Abstract
Akt is a critical regulator of cell growth, proliferation, and survival that is activated by phosphatidylinositol 3-kinase (PI3K). We investigated the effect of PI3K inhibition on activation of sterol regulatory element binding protein-2 (SREBP-2), a master regulator of cholesterol homeostasis. SREBP-2 processing increased in response to various cholesterol depletion approaches (including statin treatment) and this increase was blunted by treatment with a potent and specific inhibitor of PI3K, LY294002, or when a plasmid encoding a dominant-negative form of Akt (DN-Akt) was expressed. LY294002 also suppressed SREBP-2 processing induced by insulin-like growth factor-1. Furthermore, LY294002 treatment down-regulated SREBP-2 or -1c gene targets and decreased cholesterol and fatty acid synthesis. Fluorescence microscopy studies indicated that LY294002 disrupts transport of the SREBP escort protein, SCAP, from the endoplasmic reticulum to the Golgi. This disruption was also shown by immunofluorescence staining when DN-Akt was expressed. Taken together, our studies indicate that the PI3K/Akt pathway is involved in SREBP-2 transport to the Golgi, contributing to the control of SREBP-2 activation. Our results provide a crucial mechanistic link between the SREBP and PI3K/Akt pathways that may be reconciled teleologically because synthesis of new membrane is an absolute requirement for cell growth and proliferation.
INTRODUCTION
There is a growing body of work indicating a complex interplay between growth regulatory signaling pathways and lipid metabolism. Here we have used the 3-hydroxy-3-methylglutaryl CoA (HMG-CoA) reductase inhibitors, referred to as statins, to study the link between the Akt cell survival and proliferation pathway and the sterol-regulatory element binding protein (SREBP) transcriptional pathway of lipid homeostasis.
Statins are widely prescribed for the treatment of elevated blood cholesterol levels in patients at risk of cardiovascular disease. These agents exploit the cholesterol homeostatic mechanism of the liver cell to which they are targeted by reversibly inhibiting HMG-CoA reductase, the first rate-limiting enzyme in cholesterol biosynthesis. Deprived of newly synthesized cholesterol, the liver cell responds by up-regulating low-density lipoprotein (LDL) receptor expression through activation of the SREBP pathway (Matsuda et al., 2001). More LDL is subsequently cleared, resulting in a reduction of circulating cholesterol levels.
SREBPs are master regulators of lipid homeostasis, inducing transcription of many genes involved in lipid biosynthesis (Goldstein et al., 2006). Genes involved in fatty acid metabolism are preferentially regulated by SREBP-1c (e.g., fatty acid synthase [FAS]), whereas SREBP-2 principally regulates genes involved in cholesterol metabolism (e.g., HMG-CoA reductase) and uptake (LDL-receptor; Horton et al., 2003). SREBPs are produced as precursors embedded in the membrane of the endoplasmic reticulum (ER) bound to the cholesterol sensor, SREBP cleavage-activating protein (SCAP). When cholesterol levels are sensed to be low, SCAP escorts SREBP via COPII vesicles to the Golgi where it is processed by two proteases, designated Site-1 protease and Site-2 protease, to an active transcription factor. When cholesterol levels are sensed to be sufficient by SCAP, the SCAP/SREBP complex is held back in the ER by a retention protein (Insig-1 or -2). Therefore, exit of SCAP/SREBP from the ER is the critical point in feedback regulation of cholesterol metabolism (Nohturfft et al., 2000; Brown et al., 2002; Espenshade et al., 2002; Yang et al., 2002). Statins activate SREBP (Yang et al., 1995; Cheng et al., 1999), presumably by inhibiting cholesterol synthesis and hence reducing the regulatory pool sensed by SCAP.
There is a growing body of clinical and experimental evidence that statins exert additional benefits beyond cholesterol reduction (Laufs and Liao, 2003). For example, statins increase nitric oxide production by endothelial cells and promote angiogenesis through activation of the phosphatidylinositol 3-kinase (PI3K)/Akt pathway (Shiojima and Walsh, 2002). Akt, also called protein kinase B, is a critical regulator of PI3K-mediated cell growth and survival (Shiojima and Walsh, 2002; Song et al., 2005). Akt is recruited to the plasma membrane by its pleckstrin homology domain, which is targeted to the lipid products of PI3K. Activation of Akt then occurs through sequential phosphorylation of a threonine and serine residue. LY294002 and wortmannin are two PI3K inhibitors that inhibit Akt activation. These PI3K inhibitors also inhibit activation of SREBP-1 in response to growth factors (VEGF, platelet-derived growth factor [PDGF]) and insulin (Demoulin et al., 2004; Zhou et al., 2004; Hegarty et al., 2005), suggesting a role for PI3K/Akt in lipogenesis. Therefore, given that statins can activate both SREBP and PI3K/Akt pathways, we hypothesized that the activation of SREBP by statins may involve PI3K/Akt. We report here that the PI3K/Akt pathway has a fundamental effect on cholesterol homeostasis beyond statins, influencing SREBP-2 processing through effects on the ER-to-Golgi transport of this transcription factor.
MATERIALS AND METHODS
Materials
All solvents were analytical reagent- or HPLC-grade from Crown Scientific (Minto, NSW, Australia). Dulbecco's modified eagle's medium/Ham's Nutrient Mixture F-12 (DMEM/F12) was obtained from JRH Biosciences (Lenexa, KS). l-glutamine, newborn calf serum (NBS), fetal calf serum (FCS), and penicillin-streptomycin were obtained from Invitrogen (Carlsbad, CA). Lipoprotein-deficient NBS was prepared as described (Goldstein et al., 1983). Compactin (also called mevastatin), 2-hydroxypropyl-β-cyclodextrin, LY294002, methyl-β-cyclodextrin, mevalonate, Dulbecco's phosphate-buffered saline (PBS), protease inhibitors (or cocktail), phosphatase inhibitor cocktail, Triton X-100, essentially fatty-acid free bovine serum albumin (BSA), saponin, and brefeldin A were obtained from Sigma (St. Louis, MO). Insulin-like growth factor-1 (IGF-1) was purchased from Bio-Scientific (Gymea, NSW, Australia). Cholesterol and 25-hydroxycholesterol (cholest-5-en-3β, 25-diol; 25HC) were obtained from Steraloids (Newport, RI). Cholesterol complexed to methyl-β-cyclodextrin was prepared as described (Brown et al., 2002). [1-14C]Acetic acid, sodium salt (56 mCi/mmol), and [1-14C]oleic acid (55 mCi/mmol) were obtained from GE Healthcare (Chalfont St Giles, United Kingdom). Atorvastatin, fluvastatin, lovastatin, (p)itavastatin, pravastatin, rosuvastatin, and simvastatin were obtained from Sequoia Research Products (Oxford, United Kingdom). The squalene epoxidase inhibitor, GR144000X, was kindly donated by Glaxo-Smith Kline (Middlesex, United Kingdom). Rabbit polyclonal antibodies against phospho-Akt (Ser473) or total Akt were obtained from Cell Signaling Technology (Beverly, MA). Anti-Myc IgG-9E10 mouse monoclonal antibody (mAb; epitope corresponding to amino acids 408-439 within the C-terminal domain of c-Myc of human origin) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Chinese hamster ovary (CHO) cells and antibodies against SCAP and SREBP-2 were kind gifts of Drs. Michael S. Brown and Joseph L. Goldstein (UT Southwestern, Dallas, TX): IgG-R139, a rabbit polyclonal antibody against hamster SCAP (amino acids 54-277 and 540-707; Sakai et al., 1997); IgG-7D4, a mouse mAb against hamster SREBP-2 (amino acids 32-250; Yang et al., 1994). Mammalian expression plasmids, also generously provided by Drs. Brown and Goldstein, have been previously described: pCMV-PLAP-BP2 (513-1141; Sakai et al., 1998), pCMV-Insig-1-Myc (Yang et al., 2002), and pTK-SCAP (Nohturfft et al., 1998). The plasmids for wild-type Akt (pCMV-WT-Akt-myc) and dominant-negative Akt (pCMV-DN-Akt-myc, T308A, S473A Akt) were kindly provided by Dr. Stefanie Dimmeler (University of Frankfurt, Germany; Dimmeler et al., 1999).
Cell Culture
CHO cells were grown as a monolayer in a humidified incubator at 37°C in a 5% CO2 atmosphere. The culture media were DMEM/F12 containing penicillin (100 U/ml), streptomycin (100 μg/ml), and l-glutamine (2 mM), supplemented with various sera. The different media used were supplemented as follows: medium A, 5% (vol/vol) NBS; medium B, 5% lipoprotein-deficient NBS; medium C, 5% NBS plus 5 μg/ml cholesterol, 1 mM sodium mevalonate, 20 μM sodium oleate; medium D, 5% lipoprotein-deficient NBS plus 1 μg/ml 25HC; and medium E, 0.1% (wt/vol) BSA. Statins, 25HC, LY294002 and other test agents were added in ethanol or dimethylsulfoxide (DMSO). Within an experiment, the final concentrations of ethanol or DMSO were kept constant between conditions and did not exceed 0.25% (vol/vol).
Generation of Stably Transfected 13A/PS Cell Line
On day 0, SRD-13A cells (Rawson et al., 1999), a SCAP-deficient cell line derived from CHO-7 cells, were set up in a six-well plate at 2 × 105 cells per well in medium C. On day 1, the cells were cotransfected with pCMV-PLAP-BP2 (0.5 μg) and pTK-SCAP (0.5 μg) using FuGENE 6 transfection reagent (Roche Diagnostics, Basel, Switzerland). On day 2, the cells were switched to medium B to select for cells expressing the SCAP-containing plasmid. Fresh medium B was added every 2–3 d until colonies formed at ∼15 d. Individual colonies were cloned by limiting dilution and screened for expression of the secreted form of PLAP (see below). The most robust colonies were further tested for 16 h in medium B in the presence or absence of sterols (10 μg/ml cholesterol and 1 μg/ml 25HC). A cell line, designated 13A/PS, that showed clear and consistent sterol-regulation of PLAP secretion was maintained in medium B and used as a tool to quantify SREBP-2 processing.
