Abstract
Biochemical studies of Chlamydomonas flagellar axonemes revealed that radial spoke protein (RSP) 3 is an A-kinase anchoring protein (AKAP). To determine the physiological role of PKA anchoring in the axoneme, an RSP3 mutant, pf14, was transformed with an RSP3 gene containing a mutation in the PKA-binding domain. Analysis of several independent transformants revealed that the transformed cells exhibit an unusual phenotype: a fraction of the cells swim normally; the remainder of the cells twitch feebly or are paralyzed. The abnormal/paralyzed motility is not due to an obvious deficiency of radial spoke assembly, and the phenotype cosegregates with the mutant RSP3. We postulated that paralysis was due to failure in targeting and regulation of axonemal cAMP-dependent protein kinase (PKA). To test this, reactivation experiments of demembranated cells were performed in the absence or presence of PKA inhibitors. Importantly, motility in reactivated cell models mimicked the live cell phenotype with nearly equal fractions of motile and paralyzed cells. PKA inhibitors resulted in a twofold increase in the number of motile cells, rescuing paralysis. These results confirm that flagellar RSP3 is an AKAP and reveal that a mutation in the PKA binding domain results in unregulated axonemal PKA activity and inhibition of normal motility.
INTRODUCTION
Eukaryotic cilia and flagella are highly conserved organelles that are required for diverse and vital motile and sensory functions (Afzelius, 2004; Snell et al., 2004; Pan et al., 2005; Quarmby and Parker, 2005). Motile cilia and flagella are capable of complex, carefully coordinated movements that are important for embryonic development, fertilization, and maintenance of epithelial tracts (Silflow and Lefebvre, 2001; El Zein et al., 2003; McGrath and Brueckner, 2003). Ciliary and flagellar movement is mediated by the axoneme, a highly ordered microtubule-based structure, composed of hundreds of conserved proteins (Avidor-Reiss et al., 2004; Li et al., 2004; Pazour et al., 2005). Within the axoneme, exact spatial and temporal regulation of dynein-driven microtubule sliding is required for the production of precisely formed bends (Satir, 1968; Summers and Gibbons, 1971; Shingyoji et al., 1977; Brokaw, 1991). However, the mechanisms that regulate dynein and modulate the size and shape of the axonemal bend, parameters referred to as waveform, are poorly understood. Experimental studies of Chlamydomonas and other organisms have shown that several key structural components of the axoneme, including the inner dynein arms, central pair apparatus, and radial spokes, are important in the control of axonemal waveform, in a process involving protein phosphorylation (Brokaw et al., 1982; Brokaw and Kamiya, 1987; Porter and Sale, 2000; Kamiya, 2002; Padma et al., 2003; Smith and Yang, 2004; White et al., 2005). Here, we focus on the role of the radial spokes and an associated axonemal cAMP-dependent protein kinase (PKA), which operates to regulate dynein activity and axonemal motility.
Previous in vitro studies of isolated axonemes lacking radial spokes have revealed that an axonemal PKA controls microtubule sliding in a pathway that regulates phosphorylation of the IC138 intermediate chain of the I1 dynein complex, also known as the f-dynein (Smith and Sale, 1992; Habermacher and Sale, 1996, 1997; King and Dutcher, 1997; Yang and Sale, 2000; Smith, 2002; Hendrickson et al., 2004), which is important for the control of flagellar waveform (Brokaw and Kamiya, 1987). Specifically, the experimental evidence indicates that phosphorylation of IC138 inhibits dynein-driven microtubule sliding and that dephosphorylation, requiring PKA inhibitors, rescues microtubule sliding (Habermacher and Sale, 1997; King and Dutcher, 1997; Smith, 2002; Hendrickson et al., 2004). A subsequent study indicated that IC138 may not be a direct substrate for PKA but that PKA may instead be an upstream regulator of IC138 phosphorylation (Yang and Sale, 2000). Collectively, these results, and results from additional systems (Brokaw, 1987; Hamasaki et al., 1991; San Agustin et al., 1998; Kultgen et al., 2002; Wyatt et al., 2005), demonstrate that PKA is a structural component of the axoneme and that the axonemal PKA operates in a pathway involving the radial spokes and inner arm dynein I1 to regulate microtubule sliding.
Consistent with the presence of PKA in the Chlamydomonas axoneme, an A-kinase–anchoring protein (AKAP) has been identified as a component of the radial spokes (Gaillard et al., 2001; Yang and Yang, 2006). The radial spokes are regularly repeating axonemal structures composed of at least 23 proteins and are required for normal axonemal motility (for spoke structure, see Curry and Rosenbaum, 1993; Smith and Yang, 2004; Yang et al., 2006). Radial spoke protein (RSP) 3, originally characterized as a protein required for radial spoke assembly (Diener et al., 1993), is also an AKAP (Gaillard et al., 2001). AKAPs typically function to localize PKA in the cell through binding to the RII or RI regulatory subunits (Tasken and Aandahl, 2004; Wong and Scott, 2004). In vitro studies have shown that RSP3 binds to RII and that the RII-binding domain of Chlamydomonas RSP3 is localized to a region containing amino acids 161-178 (Figure 1). This region of RSP3 is predicted to form an amphipathic helix, the structural domain of AKAPs that serves as the site of interaction for PKA regulatory subunits, and is contained within a remarkably highly conserved domain in RSP3 orthologues (Table 1). The localization of the RSP3 RII-binding domain, here referred to as the PKA-binding domain, has been confirmed by in vitro mutagenesis studies in which amino acids valine 169 and leucine 170 were replaced by alanines, causing disruption of the amphipathic helix and resulting in a loss of PKA binding by RSP3 (Gaillard et al., 2001). Additional studies have shown that RSP3 is located near the base of the radial spoke, in proximity to I1 dynein (Piperno et al., 1981; Curry and Rosenbaum, 1993; Diener et al., 1993), and interacts with additional spoke proteins that regulate motility (Yang and Yang, 2006).
Figure 1.
RSP3 structure: amino acid substitution in the PKA-binding domain of RSP3, a protein comprised of 516 amino acids. Amino acids valine 169 and leucine 170 were substituted with alanines (V169L170→AA). The position of the RII binding domain (Gaillard et al., 2001) is shown relative to the axoneme targeting domain (Diener et al., 1993) and a putative coiled-coil domain (SMART; Schultz et al., 1998; Letunic et al., 2004). The RII-binding domain is contained within the radial_spoke_3 domain, a protein domain registered in the CDD (Marchler-Bauer et al., 2005) and present in orthologues from diverse species.
Table 1.
Percentage of amino acids conserved in RSP3 orthologues from various species compared with Chlamydomonas RSP3
| Species | % Conserved for radial_spoke_3 domain | % Conserved for PKA-binding domain |
|---|---|---|
| Anopheles gambiae | 59 | 83 |
| Trypanosoma brucei | 59 | 89 |
| Apis mellifera | 61 | 89 |
| Leishmania major | 62 | 89 |
| Canis familiaris | 66 | 89 |
| Mus musculus | 67 | 89 |
| Pan troglodytes | 67 | 89 |
| Homo sapiens | 67 | 89 |
| Gallus gallus | 68 | 89 |
| Xenopus tropicalis | 69 | 89 |
| Danio rerio | 69 | 89 |
| Ciona intestinalis | 71 | 89 |
Conserved amino acids include identical and conservatively substituted amino acids. For each species, the percentage of conserved amino acids is shown for the radial_spoke_3 domain (Marchler-Bauer et al., 2005) and for the putative PKA-binding domain in comparison with the RSP3 amino acid sequence in Chlamydomonas.
