Abstract
To analyze the compartmentation of nucleolar protein complexes, the mechanisms controlling targeting of nucleolar processing proteins onto rRNA transcription sites has been investigated. We studied the reversible disconnection of transcripts and processing proteins using digitonin-permeabilized cells in assays capable of promoting nucleolar reorganization. The assays show that the dynamics of nucleolar reformation is ATP/GTP-dependent, sensitive to temperature, and CK2-driven. We further demonstrate the role of CK2 on the rRNA-processing protein B23. Mutation of the major CK2 site on B23 induces reorganization of nucleolar components that separate from each other. This was confirmed in assays using extracts containing B23 mutated in the CK2-binding sites. We propose that phosphorylation controls the compartmentation of the rRNA-processing proteins and that CK2 is involved in this process.
INTRODUCTION
The nucleolus is a model organelle to study nuclear compartmentation (Strouboulis and Wolffe, 1996). In the nucleolus ribosomal RNAs (rRNAs) are synthesized, processed, and assembled with ribosomal proteins to form the small 40S and large 60S preribosome subunits (for a review, see Shaw and Jordan, 1995). The dynamic integration of these different processes generates a typical nucleolar organization. Three main specific components are observed by electron microscopy (Scheer and Hock, 1999): the fibrillar centers (FCs) that are light areas surrounded by a highly contrasted region, the dense fibrillar component (DFC), and the granular component (GC) in which the FCs and DFC are embedded. In the active nucleolus, the early rRNA-processing proteins associated with transcripts during elongation are localized in the central part of the nucleolus, i.e., in the DFC. The processing proteins associated with late steps of rRNA processing are localized in the external part of the nucleolus, i.e., in the GC.
The organization of active nucleoli illustrates the coordination that exists between transcription and processing mechanisms and the recruitment of the nucleolar protein complexes at specific steps of ribosome biogenesis. However the mechanisms that control the compartmentation of nucleolar protein complexes are poorly understood. With this in mind, we undertook to determine whether phosphorylation drives the connection of the processing proteins on rRNAs.
It has been established that nucleolar disorganization can be induced by DRB (5,6 dichloro-1-β-d-ribofuranosylbenzimidazole; Granick, 1975a, 1975b; Scheer et al., 1984; Haaf et al., 1991; Le Panse et al., 1999). Typically the ribosomal genes extend into the nucleoplasm forming the nucleolar necklace, each of the beads of the necklace corresponding to individual transcription sites in association with early rRNA-processing proteins (Granick, 1975a; Haaf and Ward, 1996). More recently it was demonstrated that DRB also induces the formation of masses containing late rRNA-processing proteins at a distance from the transcription sites (David-Pfeuty et al., 2001; Louvet et al., 2005). Thus, there results a separation between rRNA transcription sites and late rRNA-processing proteins, illustrating the disconnection between DFC and GC. When DRB is removed, the nucleolar processing proteins are reconnected with the rRNA transcripts and nucleolar organization is restored. We propose that this experimental approach provides a convenient tool to analyze the mechanism controlling compartmentation of late processing proteins in GC and interaction between DFC and GC.
In the present study we developed an in vitro assay using digitonin-permeabilized cells capable of promoting nucleolar reorganization after DRB removal. By loading the cytoplasm of the permeabilized cells, we investigated the parameters that favor nucleolar formation. In addition, the role of CK2 on nucleolar compartmentation of B23 (also known as nucleophosmin, numatrin or No38) was examined using overexpression of B23 mutants. B23, a phosphoprotein substrate of CK2, is a master protein in GC. It is preferentially associated with 28S pre-rRNA and its ribonuclease activity processes the internal transcribed spacer 2 of the pre-rRNA (Savkur and Olson, 1998; Itahana et al., 2003; Huang et al., 2005). In addition B23 has molecular chaperone activity (Hingorani et al., 2000) regulated by CK2 phosphorylation (Szebeni et al., 2003).
The results of these investigations indicate first that the reconnection of late processing proteins in GC is driven by ATP/GTP hydrolysis and is dependent on CK2, and second that CK2 phosphorylation of B23 plays an important role in nucleolar organization.
MATERIALS AND METHODS
Cell Culture and Fusion Proteins
Control HeLa cells and stable transfected HeLa cells were cultured without antibiotics in Eagle's minimum essential medium (MEM) supplemented with 10% fetal calf serum and 2 mM l-glutamine (Invitrogen, Carlsbad, CA), in 5% CO2 at 37°C. Cells were seeded three times a week. DRB (Sigma, St. Louis, MO) was dissolved in 95% ethanol and added for 4 h to the culture medium at a concentration of 60 μM. For ATP depletion, the cells were incubated with DMEM lacking glucose (Invitrogen) and supplemented with 6 mM 2-deoxy-d-glucose (Acros Organics), and 10 mM sodium azide (Sigma).
Cells (1 × 106/ml) in suspension were transfected using the cationic polymer reagent jetPEI (Qbiogene, Illkirch, France) or the effectene transfection reagent (Qiagen, Courtaboeuf, France).
Fusion proteins in which GFP was linked to the NH2 terminus of the proteins of interest were constructed in pEGFP-C1 (Clontech Laboratories, Palo Alto, CA). The GFP-fibrillarin and GFP-Nop52 constructs have been described (Savino et al., 1999). The wild-type GFP-B23 construct was provided by S. Huang (Chicago, IL; Chen and Huang, 2001), the substitution mutant B23-S125A was provided by M. Olson (Jackson, MI; Szebeni et al., 2003) and inserted into pEGFP-C1, and the substitution mutant GFP-B23-T199A and the deletion mutant GFP-B23-Δ186-239 we reproduced as described by Okuda et al. (2000) and Tokuyama et al. (2001) were gifts of A. Rousselet (Paris, France). Inserting B23 into pDsRed2-C1 (Clontech Laboratories) generated dsRed-B23.
