Abstract
Kettin is a large actin-binding protein with immunoglobulin-like (Ig) repeats, which is associated with the thin filaments in arthropod muscles. Here, we report identification and functional characterization of kettin in the nematode Caenorhabditis elegans. We found that one of the monoclonal antibodies that were raised against C. elegans muscle proteins specifically reacts with kettin (Ce-kettin). We determined the entire cDNA sequence of Ce-kettin that encodes a protein of 472 kDa with 31 Ig repeats. Arthropod kettins are splice variants of much larger connectin/titin-related proteins. However, the gene for Ce-kettin is independent of other connectin/titin-related genes. Ce-kettin localizes to the thin filaments near the dense bodies in both striated and nonstriated muscles. The C-terminal four Ig repeats and the adjacent non-Ig region synergistically bind to actin filaments in vitro. RNA interference of Ce-kettin caused weak disorganization of the actin filaments in body wall muscle. This phenotype was suppressed by inhibiting muscle contraction by a myosin mutation, but it was enhanced by tetramisole-induced hypercontraction. Furthermore, Ce-kettin was involved in organizing the cytoplasmic portion of the dense bodies in cooperation with α-actinin. These results suggest that kettin is an important regulator of myofibrillar organization and provides mechanical stability to the myofibrils during contraction.
INTRODUCTION
In muscle cells, the actin cytoskeleton is highly differentiated to form the myofibrils that are specialized for producing contractile forces. In addition to actin and myosin as the essential contractile components, regulatory components for the contractile activity are integrated into the structure (Squire, 1997). Despite the fact that many myofibrillar components have been identified, little is known about how these components functionally interact to assemble and maintain the myofibrils in living cells (Littlefield and Fowler, 1998; Gregorio and Antin, 2000; Clark et al., 2002). In particular, the assembly process of actin filaments is complex. During development, actin-monomer binding proteins, including profilin, and actin depolymerizing factor/cofilin regulate actin filament dynamics (Obinata, 1993; Obinata et al., 1997; Ono, 2003a, b). However, in mature myofibrils, these proteins are no longer major components of the thin filaments, and filament binding proteins, such as tropomyosin and α-actinin, and end-capping proteins, such as CapZ and tropomodulin, become the major actin-associated proteins. However, the list of actin binding proteins in muscle is still growing and cellular functions of many of these proteins are not clearly understood.
Kettin is a large protein of 500-700 kDa found in the Z-discs and the I-bands of arthropod muscles (Bullard et al., 2000; Bullard et al., 2002, 2006). Kettin directly binds to actin filaments with high-affinity (Lakey et al., 1993; Maki et al., 1995; van Straaten et al., 1999). Proteolytic removal of kettin by calpain from the Z-discs causes disintegration of the Z-discs (Lakey et al., 1993) and decrease in stiffness of the myofibrils (Kulke et al., 2001b). Kettin is one of the first proteins to colocalize with actin during the early stages of myofibrillogenesis (Ayme-Southgate et al., 2004), and genetic analysis has shown that it is essential for myofibril assembly in the fruit fly (Hakeda et al., 2000). The sequence of kettin contains 35 immunoglobulin-like (Ig) repeats separated by short linker sequences (Hakeda et al., 2000; Kolmerer et al., 2000) and is related to the connectin/titin family of giant elastic proteins (Maruyama, 1997; Gautel et al., 1999; Gregorio et al., 1999; Maruyama and Kimura, 2000; Granzier et al., 2002). However, kettin does not have fibronectin-like, elastic PEVK, or kinase domains, and seems to be a uniquely evolved member of the connectin/titin family of proteins. Importantly, recent molecular genetic studies have shown that kettin is a splice variant of connectin/titin in Drosophila (Machado and Andrew, 2000; Zhang et al., 2000) and crayfish (Fukuzawa et al., 2001). Therefore, the previously reported phenotype of the Drosophila kettin mutants (Hakeda et al., 2000) may be partly due to a defect in the D-titin gene.
Based on sequence homology, a gene coding for a kettin-like protein has been found in the nematode Caenorhabditis elegans (Hakeda et al., 2000; Kolmerer et al., 2000). Transcripts of this gene are expressed in various muscle cells, and a monoclonal antibody (mAb) against insect kettin reacts with the dense bodies in obliquely striated body wall muscle (Kolmerer et al., 2000), which is equivalent to the Z-lines in cross-striated muscle. However, the product of the C. elegans kettin-like gene has not been extensively studied at the protein level. In this study, we show that the antigen of MH44, one of monoclonal antibodies raised against C. elegans muscle proteins, is kettin (Francis and Waterston, 1985; Ono et al., 2006). Unlike arthropods, the C. elegans kettin gene is independent of other connectin/titin-related genes, and genetic manipulation of kettin can be applied without affecting them. Our functional analysis of C. elegans kettin suggests that kettin is an important regulator of actin organization and myofibril stability in muscle cells.
MATERIALS AND METHODS
Nematode Strains
Wild-type C. elegans strain N2; an RNA interference (RNAi)-sensitive strain, NL2099 rrf-3(pk1426) (Simmer et al., 2002); and a myosin heavy chain mutant unc-54(s95) (Moerman et al., 1982) were obtained from the Caenorhabditis Genetics Center (Minneapolis, MN). An α-actinin mutant, atn-1(ok84), was provided by Drs. Gary Moulder and Robert Barstead (Oklahoma Medical Research Foundation, Oklahoma City, OK). Nematodes were grown under standard conditions at 20°C (Brenner, 1974).
Immunoprecipitation and Microsequencing
During the process of purifying actin from C. elegans (Ono, 1999), high salt/ATP extracts that are enriched with the thin filament proteins were saved and used for immunoprecipitation. Briefly, frozen nematodes were thawed in a homogenizing buffer (50 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride [PMSF], pH 8.0) and homogenized by passing twice through a French pressure cell at 360-580 kg/cm2. The homogenate was centrifuged at 10,000 × g for 10 min, and the pellet was washed twice with the homogenizing buffer. The washed pellet was extracted twice with 1 volume of a high salt/ATP buffer (0.6 M KCl, 5 mM ATP, 5 mM MgCl2, 20 mM Tris-HCl, 0.2 mM EGTA, 1 mM dithiothreitol, and 1 mM PMSF, pH 8.0) and centrifuged at 10,000 × g for 10 min. The supernatant (1 ml) was dialyzed against phosphate-buffered saline (PBS) overnight at 4°C. The extract was cleared by centrifugation at 20,000 × g for 10 min, mixed with 50 μl of 10% Triton X-100 and 5 μl of the mAb MH44 (Francis and Waterston, 1985) (provided by Pamela Hoppe, Western Michigan University, Kalamazoo, MI) in ascites fluid, and incubated on ice for 2 h. Then, 30 μl of protein G-agarose beads (Pierce Chemical, Rockford, IL) that had been washed with PBS was added and incubated for 90 min at 4°C with gentle mixing. The beads were recovered by brief centrifugation at 5000 × g and washed three times with 1 ml each of PBS containing 0.5% Triton X-100 and twice with 1 ml each of PBS. The bound proteins were eluted with 50 μl of SDS-lysis buffer (2% SDS, 80 mM Tris-HCl, 5% β-mercaptoethanol. 15% glycerol, and 0.05% bromophenol blue, pH 6.8) at 97°C for 2 min.
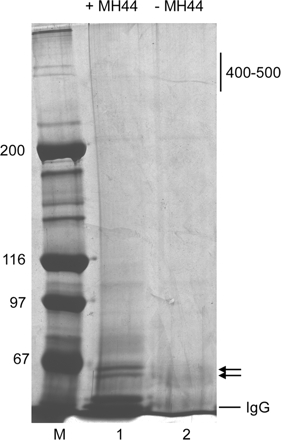
The immunoprecipitates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by silver staining (Figure 1) or Coomassie blue staining for microsequencing. The 60- to 62-kDa bands were excised together from the gel and submitted to ProtTech (Fairview Village, PA) for identification of the protein following their standard procedure. Briefly, the protein in the gel piece was digested with trypsin in 50 mM ammonium bicarbonate at pH 8.5, and the peptides were extracted by 3–5 volumes of acetonitrile, dried, and dissolved in 0.5% acetic acid. The peptides were analyzed by liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). The MS/MS data were subjected to homology search against the protein database and manually analyzed for the quality of the results.
Figure 1.
Immunoprecipitation of the MH44 antigen. Immunoprecipitates with MH44 (lane 1) and control precipitates in the absence of MH44 (lane 2) were analyzed by SDS-PAGE (5% acrylamide gel) and silver staining. There were no specific bands around 400-500 kDa, Instead, two bands of 60-62 kDa (arrows) were detected in the precipitates with MH44. These bands were subjected to microsequencing. Molecular mass markers in kilodaltons (lane M) are indicated on the left of the gel.
cDNA Cloning and Sequencing
Total C. elegans RNA was prepared from N2 using a TRI reagent (Sigma-Aldrich, St. Louis, MO). Fragments (1-2 kb) of the Ce-kettin cDNA were amplified with reverse transcriptase (RT)-PCR using a SuperScript III one-step RT-PCR with Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA) with primers listed in Supplemental Table 1. Nematode mRNAs often have the SL1 trans-spliced leader sequence at their 5′ ends. Therefore, the 5′ end of the Ce-kettin mRNA was amplified by PCR using SL1 as a forward primer (Fr-1 in Supplemental Table 1). They were cloned into a pCR-II plasmid vector using a TOPO-TA cloning kit (Invitrogen), and the sequences were determined by DNA sequencing. The sequences were manually assembled into a contiguous full-length cDNA sequence.