To determine the physiological relevance of PKA binding by RSP3 and to further test the hypothesis that RSP3 is an AKAP required for control of axonemal PKA, we performed site-directed mutagenesis of the RSP3 gene in the region coding for the PKA binding site and used the mutant gene for transformation studies and subsequent analysis of motility phenotypes. Our prediction was that the specific disruption of PKA binding by RSP3 would result in misregulation of axonemal PKA activity and abnormal flagellar motility. Our strategy was to mutate RSP3 by making alanine substitutions at residues 169 and 170 (Figure 1), which block the PKA–RSP3 interaction (Gaillard et al., 2001), and then transform pf14 cells, which are a “null” mutant for RSP3 and lack radial spokes (Diener et al., 1993). We provide evidence that disruption of the PKA-binding domain in RSP3 results in abnormal flagellar motility and that this abnormal motility can be partially rescued by the addition of PKA inhibitors. These results are consistent with a model in which RSP3 is an AKAP required for regulation of axonemal PKA and regulation of flagellar bending by the radial spokes.
MATERIALS AND METHODS
Chlamydomonas Strains and Growth Conditions
Chlamydomonas reinhardtii strains wild type (wt) (cc-125) and pf14 (lacks radial spokes) were obtained from the Chlamydomonas Center (Duke University, Durham, NC), as were the high-efficiency mating cell types cc-620 and cc-621. Pf14/nit1-305 (lacks radial spokes, deficient in a nitrate reductase gene) was obtained from Dennis Diener (Yale University, New Haven, CT). Cells were grown in liquid modified medium I, with aeration and a 14/10-h light/dark cycle (Witman, 1986).
Mutagenesis of the RSP3 Gene
Mutagenesis of an RSP3 cDNA construct encoding amino acids 104-180 was performed as described previously (Gaillard et al., 2001), such that valine 169 and leucine 170 were substituted with alanines (Figure 1). Using a plasmid containing a 3.5-kilobase (kb) HindIII-EcoRI fragment of the RSP3 gene (pRSP3-HE) (obtained from Dennis Diener) that encompasses the entire coding region of the gene, the gene sequence coding for amino acids 104-180 was excised by restriction enzyme digestion with EcoRI and SphI. This region contains part of a single exon of the RSP3 gene. The excised region was then replaced using a corresponding sequence obtained by EcoRI/SphI restriction digestion of the mutagenized RSP3 cDNA construct. DNA sequencing was then performed to ensure the accuracy of RSP3 gene mutagenesis.
Transformation with RSP3
Transformation was performed according to the method of Kindle (1990) with the following modifications. For transformation, 1 μg of either nonmutagenized pRSP3-HE plasmid or mutagenized pRSP3-HE plasmid was cotransformed with 1 μg of plasmid pMN56, which contains a 14.5-kb genomic fragment encoding nitrate reductase (Fernandez et al., 1989). In preparation for transformation, pf14/nit1-305 cells were initially grown in liquid modified medium I, and the plasmids were linearized by restriction enzyme digestion with SspI so that at least 1 kb of noncoding sequence was present on the ends of the linearized plasmids. For transformation, acid-washed glass beads (G-1152; Sigma-Aldrich, St. Louis, MO) were used and were autoclaved before use. Polyethylene glycol (PEG; Mr 6000) was substituted with PEG (Mr 8000) (P-4463; Sigma-Aldrich) and was filter sterilized before use. Glass beads, cells, PEG, and DNA were mixed together in a 15-ml conical tube, and the tube was vortexed for exactly 45 s. After vortexing, the cells were washed once with a 10× volume of SGII-NO3 liquid medium before plating.
Genetic Backcrosses
Mutant RSP3 transformant cells (mating type minus) and wt (cc-125) cells (mating type plus) were incubated in TAP medium lacking nitrogen for 3 h to induce gametogenesis (Harris, 1989). Equal numbers of cells of each mating type were then combined in a culture dish, and mating was confirmed 1–2 h later by checking for the presence of quadraflagellate cells. The cells were then plated on solid TAP medium containing 2% agar and were incubated overnight in constant light. After overnight incubation, the plates were sealed with Parafilm and wrapped in aluminum foil to block out light for 5 d. After 5 d, the plates were unwrapped and exposed to chloroform vapors for 30 s to eradicate unmated cells, and then they were returned to the dark for 1 d. The plates were removed from the dark and were then scraped with a dull razor blade to remove unmated cells. To induce germination of the zygotes, the plates were exposed to constant light and moisture for 18–24 h. Tetrads were then identified on the plates, and the tetrad progeny cells were isolated and transferred to fresh TAP plates. The motility and molecular phenotypes of the progeny were then assessed.
Analysis of Cell Motility
To assess cell motility, cells were applied to a perfusion chamber, constructed by placing two strips of double-sided tape onto a glass slide and positioning a long glass coverslip on top of the two strips of tape. The perfusion chamber was designed to hold 10–15 μl of liquid, and cells in the perfusion chamber were observed by dark-field microscopy and recorded on videotape. Each slide was observed for about 1 min, moving among distinct different fields and recording each field for ∼20 s. The focal plane used for measuring overall cell motility was defined by the presence of freely swimming or floating cells, midway between the coverslip and the slide surface: cells obviously adherent to either the slide or coverslip were disregarded. Motile cells were defined as those that were actively swimming, spinning or that had rapidly twitching flagella, whereas cells that were floating and had paralyzed or rarely twitching flagella were scored as immotile. All cells in each field (∼30 cells per field) were assessed and tabulated as motile or immotile, assessing a total of 500–600 cells for each experiment. In some cases, cells were further tabulated as swimming, spinning, regularly twitching, or paralyzed. Student's t tests were used in pairwise statistical analysis between control and experimental samples to determine whether there were statistically significant differences between the data sets.
Isolation of Axonemes
Unless otherwise stated, all chemicals were obtained from Sigma-Aldrich, and deionized H2O was used throughout. Axonemes were isolated as described previously (Witman, 1986). In brief, cells were pelleted at 1000 × g and were resuspended in HMDS buffer (10 mM HEPES, 5 mM MgSO4, 1 mM dithiothreitol [DTT], 4% sucrose, 0.1 M phenylmethylsulfonyl fluoride [PMSF], and 0.6 trypsin inhibitor unit [TIU] aprotinin, pH 7.4). Cells were then deflagellated with 0.1 M dibucaine. The dibucaine was diluted by the addition of HMDEgS buffer (10 mM HEPES, 5 mM MgSO4, 1 mM DTT, 0.5 mM EGTA, 4% sucrose, 0.1 M PMSF, and 0.6 TIU aprotinin, pH 7.4), and the cell bodies were separated from the flagella by centrifugation at 1000 × g. The flagella were further isolated by centrifugation on a 25% sucrose pad at 2600 × g using a swinging bucket rotor. The flagellar suspension was pelleted at 13,000 × g, and the flagella were then resuspended in Na low buffer (10 mM HEPES, 5 mM MgSO4, 1 mM DTT, 30 mM NaCl, 0.5 mM EDTA, 0.1 M PMSF, and 0.6 TIU aprotinin, pH 7.4). The flagella were demembranated with 0.5% NP-40 (Calbiochem, San Diego, CA), and the axonemes were pelleted at 13,000 × g. For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, axonemal protein concentration was determined by the Bradford assay using Bio-Rad Protein Assay Reagent (Bio-Rad, Hercules, CA), and axonemal protein samples were fixed for SDS-PAGE at a concentration of 4 μg/μl.
Biochemical Analyses
RII Overlays and Western Blot Analysis.
RII overlays were performed as described previously (Gaillard et al., 2001). For Western blot analysis, proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were treated with 5% nonfat milk in Tris-buffered saline (TBS), pH 7.4, to prevent nonspecific binding. The blots were incubated with anti-RSP3 serum at 1:15,000 overnight at 4°C. After washing in TBS, blots were incubated with goat anti-rabbit secondary antibodies (1:15,000) (Bio-Rad) for 1 h at 23°C. After a final series of washes in TBS, blots were developed using enhanced chemiluminescence (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Two-Dimensional (2-D) Gel Electrophoresis.