In Vitro Assay Using Permeabilized Cells and Cell Extracts
To produce permeabilized cells, HeLa cells grown on polylysine-coated slides for 24 h were treated for 4 h with 60 μM DRB. They were washed in transport buffer [20 mM HEPES, 110 mM CH3COOK, 5 mM CH3COONa, 2 mM (CH3COO)2Mg, 2 mM DTT, 0.5 mM EGTA, pH 7.2, and 1/25 of a mixture of protease inhibitors; Roche Mannheim, Mannheim, Germany] and permeabilized using 40 μg/ml digitonin (Sigma) for 5 min as described (Adam et al., 1990). The cells were incubated for 60 min at 37°C with either 1) 1/3 of transport buffer supplemented with an ATP-generating system (1 mM ATP [Fermentas Life Sciences, Burlington, Ontario, Canada], 5 mM creatine phosphate and 20 U/ml creatine phosphokinase [Calbiochem, Nottingham, United Kingdom], and 2/3 of extraction buffer; see below) or 2) transport buffer plus ATP generating system and cell extracts (1/3, 2/3, respectively). In some assays, ATP was substituted by 1 mM AMP-PNP, ADP (Sigma-Aldrich), or GTP (Fermentas Life Science) in transport buffer. The CK2 assays were performed using 500 or 1000 Units of CK2 complex (New England Biolabs, Beverly, MA) in 75 μl of a mixture (1/3, 2/3, respectively) of transport and extraction buffer (see below). Cell extracts were prepared from 50 × 106 exponentially growing HeLa cells, or HeLa permanently expressing GFP-fibrillarin and GFP-Nop52, or HeLa cells transfected with GFP-B23-S125A. The extracts were prepared 24 h after transfection using conditions ensuring that 60% of cells expressed GFP-B23-S125A. The KPM extraction buffer (50 mM KCl, 50 mM Pipes KOH, pH 7.25, 10 mM EGTA, and 1.92 mM MgCl2) was used at 4°C as described (Suprynowicz and Gerace, 1986). The suspensions were sonicated until disruption of the nuclei, centrifuged (16,000 × g for 15 min), and the supernatants were frozen in liquid nitrogen. The protein concentration was as high as 9 mg/ml as determined using the BCA protein assay reagent (Pierce, Rockford, IL).
Quantification of nucleolar reformation in permeabilized cells was established based on the presence of separated (as masses) or connected rRNA-processing proteins, or the presence of compact nucleoli. The observation of these three patterns was performed with both epifluorescence and phase-contrast microscopy (Leitz-DMRB, Deerfield, IL). Percentage values are the averages of at least 100 cells from four independent experiments per assay, and the assays were repeated 4–30 times. Standard deviations are represented as error bars.
Antibodies, Immunolabeling, and Microscopy
The antibodies directed against nucleolar proteins were human autoimmune sera against fibrillarin or UBF (rDNA transcription factor), and goat polyclonal antibodies against B23 (C19, Santa-Cruz Biotechnology, Santa Cruz, CA). The secondary antibodies conjugated with FITC or Texas red were from Jackson ImmunoResearch (West Grove, PA). Cells were fixed in 2% paraformaldehyde for 20 min at room temperature and permeabilized with 0.5% Triton X-100 for 5 min. The first antibodies were incubated for 45 min at room temperature and revealed with Texas red- or FITC-conjugated secondary antibodies. The samples were mounted with Citifluor (Canterbury, United Kingdom) and observed with a Leica microscope. Acquisitions were performed with a Micromax CCD camera (Princeton Instruments, Roper Scientific, Evry, France). In other cases optical sections were examined on a Leica SP2 AOBS confocal microscope with a 63×, 1.32 NA PlanApo lens using an Argon laser (488 nm) or a Krypton laser (568 nm) to visualize FITC or Texas red fluorescence, respectively. The images were assembled using Adobe Photoshop (San Jose, CA).
Run-on Transcription Assay and Quantification
Pol I transcription assays were performed on GFP-B23-S125A transitorily transfected cells using Br-UTP as described (Sirri et al., 2000). Br-UTP incorporation was detected by mouse anti-BrdU antibodies (Sigma); the second antibodies were Texas red conjugated. For quantification, acquisitions were performed in 16-bit gray level without autoscale with a Micromax CCD camera. Fluorescence intensities of GFP and Texas red were quantified in nucleoli. One nucleolus was considered as one region of interest and was chosen on the phase contrast image. The same region of interest was used for GFP and Texas red fluorescence, and the signals were quantified using the ImageJ software (NIH). The mean gray value and SD were recorded.
RESULTS
In Intact Cells, Nucleolar Reformation after DRB Removal Occurs in 60 min and Is ATP and Temperature Dependent
We first established the criteria allowing evaluation in situ of the process of DRB-induced nucleolar disorganization consisting in the separation of DFC from GC (Figure 1, a–c‴). The separation of transcription sites (detected by UBF) from the late processing proteins was visible after 2 h of DRB treatment (Figure 1, b–b‴) and fibrillarin remained associated with the transcripts (Louvet et al., 2005). The separation of DFC and GC identified by specific markers (respectively fibrillarin, and Nop52 or B23), occurred in 84% of DRB-treated cells after 4 h (Figure 1, c–c‴). Consequently, this duration of DRB treatment was used in further experiments. Reformation of the nucleolus was observed 20, 40, and 60 min after DRB removal; it was possible to quantify the cells in which the nucleolar components were separated and to identify two steps during nucleolar reformation (Figure 1, d–e‴). The first step was the connection between GC proteins and fibrillarin (Figure 1, d–d‴) and the second step was compaction of the nucleolus (Figure 1, e–e‴).
Figure 1.
Nucleolar reformation occurs in two steps after DRB removal. (a–a‴) Control HeLa cell showing fibrillarin localized in the central part of the nucleolus (a′), surrounded by Nop52 (a″), merge a‴. (b–b‴) Separation of UBF (rDNA transcription factor) and B23 after 2 h of DRB treatment. (c–c‴) Separation of fibrillarin and Nop52 after 4 h of DRB treatment; in the merge image c‴ green and red signals reflect the separation. (d–d‴) Connection between fibrillarin and B23, 20 min after DRB removal. (e–e‴) Compaction of the nucleolus 60 min after DRB removal. Antibodies detect Fibrillarin (αFib) or UBF; the GC proteins (Nop52 and B23) are revealed by a GFP-tag. (a, b, c, d, and e) Phase-contrast images showing the nucleolar structures. Bar, 5 μm.
The distribution of the nucleolar component 60 min after DRB removal was quantified to evaluate the efficiency of nucleolar reformation in HeLa cells: compact nucleoli were observed in 83% of the cells and nucleolar separation in none of the cells. The high percentage of nucleolar reformation demonstrates that the cells can overcome the DRB effect on nucleolar organization in 60 min. The percentage of reformation depends on the temperature because cells incubated at 33°C instead of 37°C exhibit only 37% of compact nucleoli. Interestingly, recovery was almost completely impaired by depleting the ATP pool (see Materials and Methods). The ATP pool was depleted during the last 30 min of DRB treatment and after DRB removal. After 60 min in the absence of ATP, 64% of the cells still exhibited a nucleolar separation indicating strong inhibition of nucleolar reformation.
