Abstract
Ubiquitylation of RhoA has emerged as an important aspect of both the virulence of Escherichia coli producing cytotoxic necrotizing factor (CNF) 1 toxin and the establishment of the polarity of eukaryotic cells. Owing to the molecular activity of CNF1, we have investigated the relationship between permanent activation of RhoA catalyzed by CNF1 and subsequent ubiquitylation of RhoA by Smurf1. Using Smurf1-deficient cells and by RNA interference (RNAi)-mediated Smurf1 knockdown, we demonstrate that Smurf1 is a rate-limiting and specific factor of the ubiquitin-mediated proteasomal degradation of activated RhoA. We further show that the cancer cell lines HEp-2, human embryonic kidney 293 and Vero are specifically deficient in ubiquitylation of either activated Rac, Cdc42, or Rho, respectively. In contrast, CNF1 produced the cellular depletion of all three isoforms of Rho proteins in the primary human cell types we have tested. We demonstrate that ectopic expression of Smurf1 in Vero cells, deficient for RhoA ubiquitylation, restores ubiquitylation of the activated forms of RhoA. We conclude here that Smurf1 ubiquitylates activated RhoA and that, in contrast to human primary cell types, some cancer cell lines have a lower ubiquitylation capacity of specific Rho proteins. Thus, both CNF1 and transforming growth factor-β trigger activated RhoA ubiquitylation through Smurf1 ubiquitin-ligase.
INTRODUCTION
Rho proteins belong to the Ras superfamily of GTPases (Takai et al., 2001). They organize the architecture of actin cytoskeleton to adapt cellular morphology to constraints imposed by cellular programs of differentiation, division, and migration (Etienne-Manneville and Hall, 2002; Burridge and Wennerberg, 2004). Rho proteins bind and hydrolyze GTP into GDP. They oscillate between a GDP-bound inactive form sequestered in the cytosol in association with the cellular factor RhoGDI (Del Pozo et al., 2002) and a GTP-bound active form, located in specific membrane locations, capable of binding and activating protein effectors (Burridge and Wennerberg, 2004). Transitions between both forms of Rho are regulated by guanine nucleotide exchange factors (GEFs) for activation and GTPase-activating proteins (GAPs) for inactivation (Schmidt and Hall, 2002; Moon and Zheng, 2003).
Rho proteins are the targets of a large number of bacterial toxins and bacterial effectors that are directly injected into host cell cytosol (Barbieri et al., 2002; Boquet and Lemichez, 2003). Bacterial factors activate or inactivate Rho proteins to block or mobilize Rho proteins. The cytotoxic necrotizing factor (CNF) 1 toxin activates Rho proteins but sensitizes them to ubiquitin (Ub)-mediated proteasomal degradation (Doye et al., 2002, 2006 Lerm et al., 2002). On penetration into cell cytosol, CNF1 catalyzes the activation of Rho proteins by deamidation of glutamine 63 of RhoA or the equivalent Q61 of Rac1 and Cdc42 and converts this residue into a glutamic acid (Flatau et al., 1997; Schmidt et al., 1997; Lerm et al., 1999). Mutation of this glutamine residue impairs the intrinsic and GAP-regulated GTPase activity of Rho proteins, locking them in a permanent active form bound to GTP. Nevertheless, permanently activated Rho proteins seem to be rapidly down-regulated by ubiquitylation and subsequent degradation in the proteasome. At high toxin concentration, the rate of activation/degradation of Rac supersedes that of synthesis, resulting in depletion of cellular Rac (Doye et al., 2002, Munro et al., 2004). Consequently, CNF1 produces a moderate activation of Rho proteins, which accounts for efficient bacterial internalization into cells and likely prevents host cells from producing high levels of inflammatory mediators (Munro et al., 2004).