Isoelectric focusing in the first dimension was carried out using a Bio-Rad Protean isoelectric focusing (IEF) cell and 11-cm, pI 3-10 nonlinear Ready Strips (Bio-Rad). A 50-μg sample of axonemal protein was focused for 50,000 V-h. The second dimension was performed using Bio-Rad 7.5% Criterion precast IEF gels with electrophoresis at 200 V for 1 h, 20 min. The gels were then silver stained (Merril et al., 1981).
Electron Microscopy
Specimens for electron microscopy were prepared as described previously (Mitchell and Sale, 1999). Briefly, pelleted axonemes were prepared for electron microscopy by fixation in 1% tannic acid, 1% glutaraldehyde (Ted Pella, Redding, CA), and 0.1 M Na-cacodylate for 1 h at 23°C or at 4°C overnight. The pellets were washed with 0.1 M Na-cacodylate, followed by fixation in 1% osmium tetroxide and 0.1 M Na-cacodylate for 1 h at 23°C. The pellets were rinsed in 0.1 M Na-cacodylate and were gradually dehydrated by performing sequential washes of 50, 70, 80, 95, and 100% ethanol. Fixed axonemes were mixed with a solution of 1:1 epoxy embedding medium (Eponate 12-Araldite 502; Ted Pella) to propylene oxide (Ted Pella) overnight at 23°C, with agitation. Axonemal pellets were transferred to molds containing fresh epoxy medium and were baked at 60°C for 48 h. Silver sections of the embedded axonemes were stained with lead citrate and uranyl acetate in preparation for viewing. Negatives were scanned into Adobe Photoshop (Adobe Systems, Mountain View, CA) for cropping and contrast adjustment.
Enrichment of Motile and Immotile Transformed Cells and Deflagellation and Flagellar Regeneration of Fractionated Cells
Cell Enrichment.
388 cells were gently pelleted by centrifugation at ∼500 × g for 2 min using the SS-34 rotor (Sorvall, DuPont Instruments, Newtown, CT). Tubes were then placed in light for 3–4 h to allow the motile cells to swim out of the pellet. The top (motile) and bottom (immotile) fractions were collected, and motility of the enriched motile or paralyzed cells was assessed over the next 8 h. The fractionation was performed such that, typically, >75% of the enriched motile fraction of cells were motile and <20% of the immotile fraction of cells were motile.
Flagellar Regeneration.
Enriched motile or immotile fractions of cells were centrifuged for 1 min at low speed using a clinical centrifuge. The cell pellets were then resuspended in a buffer containing 5 mM HEPES and 5% sucrose, and the cells were deflagellated by the dropwise addition of 0.5 M acetic acid. The cells were periodically checked for deflagellation, and once the flagella had been removed, the cells were quickly returned to a neutral pH by the addition of 0.5 M KOH. The cells were pelleted as described above and then resuspended in 5 ml of liquid modified medium I to allow for the regeneration of flagella. Aliquots of cells were collected at 0, 3, and 6 h, and cell motility was immediately assessed.
Gamete Motility and Temperature Sensitivity
To induce gametogenesis, wt, mutant transformant (214 and 388), and tetrad progeny cells were grown in liquid modified medium I lacking nitrogen for 3 h to overnight. After this incubation, cell motility of the gametes was measured as described previously. To test the effect of temperature on cell motility, wt, 214, 388, and tetrad progeny cells were streaked onto plates of L-medium and were incubated at 16, 23, or 32°C overnight. The cells were then scraped off of the plates, resuspended in nitrogen-deficient liquid modified medium I at the same temperatures, and incubated for 2 h. After the incubation, cell motility was assessed.
Reactivation of Motility
Cells were grown to a density of ∼1 × 106 to 1 × 107 cells/ml in liquid modified medium I. Ten milliliters of cells was placed into a 15-ml conical tube, and the cells were gently pelleted for 1 min using a clinical centrifuge. The supernatant was then removed and the cell pellet was resuspended to standard density (3 × 106 cells/ml) in a buffer containing 10 mM HEPES and 4% sucrose. Fifty microliters of resuspended cells was then removed and added to a tube containing 0.5 ml of demembranation buffer (30 mM HEPES, 5 mM MgSO4, 1 mM DTT, 1 mM EGTA, 50 mM K-acetate, 1% PEG [Mr 15,000–20,000], and 0.1% NP-40 [Calbiochem]). The cells were extracted in demembranation buffer for 30–60 s, and cessation of motility was confirmed by examination of the cells with phase contrast microscopy. One-half milliliter of reactivation buffer (30 mM HEPES, 5 mM MgSO4, 1 mM DTT, 2 mM EGTA, 50 mM K-acetate, 1% PEG [Mr 15,000–20,000], and 2 mM ATP) was then added to the tube containing demembranated cells, and reactivation of motility was immediately observed and recorded by dark-field videomicroscopy as described above. Where indicated, reactivation was performed in the presence of 50 nM PKA peptide inhibitor (PKI) or 50 nM recombinant RIIα and/or 5 μM cAMP. In some experiments (where indicated) 1 μM microcystin-LR (MC) was added before addition of PKI or RIIα and/or cAMP.
RESULTS
A Mutation in the RII-binding Domain of RSP3 Results in Decreased Flagellar Motility
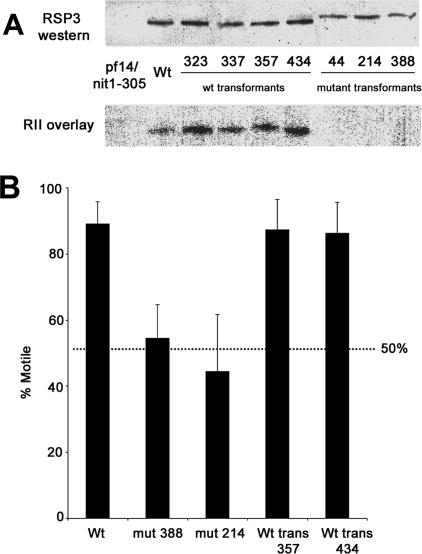
To determine the physiological role of RII binding by RSP3, the RSP3 gene was mutated in the region coding for the RII-binding domain (Figure 1). The RII-binding domain was altered as described previously for the in vitro studies of RSP3, in which valine 169 and leucine 170 were replaced by alanines. Both the mutated RSP3 gene and the wt RSP3 gene (as a control) were transformed into pf14 cells that lack axonemal RSP3 and radial spokes. Several independent transformants were obtained for each gene, and the axonemes from the transformants were analyzed for the presence of RSP3 and radial spokes. Western blot analysis of the axonemes from the transformants revealed that both the wt transformants (323, 337, 357, and 434) and the mutant transformants (44, 214, and 388) contain axonemal RSP3 (Figure 2A, top). Axonemal RSP3 of the mutant transformants migrates slightly slower on an SDS-PAGE gel compared with axonemal RSP3 of the wt transformants, consistent with the in vitro studies of RSP3 described previously (Gaillard et al., 2001), thereby providing a marker for the mutant protein. RII overlay analysis of axonemal protein from the transformants revealed that wt axonemal RSP3 binds to RII, whereas, as predicted, the mutant form of axonemal RSP3 fails to bind to RII; again, consistent with previous in vitro studies of RSP3 (Figure 2A, bottom).
Figure 2.