Fluorescence Microscopy
For immunofluorescent staining of adult body wall muscle in Figure 3, adult worms were cut in halves near the vulva by needles on poly-lysine–coated slides and permeabilized by a freeze-crack method (Epstein et al., 1993). The gonads were dissected by cutting adult hermaphrodites at the level of pharynx on poly-lysine–coated slides as described previously (Rose et al., 1997). Worm embryos were obtained by cutting gravid adults on poly-lysine–coated slides and permeabilized by a freeze-crack method. These samples were fixed by an optimal method and stained with antibodies as listed in Table 1. Immunofluorescent staining of adult body wall muscle in Figure 8 was performed by a whole-mount staining procedure as described previously (Finney and Ruvkun, 1990). When the host animals of the primary antibodies were different, they were mixed and reacted with the samples simultaneously, and followed by treatments with appropriate fluorescently labeled secondary antibodies (Table 1). We also succeeded in differentially labeling two mouse monoclonal antibodies by using secondary antibodies that distinguish IgG isotypes. MH44 is IgG2a, and mouse monoclonal antibodies of the IgG1 isotype were used for simultaneous staining with MH44. The samples were first treated with mixture of mouse IgG1 antibody and rabbit or guinea pig antibody, and then with secondary antibodies (nonspecific for IgG isotypes). They were washed with PBS and blocked with 0.1 mg/ml mouse IgG (Rockland, Gilbertsville, PA) in 1% bovine serum albumin in PBS for 10 min and reacted with MH44. Then, MH44 was visualized by Zenon Alexa Fluor mouse IgG2a labeling reagents (Invitrogen) (Table 1).
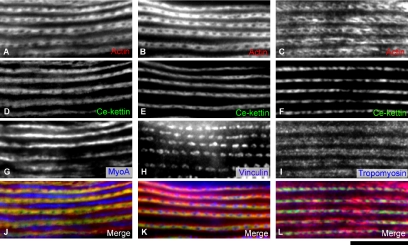
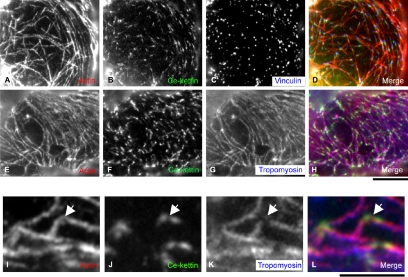
Figure 3.
Localization of Ce-kettin in adult body wall muscle. Adult body wall muscle was stained for actin (A–C), Ce-kettin (D–F), and myoA myosin heavy chain (G), vinculin (H), or tropomyosin (I). Merged images of actin (red), Ce-kettin (green), and myoA, vinculin, or tropomyosin (blue) are shown in J–L. Bar, 10 μm.
Table 1.
Fixation methods and antibodies for immunofluorescent staining
| Tissue | Protein | Fixationa | Primary antibodies | Secondary antibodiesb | Primary antibodies | Zenon-IgG2ac |
|---|---|---|---|---|---|---|
| Body wall muscle | Actin Ce-kettin MyoA | A | Rabbit anti-actin (AAN01), mouse anti-MyoA (5-6) | Cy3-DAR, A647-GAM | MH44 | A488 |
| Actin Ce-kettin Vinculin | A | Rabbit anti-actin (AAN01), mouse anti-vinculin (MH24) | Cy3-DAR, A647-GAM | MH44 | A488 | |
| Actin Ce-kettin Tropomyosin | B | Guinea pig anti-CeTM, mouse anti-actin (C4) | Cy3-DAM, A488-GAG | MH44 | A647 | |
| Ovary | Actin Ce-kettin Vinculin | A | Rabbit anti-actin (AAN01), mouse anti-vinculin (MH24) | Cy3-DAR, A647-GAM | MH44 | A488 |
| Actin Ce-kettin Tropomyosin | B | Rabbit anti-actin (AAN01) MH44, guinea pig anti-CeTM | Cy3-DAR, A647-GAM, A488-GAG | NA | NA | |
| Embryo | Actin Ce-kettin | A | Rabbit anti-actin (AAN01) MH44 | Cy3-DAR, A488-GAM | NA | NA |
NA, not applicable.
a Fixation methods: A, methanol (5 min; −20°C); and B, 4% paraformaldehyde in cytoskeleton buffer (138 mM KCl, 3 mM MgCl2, 2 mM EGTA, and 10 mM MES-KOH, pH 6.1) containing 0.32 M sucrose (30 min at room temperature), followed by methanol (5 min; −20°C).
b A488-GAG, Alexa488-labeled goat anti-guinea pig IgG (Invitrogen); A488-GAM, Alexa488-labeled goat anti-mouse IgG (Invitrogen); A647-GAM, Alexa647-labeled goat anti-mouse IgG (Invitrogen); Cy3-DAM, Cy3-labeled donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories, West Grove, PA); Cy3-DAR, Cy3-labeled donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories).
c Zenon Alexa Fluor mouse IgG2a labeling reagents. A488, Alexa488 labeled; A647, Alexa647 labeled.
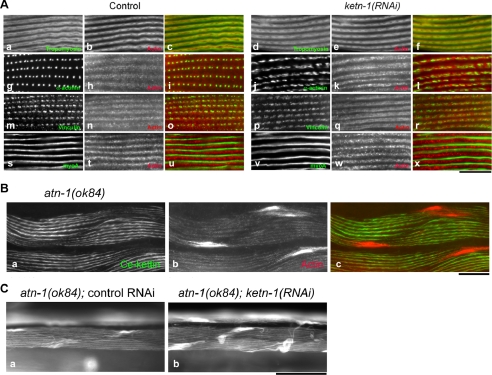
Figure 8.
Functional interaction between Ce-kettin and α-actinin in the body wall muscle. (A) Tropomyosin (a and d), α-actinin (g and j), vinculin (m and p), or myoA myosin heavy chain (s and v) was double-stained with actin (b, e, h, k, n, q, t, and w) in the body wall muscle of control or ketn-1(RNAi) adult worms. Merged images are shown (c, f, i, l, o, r, u, and x). (B) Ce-kettin (a) and actin (b) were immunolocalized in the atn-1(ok84) homozygous worms. (C) Actin filaments in the body wall muscle of the atn-1(ok84) worms with control RNAi (a) or ketn-1(RNAi) (b) were stained with tetramethylrhodamine-phalloidin. Bars, 10 μm (A), 20 μm (B), and 50 μm (C).
Guinea pig anti-tropomyosin (CeTM) antibody was described previously (Ono and Ono, 2002). Rabbit polyclonal anti-actin antibody (AAN01) was purchased from Cytoskeleton (Denver, CO). Mouse monoclonal anti-actin antibody (C4) was purchased from MP Biomedicals (Irvine, CA). Mouse monoclonal anti-myoA antibody (clone 5-6) (Miller et al., 1983) was provided by Henry Epstein (University of Texas Medical Branch at Galveston, Galveston, TX). Mouse monoclonal anti-vinculin antibody (MH24) and mouse monoclonal anti-α-actinin antibody (MH40) (Francis and Waterston, 1985) was provided by Pamela Hoppe.
Samples were viewed by epifluorescence using a Nikon Eclipse TE2000 inverted microscope with a 40 or 60× CFI Plan Fluor objective. Images were captured by a SPOT RT monochrome charge-coupled device camera (Diagnostic Instruments, Sterling Heights, MI) and processed by the IPLab imaging software (Scanalytics, Rockville, MD) and Adobe Photoshop 6.0 (Adobe Systems, Mountain View, CA).
Preparation of Recombinant Ce-Kettin Fragments
To construct an expression vector for KETN-CT1, a cDNA fragment encoding residues 3830-4250 was amplified by PCR using primers 5′-GATCGGATCCCAAGCTCCACCGACAATCTCCC and 5′-GATCGGTACCCTAGCGACTGAGTGTGAGCTTG that have added BamHI and KpnI restriction sites (underlined), respectively. The PCR product was digested by BamHI and KpnI and ligated with pQE-30 (QIAGEN, Valencia, CA) at BamHI–KpnI cloning sites. Expression of a 6xHis-tagged protein from this vector was not successful. Therefore, the cDNA fragment was excised from the vector by BamHI and SmaI and ligated with pGEX-2T (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) at BamHI–SmaI cloning sites. The cDNA insert was entirely sequenced to verify that there were no PCR-induced mutations.
