Abstract
Cholangiocellular carcinoma (CC), the second most common primary liver cancer, is associated with a poor prognosis. It has been shown that CCs harbor alterations of a number of tumor-suppressor genes and oncogenes, yet key regulators for tumorigenesis remain unknown. Here we have generated a mouse model that develops CC with high penetrance using liver-specific targeted disruption of tumor suppressors SMAD4 and PTEN. In the absence of SMAD4 and PTEN, hyperplastic foci emerge exclusively from bile ducts of mutant mice at 2 months of age and continue to grow, leading to tumor formation in all animals at 4–7 months of age. We show that CC formation follows a multistep progression of histopathological changes that are associated with significant alterations, including increased levels of phosphorylated AKT, FOXO1, GSK-3β, mTOR, and ERK and increased nuclear levels of cyclin D1. We further demonstrate that SMAD4 and PTEN regulate each other through a novel feedback mechanism to maintain an expression balance and synergistically repress CC formation. Finally, our analysis of human CC detected PTEN inactivation in a majority of p-AKT–positive CCs, while about half also lost SMAD4 expression. These findings elucidate the relationship between SMAD4 and PTEN and extend our understanding of CC formation.
Introduction
Cholangiocarcinoma, or cholangiocellular carcinoma (CC), is a malignant epithelial neoplasm with bile duct epithelial differentiation. The molecular mechanisms associated with cancer initiation and progression of this cancer remain unclear (1–3). CC accounts for approximately 15% of the total liver cancer cases in the world, with significant variations from country to country, and it is associated with poor prognosis, with most patients dying soon after diagnosis (1–4). Indeed, CC is the most common primary liver cancer–related cause of death in the United Kingdom (4). It has been shown that CC harbors alterations of a number of tumor-suppressor genes and oncogenes, including p53, p16, p27, p57, SMAD4, β-catenin, cyclin D1, ERK, Ras, AKT, and c-Myc (3, 5–12). In part because of the paucity of proper animal models, critical roles of these alterations in tumor initiation, progression, and metastasis have not been studied.
SMAD4 is a common mediator of TGF-β signals (13, 14). Deletion or mutation of SMAD4 has been detected in pancreatic cancer, colon cancer, gastric polyposis, and adenocarcinomas (reviewed in ref. 15). SMAD4 is also one of the most frequently altered tumor suppressor genes in CC (5). SMAD4 is essential for embryonic development in mice, as loss of SMAD4 results in lethality at E6–7 due to impaired extraembryonic membrane formation and decreased epiblast proliferation (16, 17). Smad4-heterozygous mice developed gastric polyposis and cancer due to haploinsufficiency (18, 19). To study SMAD4 in postnatal development, we have created mice carrying a conditional allele of Smad4 (Smad4Co) (20). This strain of mouse is useful in studying functions of SMAD4 in different organs/tissues when a tissue specific Cre expression system is used (21, 22).
Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) is mutated in a wide range of human cancers (reviewed in ref. 23). PTEN is a phosphoprotein/phospholipid dual-specificity phosphatase that antagonizes the activity of PI3K on phosphoinositide substrates (24). It has been shown that the absence of functional PTEN leads to constitutive activation of downstream components of the PI3K pathway, including AKT/PKB, a survival factor that protects various cell types against apoptosis (25). PTEN acts as a tumor suppressor in most cells by inducing G1-phase cell-cycle arrest, which has been attributed to a decrease in the level and nuclear localization of cyclin D1 (26). Currently, mutations of PTEN have not been detected in CC (27), although phosphorylated (activated) AKT was found to be expressed in 16 of 19 bile duct cancers (28) and 25 of 33 intrahepatic CCs analyzed (12). These observations suggest that PTEN may be involved in CC formation through its downstream genes. Consistent with this, inhibition of AKT activation by LY294002 significantly decreased the viability of human CC cells (10). Moreover, abnormal nuclear overexpression of cyclin D1 has been frequently observed in CC (5, 7, 11). The overexpression of cyclin D1, however, could also be caused by a number of other factors, and so the role of PTEN in CC formation remains elusive.
To investigate the roles of SMAD4 and PTEN in CC formation, we disrupted these genes in the liver using the Cre-loxP approach. We showed that the absence of SMAD4 alone failed to induce CC, while about one-third of PtenCo/CoAlb-Cre mice developed foci of CC when they were older than 1 year of age. In contrast, hyperplastic foci emerged exclusively from bile ducts of Smad4Co/CoPtenCo/CoAlb-Cre mice at 2 months of age and continued to grow, leading to CC formation in all animals at 4–7 month of age. Our analysis of human CC also detected inactivation of PTEN by epigenetic modification and loss of SMAD4 expression in 71% and 48% of p-AKT–positive tumors, respectively. These findings provide a molecular basis for the synergistic action of SMAD4 and PTEN in inhibiting CC formation.
Results
Disruption of Smad4 and Pten in liver results in tumor formation.
To disrupt Smad4 and/or Pten specifically in the liver, we crossed mice carrying a Smad4 conditional allele (Smad4Co) (20) and/or a Pten conditional allele (PtenCo) (29) with transgenic mice carrying a Cre gene under control of an albumin promoter (30). These crosses generated cohorts of mice with various genotypes, including Smad4Co/CoAlb-Cre, PtenCo/CoAlb-Cre, Smad4Co/CoPtenCo/CoAlb-Cre, Smad4+/CoPtenCo/CoAlb-Cre, and wild-type. Using PCR, we detected the predicted deletion of Pten and/or Smad4 in the livers of Smad4Co/CoAlb-Cre, PtenCo/CoAlb-Cre, and Smad4Co/CoPtenCo/CoAlb-Cre mice, and liver cancers developed in Smad4Co/CoPtenCo/CoAlb-Cre mice but not in wild-type mice (Supplemental Figure 1, A–C; supplemental material available online with this article; doi:10.1172/JCI27282DS1). Our analysis of 15 tissues/organs isolated from Smad4Co/CoPtenCo/CoAlb-Cre mice detected Cre-mediated deletion of Pten and Smad4 in liver and salivary glands (Supplemental Figure 1D).
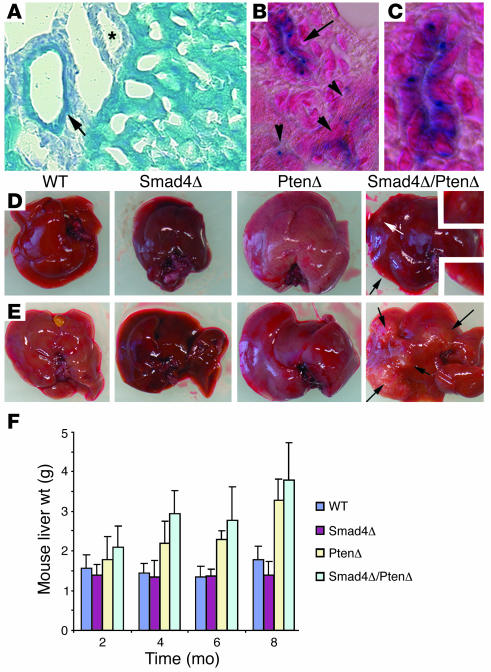
After crossing the Alb-Cre mice with a Rosa-26 reporter mouse (31), we detected β-galactosidase–positive cells in the majority of hepatocytes and bile duct epithelial cells at postnatal day (P30) (Figure 1A). Because endogenous albumin is expressed specifically in hepatocytes in adult liver, we suspected that the Cre-mediated recombination detected in the bile ducts of adult mice could be due to the albumin promoter activity during earlier developmental stages in these Alb-Cre–transgenic mice. Consistently, our analysis of mice at P15 and younger revealed β-galactosidase–positive cells in bile ducts and hepatocytes in a stochastic fashion (Figure 1, B and C).
Figure 1. Targeted disruption of Smad4 and Pten results in liver cancer.