Analysis of pf14/nit1-305 cells transformed with the RSP3 gene: Pf14/nit1-305 cells were transformed with either a wt or mutant (V169L170→AA) RSP3 gene, and independent transformants were isolated and characterized. (A) Western blot and RII overlay analysis of axonemes from wt and mutant transformants. Both wt and mutant transformants contain normal amounts of axonemal RSP3 protein. RSP3 protein found in the axonemes of mutant transformants (44, 214, and 388) migrates slightly slower on an SDS-PAGE gel, consistent with SDS-PAGE migration of the bacterially expressed, mutagenized RSP3 construct (Gaillard et al., 2001). The shift in migration provides a useful marker of the mutant RSP3. As predicted, RSP3 found in the mutant transformants also fails to bind RII in an RII overlay assay. (B) Phenotypic analysis of the transformed cells. Motility analysis, defined in Materials and Methods, of randomly selected transformants (214 and 388) shows that the mutant transformants have a significantly reduced number of motile cells compared with wt transformants (357 and 434); when normalized to wild type, cells transfected with the mutant RSP3 display ∼50% motile cells and 50% immotile cells.
Transformants were then randomly selected for further study, with particular focus on analysis of motility. To determine the degree of motility for each cell type, individual cells were scored as motile or immotile, and a percentage of motility was obtained for each cell type. Using this assay, ∼90% of wt cells were motile (Figure 2B). Similarly, ∼90% of the wt RSP3 transformants (357 and 434) were motile, confirming successful transformation rescue of pf14 (Figure 2B). In striking contrast, the mutant RSP3 transformants (214 and 388) displayed an unusual mixed motility phenotype: ∼50% of the cells were motile and ∼50% were immotile (Figure 2B). Besides a quantitative difference in motility between the wt and mutant transformants, qualitative differences in motility were also discerned. Observations of the mutant RSP3 transformants revealed that the motile cells are comprised of normally swimming cells as well as spinning or regularly twitching cells. The spinning cells were observed to be biflagellate and presumably are the result of cells having one motile flagellum and one immotile flagellum. Measurements of swimming speed and beat frequency were performed for the normally swimming mutant RSP3 transformants, revealing the same swimming speed and flagellar beat frequency as wild-type cells.
To ensure that the mutant RSP3 transformant cells are genetically identical and not a mixture of different genotype populations, the cells were subcloned several times. To do this, a single mutant RSP3 transformant cell was isolated and then allowed to multiply into a population of cells. The population of cells was then scored for motility, and in all cases the population of cells exhibited a mixed motility phenotype (nonmotile, spinning, normal swimming). Additional experiments showed that the mixed motility phenotype of the mutant RSP3 transformants is unrelated to cell culture density or temperature and is also present when the cells are gametes. Collectively, these experiments demonstrate that the mixed motility phenotype is an inherent characteristic of the mutant RSP3 transformant cells. For convenience, and because the mutant cells display ∼50% motile and 50% immotile cells, we refer to the mutant phenotype as the “50:50 phenotype.”
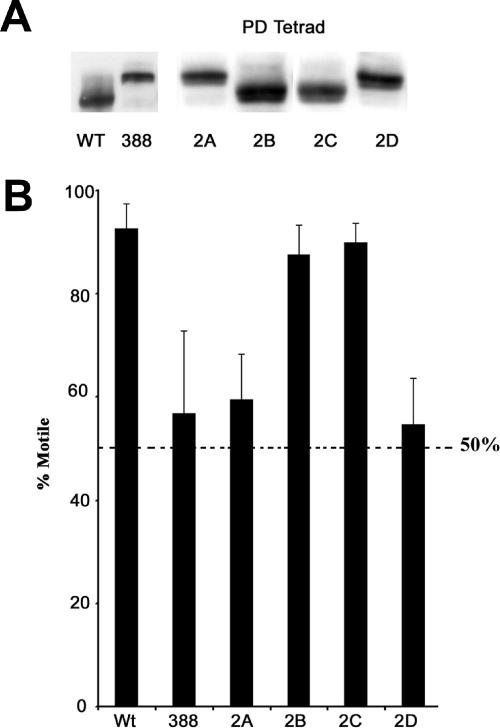
To make certain that the decreased motility of the mutant RSP3 transformants is due to the presence of the mutant RSP3 gene, 388 cells were backcrossed to wild-type cells, and tetrad analysis was performed. Each tetrad progeny was scored for motility and for the presence of RSP3 in the axoneme (assessed by Western blots). Because the mutated version of the RSP3 gene produces axonemal RSP3 protein that migrates more slowly on an SDS-PAGE gel, the presence of the mutant RSP3 gene can be easily distinguished from that of the wild-type RSP3 gene. Tetrad analysis revealed approximately equal numbers of parental ditype tetrads (PD) and nonparental ditype tetrads (NPD). Thus, the transformed mutant RSP3 gene is most likely located on a separate chromosome from the RSP3 gene found in wt cells. The motility of each of the tetrad progeny was then measured and recorded as a percentage of motility. In all cases, the 50:50 motility phenotype exclusively cosegregated with the mutant RSP3 gene, demonstrating a definitive linkage between the mutant RSP3 gene and the 50:50 motility phenotype. For example, progeny 2A and 2D, of a randomly selected PD tetrad, exhibit both decreased motility and the presence of the mutant RSP3 gene, whereas progeny 2B and 2C contain the wt RSP3 gene and have motility similar to wt cells (Figure 3, A and B).
Figure 3.
Genetic analysis of a mutant RSP3 transformant: 388 cells were mated with wt cells and tetrad analysis was performed, revealing a relatively equal occurrence of parental ditype (PD) and nonparental ditype (NPD) tetrads. (A) Western blot analysis of axonemes from a PD tetrad using an antibody to RSP3. Progeny 2B and 2C from the PD tetrad contain the wt form of RSP3 protein, whereas progeny 2A and 2D contain the mutant form of RSP3 protein, as indicated by the migratory shift of the protein band. Axonemal RSP3 from 388 cells and wt cells is shown for comparison. (B) Phenotypic analysis of the tetrad progeny. Live cell motility was assessed for each of the tetrad progeny, showing that the reduced motility phenotype exclusively cosegregates with the mutant form of axonemal RSP3 (2A and 2D).
The Mutation in the PKA-binding Domain Does Not Affect Radial Spoke Assembly
To determine whether the defective motility of the mutant RSP3 transformants is due to a defect in radial spoke assembly, axonemes from mutant RSP3 transformants were isolated and compared with isolated axonemes from wt RSP3 transformants, wt cells, and pf14 cells. Electron microscopy was performed on the axonemes, and electron microscopy images of both cross and longitudinal sections revealed that radial spokes are present in both wt (357) and mutant (388) RSP3 transformants, resembling the appearance of radial spokes in wt cells (Figure 4A). Radial spokes are clearly not present in pf14 cells, showing that transformation of the cells with the RSP3 gene (wt or mutant) is responsible for restoring radial spoke assembly (Figure 4A). Because a lack of radial spokes in the axoneme results in displacement of the central pair apparatus, the position of the central pair can be used as an assay for the presence of assembled radial spokes (see Figure 4A for reference). Axonemal cross-sections from wt cells, pf14 cells, wt RSP3 transformant cells (357 and 434), and mutant RSP3 transformant cells (214 and 388) were randomly selected, and the position of the central pair was recorded as central or displaced. The assay demonstrated that wt cells, wt RSP3 transformants (357 and 434), and mutant RSP3 transformants (214 and 388) all have axonemes that predominantly contain centered central pair apparatuses (Figure 4B), consistent with the presence of radial spokes in the axoneme. In contrast, pf14 cells have axonemes that contain mostly displaced central pair apparatuses (Figure 4B), a characteristic feature of axonemes lacking radial spokes. Axonemes were also analyzed by 2-D gel electrophoresis, focusing on radial spoke proteins 1-7, and analysis revealed that both wt (357) and mutant (388) RSP3 transformants contain wild-type levels of radial spoke proteins (Figure 4C). In contrast, pf14 axonemes lack radial spoke proteins (Figure 4C). Together, these three independent measures for the presence of radial spokes all demonstrate that the mutant RSP3 transformants contain normal amounts of radial spokes; thus, radial spoke assembly is not affected by the mutation in the PKA-binding domain of RSP3.