These results suggest that nucleolar reformation after DRB removal is at least in part, ATP and temperature dependent.
In Permeabilized Cells Nucleolar Reformation Requires ATP Hydrolysis
To investigate precisely the need for ATP in nucleolar reformation, digitonin-permeabilized cells were used. The principle of the assay is based on the permeabilization of the cell plasma membrane inducing cytoplasmic extraction while the nuclear envelope remains intact (Adam et al., 1990). The absence of nuclear diffusion was verified using antibodies directed against fibrillarin (unpublished data). We also demonstrated that permeabilization did not block rRNA transcription in the necklace (Figure 2, e–e‴). It was verified that permeabilization did not modify the percentage of the different nucleolar categories after 4 h of DRB treatment (Figure 3a).
Figure 2.
Nucleolar reformation occurs in digitonine-permeabilized cells. DRB-treated HeLa cells were permeabilized, the cytoplasm loaded with ATP-generating buffer without DRB and nucleolar reformation was examined. (a) Permeabilized control HeLa cells. (b) Permeabilized DRB-treated cells; instead of compact nucleoli, a nucleolar necklace is visible. (c) Nucleolar reformation at 37°C, 1 h after DRB removal. c1–c3, ordered nucleolar patterns: c1, separation; c2, connection; and c3, compact nucleolus. (d–d‴) The connection pattern appearing as a loosely organized nucleolus is illustrated using antibodies against fibrillarin (αFib) or GFP-Nop52. (e–e‴) In permeabilized cells after 4 h of DRB treatment rDNA transcription is detected (e′), and the Nop52 protein tagged with GFP is in masses (e″) at a distance from the transcription signal (e‴); (e) phase contrast. Bars, 10 μm.
Figure 3.
ATP/GTP hydrolysis is needed to connect nucleolar components. Quantitative evaluation of nucleolar reformation in permeabilized cells in the presence of different nucleotides. (a) After 4 h of DRB treatment, 84% of the cells exhibit separation of the nucleolar component, 9% exhibit loosely organized connected component, and 7% exhibit compact nucleoli; gray, hatched, and black columns, respectively. After 1 h having removed DRB, the percentage of the three patterns was estimated using loading buffer containing (b) ATP; (c) GTP; (d) ADP; and (e) AMP-PNP. ATP and GTP induced ∼45% reconnection of the processing proteins, whereas ADP was less efficient with only 30% connection. The nonhydrolyzable ATP analogue AMP-PNP had almost no effect compared with DRB-treated cells. Significant nucleolar reformation (compact) was not observed. (f) The table corresponds in columns 1–3 to the percentage of nucleolar organization in each condition from four different samples of one representative assay. Column 4, total number of cells observed in the four samples; column 5, total number of assays performed.
Starting with the separation of the nucleolar components, the conditions inducing nucleolar reformation were established by cytoplasm loading without DRB at 37°C for 60 min (Figure 2). Three nucleolar patterns were observed: 1) separation of the nucleolar component (Figure 2c1), 2) connection between early and late processing proteins (Figure 2, c and d–d‴), and 3) compact nucleoli (Figure 2c3). In pattern 2, fibrillarin and protein B23 or Nop52 were connected (compare connection of fibrillarin and Nop52 in permeabilized cells in Figure 2, d–d‴, and connection of fibrillarin and B23 in intact cells, Figure 1, d–d‴). With only ATP-containing buffer, separation of the nucleolar components was decreased with a corresponding increase in connected components (84–47% and 9–43%, respectively; Figure 3, a, b, and f). This reorganization was not observed in the absence of ATP (80% separation) or in the presence of DRB (84% separation) and was less efficient at 30°C (68% separation) than at 37°C (unpublished data). Thus partial reconnection of the nucleolar protein complexes obtained in the presence of ATP at 37°C indicates that the DRB effect on endogenous nucleolar proteins can be reversed. To characterize the mechanism responsible for ATP-dependent reconnection of the late processing proteins on transcription sites, permeabilized cells were supplemented with GTP, ADP, or the nonhydrolyzable ATP analogue AMP-PNP. GTP as also ATP partially restored nucleolar reconnection (Figure 3c), whereas addition of both ATP and GTP did not further enhance the reconnection (unpublished data); ADP was less efficient, and AMP-PNP had almost no effect (Figure 3, d–f). This supports the hypothesis that ATP or GTP hydrolysis is necessary to overcome DRB-induced separation of the late processing proteins from the transcription sites.
In Permeabilized Cells Nucleolar Reformation Is Enhanced by CK2
We next addressed the question of the improvement of nucleolar reformation by complementation of the ATP loading buffer with cell extracts. Nuclear targeting of the extracts was first verified. Extracts were generated from HeLa cells permanently expressing GFP-fibrillarin and GFP-Nop52 (Savino et al., 2001), and GFP was used to follow the loaded nucleolar proteins. GFP-fibrillarin and GFP-Nop52 were imported into the nucleus because the GFP signal was only detected in the nucleus (Figure 4, a′ and b′). Translocation efficiency of GFP-tagged proteins to the nuclei and nucleolar domain increased with time up to 60 min. From these observations, it seems likely that other nucleolar proteins contained in the extracts could also be imported into the nucleus. The imported GFP-fibrillarin was recruited in nucleolar structures containing endogenous fibrillarin detected by anti-fibrillarin antibodies (Figure 4a″). It is noticeable that GFP-fibrillarin is recruited in compact nucleoli, in the connected component and in the disconnected component. Similarly GFP-Nop52 colocalized with the endogenous protein B23 in compact nucleoli and in the connected component (Figure 4b″). This indicates that the nucleolar proteins loaded in the cytoplasm are associated with nucleolar reorganization. The cell extracts loaded with ATP-containing buffer increased nucleolar reformation (56% connection and 11% nucleoli) compared with ATP-containing buffer alone (compare Figure 5a with Figure 3b). A similar efficiency was found for cell extracts loaded in GTP-containing buffer (Figure 5 b). This suggests that some proteins have been removed from the nucleus during permeabilization or that the equilibrium of shuttling proteins is enhanced by the extracts loaded.
Figure 4.