(A–C) Alb-Cre activity assayed using Rosa-26 reporter mice at P30 (A) and P15 (B and C). Arrows indicate bile ducts, arrowheads indicate hepatocytes, and the asterisk marks an artery, which is not stained. The bile duct (arrow in B) is amplified in C. Magnification: ×300 (A), ×450 (B), ×900 (C). (D and E) Tumor formation in 4-month- (D) and 8-month-old (E) Smad4Co/CoPtenCo/CoAlb-Cre mice, but not in mice of other genotypes. (F) Weights of livers isolated from mice at different ages as indicated. PtenΔ, PtenCo/CoAlb-Cre mice; Smad4Δ, Smad4Co/CoAlb-Cre mice; Smad4Δ/PtenΔ, Smad4Co/CoPtenCo/CoAlb-Cre mice.
Next, we studied the development of the liver and tumorigenesis in these mutant mice. Our analysis of mice up to 10 months of age indicated that the absence of Pten or Smad4 alone does not result in tumor formation, although PtenCo/CoAlb-Cre mice developed fatty liver, as shown previously (29) and Smad4Co/CoAlb-Cre mice exhibited increased iron accumulation in the liver (32). However multiple visible tumor foci were observed in the livers of 4-month-old Smad4Co/CoPtenCo/CoAlb-Cre mice at autopsy (Figure 1D). These foci continuously increased in size (Figure 1E, arrows) as the animals aged and eventually resulted in the death of all Smad4Co/CoPtenCo/CoAlb-Cre mice before 10 months of age (Table 1). Our data also revealed that the sizes and weights of the liver were increased in both PtenCo/CoAlb-Cre mice and Smad4Co/CoPtenCo/CoAlb-Cre mice (Figure 1, D–F), primarily due to fat accumulation in the liver associated with PTEN deficiency, as documented previously (33, 34). Notably, the livers of Smad4Co/CoPtenCo/CoAlb-Cre mice were even heavier than those of PtenCo/CoAlb-Cre mice at all time points analyzed (Figure 1F). This increased weight correlated with tumor formation in the livers of the Smad4Co/CoPtenCo/CoAlb-Cre mice. We also examined the salivary glands, another organ that expresses Alb-Cre, and we detected 3 tumors in 35 Smad4Co/CoPtenCo/CoAlb-Cre mice between 6 and 10 months of age and 2 tumors in 15 PtenCo/CoAlb-Cre mice between 12 and 16 months of age (Table 1). This observation suggests that SMAD4 and PTEN do not play a significant role in suppressing tumor formation in this organ.
Table 1 .
Tumor incidence in mice of various genotypes
Smad4Co/Co PtenCo/Co Alb-Cre mice develop CC prior to their death.
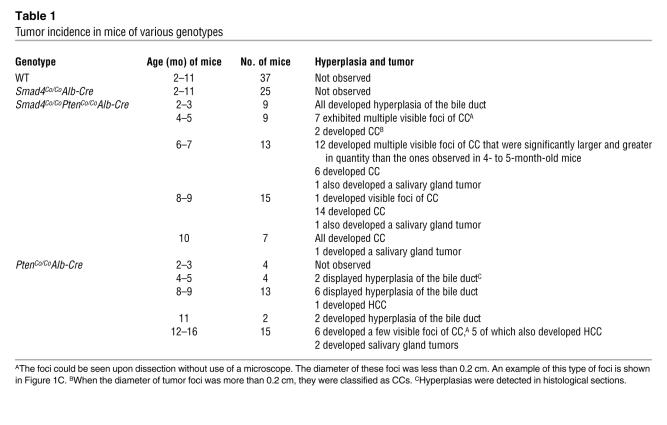
We next performed histological analysis of livers isolated from control and mutant mice. No obvious differences were detected between livers isolated from Smad4Co/CoAlb-Cre mice (Figure 2, A and B) and control mice (Figure 2, C and D) younger than 6 months of age. After 8 months of age, some of the livers in the Smad4Co/CoAlb-Cre mice showed focal accumulations of neutrophils and macrophages, but otherwise no tumors were identified in any of the mice examined (Table 1 and data not shown). Notably, livers isolated from all Smad4Co/CoPtenCo/CoAlb-Cre mice of 2 months (n = 6) and 3 months (n = 3) of age had varying degrees of hyperplasia in their bile ducts (Table 1), which were characterized by increased size, number, and altered morphology of bile duct epithelial cells (Figure 2, A and B). Multiple abnormal bile ducts were found surrounding each portal vein in Smad4Co/CoPtenCo/CoAlb-Cre livers (Figure 2B), while in control liver the hepatic portal tract was often accompanied by 1 or 2 bile ducts (Figure 2, C and D). Significantly increased branching of bile ducts was also observed in the liver of Smad4Co/CoPtenCo/CoAlb-Cre mice (Figure 2E). Our examination of 9 animals at 4–5 months of age indicated that all animals displayed multiple tumor foci on the surface of their liver that were grossly appreciable (Table 1 and Figure 1, C and D). Older animals in general had more numerous and larger liver tumors than younger ones (Table 1). Histological analysis indicated that lesions were characterized by bile duct dysplasia with a papillary growth pattern (Figure 2F), multiple tumors with varying histopathology (Figure 2, G and H), and invasive CC histologically similar to human intrahepatic CC (Figure 2I). Thus, these data suggest that the CC progression follows a multistep progression that includes hyperplasia, dysplasia, carcinoma in situ, and eventually well-established CC. No metastases were found in these mice.
Figure 2. CC formation in Smad4Co/Co PtenCo/Co Alb-Cre mice.
(A–D) Histologic analysis of livers isolated from 2-month-old Smad4Co/CoPtenCo/CoAlb-Cre (A and B), WT (C), and Smad4Co/CoAlb-Cre (D) mice. Arrows indicate bile ducts. (E) An H&E-stained liver section showing significantly increased bile duct branching in the liver of a 3-month-old Smad4Co/CoPtenCo/CoAlb-Cre mouse. The arrow and arrowhead indicate large and small branches, respectively. (F–I) Bile duct dysplasia (F), CC foci with varying histopathology (G and H), and well-developed CC (I) found in Smad4Co/CoPtenCo/CoAlb-Cre livers. Arrowheads in the insets indicate cells at the mitotic phase. (J and K) Bile duct hyperplasia (J) and CC foci in PtenCo/CoAlb-Cre (K) livers. Magnification: ×100 (A); ×200 (B–D and H); ×400 (E–G and I–K).
Ten of 23 PtenCo/CoAlb-Cre mice examined between 2 and 11 months of age displayed varying degrees of epithelial hyperplasia primarily in larger bile ducts (Figure 2J and data not shown), although no invasive CCs were found in any of the animals examined (Table 1). Of note, 6 of 15 mice examined between 12 and 16 months of age had CC foci (Figure 2K), although they were much fewer and smaller than those observed in Smad4Co/CoPtenCo/CoAlb-Cre livers. This observation suggests that the absence of PTEN alone initiates bile duct neoplasia and that bile duct neoplasia is inhibited by the presence of SMAD4, especially when the animals are young.
It was recently shown that liver-specific disruption of Pten resulted in hepatocellular carcinoma (HCC) formation late in adulthood (34). Examination of our PtenCo/CoAlb-Cre animals detected 1 HCC among 13 animals at 8–9 months of age and 5 HCCs among 15 animals between 12 and 16 months (Table 1). The fact that Smad4Co/CoPtenCo/CoAlb-Cre mice did not develop HCC before they died at 10 months of age suggests that the absence of SMAD4 did not accelerate HCC formation during this period of time.