Figure 4.
Structural analysis of axonemes from wt and mutant transformants: (A) Electron microscopy of axonemes from randomly selected wt and mutant RSP3 transformants. Cross-sections (left) and longitudinal sections (right) are shown. Both wt (357) and mutant (388) RSP3 transformants contain radial spokes in the axoneme, indicating that the amino acid substitution of RSP3 does not obviously affect radial spoke assembly. Wt and pf14/nit1-305 axonemes are shown for comparison. (B) Central pair apparatus centering assay using randomly selected wt and mutant transformants and compared with wild-type and pf14/nit1-305 axonemes. As an assay for the presence of radial spokes, electron micrograph images of axonemes were randomly scanned for cross-sections, and the position of the central pair apparatus was recorded as central (black bar) or displaced (white bar). Cross-sections of both wt (357 and 434) and mutant (214 and 388) RSP3 transformants contain mostly centered central pair apparatuses compared with pf14/nit1-305 cells that contain mostly displaced central pair apparatuses; thus, indicating that a mutation in the PKA-binding domain of RSP3 does not affect radial spoke assembly. (C) 2-D gel electrophoresis of axonemes from randomly selected wt (357) and mutant (388) RSP3 transformants compared with wild-type and pf14/nit-305 axonemes. A silver-stained subsection of the 2D gel is shown, revealing normal amounts of radial spoke proteins 1, 2, 3, 4, 5, 6, and 7; again, indicating that the RSP3 mutation does not affect radial spoke assembly. Not shown here, RSP11 also assembles in axonemes from the 388 RSP3 transformants.
Once the Flagellum Assembles, Individual Cells Do Not Switch between Motile and Immotile States
To further study the “50:50” motility phenotype and test whether individual cells can switch between motile and immotile states, mutant transformant cells were fractionated in small cultures to enrich for motile and immotile populations. The percentage of motility for the motile and immotile fractions of cells was then measured over time. The fractionation procedure was partially successful: before fractionation the cells were ∼50% motile (Figure 5, bar 1), whereas after fractionation, the motile fraction displayed ∼80–90% motility (Figure 5, bar 2) and the immotile fraction displayed ∼25–30% motility (Figure 5, bar 6). After fractionation, the cells were incubated for up to 8 h and periodically observed, and motility was measured again. The overall degree of motility for the motile and immotile fractions of cells remained unchanged over time (our unpublished data). This observation shows that individual cells do not switch between motile and immotile states once the flagellum is assembled, indicating that at the time of assembly a flagellum is either motile or immotile. Consistent with this idea, the ratio of motile cells that display motility of both flagella to those that display motility in just one flagellum (as revealed by a spinning motion) was also observed to remain constant over time.
Figure 5.
Deflagellation and flagellar regeneration of motile and immotile fractions of a mutant RSP3 transformant: 388 cells were assessed for motility (live, bar 1) and then separated into two fractions: motile (388M) and immotile (388IM). The fractionated cells were assessed for motility (pre, bars 2 and 6) and then deflagellated by pH shock. Flagella were allowed to reassemble over a period of 6 h, and motility was assessed at 0, 3, and 6 h after deflagellation (post). After flagellar reassembly, the “motile” and “immotile” fractions become heterogeneous, containing both motile and immotile cells, restoring the 50:50 phenotype.
To further test the idea that motility is determined at the time of flagellar assembly, we used a flagellar regeneration strategy. We predicted that deflagellation of the enriched motile cells (showing ∼90% motility) would restore the cells to 50% motility, and that, similarly, the “immotile” fraction (showing ∼25% motility) would be restored to 50% motility. To test this prediction, each fraction of cells was deflagellated by pH shock, and the motility of the cells was observed at 0, 3, and 6 h after deflagellation. Immediately after deflagellation, both fractions of cells were observed to be immotile, due to the absence of flagella (Figure 5, 0 h, bars 3 and 7). At 3 h after deflagellation, the motile fraction of cells exhibited ∼40% motility, whereas the immotile fraction exhibited ∼25% motility (Figure 5, 3 h, bars 4 and 8). As predicted, 6 h after deflagellation, the motile fraction of cells showed ∼60% motility and the immotile fraction ∼55% motility (Figure 5, 6 h, bars 5 and 9). Thus, after deflagellation and then regeneration of the flagella, the 50:50 motility phenotype was restored.
PKA Inhibitors Rescue Motility of Mutant RSP3 Transformant Cells
The mutation in RSP3 was designed to interrupt PKA binding (Figure 1). Therefore, we postulated that the mixed motility phenotype of the transformants, particularly the immotility (Hasegawa et al., 1987), is a consequence of misregulated axonemal PKA. To test this, and to further assess whether motility is a stable feature of each axoneme, in vitro-reactivated cell motility experiments were conducted. The motility of wt cells, wt RSP3 transformants (357), and mutant RSP3 transformants (388) was observed, and, as expected, motilities of ∼95, 95, and 55% were revealed, respectively (Figure 6A, live, bars 1, 4, and 7, respectively). Cells were then demembranated and motility was reactivated in a buffer containing 1 mM ATP. For all three cell types, the degree of motility for the reactivated cells was strikingly similar to that of the live cells (Figure 6A, compare live versus react). In particular, both the live and reactivated mutant transformants (Figure 6A, 388, bars 7 and 8) displayed the same 50:50 mixed motility phenotype for live and reactivated cells. This result further indicates that the difference between motile and immotile fractions of cells is established when the axoneme is assembled and is a stable feature of axonemal structure.
Figure 6.
Pharmacological analysis of RSP3 transformants using demembranated cell models. (A) PKI-induced rescue of motility for mutant RSP3 transformant cells. Live cell motility of wt cells, wt transformant (357) cells and mutant transformant (388) cells (live) (bars 1, 4, and 7, respectively) was measured followed by demembranation (resulting in arrest of cell motility) and reactivation of cell motility performed in the presence of either 0.5 mM ATP (react) (bars 2, 5, and 8) or ATP + 50 nM PKI (+PKI) (bars 3, 6, and 9). In each case, the percentage of motile reactivated cells matched the percentage of motile live cells, and PKI restores the motility of 388 cells to near wild-type levels (bar 9). To further analyze the 388 cells, reactivation was also performed using a buffer containing ATP + 50 nM RII (+RII) (bar 10), showing that RII also rescues the decreased motility phenotype of 388 cells. As an important control, a buffer containing ATP + 50 nM RII + 5 μM cAMP (+RII/cAMP) (bar 11), blocked RII-mediated rescue of motility. (B) Inhibition of PKI-mediated rescue of 388 cells by a phosphatase inhibitor. Live cell motility was assessed for wt and 388 cells (bars 1 and 4), followed by demembranation and reactivation of cell motility using reactivation buffer that contained ATP alone (react) (bars 2 and 5), ATP + 50 nM PKI (react + PKI) (bar 6), ATP + 1 μM microcystin + 50 nM PKI (react + MC + PKI) (bars 8 and 9), or ATP + 1 μM microcystin (react + MC) (bars 3 and 7). For the mutant RSP3 transformant (388) cells, the addition of MC blocks the PKI-induced rescue of motility.