Tagged proteins present in cell extracts are properly targeted. DRB-treated HeLa cells were permeabilized and loaded with cell extracts containing GFP-tagged nucleolar proteins for 1 h at 37°C. Note the colocalization in the same nucleolar component of exogenous (GFP-tagged proteins) and endogenous proteins detected with antibodies: (a–a″) fibrillarin; (b–b″) GFP-Nop52 and B23 detected with the corresponding antibodies. Bar, 10 μm.
Figure 5.
Cell extracts and CK2 are needed for nucleolar reformation. (a and b) ATP-containing buffer was complemented with whole-cell protein extracts. This induced the connection of the nucleolar component in more than 50% of the cells. (c and d) ATP-containing buffer complemented with CK2, respectively, 500 or 1000 U. (e and f) ATP-containing buffer complemented with whole-cell extracts and CK2 was the most efficient, leading to more than 30% compact nucleoli. Separated and connected nucleolar components and compact nucleoli are represented by gray, hatched, and black columns, respectively. (g) The table corresponds in columns 1–3 to the percentage of nucleolar organization in each condition from four different samples of one representative assay 1 h after having removed DRB. Column 4, total number of cells observed in the four samples; column 5, total number of assays performed.
The assay makes it possible to visualize the connection of nucleolar proteins to transcription sites; however the percentage of nucleolar compaction is low (<15%). As CK2 kinase is a target of DRB, it seemed possible that addition of CK2 might enhance nucleolar reformation by competition with DRB-inactivated CK2. We first examined the effect of CK2 at two concentrations in ATP-containing buffer. The effect of CK2 alone on nucleolar reformation was not significant except for a slight increase in the number of compact nucleoli (compare Figure 5, c and d, with Figure 3b). We next examined nucleolar reformation after addition of CK2 to cell extracts in ATP-containing buffer. In these conditions, the efficiency of nucleolar reformation was enhanced and dose-dependent (10% separation, 59% connection, and 31% compact nucleoli for the highest concentration; Figure 5, e–f and g). This indicates that the complementation of cell extracts with CK2 enhanced nucleolar reformation (compared Figure 5, a–b and e–f) and suggests a role of CK2 or of kinases regulated by CK2 (Miyata and Nishida, 2004) on the efficient compartmentation of the nucleolar protein complexes in functional nucleoli.
CK2 Phosphorylation Plays a Role in B23 Compartmentation
To investigate the role of CK2 on the compartmentation of processing nucleolar proteins, B23 was chosen. The sites of B23 phosphorylation in vivo have been characterized and depend on CK2 and cyclin-dependent kinases (CDKs; Chan et al., 1990; Peter et al., 1990; Okuda et al., 2000). To determine whether B23 compartmentation depends on its phosphorylation by CK2, B23 mutants were used (Figure 6A).
Figure 6.
Mutation of CK2 phosphorylation site of B23 induces reticulated nucleoli. (A) Constructs of B23: wild type (WT); Δ186-239 corresponds to the deletion mutation of the CDK sites (Okuwaki et al., 2002); S125A corresponds to the substitution mutation S125 by alanine in the region of the sequence containing the CK2-binding site (Szebeni et al., 2003); and T199A corresponds to the substitution mutation T199 by alanine (Tokuyama et al., 2001). Arrows point to the major phosphorylation sites in B23 domains: black box corresponds to the acidic region, the gray box to RNase activity, and the striped box to RNA binding activity (Hingorani et al., 2000). (B) Overexpression of GFP-B23-S125A in HeLa cells. (a–a″) Note the reticulated organization of the nucleoli visible by phase contrast (a′) only in cells expressing the GFP-B23 mutant (a); a" antibodies against B23. Bar, 10 μm.
In the B23 sequence, serine 125 is the main site of CK2 phosphorylation (Szebeni et al., 2003). The GFP-tagged B23 protein containing the amino acid substitution S125 to alanine (GFP-B23-S125A) is recruited into the nucleolus (Figure 6B, a and a″). Disruption of the nucleolus was not observed whatever the level of expression of B23-S125A. This was expected because expression of B23 deficient in CK2 phosphorylation site cannot reproduce the effect of DRB on a large number of proteins (Meggio and Pinna, 2003). We investigated the organization of nucleoli as a function of the level of expression of B23 deficient in the major CK2 phosphorylation site. In cells expressing GFP-B23-S125A to high levels, the nucleolar organization visible in phase contrast was reticulated (Figure 6B, a and a′). This reorganization is due to the mutated protein (Figure 7, a–a" and c–c″) and not to its level of expression because similar levels of GFP-B23-WT did not form reticulated nucleoli (Figure 7, b–b" and d–d″). The nucleolar components in reticulated nucleoli were not intermingled. UBF localized in space devoid of B23 most likely in the FC (Figure 7, a–a″), fibrillarin surrounded the FC (Figure 7, c and c″), and GFP-B23-S125A was excluded from the FC (Figure 7, c and c″). Therefore, reorganization of the nucleolus is induced by B23-S125A. This nucleolar reorganization is also illustrated by the decrease of rRNA transcription observed in cells expressing GFP-B23-S125A (Figure 8, a–a″), and this decrease is correlated with the level of expression of the mutant protein (Figure 8b).
Figure 7.
Expression of B23-S125A induces redistribution of UBF and fibrillarin in nucleoli. (a–a″) In the reticulated nucleoli overexpressing GFP-B23-S125A, UBF localized in the B23 depleted zone; see enlarged nucleoli in a". (b–b″) In the nucleolus expressing GFP-B23-WT, the distribution of B23 is homogeneous relative to UBF. (c–c″) In nucleoli overexpressing GFP-B23-S125A, fibrillarin distributed around the B23-depleted zone; see enlarged nucleoli in c". (d–d″) In the nucleolus expressing GFP-B23-WT, the distribution of B23 is homogeneous relative to fibrillarin. Confocal optical sections. Bars, 1 μm.
Figure 8.
Expression of GFP-B23-S125A decreases rRNA transcription. (a–a″) Cells expressing or not expressing GFP-B23-S125A. The two transfected cells (a′) exhibit a reticulated nucleolus (a). Pol l transcription revealed by Br-UTP incorporation shows a decrease of the signal in the transfected cells (a″). (b) Graph representing the transcription level as a function of GFP level in 30 nucleoli in untransfected cells (background; white triangles) and 35 nucleoli in transfected cells (gray circles); arbitrary units. Bar, 10 μm.