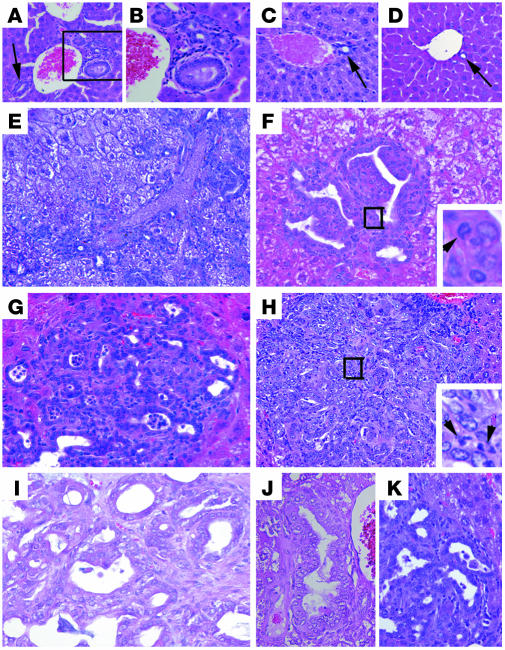
To confirm that the tumors developed from Smad4Co/CoPtenCo/CoAlb-Cre mice were indeed CCs with bile duct epithelial cell differentiation, we first labeled tissue sections with antibody A6, which recognizes oval cells and cholangiocytes in mice (35). A6 labeled all normal biliary epithelial cells in the wild-type liver (Figure 3A) and the neoplastic cells in the Smad4Co/CoPtenCo/CoAlb-Cre liver (Figure 3B), suggesting oval cell or bile duct differentiation of the tumors. Next, we used a panel of markers that have been used for CC diagnosis, including CK19 (36), AE1 (36), Mucicarmine (37), and Mucin 5AC (38). Our analysis indicated that all tumors expressed these markers (Figure 3, C–G). In contrast, labeling with an antibody to Hep Par1, a protein that is highly expressed in HCCs (36), showed that all the samples (n > 10) analyzed were negative, in comparison with the surrounding non-neoplastic hepatocytes (Figure 3, H and I, and arrow in I). These observations provide strong evidence that the loss of both SMAD4 and PTEN synergistically enhances CC formation in the liver of mutant mice.
Figure 3. Tumors developed from Smad4Co/Co PtenCo/Co Alb-Cre mice are exclusively of bile duct origin.
Molecular markers used are as indicated. (A and H) Livers from WT mice. (B–G and I) Livers from Smad4Co/CoPtenCo/CoAlb-Cre mice. The arrow in I indicates tumor cells that were Hep Par1 negative. At least 5 samples were used for each antibody. Magnification: ×300 (A–C); ×600 (D–G); ×500 (H and I).
Activation of AKT signaling pathways in Smad4Co/CoPtenCo/CoAlb-Cre tumors.
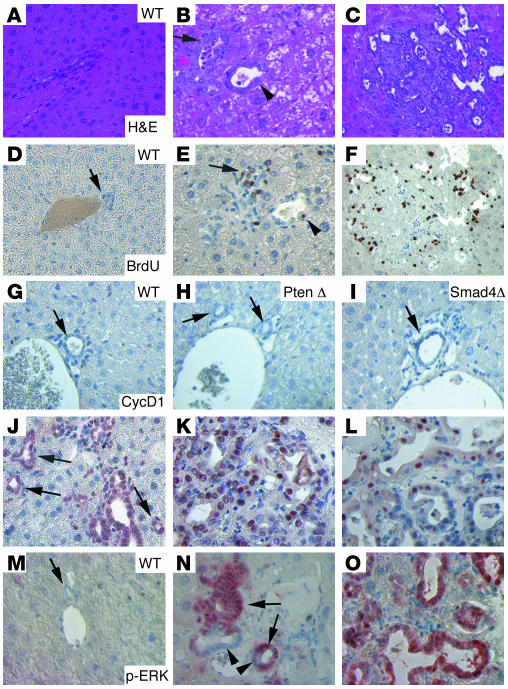
Next, we performed molecular analysis to characterize the tumors. Liver cells were normally in a quiescent state, as revealed by the lack of BrdU uptake (WT, Figure 4, A and D). In contrast, hyperplastic/neoplastic foci and tumors that developed in Smad4Co/CoPtenCo/CoAlb-Cre mice showed a markedly increased number of BrdU-positive cells (Figure 4, B, C, E, and F). This observation suggests that the loss of SMAD4 and PTEN promotes cells to exit the G1 phase, resulting in significantly increased cell proliferation. Consistent with this, hyperplastic/neoplastic foci and tumors also exhibited increased numbers of mitotic cells compared with control bile ducts (Supplemental Table 1). To identify molecular alterations that might be responsible for tumorigenesis, we first studied the expression of selected candidate genes using immunohistochemical labeling of livers isolated from 4-month-old control and mutant mice. Livers from mutant mice at this stage displayed multiple neoplastic foci and also some fully developed tumors. Previous investigations indicated that abnormal nuclear overexpression of cyclin D1 is frequently observed in human CC (5, 7, 11). We therefore performed immunohistochemical labeling using an antibody to cyclin D1. Our analysis revealed that cyclin D1 was not present at detectable levels in livers isolated from control mice (Figure 4, G–I). By contrast, we detected nuclear overexpression of cyclin D1 in hyperplastic bile ducts and even in many histologically normal-appearing bile ducts (small, with a single layer of cells; arrows in Figure 4J) in the Smad4Co/CoPtenCo/CoAlb-Cre mice. Because these bile duct were present at a high density and were not adjacent to any portal veins as normally seen in the control liver (Figure 4, G–I), we believe that they may be a part of the abnormally branched bile duct network observed in all Smad4Co/CoPtenCo/CoAlb-Cre mice. Abnormal nuclear localization of cyclin D1 was also found in CCs at all different stages, with varying frequency, ranging from 37% to 62% of cells (Figure 4, K and L). We have examined a total of 15 Smad4Co/CoPtenCo/CoAlb-Cre livers, and they all exhibited a similar pattern of cyclin D1 staining. Phosphorylation of the ERK pathway plays an important role in the induction of cyclin D1 expression (39, 40). Our analysis revealed that a majority of cells in hyperplasia (99/118, 84%; Figure 4N) and in fully developed CC (138/154, 90%; Figure 4O) exhibited high levels of phosphorylation of ERK. Of note, hyperplastic bile ducts still contained p-ERK–negative cells (arrowheads in Figure 4N), while about 95% of cells (218/230) in hyperplastic bile ducts were cyclin D1 positive (Figure 4J). This observation, i.e., that more cells are cyclin D1 positive than p-ERK positive in the hyperplasias, suggests that additional factors may also have contributed to the nuclear accumulation of cyclin D1 in this stage of tumorigenesis.
Figure 4. Molecular characterization of CCs that developed inSmad4Co/CoPtenCo/CoAlb-Cre mice.
H&E (A–C) and BrdU (D–F) images. Liver cells appeared normal in WT liver (A and D), and hyperplasic foci (B and E) and bigger tumors (C and F) developed in Smad4Co/CoPtenCo/CoAlb-Cre mice. The arrow in E indicates an area of BrdU-positive cells, and the arrowhead indicates a single BrdU-positive cell in a bile duct with moderately increased numbers of cells. Their corresponding positions are indicated in B. (G–O) Immunohistochemical staining with antibodies against cyclin D1 (CtcD1) (G–L) and p-ERK (M–O) of control liver, neoplastic foci, and bigger tumors. Arrows in J point to cyclin D1–positive bile ducts that still maintained single layers of cells. Arrowheads in N indicate p-ERK–negative cells in the single layer area of the bile duct. These cells gradually became p-ERK positive, especially when multiple layers of cells accumulated in the bile duct (arrows). Most samples were from 4-month-old mice, except for some bigger tumors (K, L, and O), which were from 8-month-old mutant mice. At least 5 samples of each genotype were used for each antibody. Unless otherwise indicated (in the upper-right corner), tumors from Smad4Co/CoPtenCo/CoAlb-Cre mice are shown. Magnification: ×500.