The result of the reactivation experiments also permitted us to further test the idea that immotility is due to misregulation of axonemal PKA in the mutant transformants. This hypothesis is based on the design of the experiment—disruption of the PKA-binding domain of RSP3 in the mutant transformants—and the fact that increased PKA activity is known to inhibit dynein activity and axonemal motility (Hasegawa et al., 1987; Howard et al., 1994; Habermacher and Sale, 1996, 1997; Smith, 2002). Therefore, we predicted that addition of PKA inhibitors, including PKI and exogenous RII, would rescue motility in the immotile fraction of reactivated cells. To test this prediction, cell models were generated and reactivation of cell motility was performed in the presence or absence of the PKA peptide inhibitor PKI (Howard et al., 1994). Strikingly, the motility of the mutant RSP3 transformant cells increased from ∼55 to ∼85% in the presence of 50 nM PKI (Figure 6A, + PKI, bar 9). In contrast, the motilities of the wt cells and wt RSP3 transformant cells remained relatively unchanged (Figure 6A, + PKI, bars 3 and 6, respectively). The motility of reactivated mutant RSP3 transformants was also increased significantly to ∼75% upon the addition of 50 nM RII (Figure 6A, + RII, bar 10; Howard et al., 1994). As a control of specificity, the RII-induced rescue of motility was suppressed by the addition of 5 μM cAMP (Figure 6A, + RII/cAMP, bar 11). Moreover, and as expected, cAMP alone had no effect on motility of 388 mutant transformant cells. Together, these experiments suggest that the motility defect exhibited by the mutant RSP3 transformant cells is caused by unregulated and inappropriately active flagellar PKA.
Rescue of Motility with PKA Inhibitors Also Requires the Activity of a Flagellar Phosphatase
On the basis of previous studies, including data showing that PKA activity is inhibitory for the motility of Chlamydomonas flagella, we postulated that rescue of motility for the mutant RSP3 transformants would also require the activity of a flagellar phosphatase (Hasegawa et al., 1987; Habermacher and Sale, 1996, 1997; Yang et al., 2000). To test this, additional experiments using reactivated cell models were performed in the presence of the phosphatase inhibitor MC (Habermacher and Sale, 1996, 1997; Yang et al., 2000). We predicted that the addition of MC would block PKI-induced rescue of motility for the mutant RSP3 transformant cells. Reactivation of wt cells and mutant RSP3 transformant cells was performed, and, as expected, the reactivated cells had motilities similar to that of the live cells. About 90% of live and reactivated wt cells were motile and ∼50% of the live and reactivated mutant RSP3 transformant cells were motile (Figure 6B, live, bars 1 and 4, respectively). As before, addition of 50 nM PKI partially rescued reactivated motility of the mutant RSP3 transformants (Figure 6B, + PKI, bar 6). However, when cell motility was reactivated in the presence of both 50 nM PKI and 1 μM MC, rescue of motility was blocked (Figure 6B, compare bar 6 with bars 8 and 9). As a control, reactivation was performed in the presence of 1 μM MC only. For both wt cells and mutant RSP3 transformant cells, MC alone had little effect on motility (Figure 6B, bars 3 and 7). Therefore, MC blocked the motility-rescuing effects of PKI, indicating that a flagellar phosphatase is required for rescue of motility.
DISCUSSION
In this study, we have revealed a physiological role for the PKA-binding domain of the radial spoke protein RSP3 and confirmed its AKAP function. Disruption of the PKA-binding domain of RSP3 results in abnormal flagellar motility, but it does not interfere with radial spoke assembly (for radial spoke structure and model mechanism, see Yang et al., 2004, 2006). Furthermore, physiological assays using cell models reveal that the abnormal flagellar motility of the RSP3 mutant cells (388) is caused, in whole or in part, by overactive, misregulated PKA. This conclusion is based on observations that both PKI and RII, selective inhibitors of PKA activity, significantly increase the level of motility for cells defective in the PKA-binding domain of RSP3, but they have no effect on wild-type cells (Figure 6). Thus, the PKA-binding domain of RSP3 is required for proper regulation of axonemal PKA, as well as for regulation of normal flagellar motility. The inhibitory effect of axonemal PKA is consistent with previous reports on in vitro reactivation of motility (Hasegawa et al., 1987) and with previous studies on global inhibition of dynein-driven microtubule sliding in axonemes from paralyzed flagellar mutants also expressing unregulated PKA (Howard et al., 1994; Habermacher and Sale, 1996, 1997; Yang et al., 2000; Smith, 2002; Hendrickson et al., 2004).
Heterogeneous Motility of the Mutant RSP3 Transformant Cells
One of the surprising results of this study is the unusual motility phenotype of mutant cells lacking the PKA-binding domain of RSP3. We found that the motility of these cells is consistently heterogeneous (50:50 phenotype): 50% of the cells are paralyzed and 50% of the cells are motile, including normally swimming cells or regularly twitching or spinning cells. The cloning of individual cells as well as genetic backcrosses have shown that this mixed motility phenotype is an inherent characteristic of the mutant RSP3 transformant cells (Figure 3B). The 50:50 phenotype was observed in multiple RPS3 mutant transformants (44, 214, and 388), and the motility phenotype was not altered in differentiated gametes nor by changes in temperature or culture conditions.
Individual RSP3 mutant transformant cells do not switch between motile and immotile states; thus, the motility phenotype is a stable feature of each cell. This conclusion is based on fractionation and flagellar regeneration experiments (Figure 5) and on reactivation experiments in which the 50:50 live cell phenotype is replicated in the reactivated cells (Figure 6). This surprising and interesting result indicates that motility or immotility is a stable feature of axonemal assembly that can be recapitulated in reactivation buffers. Because PKI significantly increases the proportion of motile 388 cells in reactivation studies (Figure 6), the inhibition of PKA seems to bypass the deficient state of assembly for the immotile fraction of axonemes containing mutant RSP3.
The 50:50 phenotype of the RSP3 mutant transformants cannot be explained by an obvious failure in assembly of the radial spokes (Figure 4). Thus, we postulate that PKA misregulation is caused specifically by a defect in radial spoke signal transduction (Yang et al., 2004). Consistent with this idea, and one of the major advances of these studies, PKA inhibitors rescue motility in the RSP3 mutant transformant cells. However, although we have demonstrated radial spoke assembly is not affected by the mutation in the PKA-binding domain of RSP3 (Figure 3), we cannot completely rule out the possibility of a minor radial spoke assembly defect due to the limitations of current structural and biochemical analysis. Recently, it has been demonstrated that RSP3 and RSP11 directly interact and that RSP11 contains an AKAP-binding domain (Yang and Yang, 2006). One prediction is that mutation in the RSP3 AKAP domain would result in failure of RSP11 to assemble in the mutant axonemes. However, the axonemes from the 388 cells contain approximately wild-type amounts of RSP11 (our unpublished data). Therefore, predictably RSP11 binds to RSP3 at sites other than the PKA-binding domain of RSP3 and/or the PKA-binding domain of RSP3 is not required for incorporation of RSP11 into the radial spokes.
Physiological Role of RSP3
We postulate that in wild-type cells, RSP3 plays a central role in regulating axonemal PKA in a pathway involving the radial spokes, by ultimately impinging on individual outer doublet microtubules to locally control inner arm dynein activity (Smith and Yang, 2004). Regulation of axonemal PKA activity by the PKA-binding domain of RSP3 is probably most important in local control of microtubule sliding and the modification of flagellar waveform, rather than for control of the flagellar beat cycle. This conclusion is based on published studies indicating a central role of the radial spokes, as well as inner arm dynein I1, for regulation of the size and shape of the flagellar bend (Brokaw et al., 1982; Brokaw and Kamiya, 1987; Smith and Yang, 2004), but not for the initiation of bend oscillation (Smith and Yang, 2004; Aoyama and Kamiya, 2005). Collectively, these studies are consistent with a model in which the radial spokes are required for proper regulation of axonemal PKA; which, based on analysis of microtubule sliding, is required for proper regulation of I1 dynein activity (Smith and Sale, 1992; Howard et al., 1994; Habermacher and Sale, 1996, 1997; King and Dutcher, 1997; Smith, 2002; Hendrickson et al., 2004). Specifically, this model predicts that a defect in the radial spokes would result in a failure to suppress axonemal PKA activity, thereby resulting in phosphorylation of the IC138 intermediate chain of I1 dynein, inappropriate inhibition of microtubule sliding and global inhibition of motility. Direct tests of this model will require characterization of PKA in Chlamydomonas with production of useful antibodies and recovery of additional informative mutant cells.