To construct an expression vector for KETN-CT2, two cDNA fragments CT2-1 and CT2-2 that overlap by 22 base pairs were amplified by RT-PCR. Primers for CT2-1 were 5′-GGTTCCGCGTGGATCCAGACAGACCAAGTTGAGACCGGC and 5′-GGGAGATTGTCGGTGGAGCTTG. Primers for CT2-2 were 5′-CAAGCTCCACCGACAATCTCCC and 5′-TCACGATGAATTCCCCTAGCGACTGAGTGTGAGCTTG. Fifteen or 16 base pairs of overlapping sequences with pGEX-2T were designed at the 5′ and 3′ ends of CT2-1 and CT2-2 (underlined), respectively. CT2-1, CT2-2, and pGEX-2T that had been cut by BamHI and SmaI were fused at their homologous ends by an In-fusion PCR cloning kit (Clontech). The cDNA insert was entirely sequenced to verify that there were no PCR-induced mutations.
To construct an expression vector for KETN-CT3, a cDNA fragment encoding residues 3554-3831 was amplified from pGEX-KETN-CT2 by PCR using primers 5′-GGTTCCGCGTGGATCCAGACAGACCAAGTTGAGACCGGC and 5′-TCACGATGAATTCCCTCAAGCTTGCTTAGATTGCCCGATCTTTG. Fifteen or 16 base pairs of overlapping sequences with pGEX-2T were added at the 5′ and 3′ ends (underlined). The PCR product and pGEX-2T that had been cut by BamHI and SmaI were fused at their homologous ends by an In-fusion PCR cloning kit (Clontech). The cDNA insert was entirely sequenced to verify that there were no PCR-induced mutations.
KETN-CT1, KETN-CT2, and KETN-CT3 were expressed in Escherichia coli as glutathione S-transferase (GST)-fusion proteins and purified by the same method. The E. coli strain BL21 (DE3) was transformed with the expression vector and cultured in M9ZB medium containing 50 μg/ml ampicillin at 37°C until absorbance at 600 nm reached 0.6 cm−1. Then, the culture was cooled to room temperature, and protein expression was induced by adding 0.1 mM isopropyl β-d-thiogalactopyranoside for 3 h at room temperature. The cells were harvested by centrifugation at 5000 × g for 10 min and disrupted by a French pressure cell at 360-580 kg/cm2 in PBS containing 0.2 mM dithiothreitol and 0.4 mM PMSF. The homogenates were centrifuged at 20,000 × g for 15 min, and the supernatants were applied to a glutathione-Uniflow column (bed volume of 1 ml) (Clontech). The columns were washed by 10 column volumes of PBS and bound proteins were eluted with 10 mM glutathione, 20 mM Tris-HCl, 0.2 mM dithiothreitol, pH 8.0. Fractions containing the GST-fusion proteins were dialyzed against 0.1 M KCl, 20 mM MES-KOH, and 0.2 mM dithiothreitol, pH 6.0, and purified further with Mono S column chromatography. The proteins were eluted from the Mono S column with a linear KCl gradient of 0.1–0.5 M and dialyzed overnight against 0.1 M KCl, 2 mM MgCl2, 20 mM HEPES-NaOH, and 0.2 mM dithiothreitol, pH 7.5. Protein concentrations were determined by a BCA Protein Assay kit (Pierce Chemical).
Actin Binding Assay
Rabbit muscle actin was prepared as described previously (Pardee and Spudich, 1982), and its binding with the kettin fragments was examined by a copelleting assay as described previously (Mohri and Ono, 2003; Mohri et al., 2004) with modifications. GST-KETN-CT1, GST-KETN-CT2, or GST-KETN-CT3 (all 0-30 μM0 was incubated with or without 5 μM F-actin in a buffer containing 0.1 M KCl, 2 mM MgCl2, 1 mM dithiothreitol, and 20 mM HEPES-NaOH, pH 7.5, for 30 min at room temperature and ultracentrifuged at 80,000 rpm for 20 min in a Beckman TLA-100 rotor. The supernatants and pellets were adjusted to the same volumes and analyzed by SDS-PAGE. Gels were stained with Coomassie brilliant blue R-250 (National Diagnostics, Atlanta, GA), scanned by a UMAX Powerlook III scanner at 300 dpi, and the band intensity was quantified by Scion Image Beta 4.02 (Scion, Frederick, MD). To quantify the amounts of the kettin fragments that bound to actin, the amounts of nonspecific sedimentation of the kettin fragments were determined from control experiments without F-actin and subtracted from the data of the assays with actin.
RNA Interference Experiments
Nematodes were treated with RNAi for ketn-1 by feeding E. coli expressing double-stranded RNA under the conditions as described previously (Ono and Ono, 2002). The RNAi clone for ketn-1 (V-2F01) was obtained from the C. elegans RNAi library from Geneservice (Cambridge, United Kingdom) (Kamath et al., 2003). Control experiments were performed with the E. coli strain HT115 (DE3) that was transformed with an empty RNAi vector L4440 (Timmons and Fire, 1998; Timmons et al., 2001). Phenotypes were analyzed in their F1 generation. Staining of worms with tetramethylrhodamine-phalloidin was performed as described previously (Ono, 2001). For tetramisole treatment, worms were harvested by M9 buffer, washed once with M9 buffer, and incubated in 10 ml M9 buffer with or without 0.01% tetramisole (MP Biomedicals) for 1 h at room temperature on a nutator. To determine brood size, F1 RNAi-treated worms were isolated at the L4 larval stage in individual plates, and the number of progeny was counted by removing the progeny from the plates until the worms cease producing progeny.
Protein Electrophoresis and Western Blot
Fifty adult worms were lysed in 20 μl of SDS-lysis buffer (2% SDS, 80 mM Tris-HCl, 5% β-mercaptoethanol, 15% glycerol, and 0.05% bromophenol blue, pH 6.8), heated at 97°C for 2 min, homogenized by brief sonication, and heated again at 97°C for 2 min. The samples were resolved by SDS-PAGE using a 4 or 5% acrylamide gel and transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore, Billerica, MA) using a Genie Blotter (Idea Scientific, Minneapolis, MN). The membrane was blocked in 5% nonfat milk in PBS containing 0.1% Tween 20 for 30 min and incubated with MH44 (1/2000-diluted ascites fluid) for 1 h followed by treatment with peroxidase-labeled goat anti-mouse IgG (Pierce Chemical). The reactivity was detected with a SuperSignal chemiluminescence reagent (Pierce Chemical). The membrane was treated with a buffer containing 2% SDS, 100 mM β-mercaptoethanol, and 62.5 mM Tris-HCl, pH 6.8, at 50°C for 30 min to remove bound probes, and reprobed with mouse monoclonal anti-myoA antibody (5-6) as a loading control.
RESULTS
Identification of the Antigen for the Monoclonal Antibody MH44 as C. elegans Kettin
Francis and Waterston (1985) generated the mAb MH44 and reported that its antigen was a 400- to 440-kDa protein and localized to the I-bands in C. elegans body wall muscle. We confirmed by Western blot that MH44 specifically reacted with this high-molecular-weight protein with very weak reactivity with smaller proteins that could be proteolytic products or unknown splice variants of the major band (Ono et al., 2006). Although MH44 has been used as a marker for the I-bands, the MH44 antigen had not been molecularly identified. To determine the molecular nature of the MH44 antigen, we isolated this protein from the crude thin filament fraction by immunoprecipitation (Figure 1). In three independent experiments, no high-molecular-weight proteins of 400-440 kDa were recovered. Instead, two 60- to 62-kDa proteins were reproducibly precipitated by MH44 (Figure 1, lane 1). We determined partial peptide sequence of these proteins by LC-MS/MS analysis and identified 27 tryptic peptides (Supplemental Table 2) that corresponded to the sequence of a putative C. elegans protein F54E2.3 (GenBank/European Molecular Biology Laboratory [EMBL]/DNA Data Bank of Japan [DDBJ] accession no. AAC78200) with a predicted Mr of 500 K, suggesting that the isolated proteins were proteolytic fragments of this large protein.
Two previous studies have reported that F54E2.3 is an orthologue of Drosophila kettin, a large Ig-repeat protein in striated muscle (Hakeda et al., 2000; Kolmerer et al., 2000). However, its gene products have been deduced from computer-predicted exon–intron structures and only partial cDNA sequences near the 3′ end of the gene have been analyzed by the Expressed Sequence Tag project. Therefore, we determined the full-length cDNA sequence (13.6 kb) of the F54E2.3 gene product from eight contiguous cDNA fragments (Supplemental Table 1) and found that this gene indeed encodes a kettin-like protein that consists of 4250 amino acids with calculated molecular weight of 471,704 (GenBank/EMBL/DDBJ accession no. AY819766).
Ce-kettin is composed of 31 Ig-repeats (Figure 2, gray region) and a unique kettin-specific sequence that is located between the 27th and 28th repeats (Figure 2). All the Ig-repeats have been accurately predicted by Kolmerer et al., (2000), and they are separated by weakly conserved linker sequences. Previous genome-wide analyses of Ig-repeat proteins in C. elegans did not annotate the Ce-kettin gene correctly (Teichmann and Chothia, 2000; Vogel et al., 2003) due to lack of information on the cDNA sequence. In addition, this gene has been predicted to generate a protein with a prion-like-Q/N-rich (PQN) domain at the N terminus and designated as pqn-43 in the WormBase (Chen et al., 2005) and the sequence database (GenBank/EMBL/DDBJ accession no. NP_503758). However, our cDNA sequence does not contain the PQN sequence in the open reading frame. Therefore, we propose that ketn-1 is an appropriate designation for the Ce-kettin gene.