We then compared the effect of mutated phosphorylation sites in B23 by either CK2 or CDK. The effect of the amino acid substitution of the Cdk1/cyclin B and Cdk2/cyclin E phosphorylation site (T199A; Okuda et al., 2000; Tokuyama et al., 2001) on the localization of B23 was examined. GFP-B23-T199A was recruited in the nucleolus whatever its level of expression and without reorganization of the nucleoli (Figure 9, a and a″). Finally, deletion of a sequence containing cell cycle–regulated sites (CDK sites comprised within amino acids 186-239) directed GFP-B23-Δ186-239 both to the nucleolus and to the nucleoplasm (unpublished data). The sequences of RNA binding and those involved in RNase activity were not deleted from the construct as shown in Figure 6A. However the deletion covered the domain of hetero-dimerization implicated in interaction of B23 with several nucleolar proteins (Fankhauser et al., 1991; Valdez et al., 1994; Li et al., 1996; Korgaonkar et al., 2005). This distribution of B23-Δ186-239 did not allow testing the effect of the truncated B23 on nucleolar organization.
Figure 9.
Expression of GFP-B23-T199A has not effect on nucleolar organization. In control cells the mutant B23 protein localizes in the nucleoli as does endogenous B23 detected by antibodies, and reticulated nucleoli are not observed. (a) GFP-B23-T199A; (a′) phase contrast; (a″) B23 antibodies. Bar, 10 μm.
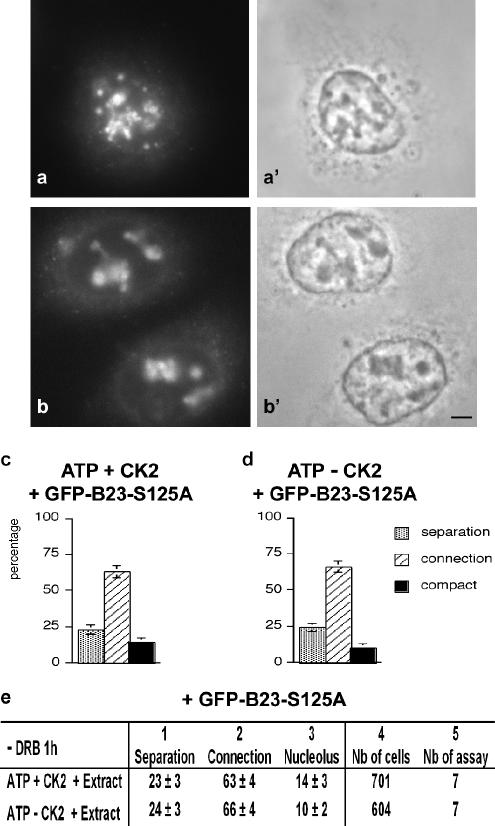
The role of the CK2 site on B23 compartmentation was also analyzed during nucleolar reformation after DRB removal. Permeabilized cells were loaded with extracts containing GFP-B23-S125A in the conditions providing 30% of compact nucleoli as defined in Figure 5f. GFP-B23-S125A was imported into the nuclei after 60 min of incubation (Figure 10, a–b′). In these cells, the three patterns of nucleolar reformation (see Figure 2; separated or connected machineries and compact nucleoli) were observed. The mutated B23 was localized in the three nucleolar patterns. The main differences observed using extracts containing mutated B23 compared with control B23 concerned machinery connection that was more heterogeneous in the former extracts. In addition to connected masses containing B23, weak labeling forming threads or large areas were visible. These areas appeared diffuse in phase-contrast microscopy (Figure 10, a′ and b′). The traffic of the mutated protein is probably modified compared with the control protein, and this could explain the heterogeneous organization of the connected machinery. Quantification of compact nucleoli versus connected machinery indicated a less efficient nucleolar compaction compared with control B23 (compare Figure 10c with Figure 5f). The absence of CK2 in the buffer seemed without effect on the percentage of connected machinery and nucleolar compaction (Figure 10, d and e). This suggests that the association of machineries during nucleolar reformation is mainly driven by the role of CK2 on B23 and not on other nucleolar proteins.
Figure 10.
Cell extracts containing a mutated site in B23 for CK2 phosphorylation, impaired nucleolar reformation. DRB-treated cells were permeabilized, the cytoplasm loaded with ATP generating buffer without DRB, and the cell extracts containing GFP-B23-S125-A, and CK2 and nucleolar reformation was examined. (a and b) Masses and structures containing mutated GFP-B23 at different stages of nucleolar machinery connection. (a′ and b′) Same cells in phase contrast showing diffuse organization of nucleolar machineries. Bar, 4 μm. (c and d) ATP-containing buffer complemented with extracts prepared from cells transfected with GFP-B23-S125A. This induced the connection of nucleolar machineries in more than 60% of the cells. ATP-containing buffer complemented (c) or not (d) with CK2 1000 U. Separated and connected nucleolar components and compact nucleoli, gray, hatched, and black columns, respectively. (e) The table corresponds in columns 1–3 to the percentage of nucleolar organization in each condition from four different samples of one representative assay. Column 4, total number of cells observed in the four samples; column 5, total number of assays performed.
DISCUSSION
After DRB removal, the late rRNA-processing proteins are recruited on transcription sites, and an active nucleolus is reassembled in 1 h. The question therefore remains as to how translocation of the processing protein is controlled. Using digitonine-permeabilized cells (Wagner et al., 2004), we demonstrate that ATP hydrolysis is necessary to reconnect the nucleolar components after DRB removal. This suggests that phosphorylation could play a role in reconnection of the processing proteins and explains why this process is temperature-dependent. In addition to ATP or GTP, nucleolar reconnection was improved by the inclusion of cell extracts, the loaded nucleolar proteins participating in nucleolar reformation. It thus appears that certain proteins have been depleted from the nucleus during permeabilization or that the equilibrium of shuttling nucleolar proteins is enhanced by the extracts added. Consequently the next step was to enhance nucleolar reformation by adding a kinase that could reverse the DRB effect. As a possible candidate, protein kinase CK2, a serine/threonine kinase requiring ATP or GTP was chosen for the following reasons. The DRB effect can be partially reversed by an excess of CK2 (Zandomeni et al., 1986), CK2 subunits are imported into the nucleus (Filhol et al., 2003), CK2 is constitutively active, and several nucleolar proteins are substrates of CK2 (Meggio and Pinna, 2003). The CK2-enriched extracts were highly efficient in facilitating the connection of processing proteins with rRNA transcripts (∼60%). We propose that CK2 activity is important for the connection of GC processing proteins with DFC (Figure 11). The fact that CK2 alone in ATP buffer did not enhance nucleolar reformation indicates that during permeabilization some proteins have been removed from the nucleus, and the extracts restore the equilibrium between cytoplasm and nucleus. The possible role of CK2 for the connection of GC with DFC is in accordance with the hypothesis that CK2 has a transversal role controlling a network of interactions (Meggio and Pinna, 2003).