To date, we have not shown a direct in vivo association of RSP3 with axonemal PKA. This is largely due to the lack of reagents available for identification of Chlamydomonas PKA subunits, and because available antibodies to PKA subunits from other species have not proven useful. Likewise, attempts to quantify levels of PKA activity among different axonemal fractions have yielded inconsistent or uninterpretable results. Thus, although there is much evidence for the presence of PKA in the Chlamydomonas axoneme (Hasegawa et al., 1987; Howard et al., 1994; Habermacher and Sale, 1996, 1997; Yang and Sale, 2000; Smith, 2002), we have little information as to the precise axonemal location of either the regulatory or catalytic PKA subunits. Because pharmacological analysis demonstrates that PKA is present in axonemes from mutant RSP3 transformant cells (Figure 6), PKA must be localized to the axoneme by interactions other than with the PKA-binding domain of RSP3 (San Agustin et al., 1998). This conclusion is consistent with those of previous studies of pf14 cells, in which PKA activity remains associated with the axoneme even when radial spokes (including RSP3) are absent (Howard et al., 1994).
An additional challenge for interpretation of this and of previous studies is that we do not know all of the PKA-mediated pathways or substrates in the axoneme; thus, we do not fully understand how misregulation of PKA leads to inhibited flagellar motility. Although there is much evidence to support the above-described model, there is no evidence to show that PKA directly phosphorylates IC138. Rather, it is possible that PKA activity is an upstream component of an inhibitory pathway that leads to the phosphorylation of IC138 by a different kinase. Accordingly, studies have shown that an axonemal CK1 is directly responsible for an inhibitory phosphorylation of IC138 (Yang and Sale, 2000). It is not yet known whether IC138 is also a direct substrate for the axonemal PKA, but independent pharmacological evidence indicates that PKA and CK1 may operate in parallel pathways to regulate microtubule sliding in the axoneme (Smith, 2002). One possibility is that axonemal PKA regulates a flagellar phosphatase required for motility. Consistent with this idea, and with previous studies of the regulation of microtubule sliding in the axoneme (Habermacher and Sale, 1996, 1997; Yang et al., 2000), rescue of motility in the RSP3 mutant cells requires an MC-sensitive flagellar phosphatase (Figure 6B). Thus, possible candidates for a PKA substrate include the MC-sensitive phosphatase or a protein modulator of phosphatase activity (Leach et al., 2003; Li et al., 2005).
RSP3 is a highly conserved protein among many diverse organisms (Table 1), and a RSP3 domain (radial_spoke_3) has recently been registered in the conserved domain database (CDD) (Marchler-Bauer et al., 2005). For Chlamydomonas, the radial_spoke_3 domain spans much of the amino acid sequence of RSP3, except for the C terminus, which is predicted to form a coiled-coil domain (Figure 1). Notably, the PKA-binding domain of RSP3 is included within the radial_spoke_3 domain and is particularly well conserved (Table 1), suggesting that RSP3 may function as an AKAP in the axonemes of many eukaryotes. Furthermore, a recent study has shown that RSP3 mRNA is up-regulated during neuronal migration of the developing mouse brain (Koukoulas et al., 2004), a process not previously known to involve the axoneme. Thus, RSP3 and its conserved radial_spoke_3 domain may be important in the physiology of other tissues, in processes other than the regulation of axonemal motility and possibly including the function of the primary cilium.
ACKNOWLEDGMENTS
We are grateful to Drs. Pinfen Yang and Chun Yang (Marquette University) for useful discussion and sharing data on pf25 before publication and to Dr. Maureen Wirschell for discussion and critical reading of the manuscript. The work was supported by National Institutes of Health Grant R37 GM-051173 and March of Dimes Grant FY04-115 (to W.S.S.) and National Institutes of Health Training Grant T32 GM-008367 (to B. C.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-02-0095) on March 29, 2006.
REFERENCES
- Afzelius B. A. Cilia-related diseases. J. Pathol. 2004;204:470–477. doi: 10.1002/path.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama S., Kamiya R. Cyclical interactions between two outer doublet microtubules in split flagellar axonemes. Biophys. J. 2005;89:3261–3268. doi: 10.1529/biophysj.105.067876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T., Maer A. M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., Zuker C. S. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- Brokaw C. J. Regulation of sperm flagellar motility by calcium and cAMP-dependent phosphorylation. J. Cell. Biochem. 1987;35:175–184. doi: 10.1002/jcb.240350302. [DOI] [PubMed] [Google Scholar]
- Brokaw C. J. Microtubule sliding in swimming sperm flagella: direct and indirect measurements on sea urchin and tunicate spermatozoa. J. Cell Biol. 1991;114:1201–1215. doi: 10.1083/jcb.114.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw C. J., Kamiya R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil. Cytoskeleton. 1987;8:68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- Brokaw C. J., Luck D. J., Huang B. Analysis of the movement of Chlamydomonas flagella: the function of the radial-spoke system is revealed by comparison of wild-type and mutant flagella. J. Cell Biol. 1982;92:722–732. doi: 10.1083/jcb.92.3.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry A. M., Rosenbaum J. L. Flagellar radial spoke: a model molecular genetic system for studying organelle assembly. Cell Motil. Cytoskeleton. 1993;24:224–232. doi: 10.1002/cm.970240403. [DOI] [PubMed] [Google Scholar]
- Diener D. R., Ang L. H., Rosenbaum J. L. Assembly of flagellar radial spoke proteins in Chlamydomonas: identification of the axoneme binding domain of radial spoke protein 3. J. Cell Biol. 1993;123:183–190. doi: 10.1083/jcb.123.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Zein L., Omran H., Bouvagnet P. Lateralization defects and ciliary dyskinesia: lessons from algae. Trends Genet. 2003;19:162–167. doi: 10.1016/S0168-9525(03)00026-X. [DOI] [PubMed] [Google Scholar]
- Fernandez E., Schnell R., Ranum L. P., Hussey S. C., Silflow C. D., Lefebvre P. A. Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 1989;86:6449–6453. doi: 10.1073/pnas.86.17.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard A. R., Diener D. R., Rosenbaum J. L., Sale W. S. Flagellar radial spoke protein 3 is an A-kinase anchoring protein (AKAP) J. Cell Biol. 2001;153:443–448. doi: 10.1083/jcb.153.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermacher G., Sale W. S. Regulation of flagellar dynein by an axonemal type-1 phosphatase in Chlamydomonas. J. Cell Sci. 1996;109:1899–1907. doi: 10.1242/jcs.109.7.1899. [DOI] [PubMed] [Google Scholar]
- Habermacher G., Sale W. S. Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J. Cell Biol. 1997;136:167–176. doi: 10.1083/jcb.136.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki T., Barkalow K., Richmond J., Satir P. cAMP-stimulated phosphorylation of an axonemal polypeptide that copurifies with the 22S dynein arm regulates microtubule translocation velocity and swimming speed in Paramecium. Proc. Natl. Acad. Sci. USA. 1991;88:7918–7922. doi: 10.1073/pnas.88.18.7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego, CA: Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- Hasegawa E., Hayashi H., Asakura S., Kamiya R. Stimulation of in vitro motility of Chlamydomonas axonemes by inhibition of cAMP-dependent phosphorylation. Cell Motil. Cytoskeleton. 1987;8:302–311. doi: 10.1002/cm.970080403. [DOI] [PubMed] [Google Scholar]