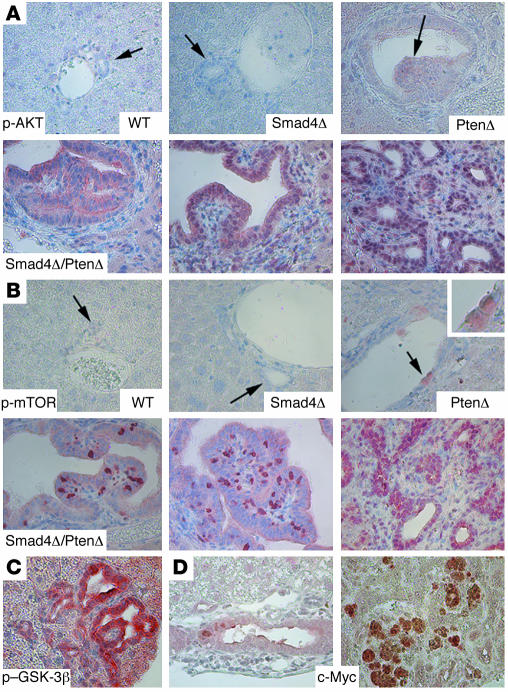
Next, we studied phosphorylation of AKT, a downstream gene of PTEN (25), in livers of control and mutant mice. Using immunohistochemical labeling, we showed that p-AKT was undetectable in wild-type and Smad mutant livers (Figure 5A), and it was slightly increased in both bile ducts and hepatocytes in Pten mutant livers (Figure 5A). The p-AKT levels were even higher in tumors (Figure 5A). Interestingly, we found that the p-AKT localization was initially both cytoplasmic and nuclear in hyperplastic bile ducts, while the nuclear component gradually increased in more advanced tumors (from left to right in the lower 3 panels; Figure 5A). Increased levels of mammalian target of rapamycin (mTOR) phosphorylation (Figure 5B) and glycogen synthase kinase-3β (GSK-3β) phosphorylation were also observed (Figure 5C and data not shown). Both mTOR and GSK-3β are major downstream targets of PTEN/PI3K/AKT signals, promoting cell cycle progression, cell growth, and proliferation. Activated GSK-3β is known to phosphorylate T286 of cyclin D1, reducing its half-life (41). Thus, the increased level of p–GSK-3β (which is an inactivated form) in CC is consistent with the abnormally increased nuclear accumulation of cyclin D1. We have also studied protein levels of some other factors that may affect cyclin D1 expression, such as pRB, p16, and p21, but did not detect a consistent pattern of change (data not shown), suggesting that they may not contribute to the increased nuclear accumulation of cyclin D1 at the early stages of tumorigenesis. Of note, we found that expression of c-Myc was present at low levels in the early stages of tumorigenesis but at higher levels in some tumors of later stages (Figure 5D), suggesting the possible involvement of this protein in later stages of tumorigenesis, when fractions of cyclin D1–positive cells are gradually reduced.
Figure 5. Activation of AKT/mTOR pathways in CCs that developed in Smad4Co/CoPtenCo/CoAlb-Cre mice.
(A) Levels of p-AKT. Genotypes are as indicted. (B) Phosphorylated mTOR (p-mTOR).The lower panels in A and B represent samples from Smad4Co/CoPtenCo/CoAlb-Cre mice. (C) Phosphorylated GSK-3β (p–GSK-3β). (D) c-Myc. Arrows in A and B indicate bile ducts. Note that the bile ducts in PtenCo/CoAlb-Cre livers were hyperplastic and abnormally enlarged. Some bile duct cells also displayed increased expression of p-AKT and p-mTOR (arrows). This phenotype was not found in WT and Smad4Co/CoAlb-Cre samples. At least 5 samples of each genotype were used for each antibody. Magnification: ×500.
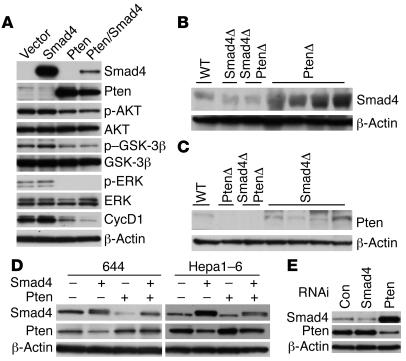
To understand further the mechanisms underlying tumorigenesis, we derived cell lines from the CCs and transfected 1 of the cell lines (858; Supplemental Figure 2, A and B) with Smad4 and/or Pten expression vectors. We found that expression of Pten alone, but not Smad4 alone, inhibited phosphorylation of AKT, GSK-3β, and ERK, as well as expression of cyclin D1 (Figure 6A). Of note, the transfection of Smad4 alone did not affect cyclin D1 expression; however, it caused a further decrease in cyclin D1 expression when cotransfected with Pten as compared with Pten transfection alone, suggesting a synergistic action between SMAD4 and PTEN in inhibiting cyclin D1 expression.
Figure 6. Analysis of cultured cells derived from liver cancers and normal livers of control mice.
(A) Transfection of Smad4 and Pten expression vectors separately or together into 858 cells. Ten micrograms of total DNA were used for each transfection in each 10-cm dish in various combinations: i.e., pcDNA vector alone, vector plus Smad4 or Pten, or Smad4 plus Pten of 5 μg each. Multiple loading controls were performed, but only 1 is shown. (B and C) Western blot analyses of SMAD4 (B) and PTEN (C) using lysates isolated from livers of various genotypes, as indicated. (D) Ectopic overexpression of Smad4 or Pten in 644 and Hepa1–6 cells decreased expression of each other. In all the above experiment, samples were harvested 48 hours after transfection. (E) Acute suppression of Smad4 or Pten in Hepa1–6 cells. Cell lysates were prepared 30 hours after transfection and subjected to Western blot analysis. Con, control.
A negative feedback loop between PTEN and SMAD4.
Our data indicate that the absence of both SMAD4 and PTEN synergistically induces CC, suggesting that the absence of one protein could be compensated by the presence of the other. To investigate this, we performed Western blot analysis on liver lysates isolated from mutant and control mice. Antibody against SMAD4 revealed a remarkable increase in SMAD4 expression in PTEN–null livers (Figure 6B). An obvious, yet less significant, increase in expression of PTEN was also observed in the SMAD4-null livers (Figure 6C). Thus, the loss of each gene was associated with an upregulation of the other.
Because the liver contains mostly hepatocytes, while only a small population of cells is from bile ducts, we suspected that such a negative inhibition between SMAD4 and PTEN could also occur in hepatocytes. To study this, we transfected Smad4 and Pten into 2 cell lines: 644, which was developed from the liver epithelial cells of a 1-month-old Smad4Co/CoPtenCo/Co mouse, and Hepa1–6, which was derived from a mouse HCC (42) (Supplemental Figure 2, E and F). Our data revealed a similar mutual inhibition effect between SMAD4 and PTEN (Figure 6, D and E). This observation suggests that the negative regulation between SMAD4 and PTEN exists in both hepatocytes and bile duct cells. To provide additional evidence for this observation, we performed RNA interference–mediated (RNAi-mediated) knockdown of Smad4 and Pten in Hepa1–6 cells. Our data revealed that acute suppression of SMAD4 resulted in increased levels of PTEN and vice verse (Figure 6E).