The relationship between transforming growth factor (TGF)-β1 and Rho signaling pathways needs to be further clarified (Jaffe and Hall, 2003, 2005). TGF-β plays an important role in epithelial cell transformation and shares with Rho proteins a capacity to regulate cellular growth and differentiation. Members of TGF-β family signal through heteromeric complex of type I and type II serine/threonine kinase receptors (Shi and Massague, 2003; Wang et al., 2003). Activated receptors trigger signaling through phosphorylation of ligand-specific SMAD proteins (Shi and Massague, 2003). Growing evidence suggests interconnections between TGF-β and Rho signaling pathways. For example, RhoA, Rac1, and Jun NH2-terminal kinase promote SMAD-mediated signaling (Atfi et al., 1997), whereas RhoB is a negative regulator of the SMAD-mediated transcription (Engel et al., 1998). Direct interconnections between both pathways, in relationship with Ub-mediated proteasomal degradation, is exemplified by the findings that RhoB degradation by the proteasome is antagonized by TGF-β (Engel et al., 1998) and that the Smad Ub regulatory factor-1 (Smurf1) bears Ub-ligase activity on wild-type RhoA (Wang et al., 2003; Ozdamar et al., 2005). Smurf1 is a HECT domain-containing protein, which ubiquitylates different cellular proteins in addition to RhoA. For example, ubiquitylation of mitogen-activated kinase kinase (MEKK) 2 by Smurf1 controls bone homeostasis (Yamashita et al., 2005). Recent findings have also established that Smurf1 Ub-mediated proteasomal degradation of RhoA in response to TGF-β is crucial during early steps of induction of the epithelial-to-mesenchymal transition (Ozdamar et al., 2005). Despite progress made, it remains to be established whether Smurf1 ubiquitylates RhoA before or after its activation and whether Smurf1 also participates in the CNF1-induced Ub-mediated degradation of RhoA (Doye et al., 2002; Jaffe and Hall, 2003; Wang et al., 2003).
Owing to the relationship between the imbalance of the activity of Rho proteins in numerous human diseases (Boettner and Van Aelst, 2002), and its participation to bacterial infection (Boquet and Lemichez, 2003), we have further investigated the relationship between activation of Rho proteins and their cellular ubiquitylation. Using Smurf1-deficient mouse embryonic fibroblast (MEF) cells we have addressed the question of whether CNF1-induced ubiquitylation of RhoA occurs through Smurf1 and consequently whether Smurf1 ubiquitylates the activated form of RhoA. We extended our observations to show that ubiquitylation of activated RhoA, Rac1, or Cdc42 is specifically impaired in some cancer cells.
MATERIALS AND METHODS
Cell Lines, Reagents, and Cellular Transfection
The cell lines used were human epithelial kidney (HEK) 293 cells (ATCC CRL-1573), Vero African green monkey epithelial kidney cells (ATCC CCL-81), HEp-2 human epithelial larynx cells (ATCC CCL-23), MCF7 human breast adenocarcinoma (ATTC HTB-22), T24 human bladder carcinoma (ATCC HTB-4), SW620 human colorectal adenocarcinoma from metastatic site lymph node (ATCC CCL-227), and 804G rat epithelial bladder cells. Human primary cells used were human umbilical vein endothelial cells (HUVECs) (PromoCell, Heidelberg, Germany), keratinocytes, and fibroblasts obtained as described previously (Turchi et al., 2003) and macrophages isolated and differentiated as described previously (Capo et al., 1998). Wild-type and Smurf1-deficient MEF cells were obtained and propagated as described previously (Yamashita et al., 2005). Anti-Rac1 and anti-Cdc42 antibodies were purchased from Transduction Laboratories (BD Biosciences, Erembodegem, Belgium); anti-RhoA has been described previously (Lang et al., 1993); anti-Smurf1 H-60 antibodies were from Santa Cruz Biotechnology (Tebu-bio, Le Perrey en Yvelines, France); anti-β actin clone (AC-74) and anti-FLAG M2 antibodies were from Sigma-Aldrich (St. Louis, MO); anti-glutathione S-transferase (GST) clone (26H1) was from Cell Signaling Technology (Beverly, MA); and anti-GFP clone (7.1 and 13.1) was from Roche Diagnostics (Mannheim, Germany). Primary antibodies were visualized using goat anti-mouse horseradish peroxidase-conjugated secondary antibodies (DakoCytomation Denmark, Glostrup, Denmark) followed by chemiluminescence detection (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). CNF1 was purified as described previously (Flatau et al., 1997). The activity of different batches of CNF1 toxin was estimated and normalized using the classical multinucleation assay in HEp-2 cells as described previously (Lemichez et al., 1997). The potency of CNF1 used in this study was such that it produces 50% of HEp-2 multinucleated cells at 10 nM. In most experiments, cells were transfected under standard electroporation condition, as described previously (Doye et al., 2002). Briefly, 5 × 106 cells were electroporated with 30 μg of plasmid DNA (monotransfection) or 15 μg of each plasmid DNA (cotransfection) except for the His-Ub pull-down experiments (see Metal-Affinity Precipitation section). In the experiment corresponding to Figure 1A, HEK293 were transfected by standard calcium phosphate method as described in the figure legend.