- Hendrickson T. W., Perrone C. A., Griffin P., Wuichet K., Mueller J., Yang P., Porter M. E., Sale W. S. IC138 is a WD-repeat dynein intermediate chain required for light chain assembly and regulation of flagellar bending. Mol. Biol. Cell. 2004;12:5431–5442. doi: 10.1091/mbc.E04-08-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard D. R., Habermacher G., Glass D. B., Smith E. F., Sale W. S. Regulation of Chlamydomonas flagellar dynein by an axonemal protein kinase. J. Cell Biol. 1994;127:1683–1692. doi: 10.1083/jcb.127.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int. Rev. Cytol. 2002;219:115–155. doi: 10.1016/s0074-7696(02)19012-7. [DOI] [PubMed] [Google Scholar]
- Kindle K. L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. J., Dutcher S. K. Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains. J. Cell Biol. 1997;136:177–191. doi: 10.1083/jcb.136.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukoulas I., Augustine C., Silkenbeumer N., Gunnersen J. M., Scott H. S., Tan S. S. Genomic organisation and nervous system expression of radial spoke protein 3. Gene. 2004;336:15–23. doi: 10.1016/j.gene.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Kultgen P. L., Byrd S. K., Ostrowski L. E., Milgram S. L. Characterization of an A-kinase anchoring protein in human ciliary axonemes. Mol. Biol. Cell. 2002;13:4156–4166. doi: 10.1091/mbc.E02-07-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach C., Shenolikar S., Brautigan D. L. Phosphorylation of phosphatase inhibitor-2 at centrosomes during mitosis. J. Biol. Chem. 2003;278:26015–26020. doi: 10.1074/jbc.M300782200. [DOI] [PubMed] [Google Scholar]
- Letunic I., Copley R. R., Schmidt S., Ciccarelli F. D., Doerks T., Schultz J., Ponting C. P., Bork P. SMART 4.0, towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. B., et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- Li M., Stefansson B., Wang W., Schaefer E. M., Brautigan D. L. Phosphorylation of the Pro-X-Thr-Pro site in phosphatase inhibitor-2 by cyclin-dependent protein kinase during M-phase of the cell cycle. Cell Signal. 2005 doi: 10.1016/j.cellsig.2005.10.020. (in press) [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., et al. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J., Brueckner M. Cilia are at the heart of vertebrate left-right asymmetry. Curr. Opin. Genet Dev. 2003;13:385–392. doi: 10.1016/s0959-437x(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Merril C. R., Dunau M. L., Goldman D. A rapid sensitive silver stain for polypeptides in polyacrylamide gels. Anal. Biochem. 1981;110:201–207. doi: 10.1016/0003-2697(81)90136-6. [DOI] [PubMed] [Google Scholar]
- Mitchell D. R., Sale W. S. Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J. Cell Biol. 1999;144:293–304. doi: 10.1083/jcb.144.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padma P., Satouh Y., Wakabayashi K., Hozumi A., Ushimaru Y., Kamiya R., Inaba K. Identification of a novel leucine-rich repeat protein as a component of flagellar radial spoke in the ascidian Ciona intestinalis. Mol. Biol. Cell. 2003;14:774–785. doi: 10.1091/mbc.02-06-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Wang Q., Snell W. J. Cilium-generated signaling and cilia-related disorders. Lab. Investig. 2005;85:452–463. doi: 10.1038/labinvest.3700253. [DOI] [PubMed] [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J., Witman G. B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Huang B., Ramanis Z., Luck D. J. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J. Cell Biol. 1981;88:73–79. doi: 10.1083/jcb.88.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M. E., Sale W. S. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 2000;151:F37–42. doi: 10.1083/jcb.151.5.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarmby L. M., Parker J. D. Cilia and the cell cycle? J Cell Biol. 2005;169:707–710. doi: 10.1083/jcb.200503053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Agustin J. T., Leszyk J. D., Nuwaysir L. M., Witman G. B. The catalytic subunit of the cAMP-dependent protein kinase of ovine sperm flagella has a unique amino-terminal sequence. J. Biol. Chem. 1998;273:24874–24883. doi: 10.1074/jbc.273.38.24874. [DOI] [PubMed] [Google Scholar]
- Satir P. Studies on cilia. 3. Further studies on the cilium tip and a “sliding filament” model of ciliary motility. J. Cell Biol. 1968;39:77–94. doi: 10.1083/jcb.39.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C. P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingyoji C., Murakami A., Takahashi K. Local reactivation of Triton-extracted flagella by iontophoretic application of ATP. Nature. 1977;265:269–270. doi: 10.1038/265269a0. [DOI] [PubMed] [Google Scholar]
- Silflow C. D., Lefebvre P. A. Assembly and motility of eukaryotic cilia and flagella. Lessons from Chlamydomonas reinhardtii. Plant Physiol. 2001;127:1500–1507. [PMC free article] [PubMed] [Google Scholar]
- Smith E. F. Regulation of flagellar dynein by the axonemal central apparatus. Cell Motil. Cytoskeleton. 2002;52:33–42. doi: 10.1002/cm.10031. [DOI] [PubMed] [Google Scholar]
- Smith E. F., Sale W. S. Regulation of dynein-driven microtubule sliding by the radial spokes in flagella. Science. 1992;257:1557–1559. doi: 10.1126/science.1387971. [DOI] [PubMed] [Google Scholar]
- Smith E. F., Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil. Cytoskeleton. 2004;57:8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell W. J., Pan J., Wang Q. Cilia and flagella revealed: from flagellar assembly in Chlamydomonas to human obesity disorders. Cell. 2004;117:693–697. doi: 10.1016/j.cell.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Summers K. E., Gibbons I. R. Adenosine triphosphate-induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc. Natl. Acad. Sci. USA. 1971;68:3092–3096. doi: 10.1073/pnas.68.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasken K., Aandahl E. M. Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol. Rev. 2004;84:137–167. doi: 10.1152/physrev.00021.2003. [DOI] [PubMed] [Google Scholar]
- White D., Aghigh S., Magder I., Cosson J., Huitorel P., Gagnon C. Two anti-radial spoke monoclonal antibodies inhibit Chlamydomonas axonemal motility by different mechanisms. J. Biol. Chem. 2005;280:14803–14810. doi: 10.1074/jbc.M414114200. [DOI] [PubMed] [Google Scholar]
- Witman G. B. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 1986;134:280–290. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]
- Wong W., Scott J. D. AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell. Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- Wyatt T. A., Forget M. A., Adams J. M., Sisson J. H. Both cAMP and cGMP are required for maximal ciliary beat stimulation in a cell-free model of bovine ciliary axonemes. Am. J. Physiol. 2005;288:L546–L551. doi: 10.1152/ajplung.00107.2004. [DOI] [PubMed] [Google Scholar]
- Yang C., Yang P. The flagellar motility of Chlamydomonas pf25 mutant lacking an AKAP-binding protein is overtly sensitive to medium conditions. Mol. Biol. Cell. 2006;17:227–238. doi: 10.1091/mbc.E05-07-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Fox L., Colbran R. J., Sale W. S. Protein phosphatases PP1 and PP2A are located in distinct positions in the Chlamydomonas flagellar axoneme. J. Cell Sci. 2000;113:91–102. doi: 10.1242/jcs.113.1.91. [DOI] [PubMed] [Google Scholar]
- Yang P., et al. Radial spoke proteins of Chlamydomonas flagella. J. Cell Sci. 2006;119:1165–1174. doi: 10.1242/jcs.02811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Sale W. S. Casein kinase I is anchored on axonemal doublet microtubules and regulates flagellar dynein phosphorylation and activity. J. Biol. Chem. 2000;275:18905–18912. doi: 10.1074/jbc.M002134200. [DOI] [PubMed] [Google Scholar]
- Yang P., Yang C., Sale W. S. Flagellar radial spoke protein 2 is a calmodulin binding protein required for motility in Chlamydomonas reinhardtii. Eukaryot. Cell. 2004;3:72–81. doi: 10.1128/EC.3.1.72-81.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]