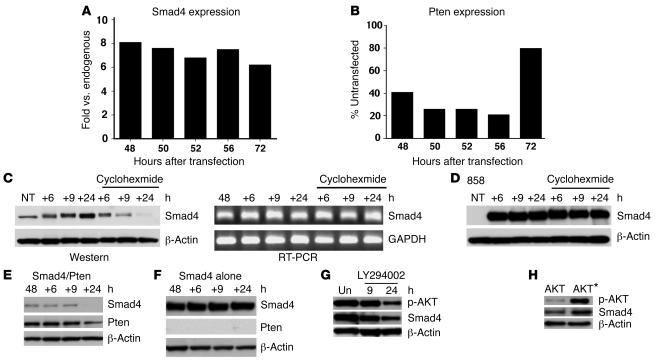
It was previously shown that TGF-β negatively regulates Pten expression at the transcription level both in vitro and in vivo (43, 44). Because SMAD4 is a common mediator of signals of the TGF-β superfamily, we hypothesized that this effect is mediated by SMAD4. To test this, we overexpressed Smad4 in 644 cells and studied its effect on Pten expression. Using real-time PCR analysis, we showed that the overexpression of Smad4 (Figure 7A) indeed resulted in decreased transcription of Pten (Figure 7B). The inhibition was more obvious in the first 56 hours after Smad4 transfection than at 72 hours. Because the cells had reached highest density and only maintained minimal proliferation activity at this point, it is possible that the inhibition efficiency of Smad4 on Pten transcription was attenuated.
Figure 7. SMAD4 and PTEN negatively regulate each other.
(A and B) Real-time PCR analysis of Smad4 (A) and Pten (B) transcription levels in 644 cells transfected with Smad4. Levels of gene expression are either expressed as fold change as compared with endogenous Smad4, which was set as 1 (A), or as percentage of untransfected Pten, which was set as 100% (B). The data shown were averaged from 3 independent transfections. (C) SMAD4 protein (left) and transcription (right) levels of 644 cells that were transfected with Smad4. (D) SMAD4 protein levels in SMAD4/PTEN double mutant (i.e., 858) cells that were transfected with Smad4. Forty-eight hours after transfection, cells were treated with cycloheximide, 100 μg/ml) or mock treated for an additional 6, 9, and 24 hours as indicated. (E and F) SMAD4 and PTEN levels in 858 cells that were transfected with Smad4 and Pten (E) or Smad4 plus vector (F). Forty-eight hours after transfection, cells were treated with cycloheximide for 6, 9, and 24 hours before being subjected to Western blot analysis. (G) SMAD4 and p-AKT levels in 858 cells that were transfected with Smad4. Forty-eight hours after transfection, cells were treated with LY294002 at 10 μM for 9 and 24 hours before being subjected to Western blot analysis. (H) Expression of the activated form of AKT* which carries changes in T308A/S473A (49), in Hepa1–6 cells increased SMAD4 protein levels (right lane) compared with WT AKT-transfected cells (left lane). NT, nontransfected cells; Un, untreated cells.
We next studied whether the expression of Pten could affect Smad4. We transfected Pten into 644 cells and found no obvious change in the transcription levels of Smad4 (data not shown), suggesting that the effect of Pten on Smad4 might be at the protein level. We therefore tested SMAD4 stability in 644 cells and Smad4Co/CoPtenCo/CoAlb-Cre cells (858 cells). After transfecting Smad4 into these cells, we monitored SMAD4 protein levels by Western blot analysis and found that SMAD4 was always present at significantly higher levels in 858 cells than in 644 cells (Figure 7, C and D), a finding consistent with the view that PTEN negatively regulates SMAD4 expression. To determine whether this effect is due to the stability of SMAD4, we treated the cells with cycloheximide 48 hours after transfection. We found that SMAD4 levels were reduced in 644 cells 9 hours after the addition of cycloheximide and diminished 24 hours later (Figure 7C, left), while Smad4 transcriptional levels were not changed (Figure 7C, right). In contrast, SMAD4 levels did not change even 24 hours after cycloheximide treatment in 858 cells (Figure 7D). Similar observations were also made in Hep1-6 cells. Because the 644 and Hep1-6 cells express PTEN while the 858 cells do not, these data suggest that the presence of PTEN promotes SMAD4 degradation, while the absence of PTEN stabilizes SMAD4.
To provide additional evidence for this hypothesis, we next cotransfected Smad4 and Pten into the 858 cells and studied SMAD4 levels prior to and after cycloheximide treatment. Consistently, our data indicated that SMAD4 disappeared 24 hours after cycloheximide treatment in the Smad4/Pten-cotransfected cells (Figure 7E), while no significant differences were observed during the same period of time in the cells transfected with Smad4 but without Pten (Figure 7F).
PTEN is known to inhibit PI3K activity and AKT phosphorylation (24). Therefore, we suspected that the effect of PTEN on SMAD4 stability might be mediated through the PI3K/AKT signaling pathway. To test this, we treated Smad4-transfected cells with LY294002, an inhibitor of PI3K activity. We found that the presence of LY294002 quickly decreased AKT phosphorylation and SMAD4 levels in these cells (Figure 7G). Conversely, the expression of a constitutively activated form of AKT resulted in increased levels of SMAD4 (Figure 7H). These data are consistent with the fact that PTEN regulates SMAD4 stability through the PI3K/AKT signaling pathway.
Inactivation of PTEN and SMAD4 in human CC.
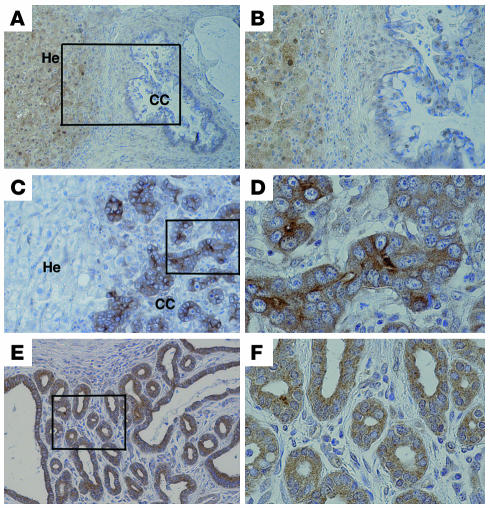
Our previous investigation indicated that 76% (25/33) of human CCs displayed increased expression of AKT (12). It was also shown that 45% (19/42) of these cancers failed to express SMAD4 protein (5). These studies implicated both AKT activation and SMAD4 inactivation in CC formation. To further address the relationship between these genes, we determined SMAD4 expression in AKT-positive samples by immunohistochemical staining using an antibody against SMAD4. Our data revealed that of 21 AKT-positive tumors analyzed, 48% (10/21) were negative for SMAD4 immunoreactivity in the tumor cells, while the neighboring hepatocytes remained positive for SMAD4 (Figure 8, A and B, and Supplemental Figure 3, A and B). Currently, genomic mutations in PTEN genes have not been found in CC (27). However, PTEN could also be inactivated by epigenetic modification through protein phosphorylation. Using an antibody against p-PTEN, we found that 71% (15/21) samples were p-PTEN positive (Figure 8, C and D). This observation suggests that PTEN inactivation is responsible for AKT activation in the majority of the CC samples we studied. Recent studies indicated that inactivation of members of the forkhead box O (FOXO) family of transcription factors, which control cell cycle and cell death, plays a critical role in malignant transformation of PTEN-negative/p-AKT–positive tumors (reviewed in ref. 45). The inactivation of FOXO proteins is characterized by protein phosphorylation and cytoplasmic retention. Therefore, we next examined phosphorylation of FOXO using an antibody that is specific for the phosphorylated form of FOXO1. Our data indicated that 76% (16/21) were positively stained and the p-FOXO1 was exclusively located in the cytoplasm (Figure 8, E and F). These data provide compelling evidence that the inactivation of PTEN and SMAD4 is a frequent event in human CC and the inactivation of FOXO by AKT signaling is involved in human CC formation. Of note, similar cytoplasmic retention of FOXO1 protein was also observed in the CC at various stages developed from our mouse model (Supplemental Figure 3, C and D). This observation reveals a similar molecular mechanism underlying tumorigenesis in our animals and the p-AKT–positive human CC.
Figure 8. Immunohistochemistry for SMAD4, p-PTEN, and p-FOXO1.
Representative images of SMAD4 immunostaining (×100 [A], ×200 [B]), p-PTEN (×200 [C], ×600 [D]), and p-FOXO (×200 [E], ×600 [F]). He, hepatocytes.