Analysis of SREBP-2 Processing by the Secreted Alkaline Phosphatase Assay
On day 0, 13A/PS cells were set up in 12-well plates in medium B at 1 × 105 cells per well. On day 2, cells were rinsed with PBS, refed fresh medium B, and incubated with various additions as described in the figure legends. After incubation for 16 h, the media were collected for determination of secreted PLAP (SEAP Gene Reporter Assay, Roche Diagnostics) and cells harvested for cell protein determination (BCA kit; Pierce, Rockford, IL; Du et al., 2004). In some experiments involving IGF-1 treatment, cells were cultured in medium E for 24 h before incubation with various additions, as described in the figure legends.
Analysis of SREBP-2 Processing by Western Blotting
SREBP-2 processing by Western blotting was analyzed as described (Adams et al., 2004) with minor modifications. On day 0, 13A/PS cells (medium B, or medium E for IGF-1 treatment experiments) or a cell line, SRD-1, overexpressing the truncated mature form of SREBP-2 (medium D; Metherall et al., 1989) were set up at 1 × 106 cells per 100-mm dish or 6 × 105 cells per 60-mm dish. On day 2, the cells were fed medium B or medium E (for IGF-1 treatment experiments) and incubated with various additions for varying times as described in the figure legends. After incubation, the cells were washed once with cold PBS, resuspended in 500 μl of buffer A (10 mM Tris-HCl, pH 7.6, 100 mM NaCl, 1% [wt/vol] SDS) containing 10 μl of Protease Inhibitor Cocktail, and then passed through a 22-gauge needle 15 times. The resultant cell lysates were then shaken for 20 min at room temperature. The protein concentration of each whole cell lysate was measured (BCA kit; Pierce, Rockford, IL) after which an aliquot (50 μg) was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE; 7.5% wt/vol) and immunoblotting (see below).
Analysis of Akt Phosphorylation
On day 0, 13A/PS cells were set up in medium B at 1 × 106 cells per 100-mm dish. On day 2, the cells were refed fresh medium B and incubated with various additions for varying times as described in the figure legends. After incubation, the cells were washed twice with cold PBS containing 0.01 volume of Phosphatase Inhibitor Cocktail and then lysed in Phosphosafe Extraction Buffer (Merck, Rahway, NJ; 100 μl) containing Protease Inhibitor Cocktail (4 μl), Phosphatase Inhibitor Cocktail (2 μl), and other protease inhibitors (25 μg/ml N-acetyl-leucinal-leucinal-norleucinal [ALLN], 1 μg/ml pepstatin A, 2 μg/ml aprotinin, 10 μg/ml leupeptin, 200 μM phenylmethylsulfonyl fluoride). The resultant cell lysates were passed through a 20-gauge needle 40 times. The protein concentration of each whole cell lysate was measured (BCA kit; Pierce) after which an aliquot (80 μg) was analyzed by SDS-PAGE (10% wt/vol) and immunoblotting (see below).
SCAP-Insig-1–binding Assay
Coimmunoprecipitaton of Insig-1 and SCAP was performed as described (Feramisco et al., 2005) with the following modifications. On day 0, SRD-13A cells were set up in medium C at 1 × 106 cells per 100-mm dish. On day 2, cells were transfected with pTK-SCAP (4.5 μg) and pCMV-Insig-1-Myc (0.9 μg) in medium A using FuGene 6 transfection reagent (Roche Diagnostics). The total amount of DNA was adjusted to 6 μg/dish with pcDNA3 and/or pTK3 mock vectors. On day 3, the cells were washed once with PBS, switched to medium B, and incubated with various additions as indicated. After incubation for 16 h, cells were rinsed twice with cold PBS and harvested for immunoprecipitation with monoclonal anti-Myc IgG-9E10 as described (Feramisco et al., 2005). Immunoprecipitated pellets and supernatants were analyzed by SDS/PAGE (10% wt/vol) and immunoblotting (see below).
Immunoblot Analysis
Samples were mixed with 0.25 volume of buffer B (250 mM Tris-HCl, pH 6.8, 10% [wt/vol] SDS, 25% (vol/vol) glycerol, 0.2% (wt/vol) bromphenol blue, and 5% (vol/vol) β-mercaptoethanol), boiled for 5 min at 95°C, and then subjected to 7.5% or 10% SDS-PAGE. After electrophoresis, the proteins were transferred to Hybond-C nitrocellulose filters (GE Healthcare). Incubations with primary antibodies were performed at 4°C using the following antibodies: 5 μg/ml IgG-7D4 (SREBP-2); 5 μg/ml IgG-R139 (SCAP); 0.4 μg/ml anti-Myc IgG-9E10 (Myc-Insig-1); 1:1000 dilution of polyclonal Phospho-Akt (Ser473) antibody (P-Akt), and 1:1000 dilution of Akt antibody (total Akt). Secondary antibodies were peroxidase-conjugated AffiniPure donkey anti-rabbit or donkey anti-mouse IgG (H+L; Jackson ImmunoResearch Laboratories, West Grove, PA) used at a 1:5000 dilution. The bound antibodies were visualized by ECL Western blotting detection reagent (GE Healthcare). The filters were exposed to Hyperfilm ECL (GE Healthcare) for periods of 2 s to 3 min. The relative intensities of bands were quantified using Sciencelab ImageGauge 4.0 Software (Fujifilm, Tokyo, Japan).
RNA Isolation and Gene Expression Analysis by Quantitative Reverse Transcriptase-PCR
On day 0, 13A/PS and CHO-7 cells were set up in six-well plates in medium B at 2.5 × 105 cells per well. On day 2, the cells were rinsed with PBS, refed fresh medium B, and incubated with various additions as described in the figure legends. After incubation for 16 h, cells were harvested for total RNA using Tri Reagent (Sigma) according to the manufacturer's instructions. Total RNA was quantified by spectrophotometry (Nanodrop ND-100 Spectrophotometer, Bio-Lab, St. Paul, MN). Reverse transcriptase-PCR was performed on 1 μg of RNA according to the manufacturer's protocol for the SuperScript III First Strand cDNA Synthesis Kit (Invitrogen). Quantitative reverse transcriptase-PCR (QRT-PCR) was performed using iQSupermix (Bio-Rad, Richmond, CA) on an ABI 7700 Sequence Detector and analyzed using ABI Prism Sequence Detector Soft-ware v1.6.3 (PE Biosystems, Foster City, CA). Primer pairs (Sigma) used for the amplification reaction of various genes from cDNAs are tabulated in Supplementary Data. PCR products were verified by sequencing. The change in gene expression levels was determined by normalizing mRNA levels of the gene of interest to the mRNA level of the housekeeping gene, porphobilinogen deaminase (PBGD). Melting curve analysis was performed to confirm the production of a single product in each reaction.
Cholesterol and Fatty Acid Synthesis Assay
On day 0, CHO-7 and 13A/PS cells were set up at a density of 4 × 105 cells/dish in 60-mm dishes in medium B. On day 2, cells were refed fresh medium B and incubated with various additions plus 1 μCi/ml [1-14C]acetic acid. After incubation for 16 h, cells were washed twice with buffer C (5 mM Tris HCl, pH 7.5, 150 mM NaCl) containing 0.2% (wt/vol) BSA and twice in buffer C alone. Cells were lysed with NaOH (0.1 M; 1 ml), and the lysates were saponified with 1 ml of 75% (wt/vol) KOH, 2 ml of ethanol plus 1 μl of butylated-hydroxytoluene (20 mM), and 20 μl of EDTA (20 mM) at 70°C for 1 h. After cooling, nonsaponifiable lipids were extracted into hexane (3 × 3 ml) and evaporated to dryness under a steam of nitrogen gas. For cholesterol synthesis, lipid extracts were redissolved in hexane (100 μl) and separated by argentation TLC using 4% (wt/vol) silver-coated Silica Gel 60 F254 plates (Merck, Whitehouse Station, NJ) and a mobile phase of heptane:ethyl acetate (2:1, vol/vol), developed four times. This method separates cholesterol from its major precursors including desmosterol. For fatty acid synthesis, fatty acids were extracted into hexane (3 × 3 ml) from the same samples after acidification with hydrochloric acid (10 M; 3 ml). Hexane extracts were evaporated, redissolved in hexane (150 μl) and separated by TLC using Silica Gel 60 F254 plates with a mobile phase of heptane:diethyl ether:glacial acetic acid (90:30:1, vol/vol/v). For visualization, TLC plates were exposed to BAS-MS imaging plate (Fujifilm) for 24 or 72 h at room temperature and scanned by FLA-5100 phosphoimager (Fujifilm).
Cholesterol Esterification Assay
On day 0, 13A/PS cells were set up at a density of 4 × 105 cells/dish in 60-mm dishes in medium B. On day 2, cells were treated with varying agents in medium B plus 1 μCi/ml [1-14C]oleic acid. After incubation for 16 h, lipids were extracted from the cell lysate as described (Du et al., 2004). The extracts were redissolved in hexane (60 μl) and separated by TLC using Silica Gel 60 F254 plates with a mobile phase of heptane:diethyl ether:glacial acetic acid (90:30:1, vol/vol/vol). For visualization, TLC plates were exposed to BAS-MS imaging plate (Fujifilm) for 72 h at room temperature and scanned by FLA-5100 phosphoimager (Fujifilm). The extent of cholesterol esterification was quantified using Sciencelab ImageGauge 4.0 software (Fujifilm).