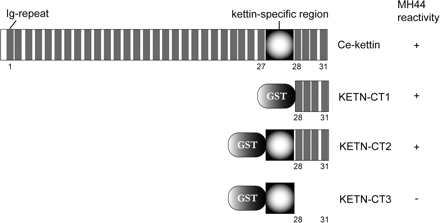
Figure 2.
Domain structure of Ce-kettin. Ig domains are shown in gray. The C-terminal fragments KETN-CT1, KETN-CT2, and KETN-CT3 were bacterially produced as fusion proteins with GST and used in in vitro assays in Figure 6. These Ce-kettin fragments were also tested for reactivity with MH44 on Western blot. The reactivity (+ or −) is shown on the right.
We found that MH44 strongly reacted with bacterially expressed C-terminal fragments of this protein, KETN-CT1 (residues 3830-4250) and KETN-CT2 (residues 3554-4250) but not with KETN-CT3 (residues 3554-3831) on Western blot (Figure 2), indicating that the epitope of MH44 is present within the four C-terminal Ig-repeats. In support of these data, the tryptic peptides of the MH44 precipitates correspond to the C-terminal region of this sequence (Supplemental Table 2).
Localization of Ce-Kettin to the Thin Filaments in Striated and Nonstriated Muscle
MH44 was previously shown to label the entire I-bands in body wall muscle (Francis and Waterston, 1985). We reexamined intracellular localization of Ce-kettin by double or triple staining with other cytoskeletal markers and found that it localized to a portion, not the entire length, of the I-bands (Figure 3). Triple staining of body wall muscle for actin (thin filaments), Ce-kettin (MH44), and myosin heavy chain (thick filaments) confirmed that Ce-kettin is associated with the thin filaments (Figure 3, A, D, G, and J). However, Ce-kettin localized to a ladder-like pattern and its localization was limited to a narrow region in the middle of the bands of actin (Figure 3, compare D–F for Ce-kettin with A–C for actin). Triple staining for actin, Ce-kettin, and vinculin (dense bodies) showed that Ce-kettin has some overlap with vinculin, but the center of the dense bodies was devoid of Ce-kettin (Figure 3, B, E, H, and K). In addition, comparison with the location of tropomyosin showed that tropomyosin localized to the outer region on the thin filaments with minimal overlap with Ce-kettin (Figure 3, C, F, I, and L). These results indicate that Ce-kettin is associated with a portion of the thin filaments near the dense bodies.
Our results on the Ce-kettin localization differ from the report by Kolmerer et al. (2000), which demonstrated that an antibody against insect kettin stained the dense bodies. Therefore, we reevaluated the specificity of the mAb MAC155 (provided by Belinda Bullard, EMBL, Heidelberg, Germany) against insect kettin that was used in Kolmerer et al. (2000). By Western blot of total C. elegans extracts, we found that MAC155 reacted with a 200-kDa protein but not with proteins of 400-500 kDa that were recognized by MH44 (our unpublished data). This result suggests that MAC155 has other reactivities with an unknown component of the dense bodies other than kettin.
In addition, Ce-kettin was expressed in nonstriated muscles, including the pharynx, the vulva, and the myoepithelial sheath of the proximal ovary (Figure 4). In particular, Ce-kettin was associated with the actin filaments in spots adjacent to the dense bodies (Figure 4, A–D) in the ovarian myoepithelial sheath that expresses the same myosin heavy chain isoform as the body wall muscle (Ardizzi and Epstein, 1987) and uses tropomyosin and troponin for regulation of contraction (Ono and Ono, 2004). Differential localization of tropomyosin and Ce-kettin on the actin filaments was more clearly observed in this nonstriated muscle (Figure 4, E–H). Tropomyosin was associated with the entire length of the actin filaments (Figure 4G), whereas Ce-kettin localized to spots that are associated with the actin filaments (Figure 4F). Observation at a higher magnification revealed that tropomyosin is not detected where Ce-kettin is located (Figure 4, I–L, arrows). These localization patterns suggest that tropomyosin and Ce-kettin bind to actin in a mutually exclusive manner.
Figure 4.
Localization of Ce-kettin in ovarian nonstriated muscle. Myoepithelial sheath of the proximal ovary was stained for actin (A, E, and I), Ce-kettin (B, F, and J), and vinculin (C) or tropomyosin (G and K). Merged images of actin (red), Ce-kettin (green), and vinculin or tropomyosin (blue) are shown in D, H, and L. High-magnification images in I–L show a representative area where differential localization of Ce-kettin and tropomyosin is clearly observed (arrows). Bars, 10 μm (A–H) and 5 μm (I–L).
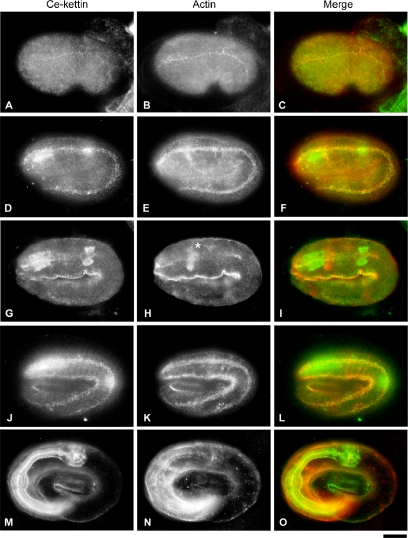
During development, expression of Ce-kettin in body wall muscle was detected as early as the comma stage (290 min after the first cleavage) (Figure 5A), and it colocalized with actin at the early stage of myofibril assembly (Figure 5, B and C). At the twofold stage (430 min), in addition to the expression in the body wall muscle (Figure 5, D–F), strong expression of Ce-kettin in the pharynx was detected (Figure 5G) before robust assembly of actin was initiated (Figure 5, G–I). At the threefold stage (520 min), expression of Ce-kettin in the body wall muscle (Figure 5, J–L) and the pharynx (Figure 5, M–O) became stronger, and its localization to the contractile apparatuses was more remarkable. These results indicate that Ce-kettin is one of the earliest proteins to be assembled into the myofibrils.
Figure 5.
Localization of Ce-kettin in embryos. Embryos at the comma (A–C), twofold (D–I), and threefold (J–O) stages were stained for Ce-kettin (A, D, G, J, and M) and actin (B, E, H, K, and N). Merged images of Ce-kettin (green) and actin (red) are shown in C, F, I, L, and O. Focus was adjusted for the body wall muscle in D–F and J–L or the pharynx in G–I and M–O. Actin-rich structure in H (asterisk) is the nerve ring. Bar, 10 μm.
Binding of the C-Terminal Kettin-specific and Ig Domains to Filamentous Actin
We reasoned that an actin binding site is present in the C terminus of Ce-kettin, because a C-terminal fragment of Ce-kettin was fractionated in the crude thin filament preparation (Figure 1). To test actin binding activity of the Ce-kettin C-terminal domains, we purified bacterially expressed C-terminal fragments of Ce-kettin and examined their ability to interact with filamentous actin. We prepared three Ce-kettin fragments as fusion proteins with GST. KETN-CT1 corresponds to residues 3830-4250 containing the four C-terminal Ig-repeats (repeats 28-31). KETN-CT2 (residues 3554-4250) has the kettin-specific sequence with unknown function in addition to Ig-repeats 28-31. KETN-CT3 (residues 3554-3831) has only the kettin-specific sequence (Figure 2).
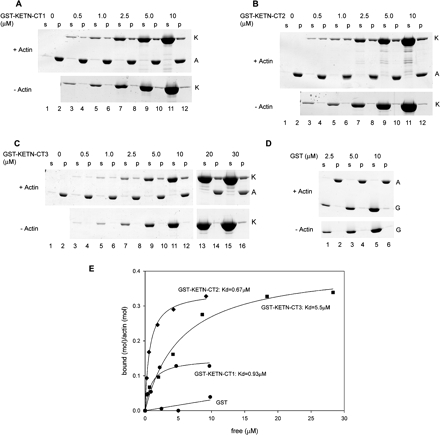
KETN-CT1, KETN-CT2, and KETN-CT3 bound to F-actin with different affinity and stoichiometry (Figure 6). In cosedimentation assays, significant portions of GST-KETN-CT1, GST-KETN-CT2, or GST-KETN-CT3, cosedimented with F-actin (Figure 6, A–C). GST alone did not cosediment with actin under the same conditions, and nonspecific trapping of GST in the pellets was very minor (Figure 6D) (Mohri et al., 2004). Quantitative analysis of the data showed that KETN-CT2 bound to F-actin with higher affinity (Kd = 0.68 ± 0.062 μM) than KETN-CT1 (Kd = 0.93 ± 0.36 μM) or KETN-CT3 (Kd = 5.5 μM ± 1.2 μM). Also interestingly, binding was saturated for KETN-CT2 or KETN-CT3 at higher stoichiometry (mol KETN-CT2/mol actin = 0.34 ± 0.0080 or ∼3:1; mol KETN-CT3/mol actin = 0.42 ± 0.029 or ∼2.4:1) than for KETN-CT1 (mol KETN-CT1/mol actin = 0.15 ± 0.016 or ∼6.7:1) (Figure 6E). These results suggest that the kettin-specific region augments the actin binding activity of the Ig-repeats.