Discussion
Despite steady increases in the worldwide incidence and mortality rate of CC over the past 3 decades (1–3), mouse models for this devastating disease are rare. Kiguchi et al. reported that overexpression of ErbB-2 in the basal layer of biliary epithelium led to gallbladder carcinoma formation in 100% of transgenic mice by 3 months of age, and at the same time 25% of the mice also developed CC (46). However, CC formation in this animal has not been characterized, perhaps due to its high penetrance of gallbladder carcinomas. In this study, we studied functions of Smad4 and Pten in liver development and neoplasia, using liver-specific disruption of these genes in the mouse. We showed that absence of SMAD4 alone failed to cause CC, although some PtenCo/CoAlb-Cre mice exhibited CC foci between 12 and 16 months of age. In contrast, neoplastic foci emerged from bile ducts of Smad4Co/CoPtenCo/CoAlb-Cre mice at 2 months of age and continued to grow, leading to tumor foci and/or CC formation in all animals at 4–5 months of age. This observation uncovers a synergistic role of PTEN and SMAD4 in repressing CC formation. The mechanism underlying the synergistic action of these genes is concomitant repression of Pten transcription by SMAD4 and promotion of SMAD4 degradation by PTEN. This reciprocal negative feedback loop ensures that the absence of one gene triggers upregulation of the other and may have the following significance. First, in the abnormal situation when mutations occur, the effect of the absence of one tumor suppressor can be compensated by the increased expression of the other, and, therefore, tumorigenesis can be repressed.
Second, this negative regulation loop may serve as a mechanism to ensure that the expression levels of these genes are not disproportionally high in relation to each other during the normal developmental process. This is perhaps because both SMAD4 and PTEN have profound inhibitory effects on cell proliferation. It has been shown that targeted disruption of murine Pten results in hyperproliferation prior to the embryonic lethality at E9.5 (47). Mice carrying tissue-specific disruption of PTEN or mice heterozygous for a PTEN-null mutation exhibited increased cell proliferation and were highly susceptible to spontaneous tumor formation (29, 48). Similarly, mice heterozygous for SMAD4-null mutation or mice carrying conditional knockout SMAD4 in mammary epithelium also developed tumors characterized by increased cell proliferation (18, 21). It has been shown that PTEN-deficient cells exhibit elevated PKB/AKT activity and accelerated G1/S progression that may be caused by increased nuclear accumulation of cyclin D1 (26, 47). On the other hand, the pathway through which SMAD4 affects cell proliferation is less clear, but increased expression of cyclin D1 has been reported in SMAD4-deficient tumor cells (18). These observations suggest that cyclin D1 may be a common target of SMAD4 and PTEN in inhibiting cell proliferation. Our data indicated that cotransfection of Smad4 and Pten indeed results in more dramatic suppression of cyclin D1 than transfection of these genes separately.
It was recently shown that a liver-specific disruption of Pten resulted in HCC in 45% (9/19) of mice at 40–44 weeks and 67% (8/12) of mice at 74–78 weeks of age (34). Examination of our PtenCo/CoAlb-Cre animals indicated that 8% (1/13) developed HCC at 8–9 months (34–38 weeks) and 33% (5/15) at 12–16 months (52–65 weeks) (Table 1). The average genetic background of the mice in that study was 75% B6C57/25% 129, while the background of our mice was, on average, 37.5% 129/25% FVB/25% Balb/c/12.5% Black Swiss. The difference in the genetic background could be a major factor underlying the different HCC frequencies observed in these mice. Consistently, our examination of 12 PtenCo/CoAlb-Cre mice, which were in a genetic background of 87.5% B6C57/12.5% 129, revealed that 91% (11/12) developed HCC between 12 and 20 months of age. This observation indicated that when the animals have a similar genetic background, they also have similar frequencies of HCC formation. Of note, some PtenCo/CoAlb-Cre mice in both genetic backgrounds we studied developed CC foci and/or CC formation after 1 year of age. This includes 6/15 mice listed in Table 1 and 5/12 mice in the genetic background of 87.5% B6C57/12.5% 129 (data not shown), while no documentation about CC formation was presented in the study by Horie et al. (34). Because the mice in Horie’s study (75% B6C57/25% 129) share a similar genetic background with that of our mice (87.5% B6C57/12.5% 129), they might also develop CC at an older age, although this remained to be confirmed. We would also like to indicate that our Smad4Co/CoPtenCo/CoAlb-Cre mice may not be a pure CC model, as we believe that if these animals could live slightly longer, they would develop both CC and HCC.
Our analysis of mutant mice showed that Alb-Cre is expressed in both bile ducts and hepatocytes (Figure 1, A and B). We also showed that the regulation of the loop between SMAD4 and PTEN occurs in both hepatocytes and bile duct cells (Figures 6 and 7). However, 100% of Smad4Co/CoPtenCo/CoAlb-Cre animals developed CC at 4–7 months of age, and none of them developed HCC during the same period of time. Although HCC might potentially occur in an older population of Smad4Co/CoPtenCo/CoAlb-Cre mice, this did not happen till the death of all animals at 10 months of age. This observation suggests that bile duct cells are more sensitive to the loss of both SMAD4 and PTEN than hepatocytes for malignant transformation. The molecular mechanism underlying this differential response to the loss of SMAD4 and PTEN in CC and HCC formation is currently unclear and will be addressed in future studies.
Human CCs harbor alterations of a number of tumor suppressor genes, including p53, p16, p27, p57, SMAD4, and oncogenes such as β-catenin, cyclin D1, ERK, Ras, AKT, and c-Myc (3, 5–12). These alterations vary significantly among individuals, perhaps due to the heterogeneous nature of the human. We have sequenced Kras (exons 1 and 2), p53 (exons 2–9), and the promoter of the Ink4a/p16 gene, and we did not detect any mutations in these genes. We have also examined the methylation status of the Ink4a/p16 promoter, and we did not find any alterations compared with wild-type control. These findings suggest that once both SMAD4 and PTEN are mutated in early stages of liver development, CC formation does not require mutations of Kras, Ink4a/ARF, and p53 in our mice with a defined genetic background. Instead, we detected, at the earliest stages of tumorigenesis, activation of AKT, mTOR, ERK, and cyclin D1 and inactivation of FOXO1, some of which have been implicated in human CC formation. Because the CC in Smad4Co/CoPtenCo/CoAlb-Cre mice developed at defined time points with high penetrance and activation of AKT has been linked to a majority of human CCs, this animal model should be useful in identifying markers associated with the earliest changes during CC initiation and the mechanism(s) underlying tumor progression, as well as for developing techniques for drug screening, chemoprevention, and therapeutic treatment of the disease.
Methods
Mice.
Mice carrying Smad4 (20) and Pten (29) conditional knockout alleles were crossed with an Alb-Cre–transgenic mouse (30). After multiple rounds of crossing, we have generated mice with the following genotypes: Smad4Co/CoPtenCo/CoAlb-Cre, Smad4+/CoPtenCo/CoAlb-Cre, PtenCo/CoAlb-Cre, Smad4Co/CoAlb-Cre, and wild type. Genotyping of Smad4Co and PtenCo mice were as described previously (20, 29). The Alb-Cre–transgenic mice were genotyped using 2 primers for Cre: Cre-1 (5′-CCTGTTTTGCACGTTCACCG-3′) and Cre-3 (5′-ATGCTTCTGTCCGTTTGCCG-3′). Rosa-26–β-gal reporter line (R26R) (31) was purchased from the Jackson Laboratory and genotyped using primers for the lacZ gene: Lac-1 (5′-GATCCGCGCTGGCTACCGGC-3′) and Lac-2 (5′-GGATACTGACGAAACGCCTGCC-3′). All experiments were approved by the Animal Care and Use Committee of the NIDDK.
Cell culture and treatment.