Fluorescence Microscopy
On day 0, CHO/pGFP-SCAP cells (Nohturfft et al., 2000), a cell line stably expressing GFP-SCAP, were set up in six-well plates containing sterile 19 × 19-mm glass coverslips at 2 × 105 cells per well in medium B. On day 1, cells were washed with PBS, refed 2 ml of medium B, and incubated with various additions for 4 h as indicated in the figure legends. In some experiments involving IGF-1 treatment, cells were cultured in medium E for 24 h before incubation with various additions. After incubation, cells were rinsed once with PBS and fixed with 1 ml of 3% (vol/vol) formaldehyde/PBS for 10 min at room temperature. Cells were then rinsed twice with PBS and mounted on glass slides with mounting media containing anti-fading reagent (Biomeda, Foster City, CA). Images were obtained using a Leica TCS SP laser scanning spectral confocal microscope (Deerfield, IL). GFP was excited with the 488-nm laser lines from an argon laser. Confocal image stacks were edited using Leica Confocal Software, LCS Lite.
Immunofluorescence Staining
On day 0, CHO/pGFP-SCAP cells were set up as above. On day 1, cells were transfected with myc-tagged wild-type Akt (WT-Akt) or dominant-negative Akt (DN-Akt) plasmids (1 μg/well) by using FuGene 6 transfection reagent according to the manufacturer's instructions (Roche Diagnostics). On day 2, cells were refed 2 ml of medium B per well and incubated with compactin (5 μM) for 4 h. After incubation, the media were aspirated, and the cells were fixed with 1 ml of 3% (vol/vol) formaldehyde for 10 min at room temperature. Cells were rinsed with PBS (three times for 5 min) and then permeabilized with 0.1% (vol/vol) Triton X-100 in PBS (1 ml, 2 min). After washing with PBS (three times for 5 min), cells were incubated with 10% (vol/vol) FCS in PBS for 30 min at room temperature. Cells on the coverslips were then incubated with primary antibody against myc-tag (IgG 9E10, 1.8 μg/ml in 10% FCS in PBS plus 0.1% [wt/vol] of saponin) for 16 h at 4°C. After washing with PBS (three times for 10 min), cells were incubated with 5 μg/ml Alexa Fluro-568–conjugated secondary antibody (Molecular Probes, Eugene, OR) for 1 h at room temperature. Cells were washed with PBS (three times for 10 min) and then mounted on glass slides with mounting media containing anti-fading reagent (Biomeda, Foster City, CA). Images were obtained using a Leica TCS SP laser scanning spectral confocal microscope. Confocal image stacks were edited using Leica confocal software, LCS Lite.
Data Presentation
Data are presented as mean + SEM. All results are representative of at least two separate experiments.
RESULTS
Characterization of a CHO Stable Cell Line That Releases a PLAP Reporter When SREBP-2 Is Processed
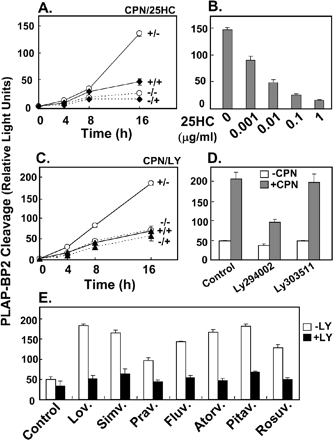
To measure SREBP-2 processing, we adopted a reporter assay devised by Sakai et al. (1998), which allows higher sample throughput and ready quantification. We created a stable cell line that expresses the carboxy-terminal half of human SREBP-2 fused to the secreted form of human placental alkaline phosphatase (PLAP-BP2). This cell line, designated 13A/PS, was highly responsive to changes in sterol status. Thus, more PLAP was secreted in the presence of the archetypal statin, compactin, whereas less PLAP was secreted in the presence of 25-HC, a commonly used oxysterol that is a potent inhibitor of SREBP-2 processing (Figure 1A). A 16-h incubation was required to observe robust effects on PLAP secretion (Figure 1A). Figure 1B shows that PLAP-BP2 processing decreased in response to nanomolar concentrations of added 25HC (0.001 mg/ml = 2.5 nM). We next determined if inhibition of PI3K/Akt affects SREBP-2 processing using a potent and specific cell-permeable inhibitor of PI3K, LY294002. Figure 1C shows that LY294002 inhibited PLAP secretion to a similar extent as 25HC during a 16-h incubation (cf. Figure 1A). By contrast, its inactive analogue, LY303511, had no effect (Figure 1D). Another commonly used PI3K inhibitor, wortmannin, also decreased compactin-induced PLAP-BP2 processing (unpublished data). To ensure that the effect of PI3K inhibition was not restricted to compactin, we tested a panel of statins, many of which are in current clinical use. For each statin, addition of LY294002 reduced PLAP-BP2 processing to basal levels (Figure 1E). These data indicate that inhibition of PI3K negates the statin-induced activation of SREBP-2.
Figure 1.
Processing of a PLAP-BP2 fusion protein increases with compactin treatment and decreases with 25-hydroxycholesterol or LY294002 treatment. (A) 13A/PS cells, a cell line stably expressing PLAP-BP2, were incubated for 2, 4, 8, or 16 h in medium B in the absence (dashed lines) or presence (solid lines) of compactin (CPN, 5 μM), and/or in the absence (○) or presence (♦) of 25-hydroxycholesterol (25HC, 1 μg/ml). (B) 13A/PS cells were incubated for 16 h in medium B with varying concentrations of 25HC (in μg/ml) in the presence of compactin (5 μM). (C) 13A/PS cells were incubated for 2, 4, 8, or 16 h in medium B in the absence (dashed lines) or presence (solid lines) of compactin (CPN, 5 μM) and/or in the absence (○) or presence (▴) of LY294002 (LY, 20 μM). (D) 13A/PS cells were incubated for 16 h in medium B in the absence (blank) or presence (shade) of compactin (CPN, 5 μM) and/or in the absence (control) or presence of LY294002 (20 μM), LY303511 (20 μM). (E) 13A/PS cells were incubated for 16 h in medium B in the absence (Control) or presence of various statins (1 μM: lovastatin, simvastatin, pravastatin, fluvastatin, atorvastatin, pitavastatin, rosuvastatin) and/or in the absence or presence of LY294002 (LY, 20 μM). For all experiments, medium was assayed for PLAP secretion by luminometry, and cell protein was determined. Values are means + SEM (from n = 3 replicate cultures).
PLAP-BP2 Processing Is Induced by Cholesterol Depletion and Reduced by PI3K Inhibition
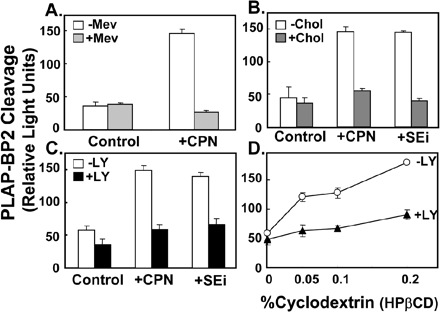
The ability of statins to increase SREBP-2 processing has been attributed to its inhibition of cholesterol synthesis and consequent lowering of cellular cholesterol levels (Yang et al., 1995; Cheng et al., 1999). However, statins also inhibit isoprenoid formation, which may impact on PI3K/Akt signaling (Laufs and Liao, 2003). To further investigate how statins stimulate SREBP-2 processing, we tested whether the effect could be inhibited by addition of mevalonate, the product of HMG-CoA reductase. Exogenous mevalonate suppressed PLAP-BP2 processing stimulated by compactin treatment (Figure 2A). We then tested if inhibition of a later step in cholesterol synthesis, after isoprenoid synthesis, also increases SREBP-2 processing. Inhibition of squalene epoxidase by GR144000X had a similar stimulatory effect on PLAP secretion as compactin (Figure 2B). Cholesterol delivered in complex with methyl-β-cyclodextrin overcame the effect of the two cholesterol synthesis inhibitors (Figure 2B). Addition of the PI3K/Akt inhibitor, LY294002, similarly decreased PLAP secretion stimulated by the squalene epoxidase inhibitor, GR144000X (Figure 2C). We then tested if depleting cholesterol in a statin-independent way increased SREBP-2 processing. As observed with compactin treatment, cholesterol depletion using hydroxypropyl-β-cyclodextrin increased PLAP secretion, which was also reversed by LY294002 treatment (Figure 2D). Taken together, our results first support previous work suggesting that statins increase SREBP-2 processing by reducing cellular cholesterol status (Yang et al., 1995; Cheng et al., 1999) and second indicate that the PI3K/Akt pathway is involved in SREBP-2 processing.
Figure 2.
Effects of cellular cholesterol status and LY294002 treatment on processing of a PLAP-BP2 fusion protein. (A) 13A/PS cells were incubated for 16 h in medium B in the absence or presence of compactin (CPN, 5 μM) and were coincubated in the absence or presence of mevalonate (Mev, 5 mM). (B) 13A/PS cells were incubated for 16 h in medium B in the absence or presence of compactin (CPN, 5 μM) and/or an inhibitor of squalene epoxidase (SEi, GR144000X, 5 μM). Cells were coincubated in the absence or presence of cholesterol complexed to methyl β-cyclodextrin (chol, 20 μg/ml). (C) 13A/PS cells were incubated for 16 h in medium B in the absence or presence of compactin (CPN, 5 μM) and/or an inhibitor of squalene epoxidase (SEi, GR144000X, 5 μM). Cells were coincubated in the absence or presence of LY294002 (LY, 20 μM). (D) 13A/PS cells were incubated for 16 h in medium B containing varying concentrations of hydroxypropyl-β-cyclodextrin (HPβCD, wt/vol) to deplete cellular cholesterol levels in the absence or presence of LY294002 (LY, 20 μM). For all experiments, medium was assayed for PLAP secretion by luminometry and cell protein determined. Values are means + SEM (n = 3 replicate cultures).