Figure 6.
Binding of the C-terminal fragments of Ce-kettin with actin filaments in vitro. Various concentrations of GST-KETN-CT1 (A), GST-KETN-CT2 (B), GST-KETN-CT3 (C), or GST (D) were incubated with or without 5 μM F-actin, and their interactions were examined by copelleting assays. The samples were fractionated into supernatants (s) and pellets (p) and analyzed by SDS-PAGE. Positions of actin (A), Ce-kettin fragments (K), and GST (G) are indicated on the right of the gels. The data were quantified by densitometric analysis (E).
RNA Interference of Ce-Kettin Causes Disorganization of Actin Filaments in a Contraction-dependent Manner
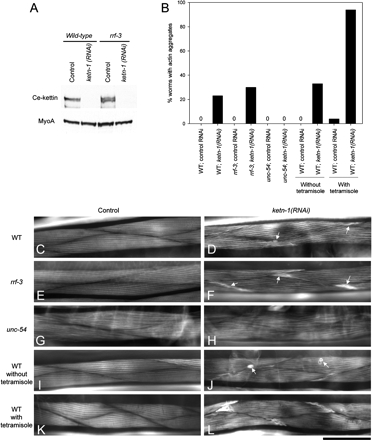
To examine the in vivo function of Ce-kettin, Ce-kettin was knocked down by RNAi and the resultant phenotype examined. RNAi of Ce-kettin [ketn-1(RNAi)] by the feeding method effectively reduced Ce-kettin to undetectable levels on Western blot in wild-type background (Figure 7A). The ketn-1(RNAi)–treated worms moved slightly slower than control worms (our unpublished data), suggesting that ketn-1(RNAi) caused a defect in body wall muscle contractility. Staining of the muscle actin filaments with fluorescently labeled phalloidin revealed that the ketn-1(RNAi)–treated worms had alterations in the actin filaments in the body wall muscle (Figure 7, C–F). Small round actin aggregates or unusual accumulations of actin were detected in 23% (n = 100) of the ketn-1(RNAi)–treated worms (Figure 7D, arrows), whereas no worms exhibited these phenotypes in control experiments (n = 100) (Figure 7, B and C). Some genes are not efficiently affected by RNAi in wild-type background but are susceptible to RNAi in an rrf-3 mutant background (Simmer et al., 2002, 2003). However, in the rrf-3 background, the phenotypes were only slightly enhanced. The rrf-3; ketn-1(RNAi) worms also had small round actin aggregates or unusual accumulations of actin in the body wall muscle in 30% of treated animals (n = 100) (Figure 7, B and F), whereas no control rrf-3 worms exhibited these phenotypes (n = 100) (Figure 7, B and E). Therefore, the low penetrance of the ketn-1(RNAi) phenotype is not simply due to the efficiency of RNAi. Actin organization in the pharynx and ovarian nonstriated muscle was not significantly altered by the RNAi treatments (our unpublished data). However, brood size was reduced by ketn-1(RNAi) in the rrf-3 background [83.4 ± 18 in control RNAi and 47.2 ± 6.8 in ketn-1(RNAi), n = 5], but not in wild-type background [280 ± 30 in control RNAi and 281 ± 28 in ketn-1(RNAi), n = 5], suggesting that the reproductive system, in particular the activity of the ovarian myoepithelial sheath, uterine muscle, or vulval muscle, might be partially impaired. In the control experiments, rrf-3 mutants produced fewer progeny than wild type, suggesting that rrf-3 is less tolerant to functional alterations in the reproductive system. These results strongly suggest that Ce-kettin is involved in assembly and/or maintenance of actin filaments in muscle cells.
Figure 7.
Disorganization of actin filaments in adult body wall muscle by RNA interference of Ce-kettin. (A) Protein levels of Ce-kettin in control or ketn-1 (RNAi) worms (50 worms per lane) in wild-type or rrf-3 background were examined by Western blot with MH44. The same membrane was reprobed with anti-myoA antibody to confirm equal loading of the proteins. (B) Frequency of formation of actin aggregates. Worms were stained with tetramethylrhodamine-phalloidin, and percentages of adult worms (n = 100) with actin aggregates in the body wall muscle were determined. (C–H) Actin organization in the body wall muscle of control RNAi (C, E, G, I, and K) or ketn-1 (RNAi) (D, F, H, J, and L) worms in wild-type (C, D, and I–L), rrf-3 (E and F), or unc-54 (G and H) backgrounds was examined by staining with tetramethylrhodamine-phalloidin. Worms in I–L were incubated in M9 buffer for 1 h in the absence (I and J) or presence (K and L) of 0.01% tetramisole before fixation. In ketn-1 (RNAi) worms, small round aggregates and unusual actin accumulations at the edges of muscle cells (arrows in D, F, and J) were detected, and this phenotype was enhanced by tetramisole-induced hypercontraction (L). Bar, 50 μm.
Although disorganized actin filaments after ketn-1(RNAi) were detected at low penetrance, we found that this phenotype is sensitive to the state of muscle contraction. unc-54(s95) is a mutation in the head domain of the UNC-54 myosin heavy chain that reduces muscle contraction without affecting overall structure of the myofibrils (Moerman et al., 1982). In the unc-54(s95) homozygous background, ketn-1(RNAi) did not cause formation of actin aggregates (Figure 7, B and H), suggesting that reduced muscle contraction suppressed disruption of actin filaments. In contrast, when hypercontraction was induced by tetramisole, an agonist of acetylcholine receptors, larger actin aggregates were formed more frequently at the cell periphery in 94% (n = 100) of the ketn-1(RNAi) worms (Figure 7B and compare Figure 7, J and L). These results suggest that disorganization of actin filaments in the ketn-1(RNAi) worms is caused by mechanical disruption of the actin filaments during contraction and that Ce-kettin provides stability to the myofibrils.
Ce-Kettin Regulates Localization of Tropomyosin and α-Actinin
In vitro studies have shown that insect kettin binds to α-actinin, whereas it competes with tropomyosin for binding to actin (Lakey et al., 1993; van Straaten et al., 1999), but their functional relationship in vivo is not understood. We examined how Ce-kettin interacts with tropomyosin and α-actinin in the C. elegans body wall muscle. As shown in Figure 3, tropomyosin is normally localized to the outer region on the thin filaments. In worms treated with control RNAi, the normal localization of tropomyosin as double lines was observed (Figure 8A, a). However, in ketn-1(RNAi) worms, tropomyosin also localized to regions where Ce-kettin normally localizes, resulting in the ladder-like localization (Figure 8A, d). This result strongly suggests that Ce-kettin and tropomyosin compete for binding to actin in vivo.
Normal organization of α-actinin was dependent on Ce-kettin. α-Actinin was deposited at the dense bodies in control worms (Figure 8A, g), whereas, in ketn-1(RNAi) worms, the pattern of α-actinin was disturbed and the shape of the accumulations of α-actinin became highly irregular (Figure 8A, j). In contrast, the pattern of vinculin at the dense bodies was only slightly affected by ketn-1(RNAi) (Figure 8A, compare m and p). Vinculin is at the base of the dense bodies near the plasma membrane, whereas α-actinin localizes at the cytoplasmic portion (Francis and Waterston, 1985). Therefore, Ce-kettin is specifically involved in the organization of the cytoplasmic region of the dense bodies. The striated pattern of the myoA myosin heavy chain was unaffected by ketn-1(RNAi) (Figure 8A, compare s and v), supporting the specific function of Ce-kettin in the thin filaments and the dense bodies.
The functional relationship between Ce-kettin and α-actinin was further examined in an α-actinin mutant (Figure 8, B and C). atn-1(ok84) has a 1.1-kb deletion in the single gene for α-actinin atn-1 (Barstead et al., 1991), is homozygous viable, and shows only minor alterations in the striated arrangement of actin filaments and small aggregations of actin at the edges of the body wall muscle cells (Moulder and Barstead, personal communication) (Figure 8B, b). Position of the atn-1(ok84) deletion suggests that a truncated α-actinin protein containing the N-terminal actin binding domain could be expressed, but the presence of such a truncated protein has not been confirmed (Moulder and Barstead, personal communication). In the α-actinin mutant, Ce-kettin colocalized with actin to the striated myofibrils, but the pattern of Ce-kettin was continuous (Figure 8B, a) rather than ladder-like (Figure 3). This is probably because the cytoplasmic portion of the dense bodies may not be well defined, in which α-actinin is the major component. Ce-kettin was not associated with the actin aggregates in the α-actinin mutant (Figure 8B, a–c). Furthermore, when the α-actinin mutant was treated with ketn-1(RNAi), disorganization of the actin filaments was enhanced and larger actin aggregates were often detected (Figure 8C, compare a and b). These results suggest that Ce-kettin and α-actinin cooperate to organize the dense bodies and the thin filaments.