Figure 11.

Nucleolar reformation after DRB removal requires ATP hydrolysis and is driven by CK2. DRB induced the separation of nucleolar transcription (circle) from late processing proteins (square). After DRB removal the nucleolar components first reconnect before being compacted into a nucleolus. The efficiency of ATP, GTP, and CK2 on connection of nucleolar components is high (black bent arrow), whereas the efficiency of CK2 alone on nucleolar compaction is moderate (gray bent arrow).
Among the nucleolar proteins known to be phosphorylated in vivo by CK2, the possibility was raised that phosphorylation of B23 might play a role on compartmentation of the protein. This hypothesis is based on the fact that depending on the means used to destroy nucleolar compartmentation, the behavior of B23 varies. Indeed B23 is homogeneously distributed throughout the nucleoplasm by blocking rDNA transcription using actinomycin D (Yung et al., 1990; Zatsepina et al., 1997), by competition for rRNAs with FRGY proteins (Gonda et al., 2003), and by several anticancer drugs that interfere with nucleolar activity (Chan et al., 1996). On the contrary when nucleolar disorganization is induced by kinase inhibitors (DRB or roscovitine), B23 remains in granular masses disconnected of DFC (David-Pfeuty et al., 2001; Sirri et al., 2002; Louvet et al., 2005). This suggests at least a partial role of kinases on DFC and GC connection. It was demonstrated that GTP is required to bring B23 into the nucleolus, whereas ATP is necessary for translocation to the nucleoplasm (Finch et al., 1993; Finch and Chan, 1996). Because B23 is not a GTP-binding protein, this indicates that nucleolar targeting of B23 is controlled by a GTP-dependent mechanism. This mechanism could be related to CK2 because its kinase activity needs ATP or GTP. In the present study we observed reorganization of the nucleolus by overexpression of GFP-B23-S125A, a substitution that impairs CK2 phosphorylation of B23 (Szebeni et al., 2003). At interphase, phosphorylation of B23 is regulated by CK2 (Chan et al., 1990). The modifications induced by B23-S125A could be part of the process leading to disconnection of the processing nucleolar proteins from transcription because the nucleolar components are no longer intermingled. This nucleolar reorganization could result from defects in the dissociation of B23-substrate complexes because phosphorylation of B23 by CK2 promotes this dissociation (Szebeni et al., 2003). Similarly the perturbations of the nucleolar reformation induced by B23 mutated in its CK2 phosphorylation site in permeabilized cells could be of the same origin by modifying the B23 traffic.
The B23 mutated CK2 site-induced reorganization of the nucleolar component but not disruption of the nucleolus, as did DRB. On the basis of the present observations, we propose that the B23 mutant participates in reorganization of the nucleolar component, part of the process leading to nucleolar disruption. However the B23 mutant had no effect on rDNA compaction as opposed to DRB that triggers an event necessary for nucleolar disruption. It is well established that DRB induces rDNA extension in the nucleoplasm forming the nucleolar necklace (Scheer and Rose, 1984; Haaf et al., 1991). In permeabilized cells, 31% of compact nucleoli were formed with CK2-enriched extracts, indicating a role for these enriched extracts on rDNA compaction (Figure 11). This output value is significant in permeabilized cell assays. However, it does not necessarily exclude that in addition to CK2, other partners are necessary to overcome rDNA decompaction.
During interphase, modifications or blockage of ribosome production induce reorganization or dispersion of the nucleolar components. Nucleolar segregation is the most typical reorganization phenomenon resulting from blockage of rDNA transcription (Hadjiolov, 1985; Puvion-Dutilleul et al., 1992; Scheer et al., 1993; Dousset et al., 2000; Gébrane-Younès et al., 2005). In this case most of the nucleolar protein complexes remain closely linked, regrouped by category of functions, i.e., transcription, early processing, or late processing. Nucleolar segregation suggests that transcription is necessary to recruit and maintain the sequential order of protein association necessary to build the primary 90S preribosomal complexes and then process the 40S and 60S ribosome subunits (Harnpicharnchai et al., 2001; Fatica and Tollervey, 2002; Fromont-Racine et al., 2003; Saveanu et al., 2003).
In addition other interactions control the compartmentation of nucleolar proteins in the absence of ribosome biogenesis because nucleolar components are maintained and reorganized (Hadjiolov, 1985; Chan et al., 1996; Rubbi and Milner, 2003; Gébrane-Younès et al., 2005). In the case of the separation of transcription and processing masses, the reorganization indicates that the presence of rRNAs is not sufficient to attract processing proteins and that processing proteins can form nuclear structures independently of the transcripts. The disconnection of the rRNA-processing proteins from rRNA transcripts is induced by kinase inhibitors, suggesting that kinase(s) is (are) involved in the connection of the nucleolar component in active nucleoli. Here, we demonstrate the role of CK2 on the compartmentation of the nucleolar processing proteins. Components of CK2 have already been identified in several complexes containing preribosomal proteins in yeast (Gavin et al., 2002; Ho et al., 2002; De Marchis et al., 2005), and the regulation of ribosome biogenesis by CK2 is well documented (for a review see Meggio and Pinna, 2003). The nucleolar CK2 substrates, B23, nucleolin, Nop140, UBF, and pol I are localized in different nucleolar components, and CK2 phosphorylation could have a global role on nucleolar activity.
ACKNOWLEDGMENTS
We thank S. Huang, M.O.J. Olson, and A. Rousselet for the kind gifts of probes; S. Chamot for cell culture; M. Barre for help with the illustrations; and A.-L. Haenni for critical reading of the manuscript. This work was supported in part by grants from the Centre National de la Recherche Scientifique to UMR 7592, and from the Association pour la Recherche sur le Cancer (ARC, Contract 3303).