Statin-induced SREBP-2 Processing Is Acutely Reduced by the PI3K Inhibitor, LY294002
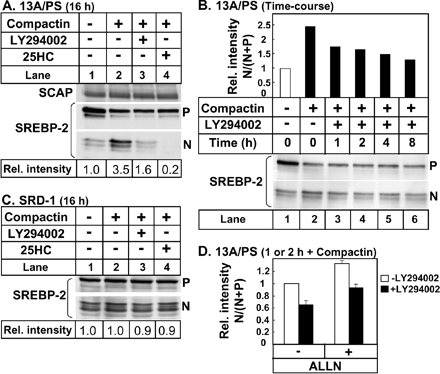
To confirm that the effect of LY294002 occurred at the level of SREBP-2 processing, we blotted whole cell lysates from 13A/PS cells with an mAb directed against the N-teminal domain of SREBP-2 (Yang et al., 1995). In agreement with previous studies (Yang et al., 1995; Cheng et al., 1999), compactin treatment increased mature (nuclear) SREBP-2 (Figure 3A). Treatment with the PI3K inhibitor, LY294002, reduced mature SREBP-2, although to a lesser extent when compared with 25HC treatment. SCAP protein levels were unaffected by all treatments (Figure 3A). Time-course experiments showed that LY294002 decreased statin-induced SREBP-2 processing acutely. After an hour of LY294002 treatment, the relative proportion of mature SREBP-2 decreased by ∼30% (Figure 3B, lane 3). To address if LY294002 affects the stability of mature SREBP-2, we performed an overnight incubation in SRD-1 cells that overexpress the truncated mature form of SREBP-2 (Metherall et al., 1989). The stability of mature SREBP-2 in SRD-1 cells was unaffected by either treatment with LY294002 or 25HC (Figure 3C). We confirmed this in a shorter time-course experiment in 13A/PS cells, by coincubating with the cysteine protease inhibitor, ALLN, to block degradation of nuclear SREBP-2 (Wang et al., 1994; Yang et al., 1995; Hegarty et al., 2005). Statin-induced SREBP-2 processing was reduced to a similar extent by LY294002-treatment, either in the absence or presence of ALLN (Figure 3D). The nuclear portion of SREBP-2 is missing in the PLAP-BP2 construct used in earlier experiments, and yet cleavage of PLAP-BP2 induced by statins was still inhibited by LY294002 (Figures 1 and 2). Taken together, these results indicate that LY294002-treatment decreases SREBP-2 processing and not nuclear SREBP-2 protein stability.
Figure 3.
SREBP-2 processing is inhibited acutely by LY294002 treatment without affecting stability of nuclear SREBP-2. (A) 13A/PS cells were incubated for 16 h in medium B in the absence or presence of compactin (5 μM) and were coincubated in the absence or presence of LY294002 (20 μM) or 25HC (1 μg/ml). After immunoblotting for SREBP-2, the filter was stripped and reprobed for SCAP. (B) 13A/PS cells were incubated in medium B in the absence (lane 1) or presence (lanes 2–6) of compactin (5 μM). After 16 h, cells were refed fresh medium B, continuing compactin treatment in the absence or presence of LY294002 (20 μM) for varying times (0–8 h). (C) SRD-1 cells that overexpress the active (nuclear) form of SREBP-2 were incubated for 16 h in medium B in the absence or presence of compactin (5 μM) and were coincubated in the absence or presence of LY294002 (20 μM) or 25HC (1 μg/ml). (D) 13A/PS cells were incubated in medium B containing compactin (5 μM). After 16 h, cells were refed fresh medium B containing compactin and coincubated for 1 or 2 h in the absence or presence of LY294002 (20 μM) and/or the cysteine protease inhibitor, ALLN (25 μg/ml). For D, values are mean + half-range for the 1- and 2-h time points, where the conditions lacking both LY294002 and ALLN were set at 1. Total cell lysates (containing 50 μg protein) were subjected to 7.5% SDS-PAGE and immunoblot analysis with anti-SCAP IgG-R139 or anti-SREBP-2 IgG-7D4 as described in Materials and Methods. P and N denote the precursor and cleaved nuclear forms of SREBP-2, respectively. Relative intensity of the N band relative to the total (N+P bands) was quantified by densitometry and presented relative to the vehicle-treated control in each panel. All results shown are representative of at least two separate experiments.
LY294002 Has Downstream Effects on SREBP-Target Gene Expression and Function
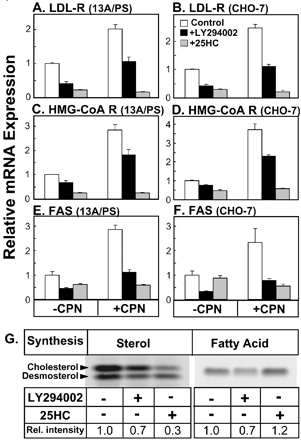
Having established that inhibition of PI3K/Akt decreases SREBP-2 processing, we determined if LY294002 treatment influences expression of SREBP-2 gene targets. 25HC, well known to down-regulate SREBP target gene expression (Yang et al., 1994), was included as a positive control. LY294002 treatment decreased expression of LDL-R, a key cholesterol uptake mechanism, in the presence or absence of compactin (Figure 4, A and B). Expression of the rate-limiting cholesterol biosynthetic enzyme, HMG-CoA reductase, was also reduced with LY294002 treatment (Figure 4, C and D). Again, the effect of LY294002 was not as marked as that observed with 25HC treatment. Similar results were obtained in the stable cell line, 13A/PS (Figure 4, A and C), and the wild-type parental cell line, CHO-7 (Figure 4, B and D). LDL-R and HMG-CoA reductase are primarily regulated by SREBP-2 (Horton et al., 2003). LY294002 treatment also reduced expression of an SREBP-1c gene target, FAS (Figure 4, E and F). In the absence of compactin, LY294002 treatment reduced FAS gene expression to a greater extent than 25HC. This may be due to FAS also being regulated by the liver X receptor and 25HC being a weak agonist for this nuclear receptor (DeBose-Boyd et al., 2001). To determine if the LY294002-mediated decrease in transcription of key lipid biosynthetic genes translates to decreased lipid synthesis, CHO-7 cells were metabolically labeled with [14C]acetic acid in the absence of compactin, and the extent of cholesterol and fatty acid synthesis was determined. LY294002 treatment decreased synthesis of both cholesterol and fatty acids by ∼30% (Figure 4G). Therefore, inhibition of LY294002 on SREBP-2 processing has downstream consequences, down-regulating SREBP-2 (and -1c) target gene expression and decreasing lipid synthesis.
Figure 4.
LY294002 treatment has downstream effects on SREBP-target genes and lipid synthesis. 13A/PS (A, C, and E) or CHO-7 (B, D, F, and G) cells were incubated for 16 h in medium B in the absence or presence of compactin (CPN, 5 μM) and were coincubated in the absence or presence of LY294002 (20 μM) or 25HC (1 μg/ml). mRNA levels for (A and B) LDL-R, (C and D) HMG-CoA Reductase, or (E and F) FAS were measured using QRT-PCR, normalized to PBGD mRNA levels. Data are presented relative to vehicle-treated controls and are means + SEM (n = 3 replicate cultures). (G) CHO-7 cells were incubated for 16 h in medium B containing [14C]acetic acid in the absence or presence of LY294002 (20 μM) or 25HC (1 μg/ml). As described in Materials and Methods, lipid extracts (from equivalent cell proteins) were separated by TLC, and bands corresponding to authentic standards were visualized by phosphoimager (24 h- and 72-h exposure, for fatty acid and sterol synthesis, respectively) and presented relative to the vehicle-treated control in each panel.
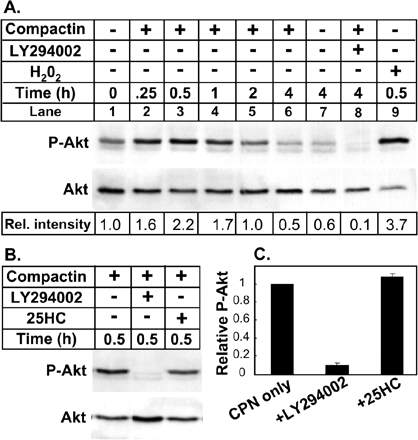
Statin-induced Akt Phosphorylation Is Abolished by Treatment with LY294002 But Not with 25HC
Statins have been reported to affect Akt activation. Most studies report that statins increase Akt phosphorylation (e.g., Kureishi et al., 2000), but there are also reports of no effect or a reduction in Akt phosphorylation (e.g., Zhuang et al., 2005). To examine the effect of compactin on Akt activation in our CHO-based cells, immunoblot analyses were performed on lysates from treated 13A/PS cells using an antibody that is specific for Akt phosphorylated at serine residue 473. Increased Akt phosphorylation was detected as early as 15 min after exposure to compactin and peaked at ∼30 min (Figure 5A, lane 3). The extent of Akt phosphorylation declined to untreated levels by 4 h (Figure 5A, lane 6 vs. lane 7). Treatment with the PI3K inhibitor, LY294002, ablated Akt phosphorylation in the presence (Figure 5A, lane 8) or absence of compactin (unpublished data). Treatment with hydrogen peroxide increases Akt phosphorylation acutely (Thomas et al., 2002) and was included as a positive control (Figure 5A, lane 9). The precise mechanism whereby 25HC induces SCAP-Insig-1 binding to suppress SREBP-2 processing remains obscure. We tested if this oxysterol also affects Akt phosphorylation. Figure 5B shows a representative immunoblot, and the relative extent of Akt phosphorylation was quantified from three separate experiments in Figure 5C. Figure 5, B and C, clearly demonstrates that 25HC had no effect on compactin-mediated Akt phosphorylation, indicating that the mechanism whereby 25HC suppresses SREBP-2 processing does not involve Akt phosphorylation.