To derive cell lines from CC and normal livers, we minced tumor tissue and livers of various genotypes in DMEM (Invitrogen Corp.) containing 10 μg/ml of bacterial collagenase type III. After overnight digestion, cells were washed with PBS and cultured in DMEM containing 10% FBS, glutamine, antibiotics, insulin (5 μg/ml), and EGF (10 μg/ml). Cultures derived from CCs frequently exhibited numerous foci of epithelial differentiation (Supplemental Figure 2A), which were expanded using cloning rings. A total of 20 cell lines that were derived from CCs and appeared to be epithelial cells (Supplemental Figure 2B) and expressed CK19 and AE1 (data not shown) were established. These cells did not express SMAD4 and PTEN (Supplemental Figure 2C) and exhibited minimal levels of genes immediately downstream of TGF-β, such as Smad6 and Smad7 (Supplemental Figure 2D). Cultures from the normal liver of Smad4Co/CoPtenCo/Cocontrol mice did not form obvious foci. After continuous expansion of areas with epithelial-like cells using cloning rings, 2 cell lines (i.e., 644 and 644-2) were established from the liver of 1-month-old Smad4Co/CoPtenCo/Comice through spontaneous immortalization during a prolonged period of culture. One cell line (i.e., 644) was further characterized and showed epithelial-like morphology (Supplemental Figure 2E). Hepa1–6 (Supplemental Figure 2F), a cell line derived from a mouse HCC (42), was cultured in DMEM with 10% FBS, glutamine, and antibiotics. Plasmids bearing Smad4, Pten (gifts from Y. Zhang, NIH), activated AKT (49), or GFP expression unit were transfected into these cells 24 hours after plating (about 80% confluence) using Lipofectamine 2000 (Invitrogen Corp.). In some cases, cycloheximide (Sigma-Aldrich) at a concentration of 100 μg/ml was added 48 hours after transfection to block protein synthesis. RNAi specific for Smad4 (SC-29485; Santa Cruz Biotechnology Inc.), Pten (SC-36326; Santa Cruz Biotechnology Inc.), and control (SC-37007; Santa Cruz Biotechnology Inc.) at a concentration of 0.2 μM were transfected into 644 and Hepa1–6 cells using Oligofectamine (Invitrogen Corp.).
Real-time PCR analysis.
Total RNA was extracted from cells using RNA STAT-60 (Tel-Test Inc.). Real-time PCR was performed with ABI PRISM 7000 Sequence Detection System (Applied Biosystems), using the following primers: Smad4-1, 5′-GACAGTGTCTGTGTGAATCC-3′; Smad4-2, 5′-TACTTGGCGGGTGTTGGATG-3′; Pten-1, 5′-TGCAGAAAGACTTGAAGGTG-3′; Pten-2, 5′-ATAAGTTCTAGCTGTGGTGG-3′; GAPDH-1, 5′-ACAGCCGCATCTTCTTGTGC-3′; GAPDH-2, 5′-TTTGATGTTAGTGGGGTCTCGC-3′.
Immunoblot analysis.
Western blot analyses were performed by standard procedures using ECL detection (Amersham Biosciences). The following primary antibodies were used: SMAD4 and cyclin D1 (Santa Cruz Biotechnology Inc.), β-actin (Sigma-Aldrich), PTEN, AKT, p-AKT (Ser473), GSK-3β, p–GSK-3β, and p-ERK1/2 (Cell Signaling Technology). HRP-conjugated anti-rabbit and anti-mouse antibodies (Kirkegaard & Perry Laboratories Inc.) were used as secondary antibodies.
Histology and immunohistochemical staining.
Paraffin sections of 4–5 μm were prepared for H&E and antibody staining. The following antibodies were used for immunohistochemical staining for mouse specimens: A6 antibody (a gift from Valentina M. Factor, NIH), AE-1 (Signet Laboratories), CK19 (Dako), Hep Par1 (Dako), c-Myc (Santa Cruz Biotechnology Inc.), Mucin 5AC (Novocastra), FOXO1 (catalog 9461; Cell Signaling Technology), and BrdU (BrdU staining kit; Zymed Laboratories Inc.), p-AKT (Ser473), p–GSK-3β (Ser9), p-ERK1/2, and p-mTOR (Ser2448) (Cell Signaling Technology).
For human CC specimens, heat antigen retrieval was performed in 1 mM EDTA (pH 8.0) for 40 minutes using a commercial vegetable steamer. The following primary antibodies were used for overnight incubation at 4°C: SMAD4 (1:50; SC-7966, Santa Cruz Biotechnology Inc.), p-PTEN (1:50; catalog 9551, Cell Signaling Technology), and p-FOXO (1:25; catalog 9461, Cell Signaling Technology). For SMAD4, cells staining more weakly and strongly than hepatocytes were considered negative and positive, respectively. For p-PTEN and p-FOXO1, cells that were stained were considered positive and those that did not were considered negative.
Supplementary Material
Acknowledgments
We thank V.M. Factor for the A6 antibody, S.P. Monga for the Hepa1–6 cell line, S.W. Michnick for the activated form of AKT, and Ying Zhang and members of the Deng lab for critically reading the manuscript. We are grateful for Ralph H. Hruban for his kind help in evaluating histopathology of CC and editing this manuscript. This research was supported by the Intramural Research Program of the NIDDK, NIH.
Footnotes
Nonstandard abbreviations used: CC, cholangiocellular carcinoma; FOXO, forkhead box O; GSK-3β, glycogen synthase kinase-3β; HCC, hepatocellular carcinoma; mTOR, mammalian target of rapamycin; P30, postnatal day 30; PTEN, phosphatase and tensin homolog deleted on chromosome 1.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 116:1843–1852 (2006). doi:10.1172/JCI272.