DISCUSSION
In this study, we identified the C. elegans kettin gene and the gene product as the antigen for the mAb MH44 that has been used as a marker for the I-bands in the C. elegans muscle. Comparisons of the localization pattern of Ce-kettin with that of other thin filament proteins revealed that Ce-kettin is associated with the thin filaments near the dense bodies, the homologous structures to the Z-discs in cross-striated muscles. Biochemical analysis showed that the four C-terminal Ig domains of Ce-kettin directly bind to actin filaments in vitro and that the kettin-specific sequence immediately N-terminal to these Ig domains enhances its actin binding activity. RNAi of Ce-kettin caused disorganization of actin filaments in body wall muscle. This phenotype was suppressed by reducing muscle contraction, but enhanced by hypercontraction. RNAi of Ce-kettin also disturbed organization of α-actinin at the dense bodies and enhanced the actin disorganization phenotype in an α-actinin mutant. These results suggest that Ce-kettin is important for maintaining organized architecture of the actin filaments and dense bodies and provides mechanical stability to the contractile apparatuses in muscle cells.
Localization of Ce-kettin on the thin filaments was proximal to the dense bodies and was similar to that of kettins from insects (Lakey et al., 1990, 1993; van Straaten et al., 1999) and crayfish (Maki et al., 1995; Fukuzawa et al., 2001), which are localized to the side of the Z-discs. In addition, the N terminus of insect kettin is located within the Z-discs (van Straaten et al., 1999). Because MH44 recognizes the C terminus of Ce-kettin, the N terminus of Ce-kettin is likely to be embedded in the dense bodies. This limited localization of kettin is probably due to competition with tropomyosin for binding to actin (van Straaten et al., 1999), because Ce-tropomyosin is more distally localized on the thin filaments (Figures 3 and 4). We also found that Ce-kettin localizes to the proximal region of the thin filaments in the ovarian nonstriated muscle (Figure 4). This is the first demonstration of kettin as a component of nonstriated myofibrils. Although the C. elegans ovarian muscle is nonstriated, it shares some physiological properties with striated muscle and uses the tropomyosin–troponin system for its contraction (Myers et al., 1996; Ono and Ono, 2004). Therefore, Ce-kettin may play common cellular roles in both striated and nonstriated muscles.
We showed that the four C-terminal Ig-repeats directly bind to actin filaments and that the adjacent kettin-specific region enhances its actin binding. Previously, the full-length kettin has been demonstrated to bind to actin filaments with high affinity (Maki et al., 1995; van Straaten et al., 1999), and the stoichiometry has suggested that one Ig-repeat may bind one actin monomer in the filament (van Straaten et al., 1999). This was also supported by the report that a single Ig-repeat of kettin is capable of binding to actin filaments (Lakey et al., 1993). However, our results show that the four C-terminal Ig-repeats (KETN-CT1) bind to actin at lower stoichiometry than one repeat per one actin monomer, and, interestingly, that the adjacent non-Ig region augments both affinity and stoichiometry for binding to actin. This suggests that the Ig-repeats and non-Ig region of the molecule cooperatively bind to actin filaments. Similarly, a single Ig-domain of myotilin is sufficient for actin binding, but flanking non-Ig sequences also play important roles in actin binding (von Nandelstadh et al., 2005). The full-length kettin binds to actin with a nanomolar affinity (van Straaten et al., 1999), whereas our C-terminal fragments binds to actin with a micromolar affinity, suggesting that other Ig-repeats and non-Ig sequence may cooperate to achieve tight binding to actin filaments with proper stoichiometry.
The Ce-kettin gene ketn-1 is independent of genes coding for other connectin/titin-related Ig-repeat proteins. ketn-1 (RNAi) caused disorganization of actin filaments in muscle. This phenotype is different from the defects in regulation of muscle contraction in the mutants of unc-22 coding for twitchin (Benian et al., 1993) and the defects in M-line assembly in the mutants of unc-89 coding for UNC-89, a protein similar to obscurin (Benian et al., 1996; Small et al., 2004). Therefore, the RNAi phenotype strongly suggests that Ce-kettin has specific function in assembly and/or maintenance of actin filaments in muscle. However, the phenotype is relatively weak with low penetrance under normal culture conditions, and the extent of actin disorganization was not as drastic as that observed in mutants of actin depolymerizing factor/cofilin (UNC-60B) (Ono et al., 1999, 2003), actin-interacting protein 1 (UNC-78) (Ono, 2001), or a calponin-like protein (UNC-87) (Goetinck and Waterston, 1994a,b), or in worms that are treated with RNAi of tropomyosin (Ono and Ono, 2002). This suggests that kettin might have partially redundant function with other muscle proteins. Ce-titins are candidates for functionally redundant proteins with Ce-kettin. The N-terminal portion of Ce-titins localizes to the I-bands (Flaherty et al., 2002), but mutant or RNAi phenotypes for the Ce-titin gene have not been clearly defined. Vertebrate connectin/titin directly binds to actin filaments (Kimura et al., 1984; Maruyama et al., 1987) via the PEVK domain in the I-band part (Kulke et al., 2001a; Yamasaki et al., 2001; Linke et al., 2002; Nagy et al., 2004) and is involved in maintenance of the thin filament structure (Linke et al., 1999). Thus, vertebrate connectin/titin may have a similar actin regulatory function to kettin by using a different domain from kettin for binding to actin filaments.
We demonstrated that the organization of α-actinin at the dense bodies was disturbed by depletion of Ce-kettin. This suggests that Ce-kettin may directly interact with α-actinin and regulate its assembly, or that Ce-kettin may organize actin filaments near the barbed ends, possibly by bundling, and facilitate efficient association with the dense bodies. α-Actinin and insect kettin have been shown to bind to actin simultaneously (van Straaten et al., 1999). Thus, these two proteins may cooperate to strengthen the anchorage between thin filaments and dense bodies and provide mechanical stability. Insect kettin also binds to myosin and is involved in stiffness of myofibrils (Kulke et al., 2001b). Although we have not examined interaction between Ce-kettin and myosin, it is also a possible mechanism to stabilize the myofibrils.
Kettin is a unique member of the connectin/titin family of Ig-repeat proteins and is found only in invertebrates (Bullard et al., 2002). However, kettin might be functionally homologous to palladin (Parast and Otey, 2000), myopalladin (Bang et al., 2001), and myotilin (Salmikangas et al., 1999), which have two to five Ig-repeats and localize to the Z-lines of vertebrate striated muscle. These proteins are critical for actin filament reorganization in nonmuscle cells (Boukhelifa et al., 2001, 2003; Salmikangas et al., 2003; Otey et al., 2005) and myofibril assembly in muscle cells (Bang et al., 2001; Salmikangas et al., 2003; Otey et al., 2005). In particular, myotilin directly binds to actin filaments, and its mutations in the human gene are associated with limb girdle muscular dystrophy 1A (Salmikangas et al., 1999) and myofibrillar myopathy (Selcen and Engel, 2004), which are termed myotilinopathies (Goebel, 2005; Olive et al., 2005). Therefore, C. elegans could be an excellent model to study functions of actin binding proteins with Ig-repeats in muscle and their interaction with other myofibrillar proteins.
Supplementary Material
ACKNOWLEDGMENTS
We thank Belinda Bullard and Taylor Allen for communicating studies on C. elegans kettin and helpful discussion; Sumiko Kimura for helpful discussion; Guy Benian for comments on the manuscript; Henry Epstein for anti-myoA antibody; Gary Moulder and Robert Barstead for an α-actinin mutant; and Pamela Hoppe for MH24, MH40, and MH44. Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institute of Health National Center for Research Resources. This work was supported by National Institutes of Health Grant R01 AR48615 (to S. O.).