Figure 5.
Statin-induced Akt phosphorylation is abolished by treatment with LY294002 but not with 25HC. (A) 13A/PS cells were incubated for varying times (0–4 h) in medium B in the absence or presence of compactin (5 μM) and were coincubated in the absence or presence of LY294002 (20 μM) or hydrogen peroxide (500 μM). (B and C) 13A/PS cells were incubated for 30 min in medium B in the presence of compactin (5 μM) and were coincubated in the absence or presence of LY294002 (20 μM) or 25HC (1 μg/ml). Panel C shows the relative intensity of the P-Akt band relative to the total Akt band (means + SEM) from three separate experiments. There is no error bar in the compactin only control in C, because this condition was set at 1 for all three separate experiments. Whole cell lysates (containing 80 μg protein) were subjected to 10% SDS-PAGE and immunoblot analysis with anti-Phospho-Akt (Ser473; P-Akt) or anti-Total Akt as described in Materials and Methods. Relative intensity of the P-Akt band relative to the total Akt band was quantified by densitometry and presented relative to control in each panel.
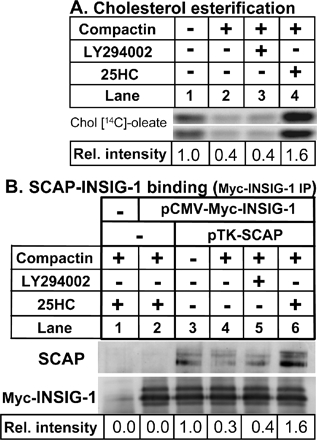
LY294002 Does Not Stimulate Cholesterol Delivery to the ER and/or Increase SCAP-Insig-1 Binding
Acyl-CoA:cholesterol acyl-transferase (ACAT) is an ER-resident enzyme that catalyzes the esterification of cholesterol in response to an expanded pool of cholesterol (Lange and Steck, 1997). We investigated if LY294002 increases cholesterol delivery to the ER by measuring the extent of cholesteryl [14C]oleate formation when 13A/PS cells were incubated with [14C]oleic acid. 25HC is well documented to stimulate cholesterol esterification (e.g., Du et al., 2004). Unlike 25HC, LY294002 did not affect cholesterol esterification (Figure 6A). Recent work (Adams et al., 2004) suggests that 25HC-induced binding of the SREBP escort protein, SCAP, to the ER-retention protein, Insig-1, is independent of cholesterol delivery to the ER. If LY294002 were to increase cholesterol delivery to the ER regulatory pool or work by an alternative mechanism like 25HC, we would predict that LY294002 should also increase SCAP-Insig-1 binding. The extent of SCAP-Insig-1 binding can be determined by immunoprecipitation of Myc-Insig-1 when Myc-tagged Insig-1 and SCAP are coexpressed in SRD-13A cells (Feramisco et al., 2005). When Myc-Insig-1 was expressed alone, no SCAP was pulled down by Myc-Insig-1 immunoprecipitation since this cell line lacks SCAP (Figure 6B, lane 2). The greatest SCAP-Insig-1 binding was observed when cells were treated with 25HC (Figure 6B, lane 6), followed by the condition lacking compactin (Figure 6B, lane 3). The least SCAP-Insig-1 binding was observed with compactin alone (Figure 6B, lane 4) or in combination with LY294002 treatment (Figure 6B, lane 5). Therefore, these studies indicate that LY294002 does not enhance cholesterol delivery to the ER or retention of the SCAP/SREBP-2 complex by Insig-1 in the ER.
Figure 6.
LY294002 does not stimulate cholesterol delivery to the ER and/or increase SCAP-Insig-1 binding. (A) 13A/PS cells were incubated for 16 h in medium B containing [14C]oleic acid in the absence or presence of compactin (5 μM) and were coincubated in the absence or presence of LY294002 (20 μM) or 25HC (1 μg/ml). Lipid extracts (from equivalent cell proteins) were separated by TLC and bands corresponding to authentic cholesteryl oleate standard were visualized by phosphoimager. Shown are two sets prepared from duplicate cultures. Relative intensities of the bands were quantified by densitometry and averaged for the two sets. (B) Coimmunoprecipitation of Insig-1 and SCAP. SRD-13A cells set up in 100-mm dishes were transfected for 24 h with the following plasmids: pcDNA3 alone (Mock, lane 1), pCMV-Insig-1-Myc (0.9 μg, lanes 2–6,) and pTK-SCAP (4.5 μg, lanes 3–6). The total amount of DNA was adjusted to 6 μg/dish with pcDNA3 and/or pTK3 mock vectors. Cells were incubated for 16 h in medium B in the absence or presence of compactin (5 μM) and were coincubated in the absence or presence of LY294002 (20 μM) or 25HC (1 μg/ml). Cells were then harvested for immunoprecipitation with anti-Myc IgG-9E10 and subjected to 10% SDS-block and immunoblot analysis with anti-SCAP IgG-R139 or anti-Myc IgG-9E10 as described in Materials and Methods. Immunoblot analysis showed no Myc-Insig-1 and little SCAP in the immunoprecipitation supernatants (unpublished data), consistent with the article in which the method was originally described (Feramisco et al., 2005). Relative intensity was quantified by densitometry and presented relative to the vehicle-treated control. All results shown are representative of at least two separate experiments.
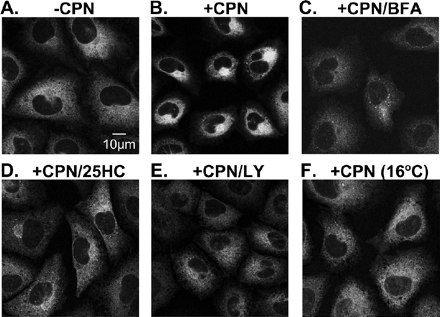
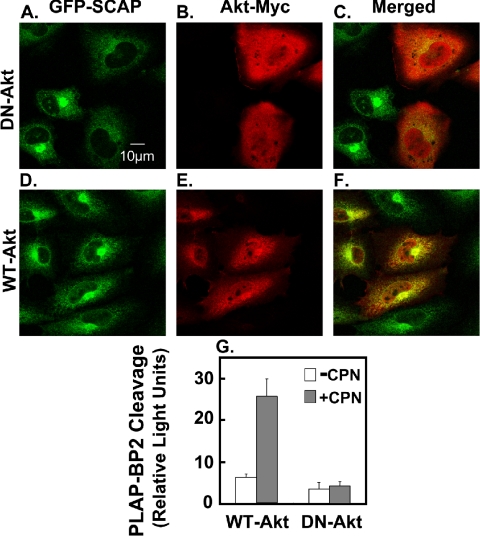
Akt Inactivation Disrupts ER-to-Golgi Transport of GFP-SCAP
To determine if Akt inactivation influences the movement of SCAP, we performed direct fluorescence and immunofluorescence microscopy on SCAP-null cells expressing GFP-SCAP. These cells (CHO/pGFP-SCAP) have previously been used to show that SCAP moves from ER-to-Golgi in a sterol-dependent manner (Nohturfft et al., 2000). In the absence of compactin, GFP-SCAP had a diffuse, reticular pattern (Figure 7A) that is typical of ER localization. After 4 h of compactin treatment, there was bright juxtanuclear fluorescence (Figure 7B), representing transport of GFP-SCAP to the Golgi (Nohturfft et al., 2000). Thus, this discrete Golgi localization was abolished when cells were coincubated with brefeldin A (Figure 7C), an agent which causes Golgi membranes to fuse with the ER (Sciaky et al., 1997). Coincubation with LY294002 produced less intense and less discrete juxtanuclear fluorescence (Figure 7E), which was distinct from the more diffuse ER fluorescence patterns evident when ER-to-Golgi transport was inhibited either by treatment with 25HC (Figure 7D) or a 16°C temperature block (Figure 7F). These observations indicate that LY294002 may interfere with ER-to-Golgi transport of the SREBP/SCAP complex. We then tested if a dominant-negative form of Akt (where the two phosphorylation sites were replaced with alanines: T308A, S473A; Dimmeler et al., 1999) had a similar effect on compactin-induced GFP-SCAP transport as pharmacological inhibition of PI3K by LY294002. When cells expressed this dominant-negative Akt (myc-tagged), GFP-SCAP transport to the Golgi was also disrupted (Figure 8, A–C). By contrast, nonexpressing cells in the same field still showed pronounced Golgi localization of GFP-SCAP. In addition, when cells expressed wild-type Akt, GFP-SCAP transport was unaffected (Figure 8, D–F). To further demonstrate that Akt is involved in SREBP processing, we transiently transfected the SCAP-null SRD-13A cells with plasmids encoding SCAP, the PLAP-BP2 reporter together with either wild-type Akt or dominant-negative Akt plasmids. Compactin stimulated PLAP secretion when wild-type Akt was transfected, whereas when dominant-negative Akt was transfected, the statin effect was completely abolished (Figure 8G). These observations suggest that inactivation of Akt by either PI3K inhibition (LY294002) or dominant-negative Akt transfection disrupts ER-to-Golgi transport of GFP-SCAP and therefore inhibits SREBP processing.