References
- 1.Okuda K., Nakanuma Y., Miyazaki M.2002. . Cholangiocarcinoma: recent progress. Part 2: molecular pathology and treatment. J. Gastroenterol. Hepatol. 171056–1063. [DOI] [PubMed] [Google Scholar]
- 2.Olnes M.J., Erlich R. A review and update on cholangiocarcinoma. Oncology. 2004;66:167–179. doi: 10.1159/000077991. [DOI] [PubMed] [Google Scholar]
- 3.Sirica A.E. Cholangiocarcinoma: molecular targeting strategies for chemoprevention and therapy. Hepatology. 2005;41:5–15. doi: 10.1002/hep.20537. [DOI] [PubMed] [Google Scholar]
- 4.Taylor-Robinson S.D., et al. Increase in mortality rates from intrahepatic cholangiocarcinoma in England and Wales 1968-1998. Gut. 2001;48:816–820. doi: 10.1136/gut.48.6.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang Y.K., Kim W.H., Jang J.J. Expression of G1-S modulators (p53, p16, p27, cyclin D1, Rb) and Smad4/Dpc4 in intrahepatic cholangiocarcinoma. Hum. Pathol. 2002;33:877–883. doi: 10.1053/hupa.2002.127444. [DOI] [PubMed] [Google Scholar]
- 6.Ito Y., et al. Expression of p57/Kip2 protein in extrahepatic bile duct carcinoma and intrahepatic cholangiocellular carcinoma. Liver. 2002;22:145–149. doi: 10.1034/j.1600-0676.2002.01532.x. [DOI] [PubMed] [Google Scholar]
- 7.Sugimachi K., et al. The role of overexpression and gene amplification of cyclin D1 in intrahepatic cholangiocarcinoma. J. Hepatol. 2001;35:74–79. doi: 10.1016/s0168-8278(01)00079-4. [DOI] [PubMed] [Google Scholar]
- 8.Sugimachi K., et al. Altered expression of beta-catenin without genetic mutation in intrahepatic cholangiocarcinoma. Mod. Pathol. 2001;14:900–905. doi: 10.1038/modpathol.3880409. [DOI] [PubMed] [Google Scholar]
- 9.Tannapfel A., et al. Mutations of the BRAF gene in cholangiocarcinoma but not in hepatocellular carcinoma. Gut. 2003;52:706–712. doi: 10.1136/gut.52.5.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu T., Leng J., Han C., Demetris A.J. The cyclooxygenase-2 inhibitor celecoxib blocks phosphorylation of Akt and induces apoptosis in human cholangiocarcinoma cells. Mol. Cancer Ther. 2004;3:299–307. [PubMed] [Google Scholar]
- 11.Tokumoto N., et al. Immunohistochemical and mutational analyses of Wnt signaling components and target genes in intrahepatic cholangiocarcinomas. Int. J. Oncol. 2005;27:973–980. [PubMed] [Google Scholar]
- 12.Kobayashi S., Werneburg N.W., Bronk S.F., Kaufmann S.H., Gores G.J. Interleukin-6 contributes to Mcl-1 up-regulation and TRAIL resistance via an Akt-signaling pathway in cholangiocarcinoma cells. Gastroenterology. 2005;128:2054–2065. doi: 10.1053/j.gastro.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Massague J. TGF-beta signal transduction. Annu. Rev. Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 14.Heldin C.H., Miyazono K., ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 15.Miyaki M., Kuroki T. Role of Smad4 (DPC4) inactivation in human cancer. Biochem. Biophys. Res. Commun. 2003;306:799–804. doi: 10.1016/s0006-291x(03)01066-0. [DOI] [PubMed] [Google Scholar]
- 16.Sirard C., et al. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev. 1998;12:107–119. doi: 10.1101/gad.12.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang X., Li C., Xu X., Deng C. The tumor suppressor SMAD4/DPC4 is essential for epiblast proliferation and mesoderm induction in mice. . Proc. Natl. Acad. Sci. U. S. A. 1998;95:3667–3672. doi: 10.1073/pnas.95.7.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X., et al. Haploid loss of the tumor suppressor Smad4/Dpc4 initiates gastric polyposis and cancer in mice. Oncogene. 2000;19:1868–1874. doi: 10.1038/sj.onc.1203504. [DOI] [PubMed] [Google Scholar]
- 19.Takaku K., et al. Gastric and duodenal polyps in Smad4 (Dpc4) knockout mice. Cancer Res. 1999;59:6113–6117. [PubMed] [Google Scholar]
- 20.Yang X., Li C., Herrera P.L., Deng C.X. Generation of Smad4/Dpc4 conditional knockout mice. . Genesis. 2002;32:80–81. doi: 10.1002/gene.10029. [DOI] [PubMed] [Google Scholar]
- 21.Li W., et al. Squamous cell carcinoma and mammary abscess formation through squamous metaplasia in Smad4/Dpc4 conditional knockout mice. Development. 2003;130:6143–6153. doi: 10.1242/dev.00820. [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y.X., et al. Cerebellar deficits and hyperactivity in mice lacking Smad4. J. Biol. Chem. 2003;278:42313–42320. doi: 10.1074/jbc.M308287200. [DOI] [PubMed] [Google Scholar]
- 23.Sansal I., Sellers W.R. The biology and clinical relevance of the PTEN tumor suppressor pathway. J. Clin. Oncol. 2004;22:2954–2963. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 24.Maehama T., Dixon J.E. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 25.Downward J. Mechanisms and consequences of activation of protein kinase B/Akt. Curr. Opin. Cell Biol. 1998;10:262–267. doi: 10.1016/s0955-0674(98)80149-x. [DOI] [PubMed] [Google Scholar]
- 26.Radu A., Neubauer V., Akagi T., Hanafusa H., Georgescu M.M. PTEN induces cell cycle arrest by decreasing the level and nuclear localization of cyclin D1. Mol. Cell. Biol. 2003;23:6139–6149. doi: 10.1128/MCB.23.17.6139-6149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pineau P., et al. Homozygous deletion scanning in hepatobiliary tumor cell lines reveals alternative pathways for liver carcinogenesis. Hepatology. 2003;37:852–861. doi: 10.1053/jhep.2003.50138. [DOI] [PubMed] [Google Scholar]
- 28.Tanno S., et al. Serine/threonine kinase AKT is frequently activated in human bile duct cancer and is associated with increased radioresistance. Cancer Res. 2004;64:3486–3490. doi: 10.1158/0008-5472.CAN-03-1788. [DOI] [PubMed] [Google Scholar]
- 29.Groszer M., et al. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- 30.Yakar S., et al. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 32.Wang R.H., et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Stiles B., et al. Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2082–2087. doi: 10.1073/pnas.0308617100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horie Y., et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J. Clin. Invest. 2004;113:1774–1783. doi: 10.1172/JCI200420513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engelhardt N.V., et al. Common antigen of oval and biliary epithelial cells (A6) is a differentiation marker of epithelial and erythroid cell lineages in early development of the mouse. Differentiation. 1993;55:19–26. doi: 10.1111/j.1432-0436.1993.tb00029.x. [DOI] [PubMed] [Google Scholar]
- 36.Lau S.K., Prakash S., Geller S.A., Alsabeh R. Comparative immunohistochemical profile of hepatocellular carcinoma, cholangiocarcinoma, and metastatic adenocarcinoma. Hum. Pathol. 2002;33:1175–1181. doi: 10.1053/hupa.2002.130104. [DOI] [PubMed] [Google Scholar]
- 37.Chaudhary H.B., Bhanot P., Logrono R. Phenotypic diversity of intrahepatic and extrahepatic cholangiocarcinoma on aspiration cytology and core needle biopsy. Cancer. 2005;105:220–228. doi: 10.1002/cncr.21155. [DOI] [PubMed] [Google Scholar]
- 38.Wongkham S., et al. Serum MUC5AC mucin as a potential marker for cholangiocarcinoma. . Cancer Lett. 2003;195:93–99. doi: 10.1016/s0304-3835(02)00691-2. [DOI] [PubMed] [Google Scholar]
- 39.Fassett J.T., Tobolt D., Nelsen C.J., Albrecht J.H., Hansen L.K. The role of collagen structure in mitogen stimulation of ERK, cyclin D1 expression, and G1-S progression in rat hepatocytes. J. Biol. Chem. 2003;278:31691–31700. doi: 10.1074/jbc.M300899200. [DOI] [PubMed] [Google Scholar]
- 40.Kase S., et al. Phosphorylation of extracellular signal-regulated kinase and p27(KIP1) after retinal detachment. Graefes Arch. Clin. Exp. Ophthalmol. 2005;244:352–358. doi: 10.1007/s00417-005-0016-5. [DOI] [PubMed] [Google Scholar]
- 41.Diehl J.A., Cheng M., Roussel M.F., Sherr C.J. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monga S.P., et al. Hepatocyte growth factor induces Wnt-independent nuclear translocation of beta-catenin after Met-beta-catenin dissociation in hepatocytes. Cancer Res. 2002;62:2064–2071. [PubMed] [Google Scholar]
- 43.Ebert M.P., et al. Reduced PTEN expression in the pancreas overexpressing transforming growth factor-beta 1. Br. J. Cancer. 2002;86:257–262. doi: 10.1038/sj.bjc.6600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li D.M., Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 45.Burgering B.M., Medema R.H. Decisions on life and death: FOXO Forkhead transcription factors are in command when PKB/Akt is off duty. . J. Leukoc. Biol. 2003;73:689–701. doi: 10.1189/jlb.1202629. [DOI] [PubMed] [Google Scholar]
- 46.Kiguchi K., et al. Constitutive expression of ErbB-2 in gallbladder epithelium results in development of adenocarcinoma. Cancer Res. 2001;61:6971–6976. [PubMed] [Google Scholar]
- 47.Stambolic V., et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 48.Backman S.A., et al. Early onset of neoplasia in the prostate and skin of mice with tissue-specific deletion of Pten. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1725–1730. doi: 10.1073/pnas.0308217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Remy I., Montmarquette A., Michnick S.W. PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat. Cell Biol. 2004;6:358–365. doi: 10.1038/ncb1113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.