Abbreviations used:
- GST
glutathione S-transferase
- Ig
immunoglobulin-like
- RNAi
RNA interference.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-02-0114) on April 5, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Ardizzi J. P., Epstein H. F. Immunochemical localization of myosin heavy chain isoforms and paramyosin in developmentally and structurally diverse muscle cell types of the nematode Caenorhabditis elegans. J. Cell Biol. 1987;105:2763–2770. doi: 10.1083/jcb.105.6.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayme-Southgate A., Bounaix C., Riebe T. E., Southgate R. Assembly of the giant protein projectin during myofibrillogenesis in Drosophila indirect flight muscles. BMC Cell Biol. 2004;5:17. doi: 10.1186/1471-2121-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang M. L., Mudry R. E., McElhinny A. S., Trombitas K., Geach A. J., Yamasaki R., Sorimachi H., Granzier H., Gregorio C. C., Labeit S. Myopalladin, a novel 145-kilodalton sarcomeric protein with multiple roles in Z-disc and I-band protein assemblies. J. Cell Biol. 2001;153:413–427. doi: 10.1083/jcb.153.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barstead R. J., Kleiman L., Waterston R. H. Cloning, sequencing, and mapping of an alpha-actinin gene from the nematode Caenorhabditis elegans. Cell Motil. Cytoskeleton. 1991;20:69–78. doi: 10.1002/cm.970200108. [DOI] [PubMed] [Google Scholar]
- Benian G. M., L'Hernault S. W., Morris M. E. Additional sequence complexity in the muscle gene, unc-22, and its encoded protein, twitchin, of Caenorhabditis elegans. Genetics. 1993;134:1097–1104. doi: 10.1093/genetics/134.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benian G. M., Tinley T. L., Tang X., Borodovsky M. The Caenorhabditis elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J. Cell Biol. 1996;132:835–848. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukhelifa M., Hwang S. J., Valtschanoff J. G., Meeker R. B., Rustioni A., Otey C. A. A critical role for palladin in astrocyte morphology and response to injury. Mol. Cell Neurosci. 2003;23:661–668. doi: 10.1016/s1044-7431(03)00127-1. [DOI] [PubMed] [Google Scholar]
- Boukhelifa M., Parast M. M., Valtschanoff J. G., LaMantia A. S., Meeker R. B., Otey C. A. A role for the cytoskeleton-associated protein palladin in neurite outgrowth. Mol. Biol. Cell. 2001;12:2721–2729. doi: 10.1091/mbc.12.9.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard B., Burkart C., Labeit S., Leonard K. The function of elastic proteins in the oscillatory contraction of insect flight muscle. J. Muscle Res. Cell Motil. 2006 doi: 10.1007/s10974-005-9032-7. (in press) [DOI] [PubMed] [Google Scholar]
- Bullard B., Goulding D., Ferguson C., Leonard K. Links in the chain: the contribution of kettin to the elasticity of insect muscles. Adv. Exp. Med. Biol. 2000;481:207–218. doi: 10.1007/978-1-4615-4267-4_12. discussion 219–220. [DOI] [PubMed]
- Bullard B., Linke W. A., Leonard K. Varieties of elastic protein in invertebrate muscles. J. Muscle Res. Cell Motil. 2002;23:435–447. doi: 10.1023/a:1023454305437. [DOI] [PubMed] [Google Scholar]
- Chen N., et al. WormBase: a comprehensive data resource for Caenorhabditis biology and genomics. Nucleic Acids Res. 2005;33:D383–389. doi: 10.1093/nar/gki066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. A., McElhinny A. S., Beckerle M. C., Gregorio C. C. Striated muscle cytoarchitecture: an intricate web of form and function. Annu. Rev. Cell Dev. Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- Epstein H. F., Casey D. L., Ortiz I. Myosin and paramyosin of Caenorhabditis elegans embryos assemble into nascent structures distinct from thick filaments and multi-filament assemblages. J. Cell Biol. 1993;122:845–858. doi: 10.1083/jcb.122.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney M., Ruvkun G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- Flaherty D. B., Gernert K. M., Shmeleva N., Tang X., Mercer K. B., Borodovsky M., Benian G. M. Titins in C. elegans with unusual features: coiled-coil domains, novel regulation of kinase activity and two new possible elastic regions. J. Mol. Biol. 2002;323:533–549. doi: 10.1016/s0022-2836(02)00970-1. [DOI] [PubMed] [Google Scholar]
- Francis G. R., Waterston R. H. Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol. 1985;101:1532–1549. doi: 10.1083/jcb.101.4.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa A., Shimamura J., Takemori S., Kanzawa N., Yamaguchi M., Sun P., Maruyama K., Kimura S. Invertebrate connectin spans as much as 3.5 micron in the giant sarcomeres of crayfish claw muscle. EMBO J. 2001;20:4826–4835. doi: 10.1093/emboj/20.17.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautel M., Mues A., Young P. Control of sarcomeric assembly: the flow of information on titin. Rev. Physiol. Biochem. Pharmacol. 1999;138:97–137. doi: 10.1007/BFb0119625. [DOI] [PubMed] [Google Scholar]
- Goebel H. H. Congenital myopathies in the new millennium. J. Child Neurol. 2005;20:94–101. doi: 10.1177/08830738050200020201. [DOI] [PubMed] [Google Scholar]
- Goetinck S., Waterston R. H. The Caenorhabditis elegans muscle-affecting gene unc-87 encodes a novel thin filament-associated protein. J. Cell Biol. 1994a;127:79–93. doi: 10.1083/jcb.127.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetinck S., Waterston R. H. The Caenorhabditis elegans UNC-87 protein is essential for maintenance, but not assembly, of bodywall muscle. J. Cell Biol. 1994b;127:71–78. doi: 10.1083/jcb.127.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzier H., Labeit D., Wu Y., Labeit S. Titin as a modular spring: emerging mechanisms for elasticity control by titin in cardiac physiology and pathophysiology. J. Muscle Res. Cell Motil. 2002;23:457–471. doi: 10.1023/a:1023458406346. [DOI] [PubMed] [Google Scholar]
- Gregorio C. C., Antin P. B. To the heart of myofibril assembly. Trends Cell Biol. 2000;10:355–362. doi: 10.1016/s0962-8924(00)01793-1. [DOI] [PubMed] [Google Scholar]
- Gregorio C. C., Granzier H., Sorimachi H., Labeit S. Muscle assembly: a titanic achievement? Curr. Opin. Cell Biol. 1999;11:18–25. doi: 10.1016/s0955-0674(99)80003-9. [DOI] [PubMed] [Google Scholar]
- Hakeda S., Endo S., Saigo K. Requirements of Kettin, a giant muscle protein highly conserved in overall structure in evolution, for normal muscle function, viability, and flight activity of Drosophila. J. Cell Biol. 2000;148:101–114. doi: 10.1083/jcb.148.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kimura S., Maruyama K., Huang Y. P. Interactions of muscle beta-connectin with myosin, actin, and actomyosin at low ionic strengths. J. Biochem. 1984;96:499–506. doi: 10.1093/oxfordjournals.jbchem.a134862. [DOI] [PubMed] [Google Scholar]
- Kolmerer B., et al. Sequence and expression of the kettin gene in Drosophila melanogaster and Caenorhabditis elegans. J. Mol. Biol. 2000;296:435–448. doi: 10.1006/jmbi.1999.3461. [DOI] [PubMed] [Google Scholar]
- Kulke M., Fujita-Becker S., Rostkova E., Neagoe C., Labeit D., Manstein D. J., Gautel M., Linke W. A. Interaction between PEVK-titin and actin filaments: origin of a viscous force component in cardiac myofibrils. Circ. Res. 2001a;89:874–881. doi: 10.1161/hh2201.099453. [DOI] [PubMed] [Google Scholar]
- Kulke M., Neagoe C., Kolmerer B., Minajeva A., Hinssen H., Bullard B., Linke W. A. Kettin, a major source of myofibrillar stiffness in Drosophila indirect flight muscle. J. Cell Biol. 2001b;154:1045–1057. doi: 10.1083/jcb.200104016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakey A., Ferguson C., Labeit S., Reedy M., Larkins A., Butcher G., Leonard K., Bullard B. Identification and localization of high molecular weight proteins in insect flight and leg muscle. EMBO J. 1990;9:3459–3467. doi: 10.1002/j.1460-2075.1990.tb07554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakey A., Labeit S., Gautel M., Ferguson C., Barlow D. P., Leonard K., Bullard B. Kettin, a large modular protein in the Z-disc of insect muscles. EMBO J. 1993;12:2863–2871. doi: 10.1002/j.1460-2075.1993.tb05948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke W. A., Kulke M., Li H., Fujita-Becker S., Neagoe C., Manstein D. J., Gautel M., Fernandez J. M. PEVK domain of titin: an entropic spring with actin-binding properties. J. Struct. Biol. 2002;137:194–205. doi: 10.1006/jsbi.2002.4468. [DOI] [PubMed] [Google Scholar]
- Linke W. A., Rudy D. E., Centner T., Gautel M., Witt C., Labeit S., Gregorio C. C. I-band titin in cardiac muscle is a three-element molecular spring and is critical for maintaining thin filament structure. J. Cell Biol. 1999;146:631–644. doi: 10.1083/jcb.146.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlefield R., Fowler V. M. Defining actin filament length in striated muscle: rulers and caps or dynamic stability? Annu. Rev. Cell Dev. Biol. 1998;14:487–525. doi: 10.1146/annurev.cellbio.14.1.487. [DOI] [PubMed] [Google Scholar]