Figure 7.
LY294002 disrupts ER-to-Golgi transport of GFP-SCAP in CHO Cells. Stably transfected CHO/pGFP-SCAP cells were set up on glass cover slips as described in Materials and Methods. On day 1, cells were cultured in medium B for 4 h in the absence (A) or presence (B–F) of compactin (5 μM) and were coincubated with (C) brefeldin A (20 μM), (D) 25HC (1 μg/ml), or (E) LY294002 (20 μM). All incubations were at 37°C except for F, which was at 16°C. Representative fields are shown for each condition from repeated (n = 5) experiments. Parental cells (SRD-13A) lacking GFP-SCAP showed no fluorescence under these conditions (unpublished data).
Figure 8.
Dominant-negative Akt also disrupts ER-to-Golgi transport of GFP-SCAP in CHO cells and inhibits SREBP-2 processing. (A–F) Stably transfected CHO/pGFP-SCAP cells were set up on coverslips as described in Materials and Methods. On day 1, cells were transfected with dominant-negative Akt (DN-Akt; A–C) or wild type-Akt (WT-Akt; D–F). On day 2, cells were cultured in medium B for 4 h in the presence of compactin (5 μM). Immunofluorescence staining was performed as described in Materials and Methods. Representative fields are shown for each condition from repeated (n = 5) experiments. Parental cells (SRD-13A) lacking GFP-SCAP showed no fluorescence under these conditions (unpublished data). (G) SCAP-null SRD-13A cells were transiently transfected with pCMV-PLAP-BP2 (0.25 μg), pTK-SCAP (0.05 μg), DN-Akt (0.5 μg), or WT-Akt (0.5 μg) in medium A for 24 h. Cells were incubated in medium B for 16 h in the presence or absence of compactin (CPN, 5 μM). Medium was assayed for PLAP secretion by luminometry, and cell protein was determined. Values are means + SEM and are representative of three separate experiments.
IGF-1 Induces SREBP Processing and ER-to-Golgi Transport of GFP-SCAP, Which Is Inhibited by LY294002 and 25HC
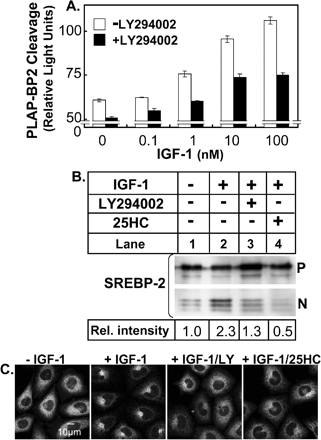
Various growth factors signal cell growth and proliferation via the Akt pathway whereas the statin model used thus far may be more related to cholesterol homeostasis than cell growth or proliferation. To determine if growth factors also induce SREBP processing and transport of GFP-SCAP to the Golgi, we used IGF-1 because CHO cells express the IGF-1 receptor endogenously (Luo et al., 2005). Treatment of 13A/PS cells with IGF-1 increased PLAP-BP2 processing in a dose-dependent manner that was blunted by addition of LY294002 (Figure 9A). Western blotting analysis confirmed IGF-1's ability to increase SREBP-2 processing that was inhibited by treatment with LY294002 or 25HC. (Figure 9B). Moreover, experiments in CHO/pGFP-SCAP cells (Figure 9C) show that IGF-1-treatment induces GFP-SCAP movement to the Golgi that again could be blocked by treatment with LY294002 or 25HC. Therefore, IGF-1, a proliferative stimulus working through Akt (Luo et al., 2005), induces ER-to-Golgi transport of GFP-SCAP and thereby stimulates SREBP processing.
Figure 9.
IGF-1 induces SREBP processing and ER-to-Golgi transport of GFP-SCAP, which is inhibited by LY294002 and 25-hydroxycholesterol. (A) 13A/PS cells were cultured in medium E for 24 h before the incubation for 16 h in medium E containing various concentrations of IGF-1 in the absence or presence of LY294002 (20 μM). Medium was assayed for PLAP secretion by luminometry, and cell protein was determined. Values are means + SEM (from n = 3 replicate cultures, representative of 3 separate experiments). (B) 13A/PS cells were cultured in medium E for 24 h before the incubation for 16 h in medium E in the absence or presence of IGF-1 (50 nM) and were coincubated in the absence or presence of LY294002 (20 μM) or 25HC (1 μg/ml). Total cell lysates (containing 50 μg protein) were subjected to 7.5% SDS-PAGE and immunoblot analysis with anti-SREBP-2 IgG-7D4 as described in Materials and Methods. P and N denote the precursor and cleaved nuclear forms of SREBP-2, respectively. Relative intensity of the N band relative to the total (N+P bands) was quantified by densitometry and presented relative to the vehicle-treated control (lane 1). (C) Stably transfected CHO/pGFP-SCAP cells were set up on glass coverslips as described in Materials and Methods. On day 1, cells were cultured in medium E for 24 h. On day 2, cells were switched to fresh medium E and treated for 16 h in the absence or presence of IGF-1 (50 nM) and were coincubated with LY294002 (20 μM) or 25HC (1 μg/ml). Representative fields are shown for each condition from repeated (n = 3) experiments. Parental cells (SRD-13A) lacking GFP-SCAP showed no fluorescence under these conditions (unpublished data).
DISCUSSION
SREBP-2 is a master regulator of cholesterol homeostasis, whereas the PI3K/Akt pathway is crucial in regulating cell growth, proliferation, and survival. Our studies provide mechanistic insights that link the SREBP-2 and PI3K/Akt pathways. On the basis of our studies using a PLAP-BP2 expressing cell line (Figures 1 and 2) and confirmed by immunoblot analysis (Figure 3), we found that the selective PI3K inhibitor, LY294002, inhibits SREBP-2 processing rather than affecting protein stability of nuclear SREBP-2. Various compounds, including sterols, inhibit SREBP processing by altering SCAP conformation and/or increasing SCAP-Insig-1 binding in the ER (Brown et al., 2002; Yang et al., 2002; Adams et al., 2003). We showed that LY294002 does not increase cholesterol delivery to the ER (as assessed by cholesterol esterification) nor enhance SCAP-Insig-1 binding (Figure 6). Fluorescence microscopy studies in GFP-SCAP–expressing cells indicated that inhibition of PI3K/Akt disrupts ER-to-Golgi transport (Figures 7 and 8), whereas stimulation of PI3K/Akt pathway by the proliferative stimulus, IGF-1, induced ER-to-Golgi transport of GFP-SCAP (Figure 9). Our data are consistent with an Akt-dependent step in transport of SCAP/SREBP-2 from the ER to the Golgi, contributing to the control of the proteolytic cleavage of SREBP-2 to the active form.
ER-to-Golgi transport of proteins initially involves cargo selection and concentration into specific regions of the ER by the action of the COPII coat complex (Watanabe and Riezman, 2004). In sterol-depleted conditions, COPII proteins bind to the SCAP/SREBP complex when added to isolated membranes in vitro (Espenshade et al., 2002; Sun et al., 2005). Protein phosphorylation, known to modulate many membrane traffic processes throughout the cell, has also been implicated in transport between the ER and Golgi (Palmer et al., 2005). Hence, there is evidence that certain proteins involved in COPII-dependent export from the ER can be phosphorylated, including COPII components themselves (Vaughan et al., 2002; Dudognon et al., 2004). Furthermore, certain kinases have been implicated in ER-to-Golgi traffic, including protein kinase A (Aridor and Balch, 2000; Lee and Linstedt, 2000), diacylglycerol kinase delta (Nagaya et al., 2002), and PCTAIRE kinases, a relatively uncharacterized branch of the cyclin-dependent kinase family (Palmer et al., 2005). Ours is the first study to provide evidence that PI3K/Akt is also involved in ER-to-Golgi transport. However, the precise target(s) of Akt remains elusive. Our immunofluorescence studies (Figure 8) suggest that GFP-SCAP may colocalize to some extent with Akt. This was more evident for wild-type Akt (Figure 8F) than for dominant-negative Akt (Figure 8C). However, Porstmann et al. (2005) were unable to identify high-stringency Akt phosphorylation sites in SREBP proteins, SCAP, Insig or site-1 and -2 proteases. Moreover, we have been unable to demonstrate any direct interaction of Akt and SCAP using a coimmunoprecipitation approach (our unpublished data). Further studies are needed to identify the precise target(s) of Akt in ER-to-Golgi transport and to determine if an Akt-dependent step regulates ER-to-Golgi transport of other cargoes besides SCAP/SREBP-2.
Our studies indicate that inhibition of PI3K/Akt has a general and fundamental effect on cellular lipid homeostasis. We showed that first, LY294002 blunted the increase in SREBP-2 processing regardless of how cellular cholesterol status was depleted or in response to a growth factor, IGF-1. Second, LY294002 treatment down-regulated the SREBP-2 or -1c gene targets, LDL-R, HMG-CoA reductase, and FAS, and also decreased cholesterol and fatty acid synthesis. Therefore, changes in the activity of the PI3K/Akt pathway influences SREBP-2 processing and this has downstream effects on cellular lipid homeostasis.