Abbreviations used:
- AMP-PNP
5′-adenylylimidodiphosphate
- CDK
cyclin-dependent kinase
- CK2
casein kinase 2
- DFC
dense fibrillar component
- DRB
5,6 dichloro-1-β-d-ribofuranosylbenzimidazole
- FC
fibrillar component
- GC
granular component
- GFP
green fluorescent protein
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-10-0923) on March 15, 2006.
REFERENCES
- Adam S. A., Marr R. S., Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. K., Liu Q. R., Durban E. The major phosphorylation site of nucleophosmin (B23) is phosphorylated by a nuclear kinase II. Biochem. J. 1990;270:549–552. doi: 10.1042/bj2700549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P. K., Qi Y., Amley J., Koller C. A. Quantitation of the nucleophosmin/B23-translocation using imaging analysis. Cancer Lett. 1996;100:191–197. doi: 10.1016/0304-3835(95)04100-1. [DOI] [PubMed] [Google Scholar]
- Chen D., Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 2001;153:169–176. doi: 10.1083/jcb.153.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Pfeuty T., Nouvian-Dooghe Y., Sirri V., Roussel P., Hernandez-Verdun D. Common and reversible regulation of wild-type p53 function and of ribosomal biogenesis by protein kinases in human cells. Oncogene. 2001;20:5951–5963. doi: 10.1038/sj.onc.1204741. [DOI] [PubMed] [Google Scholar]
- De Marchis M. L., Giorgi A., Schinina M. E., Bozzoni I., Fatica A. Rrp15p, a novel component of pre-ribosomal particles required for 60S ribosome subunit maturation. RNA. 2005;11:495–502. doi: 10.1261/rna.7200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dousset T., Wang C., Verheggen C., Chen D., Hernandez-Verdun D., Huang S. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol. Biol. Cell. 2000;11:2705–2717. doi: 10.1091/mbc.11.8.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Izaurralde E., Adachi Y., Wingfield P., Laemmli U. K. Specific complex of human immunodeficiency virus type 1 rev and nucleolar B23 proteins: dissociation by the Rev response element. Mol. Cell. Biol. 1991;11:2567–2575. doi: 10.1128/mcb.11.5.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A., Tollervey D. Making ribosomes. Curr. Opin. Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- Filhol O., Nueda A., Martel V., Gerber-Scokaert D., Benitez M. J., Souchier C., Saoudi Y., Cochet C. Live-cell fluorescence imaging reveals the dynamics of protein kinase CK2 individual subunits. Mol. Cell. Biol. 2003;23:975–987. doi: 10.1128/MCB.23.3.975-987.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch R. A., Chan P. K. ATP depletion affects NPM translocation and exportation of rRNA from nuclei. Biochem. Biophys. Res. Commun. 1996;222:553–558. doi: 10.1006/bbrc.1996.0782. [DOI] [PubMed] [Google Scholar]
- Finch R. A., Revankar G. R., Chan P. K. Nucleolar localization of nucleophosmin/B23 requires GTP. J. Biol. Chem. 1993;268:5823–5827. [PubMed] [Google Scholar]
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- Gavin A. C., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gébrane-Younès J., Sirri V., Junéra H. R., Roussel P., Hernandez-Verdun D. Nucleolus: an essential nuclear domain. In: Diekmann P.H.a.S., editor. Visions of the Cell Nucleus. Stevenson Ranch, CA: 2005. pp. 120–135. [Google Scholar]
- Gonda K., Fowler J., Katoku-kikyo N., Haroldson J., Wudel J., Kikyo N. Reversible disassembly of somatic nucleoli by the germ cell proteins FRGY2a and FRGY2b. Nat. Cell Biol. 2003;5:205–210. doi: 10.1038/ncb939. [DOI] [PubMed] [Google Scholar]
- Granick D. Nucleolar necklaces in chick embryo fibroblast cells. I. Formation of necklaces by dichlororibobenzimidazole and other adenosine analogues that decrease RNA synthesis and degrade preribosomes. J. Cell Biol. 1975a;65:398–417. doi: 10.1083/jcb.65.2.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granick D. Nucleolar necklaces in chick embryo fibroblast cells. II. Microscope observations of the effect of adenosine analogues on nucleolar necklace formation. J. Cell Biol. 1975b;65:418–427. doi: 10.1083/jcb.65.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T., Hayman D. L., Schmid M. Quantitative determination of rDNA transcription units in vertebrate cells. Exp. Cell Res. 1991;193:78–86. doi: 10.1016/0014-4827(91)90540-b. [DOI] [PubMed] [Google Scholar]
- Haaf T., Ward D. C. Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp. Cell Res. 1996;224:163–173. doi: 10.1006/excr.1996.0124. [DOI] [PubMed] [Google Scholar]
- Hadjiolov A. A. The Nucleolus and Ribosome Biogenesis. New York: Springer-Verlag; 1985. [Google Scholar]
- Harnpicharnchai P., et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- Hingorani K., Szebeni A., Olson M. O. Mapping the functional domains of nucleolar protein B23. J. Biol. Chem. 2000;275:24451–24457. doi: 10.1074/jbc.M003278200. [DOI] [PubMed] [Google Scholar]
- Ho Y., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Huang N., Negi S., Szebeni A., Olson M. O. Protein NPM3 interacts with the multifunctional nucleolar protein B23/NPM and inhibits ribosome biogenesis. J. Biol. Chem. 2005;280:5496–5502. doi: 10.1074/jbc.M407856200. [DOI] [PubMed] [Google Scholar]
- Itahana K., Bhat K. P., Jin A., Itahana Y., Hawke D., Kobayashi R., Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- Korgaonkar C., Hagen J., Tompkins V., Frazier A. A., Allamargot C., Quelle F. W., Quelle D. E. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol. Cell. Biol. 2005;25:1258–1271. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Panse S., Masson C., Héliot L., Chassery J.-M., Junéra H. R., Hernandez-Verdun D. 3-D Organization of single ribosomal transcription units after DRB inhibition of RNA polymerase II transcription. J. Cell Sci. 1999;112:2145–2154. doi: 10.1242/jcs.112.13.2145. [DOI] [PubMed] [Google Scholar]
- Li Y. P., Busch R. K., Valdez B. C., Busch H. C23 interacts with B23, a putative nucleolar-localization-signal-binding protein. Eur. J. Biochem. 1996;237:153–158. doi: 10.1111/j.1432-1033.1996.0153n.x. [DOI] [PubMed] [Google Scholar]