- Machado C., Andrew D. J. D-Titin: a giant protein with dual roles in chromosomes and muscles. J. Cell Biol. 2000;151:639–652. doi: 10.1083/jcb.151.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki S., Ohtani Y., Kimura S., Maruyama K. Isolation and characterization of a kettin-like protein from crayfish claw muscle. J. Muscle Res. Cell Motil. 1995;16:579–585. doi: 10.1007/BF00130239. [DOI] [PubMed] [Google Scholar]
- Maruyama K. Connectin/titin, giant elastic protein of muscle. FASEB J. 1997;11:341–345. doi: 10.1096/fasebj.11.5.9141500. [DOI] [PubMed] [Google Scholar]
- Maruyama K., Hu D. H., Suzuki T., Kimura S. Binding of actin filaments to connectin. J. Biochem. 1987;101:1339–1346. doi: 10.1093/oxfordjournals.jbchem.a122001. [DOI] [PubMed] [Google Scholar]
- Maruyama K., Kimura S. Connectin: from regular to giant sizes of sarcomeres. Adv. Exp. Med. Biol. 2000;481:25–33. doi: 10.1007/978-1-4615-4267-4_2. [DOI] [PubMed] [Google Scholar]
- Miller D. M., Ortiz I., Berliner G. C., Epstein H. F. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- Moerman D. G., Plurad S., Waterston R. H., Baillie D. L. Mutations in the unc-54 myosin heavy chain gene of Caenorhabditis elegans that alter contractility but not muscle structure. Cell. 1982;29:773–781. doi: 10.1016/0092-8674(82)90439-1. [DOI] [PubMed] [Google Scholar]
- Mohri K., Ono S. Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J. Cell Sci. 2003;116:4107–4118. doi: 10.1242/jcs.00717. [DOI] [PubMed] [Google Scholar]
- Mohri K., Vorobiev S., Fedorov A. A., Almo S. C., Ono S. Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 2004;279:31697–31707. doi: 10.1074/jbc.M403351200. [DOI] [PubMed] [Google Scholar]
- Myers C. D., Goh P. Y., Allen T. S., Bucher E. A., Bogaert T. Developmental genetic analysis of troponin T mutations in striated and nonstriated muscle cells of Caenorhabditis elegans. J. Cell Biol. 1996;132:1061–1077. doi: 10.1083/jcb.132.6.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Cacciafesta P., Grama L., Kengyel A., Malnasi-Csizmadia A., Kellermayer M. S. Differential actin binding along the PEVK domain of skeletal muscle titin. J. Cell Sci. 2004;117:5781–5789. doi: 10.1242/jcs.01501. [DOI] [PubMed] [Google Scholar]
- Obinata T. Contractile proteins and myofibrillogenesis. Int. Rev. Cytol. 1993;143:153–189. doi: 10.1016/s0074-7696(08)61875-6. [DOI] [PubMed] [Google Scholar]
- Obinata T., Nagaoka-Yasuda R., Ono S., Kusano K., Mohri K., Ohtaka Y., Yamashiro S., Okada K., Abe H. Low molecular-weight G-actin binding proteins involved in the regulation of actin assembly during myofibrillogenesis. Cell Struct. Funct. 1997;22:181–189. doi: 10.1247/csf.22.181. [DOI] [PubMed] [Google Scholar]
- Olive M., Goldfarb L. G., Shatunov A., Fischer D., Ferrer I. Myotilinopathy: refining the clinical and myopathological phenotype. Brain. 2005;128:2315–2326. doi: 10.1093/brain/awh576. [DOI] [PubMed] [Google Scholar]
- Ono S. Purification and biochemical characterization of actin from Caenorhabditis elegans: its difference from rabbit muscle actin in the interaction with nematode ADF/cofilin. Cell Motil. Cytoskeleton. 1999;43:128–136. doi: 10.1002/(SICI)1097-0169(1999)43:2<128::AID-CM4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Ono S. The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol. 2001;152:1313–1319. doi: 10.1083/jcb.152.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. Function of ADF/cofilin in muscle cells: an important regulator of actin cytoskeletal dynamics in myofibril assembly and muscle diseases. In: Fagan J., Davidson J. N., Shimizu N., editors. Recent Developments in Cell Research. vol. 1. Kerala, India: Research Signpost; 2003a. pp. 31–44. [Google Scholar]
- Ono S. Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1, new blades for twisted filaments. Biochemistry. 2003b;42:13363–13370. doi: 10.1021/bi034600x. [DOI] [PubMed] [Google Scholar]
- Ono S., Baillie D. L., Benian G. M. UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J. Cell Biol. 1999;145:491–502. doi: 10.1083/jcb.145.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., Mohri K., Ono K. Molecular and biochemical characterization of kettin in Caenorhabditis elegans. J. Muscle Res. Cell Motil. 2006 doi: 10.1007/s10974-005-9028-3. [DOI] [PubMed] [Google Scholar]
- Ono S., Ono K. Tropomyosin inhibits ADF/cofilin–dependent actin filament dynamics. J. Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K., Ono S. Tropomyosin and troponin are required for ovarian contraction in the Caenorhabditis elegans reproductive system. Mol. Biol. Cell. 2004;15:2782–2793. doi: 10.1091/mbc.E04-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K., Parast M., Alberico C., Benian G. M., Ono S. Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J. Cell Sci. 2003;116:2073–2085. doi: 10.1242/jcs.00421. [DOI] [PubMed] [Google Scholar]
- Otey C. A., Rachlin A., Moza M., Arneman D., Carpen O. The palladin/myotilin/myopalladin family of actin-associated scaffolds. Int. Rev. Cytol. 2005;246:31–58. doi: 10.1016/S0074-7696(05)46002-7. [DOI] [PubMed] [Google Scholar]
- Parast M. M., Otey C. A. Characterization of palladin, a novel protein localized to stress fibers and cell adhesions. J. Cell Biol. 2000;150:643–656. doi: 10.1083/jcb.150.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee J. D., Spudich J. A. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- Rose K. L., Winfrey V. P., Hoffman L. H., Hall D. H., Furuta T., Greenstein D. The POU gene ceh-18 promotes gonadal sheath cell differentiation and function required for meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 1997;192:59–77. doi: 10.1006/dbio.1997.8728. [DOI] [PubMed] [Google Scholar]
- Salmikangas P., Mykkanen O. M., Gronholm M., Heiska L., Kere J., Carpen O. Myotilin, a novel sarcomeric protein with two Ig-like domains, is encoded by a candidate gene for limb-girdle muscular dystrophy. Hum. Mol. Genet. 1999;8:1329–1336. doi: 10.1093/hmg/8.7.1329. [DOI] [PubMed] [Google Scholar]
- Salmikangas P., van der Ven P. F., Lalowski M., Taivainen A., Zhao F., Suila H., Schroder R., Lappalainen P., Furst D. O., Carpen O. Myotilin, the limb-girdle muscular dystrophy 1A (LGMD1A) protein, cross-links actin filaments and controls sarcomere assembly. Hum. Mol. Genet. 2003;12:189–203. doi: 10.1093/hmg/ddg020. [DOI] [PubMed] [Google Scholar]
- Selcen D., Engel A. G. Mutations in myotilin cause myofibrillar myopathy. Neurology. 2004;62:1363–1371. doi: 10.1212/01.wnl.0000123576.74801.75. [DOI] [PubMed] [Google Scholar]
- Simmer F., Moorman C., Van Der Linden A. M., Kuijk E., Van Den Berghe P. V., Kamath R., Fraser A. G., Ahringer J., Plasterk R. H. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S. P., Nonet M. L., Fire A., Ahringer J., Plasterk R. H. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Small T. M., Gernert K. M., Flaherty D. B., Mercer K. B., Borodovsky M., Benian G. M. Three new isoforms of Caenorhabditis elegans UNC-89 containing MLCK-like protein kinase domains. J. Mol. Biol. 2004;342:91–108. doi: 10.1016/j.jmb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Squire J. M. Architecture and function in the muscle sarcomere. Curr. Opin. Struct. Biol. 1997;7:247–257. doi: 10.1016/s0959-440x(97)80033-4. [DOI] [PubMed] [Google Scholar]
- Teichmann S. A., Chothia C. Ig superfamily proteins in Caenorhabditis elegans. J. Mol. Biol. 2000;296:1367–1383. doi: 10.1006/jmbi.1999.3497. [DOI] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Timmons L., Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- van Straaten M., Goulding D., Kolmerer B., Labeit S., Clayton J., Leonard K., Bullard B. Association of kettin with actin in the Z-disc of insect flight muscle. J. Mol. Biol. 1999;285:1549–1562. doi: 10.1006/jmbi.1998.2386. [DOI] [PubMed] [Google Scholar]
- Vogel C., Teichmann S. A., Chothia C. The Ig superfamily in Drosophila melanogaster and Caenorhabditis elegans and the evolution of complexity. Development. 2003;130:6317–6328. doi: 10.1242/dev.00848. [DOI] [PubMed] [Google Scholar]
- von Nandelstadh P., Gronholm M., Moza M., Lamberg A., Savilahti H., Carpen O. Actin-organising properties of the muscular dystrophy protein myotilin. Exp. Cell Res. 2005;310:131–139. doi: 10.1016/j.yexcr.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Yamasaki R., et al. Titin-actin interaction in mouse myocardium: passive tension modulation and its regulation by calcium/S100A1. Biophys. J. 2001;81:2297–2313. doi: 10.1016/S0006-3495(01)75876-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Featherstone D., Davis W., Rushton E., Broadie K. Drosophila D-titin is required for myoblast fusion and skeletal muscle striation. J. Cell Sci. 2000;113:3103–3115. doi: 10.1242/jcs.113.17.3103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.