Our results confirm previous studies that have linked the Akt and SREBP pathways (Demoulin et al., 2004; Zhou et al., 2004; Hegarty et al., 2005; Porstmann et al., 2005). For example, pharmacological inhibition of PI3K/Akt decreased mature SREBP-1 in response to growth factors or insulin (Demoulin et al., 2004; Zhou et al., 2004; Hegarty et al., 2005). In addition, Zhou et al. (2004) showed that a constitutively active form of Akt activated the LDL-R and FAS promoters in concert with VEGF, whereas dominant-negative Akt blocked VEGF-induced activation of these promoters. Our studies extend these findings by clearly demonstrating a role for PI3K/Akt in cellular cholesterol metabolism via effects on SCAP transport from the ER to the Golgi and hence SREBP-2 activation.
We have focused on the effects of PI3K inhibition on SREBP-2 processing because this transcription factor mainly regulates cholesterol metabolism (Horton et al., 2003) and previous studies have mostly focused on SREBP-1. Akt is also likely to influence transport of SREBP-1 from the ER to the Golgi and consequent activation, because LY294002 also decreased fatty acid synthesis and FAS gene expression (Figure 4). FAS, like other genes involved in fatty acid synthesis, is preferentially regulated by SREBP-1c (Horton et al., 2003).
Our data are consistent with previous assertions (Yang et al., 1995; Cheng et al., 1999) that statins increase SREBP-2 processing by depleting cellular cholesterol because inhibition at a later step in the pathway (catalyzed by squalene epoxidase) had a similar effect as did depletion of cellular cholesterol by incubating with low levels of hydroxypropyl-β-cyclodextrin (Figure 2). Moreover, addition of cholesterol reversed the increase in SREBP-2 processing observed when cells were treated with statin or a squalene epoxidase inhibitor. Furthermore, statins reduced cholesterol levels in the ER regulatory pool, as evidenced by less cholesterol esterification by the ER resident protein, ACAT, and decreased SCAP-Insig-1 binding (Figure 6). Therefore, our studies add support to previous suggestions that statins increase SREBP-2 processing by reducing the local concentration of cholesterol in the ER.
The PI3K/Akt pathway is best known for its role in promoting cell growth, proliferation, and survival through increased glucose utilization and prevention of apoptosis (Whiteman et al., 2002; Song et al., 2005). Our results with the growth factor, IGF-1, indicate that a proliferative stimulus, working through the PI3K/Akt pathway, induces ER-to-Golgi movement of SCAP, and activates SREBP-2 processing (Figure 9). Synthesis of new membrane is an absolute requirement for cell growth and proliferation, and hence a connection between the SREBP and Akt pathways makes sense from a teleological perspective. Indeed, Goldstein and Brown (Goldstein et al., 2002) have used mutant CHO cell lines to demonstrate that an intact SREBP pathway is required for long-term proliferation. Nohturfft and colleagues recently reported (Castoreno et al., 2005) that phagocytosis-induced membrane biogenesis was initiated by activation of SREBP-1a and SREBP-2 without lipid deprivation. In light of our current data, it would be interesting to determine if the Akt signaling pathway is involved in phagocytosis-induced membrane biogenesis.
The oxysterol, 25HC, was used as a positive control in our experiments, because it is a potent suppressor of SREBP processing. However, its precise mechanism of action remains unknown. Recent work (Adams et al., 2004) suggests that 25HC induces SCAP-Insig-1 binding without altering the conformation of SCAP and proposes the existence of a 25HC sensing protein. Our work indicates that 25HC's potent suppression of SREBP processing is unlikely to involve the PI3K/Akt signaling pathway because 25HC had no effect on Akt phosphorylation (Figure 5B).
In terms of implications of our results in vivo, it is worth noting that activation of Akt by PI3K is associated with several human cancers (Osaki et al., 2004). It is not known if overall cholesterol metabolism is normal in patients with mutations in the PI3K/Akt pathway, but overactive fatty acid and cholesterol synthesis have been described for various types of tumors (reviewed in Freeman and Solomon, 2004; Menendez and Lupu, 2004). Similarly, it is not clear if cholesterol metabolism is influenced in mice in which the various Akt isoforms have been deleted. Interestingly however, constitutively active Akt targeted to liver in mice increased LDL-R expression (Ono et al., 2003), which is consistent with our finding that inhibition of PI3K/Akt down-regulated LDL-R expression in CHO cells (Figure 4, A and B). As shown by us (Figure 5A) and others previously (e.g., Kureishi et al., 2000), statins can increase Akt activation, but this is unlikely to be a concern in terms of human cancer risk, because the consensus from large well-controlled trials is that statin therapy does not increase rates of fatal or nonfatal cancers. Indeed, recent studies indicate that statin use may even reduce the risk of certain cancers (Poynter et al., 2005). Further work is required to determine precisely what role SREBP activation plays in cell transformation and tumor development.
In summary, our studies indicate that there is an Akt-dependent step in SCAP/SREBP transport to the Golgi, adding another input into SREBP activation beside the familiar lipid end products. Our results also highlight a fundamental connection between the SREBP and PI3K/Akt pathways, which may be reconciled teleologically since synthesis of new membrane is an absolute requirement for cell growth and proliferation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Michael S. Brown, Joseph L. Goldstein, and Stefanie Dimmeler for generously sharing their valuable tools. We are also grateful to Yen Pham for her excellent technical assistance in the initial stages of this project and to Ingrid Gelissen for critically reading this manuscript. This work was supported by grants from the National Heart Foundation of Australia (G03S1178) and the Australian Research Council (DP0558041).
Abbreviations used:
- 25HC
25-hydroxycholesterol
- CHO
Chinese hamster ovary
- ER
endoplasmic reticulum
- FAS
fatty acid synthase
- GFP
green fluorescent protein
- HMG-CoA reductase
3-hydroxy-3-methylglutaryl co-enzyme A reductase
- IGF-1
insulin-like growth factor-1
- LDL
low-density lipoprotein
- NBS
newborn calf serum
- PI3K
phosphatidylinositol 3-kinase
- PLAP
placental alkaline phosphatase
- QRT-PCR
quantitative reverse transcriptase-polymerase chain reaction
- SCAP
SREBP cleavage-activating protein
- SREBP
sterol-regulatory element-binding protein
- TLC
thin-layer chromatography.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-11-1094) on March 29, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Adams C. M., Goldstein J. L., Brown M. S. Cholesterol-induced conformational change in SCAP enhanced by Insig proteins and mimicked by cationic amphiphiles. Proc. Natl. Acad. Sci. USA. 2003;100:10647–10652. doi: 10.1073/pnas.1534833100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams C. M., Reitz J., De Brabander J. K., Feramisco J. D., Li L., Brown M. S., Goldstein J. L. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J. Biol. Chem. 2004;279:52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- Aridor M., Balch W. E. Kinase signaling initiates coat complex II (COPII) recruitment and export from the mammalian endoplasmic reticulum. J. Biol. Chem. 2000;275:35673–35676. doi: 10.1074/jbc.C000449200. [DOI] [PubMed] [Google Scholar]
- Brown A. J., Sun L., Feramisco J. D., Brown M. S., Goldstein J. L. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell. 2002;10:237–245. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- Castoreno A. B., Wang Y., Stockinger W., Jarzylo L. A., Du H., Pagnon J. C., Shieh E. C., Nohturfft A. Transcriptional regulation of phagocytosis-induced membrane biogenesis by sterol regulatory element binding proteins. Proc. Natl. Acad. Sci. USA. 2005;102:13129–13134. doi: 10.1073/pnas.0506716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D., Espenshade P. J., Slaughter C. A., Jaen J. C., Brown M. S., Goldstein J. L. Secreted site-1 protease cleaves peptides corresponding to luminal loop of sterol regulatory element-binding proteins. J. Biol. Chem. 1999;274:22805–22812. doi: 10.1074/jbc.274.32.22805. [DOI] [PubMed] [Google Scholar]
- DeBose-Boyd R. A., Ou J., Goldstein J. L., Brown M. S. Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc. Natl. Acad. Sci. USA. 2001;98:1477–1482. doi: 10.1073/pnas.98.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demoulin J. B., Ericsson J., Kallin A., Rorsman C., Ronnstrand L., Heldin C. H. PDGF stimulates membrane lipid synthesis through activation of phosphatidylinositol 3-kinase and sterol regulatory element-binding proteins. J. Biol. Chem. 2004;279:35392–35402. doi: 10.1074/jbc.M405924200. [DOI] [PubMed] [Google Scholar]
- Dimmeler S., Fleming I., Fisslthaler B., Hermann C., Busse R., Zeiher A. M. Activation of nitric oxide synthase in endothelial cells by Akt-dependent phosphorylation. Nature. 1999;399:601–605. doi: 10.1038/21224. [DOI] [PubMed] [Google Scholar]
- Du X., Pham Y. H., Brown A. J. Effects of 25-hydroxycholesterol on cholesterol esterification and SREBP processing are dissociable: implications for cholesterol movement to the regulatory pool in the endoplasmic reticulum. J. Biol. Chem. 2004;279:47010–47016. doi: 10.1074/jbc.M408690200. [DOI] [PubMed] [Google Scholar]