- Louvet E., Junera H. R., Le Panse S., Hernandez-Verdun D. Dynamics and compartmentation of the nucleolar processing machinery. Exp. Cell Res. 2005;304:457–470. doi: 10.1016/j.yexcr.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Meggio F., Pinna L. A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- Miyata Y., Nishida E. CK2 controls multiple protein kinases by phosphorylating a kinase-targeting molecular chaperone, Cdc37. Mol. Cell. Biol. 2004;24:4065–4074. doi: 10.1128/MCB.24.9.4065-4074.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda M., et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Okuwaki M., Tsujimoto M., Nagata K. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol. Biol. Cell. 2002;13:2016–2030. doi: 10.1091/mbc.02-03-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M., Nakagawa J., Dorée M., Labbé J.-C., Nigg E. A. Identification of major nucleolar proteins as candidate mitotic substrates of cdc2 kinase. Cell. 1990;60:791–801. doi: 10.1016/0092-8674(90)90093-t. [DOI] [PubMed] [Google Scholar]
- Puvion-Dutilleul F., Mazan S., Nicoloso M., Pichard E., Bachellerie J. P., Puvion E. Alterations of nucleolar ultrastructure and ribosome biogenesis by actinomycin D. Implications for U3 snRNP function. Eur. J. Cell Biol. 1992;58:149–162. [PubMed] [Google Scholar]
- Rubbi C. P., Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu C., Namane A., Gleizes P. E., Lebreton A., Rousselle J. C., Noaillac-Depeyre J., Gas N., Jacquier A., Fromont-Racine M. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino T. M., Bastos R., Jansen E., Hernandez-Verdun D. The nucleolar antigen Nop52, the human homologue of the yeast ribosomal RNA processing RRP1, is recruited at late stages of nucleologenesis. J. Cell Sci. 1999;112:1889–1900. doi: 10.1242/jcs.112.12.1889. [DOI] [PubMed] [Google Scholar]
- Savino T. M., Gébrane-Younès J., De Mey J., Sibarita J.-B., Hernandez-Verdun D. Nucleolar assembly of the rRNA processing machinery in living cells. J. Cell Biol. 2001;153:1097–1110. doi: 10.1083/jcb.153.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur R. S., Olson M.O.J. Preferential cleavage in pre-ribosomal RNA by protein B23 endoribonuclease. Nucleic Acids Res. 1998;26:4508–4515. doi: 10.1093/nar/26.19.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U., Hock R. Structure and function of the nucleolus. Curr. Opin. Cell Biol. 1999;11:385–390. doi: 10.1016/S0955-0674(99)80054-4. [DOI] [PubMed] [Google Scholar]
- Scheer U., Hügle B., Hazan R., Rose K. M. Drug-induced dispersal of transcribed rRNA genes and transcriptional products: immunolocalization and silver staining of different nucleolar components in rat cells treated with 5,6-Dichloro-β-D-ribofuranosylbenzimidazole. J. Cell Biol. 1984;99:672–679. doi: 10.1083/jcb.99.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U., Rose K. M. Localisation of RNA polymerase I in interphase cells and mitotic chromosomes by light and electron microscopic immunocytochemistry. Proc. Natl. Acad. Sci. USA. 1984;81:1431–1435. doi: 10.1073/pnas.81.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U., Thiry M., Goessens G. Structure, function and assembly of the nucleolus. Trends Cell Biol. 1993;3:236–241. doi: 10.1016/0962-8924(93)90123-i. [DOI] [PubMed] [Google Scholar]
- Shaw P. J., Jordan E. G. The nucleolus. Annu. Rev. Cell Dev. Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Sirri V., Hernandez-Verdun D., Roussel P. Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J. Cell Biol. 2002;156:969–981. doi: 10.1083/jcb.200201024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirri V., Roussel P., Hernandez-Verdun D. In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J. Cell Biol. 2000;148:259–270. doi: 10.1083/jcb.148.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strouboulis J., Wolffe A. P. Functional compartmentalization of the nucleus. J. Cell Sci. 1996;109:1991–2000. doi: 10.1242/jcs.109.8.1991. [DOI] [PubMed] [Google Scholar]
- Suprynowicz F. A., Gerace L. A fractionated cell-free system for analysis of prophase nuclear disassembly. J. Cell Biol. 1986;103:2073–2081. doi: 10.1083/jcb.103.6.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebeni A., Hingorani K., Negi S., Olson M.O.J. Role of protein kinase CK2 phosphorylation in the molecular chaperone activity of nucleolar protein B23. J. Biol. Chem. 2003;278:9107–9115. doi: 10.1074/jbc.M204411200. [DOI] [PubMed] [Google Scholar]
- Tokuyama Y., Horn H. F., Kawamura K., Tarapore P., Fukasawa K. Specific phosphorylation of nucleophosmin on Thr (199) by cyclin-dependent kinase 2-cyclin E and its role in centrosome duplication. J. Biol. Chem. 2001;276:21529–21537. doi: 10.1074/jbc.M100014200. [DOI] [PubMed] [Google Scholar]
- Valdez B. C., Perlaky L., Henning D., Saijo Y., Chan P. K., Busch H. Identification of the nuclear and nucleolar localization signals of the protein p120. Interaction with translocation protein B23. J. Biol. Chem. 1994;269:23776–23783. [PubMed] [Google Scholar]
- Wagner S., Chiosea S., Ivshina M., Nickerson J. A. In vitro FRAP reveals the ATP-dependent nuclear mobilization of the exon junction complex protein SRm160. J. Cell Biol. 2004;164:843–850. doi: 10.1083/jcb.200307002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung B. Y., Bor A. M., Chan P. K. Short exposure to actinomycin D induces “reversible” translocation of protein B23 as well as “reversible” inhibition of cell growth and RNA synthesis in HeLa cells. Cancer Res. 1990;50:5987–5991. [PubMed] [Google Scholar]
- Zandomeni R., Zandomeni M. C., Shugar D., Weimann R. Casein kinase type II is involved in the inhibition by 5,6 dichloro 1 b D ribofuranosylbenzimidazole of specific RNA polymerase II transcription. J. Biol. Chem. 1986;261:3414–3419. [PubMed] [Google Scholar]
- Zatsepina O. V., Todorov I. T., Philipova R. N., Krachmarov C. P., Trendelenburg M. F., Jordan E. G. Cell cycle-dependent translocations of a major nucleolar phosphoprotein, B23, and some characteristics of its variants. Eur. J. Cell Biol. 1997;73:58–70. [PubMed] [Google Scholar]