- Dudognon P., Maeder-Garavaglia C., Carpentier J. L., Paccaud J. P. Regulation of a COPII component by cytosolic O-glycosylation during mitosis. FEBS Lett. 2004;561:44–50. doi: 10.1016/S0014-5793(04)00109-7. [DOI] [PubMed] [Google Scholar]
- Espenshade P. J., Li W. P., Yabe D. Sterols block binding of COPII proteins to SCAP, thereby controlling SCAP sorting in ER. Proc. Natl. Acad. Sci. USA. 2002;99:11694–11699. doi: 10.1073/pnas.182412799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feramisco J. D., Radhakrishnan A., Ikeda Y., Reitz J., Brown M. S., Goldstein J. L. Intramembrane aspartic acid in SCAP protein governs cholesterol-induced conformational change. Proc. Natl. Acad. Sci. USA. 2005;102:3242–3247. doi: 10.1073/pnas.0500206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. R., Solomon K. R. Cholesterol and prostate cancer. J. Cell. Biochem. 2004;91:54–69. doi: 10.1002/jcb.10724. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Basu S. K., Brown M. S. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., DeBose-Boyd R. A., Brown M. S. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Rawson R. B., Brown M. S. Mutant mammalian cells as tools to delineate the sterol regulatory element-binding protein pathway for feedback regulation of lipid synthesis. Arch. Biochem. Biophys. 2002;397:139–148. doi: 10.1006/abbi.2001.2615. [DOI] [PubMed] [Google Scholar]
- Hegarty B. D., Bobard A., Hainault I., Ferre P., Bossard P., Foufelle F. Distinct roles of insulin and liver X receptor in the induction and cleavage of sterol regulatory element-binding protein-1c. Proc. Natl. Acad. Sci. USA. 2005;102:791–796. doi: 10.1073/pnas.0405067102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S.W., Brown M. S., Goldstein J. L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kureishi Y., Luo Z., Shiojima I., Bialik A., Fulton D., Lefer D. J., Sessa W. C., Walsh K. The HMG-CoA reductase inhibitor simvastatin activates the protein kinase Akt and promotes angiogenesis in normocholesterolemic animals. Nat. Med. 2000;6:1004–1010. doi: 10.1038/79510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange Y., Steck T. L. Quantitation of the pool of cholesterol associated with acyl-CoA:cholesterol acyltransferase in human fibroblasts. J. Biol. Chem. 1997;272:13103–13108. doi: 10.1074/jbc.272.20.13103. [DOI] [PubMed] [Google Scholar]
- Laufs U., Liao J. K. Isoprenoid metabolism and the pleiotropic effects of statins. Curr. Atheroscler. Rep. 2003;5:372–378. doi: 10.1007/s11883-003-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H., Linstedt A. D. Potential role for protein kinases in regulation of bidirectional endoplasmic reticulum-to-Golgi transport revealed by protein kinase inhibitor H89. Mol. Biol. Cell. 2000;11:2577–2590. doi: 10.1091/mbc.11.8.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Field S. J., Lee J. Y., Engelman J. A., Cantley L. C. The p85 regulatory subunit of phosphoinositide 3-kinase down-regulates IRS-1 signaling via the formation of a sequestration complex. J. Cell Biol. 2005;170:455–464. doi: 10.1083/jcb.200503088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M., Korn B. S., Hammer R. E., Moon Y. A., Komuro R., Horton J. D., Goldstein J. L., Brown M. S., Shimomura I. SREBP cleavage-activating protein (SCAP) is required for increased lipid synthesis in liver induced by cholesterol deprivation and insulin elevation. Genes Dev. 2001;15:1206–1216. doi: 10.1101/gad.891301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez J. A., Lupu R. Fatty acid synthase-catalyzed de novo fatty acid biosynthesis: from anabolic-energy-storage pathway in normal tissues to jack-of-all-trades in cancer cells. Arch. Immunol. Ther. Exp. (Warsz) 2004;52:414–426. [PubMed] [Google Scholar]
- Metherall J. E., Goldstein J. L., Luskey K. L., Brown M. S. Loss of transcriptional repression of three sterol-regulated genes in mutant hamster cells. J. Biol. Chem. 1989;264:15634–15641. [PubMed] [Google Scholar]
- Nagaya H., Wada I., Jia Y. J., Kanoh H. Diacylglycerol kinase delta suppresses ER-to–Golgi traffic via its SAM and PH domains. Mol. Biol. Cell. 2002;13:302–316. doi: 10.1091/mbc.01-05-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A., Brown M. S., Goldstein J. L. Sterols regulate processing of carbohydrate chains of wild-type SREBP cleavage-activating protein (SCAP), but not sterol-resistant mutants Y298C or D443N. Proc. Natl. Acad. Sci. USA. 1998;95:12848–12853. doi: 10.1073/pnas.95.22.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A., Yabe D., Goldstein J. L., Brown M. S., Espenshade P. J. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- Ono H., et al. Hepatic Akt activation induces marked hypoglycemia, hepatomegaly, and hypertriglyceridemia with sterol regulatory element binding protein involvement. Diabetes. 2003;52:2905–2913. doi: 10.2337/diabetes.52.12.2905. [DOI] [PubMed] [Google Scholar]
- Osaki M., Oshimura M., Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis. 2004;9:667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- Palmer K. J., Konkel J. E., Stephens D. J. PCTAIRE protein kinases interact directly with the COPII complex and modulate secretory cargo transport. J. Cell Sci. 2005;118:3839–3847. doi: 10.1242/jcs.02496. [DOI] [PubMed] [Google Scholar]
- Porstmann T., Griffiths B., Chung Y. L., Delpuech O., Griffiths J. R., Downward J., Schulze A. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene. 2005;24:6465–6481. doi: 10.1038/sj.onc.1208802. [DOI] [PubMed] [Google Scholar]
- Poynter J. N., Gruber S. B., Higgins P. D., Almog R., Bonner J. D., Rennert H. S., Low M., Greenson J. K., Rennert G. Statins and the risk of colorectal cancer. N. Engl. J. Med. 2005;352:2184–2192. doi: 10.1056/NEJMoa043792. [DOI] [PubMed] [Google Scholar]
- Rawson R. B., DeBose-Boyd R., Goldstein J. L., Brown M. S. Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J. Biol. Chem. 1999;274:28549–28556. doi: 10.1074/jbc.274.40.28549. [DOI] [PubMed] [Google Scholar]
- Sakai J., Nohturfft A., Cheng D., Ho Y. K., Brown M. S., Goldstein J. L. Identification of complexes between the COOH-terminal domains of sterol regulatory element-binding proteins (SREBPs) and SREBP cleavage-activating protein. J. Biol. Chem. 1997;272:20213–20221. doi: 10.1074/jbc.272.32.20213. [DOI] [PubMed] [Google Scholar]
- Sakai J., Rawson R. B., Espenshade P. J., Cheng D., Seegmiller A. C., Goldstein J. L., Brown M. S. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol. Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- Sciaky N., Presley J., Smith C., Zaal K. J., Cole N., Moreira J. E., Terasaki M., Siggia E., Lippincott-Schwartz J. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J. Cell Biol. 1997;139:1137–1155. doi: 10.1083/jcb.139.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiojima I., Walsh K. Role of Akt signaling in vascular homeostasis and angiogenesis. Circ. Res. 2002;90:1243–1250. doi: 10.1161/01.res.0000022200.71892.9f. [DOI] [PubMed] [Google Scholar]
- Song G., Ouyang G., Bao S. The activation of Akt/PKB signaling pathway and cell survival. J. Cell Mol. Med. 2005;9:59–71. doi: 10.1111/j.1582-4934.2005.tb00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. P., Li L., Goldstein J. L., Brown M. S. Insig required for sterol-mediated inhibition of Scap/SREBP binding to COPII proteins in vitro. J. Biol. Chem. 2005;280:26483–26490. doi: 10.1074/jbc.M504041200. [DOI] [PubMed] [Google Scholar]
- Thomas S. R., Chen K., Keaney J. F., Jr. Hydrogen peroxide activates endothelial nitric-oxide synthase through coordinated phosphorylation and dephosphorylation via a phosphoinositide 3-kinase-dependent signaling pathway. J. Biol. Chem. 2002;277:6017–6024. doi: 10.1074/jbc.M109107200. [DOI] [PubMed] [Google Scholar]
- Vaughan P. S., Miura P., Henderson M., Byrne B., Vaughan K. T. A role for regulated binding of p150(Glued) to microtubule plus ends in organelle transport. J. Cell Biol. 2002;158:305–319. doi: 10.1083/jcb.200201029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Sato R., Brown M. S., Hua X., Goldstein J. L. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Watanabe R., Riezman H. Differential ER exit in yeast and mammalian cells. Curr. Opin. Cell Biol. 2004;16:350–355. doi: 10.1016/j.ceb.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Whiteman E. L., Cho H., Birnbaum M. J. Role of Akt/protein kinase B in metabolism. Trends Endocrinol. Metab. 2002;13:444–451. doi: 10.1016/s1043-2760(02)00662-8. [DOI] [PubMed] [Google Scholar]
- Yang J., Brown M. S., Ho Y. K., Goldstein J. L. Three different rearrangements in a single intron truncate sterol regulatory element binding protein-2 and produce sterol-resistant phenotype in three cell lines. Role of introns in protein evolution. J. Biol. Chem. 1995;270:12152–12161. doi: 10.1074/jbc.270.20.12152. [DOI] [PubMed] [Google Scholar]
- Yang J., Sato R., Goldstein J. L., Brown M. S. Sterol-resistant transcription in CHO cells caused by gene rearrangement that truncates SREBP-2. Genes Dev. 1994;8:1910–1919. doi: 10.1101/gad.8.16.1910. [DOI] [PubMed] [Google Scholar]
- Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., Brown M. S. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Zhou R. H., Yao M., Lee T. S., Zhu Y., Martins-Green M., Shyy J. Y. Vascular endothelial growth factor activation of sterol regulatory element binding protein: a potential role in angiogenesis. Circ. Res. 2004;95:471–478. doi: 10.1161/01.RES.0000139956.42923.4A. [DOI] [PubMed] [Google Scholar]
- Zhuang L., Kim J., Adam R. M., Solomon K. R., Freeman M. R. Cholesterol targeting alters lipid raft composition and cell survival in prostate cancer cells and xenografts. J. Clin. Invest. 2005;115:959–968. doi: 10.1172/JCI200519935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.