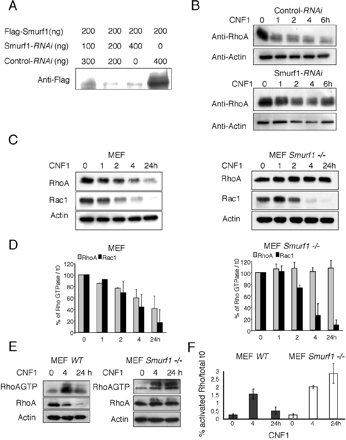
Figure 1.
Depletion of Smurf1 impairs the CNF1-induced cellular depletion of RhoA. (A) Immunoblot anti-FLAG. Smurf1-RNAi expression impairs the ectopic expression of Smurf1. HEK293 were cotransfected using calcium phosphate with 200 ng of FLAG-Smurf1 expression plasmid and different doses of expression plasmids of either Smurf1-RNAi or control-RNAi. Cells were lysed 24 h after transfection. (B) Immunoblots showing the kinetics of CNF1-induced RhoA depletion in HEK293 cells transfected with either Smurf1-RNAi or control-RNAi expression plasmids. Cells were transfected 24 h before addition of 10−9 M CNF1. Equal protein loading was verified by anti-actin immunoblotting. (C) Immunoblots showing the kinetics of CNF1-induced RhoA and Rac depletion in either Wild-type or Smurf1-deficient MEF cells intoxicated with 10−9 M CNF1. Equal protein loading was verified by anti-actin immunoblotting. (D) Quantification of the CNF1-induced RhoA and Rac depletion (mean values of 2 independent experiments). (E) Immunoblots showing the kinetic of CNF1-induced activation of RhoA in both wild-type and Smurf1-deficient MEF cells. Cells were treated with 10−9 M CNF1 and processed for GST-Rhotekin pull-down (labeled RhoAGTP). Total RhoA protein used in the assay was analyzed on 2% of the total cell lysates (labeled RhoA). Equal amounts of protein used in the assays were verified by anti-actin immunoblotting. (F) Quantification of CNF1-induced activation of Rho in wild-type and Smurf1-deficient MEF cells (mean values of 2 independent experiments).
DNA Constructs and Quantitative Reverse Transcription (RT)-PCR Conditions
For GST-Rho pull-down experiments of Smurf1, the cDNAs encoding RhoA and RhoA mutants (L63 and N19) were subcloned as BamH1/EcoR1 fragments into pLef mammalian GST-expressing vector (Rudert et al., 1996). For RNAi-mediated cellular knock-down experiments, Smurf1-RNAi sequence (5′-GCGTTTGGATCTATGCAAAC) or control-RNAi sequence (5′-ACGGTGGGAACATGACGATT) were cloned in the pSIRNA vector (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. For immunofluorescence experiments, pEGFP-C2 (Clontech, Mountain View, CA) was used to visualize transfected cells. pRBG4-6HisUb, used in metal-affinity precipitation experiments, was a gift from Dr. R. R. Kopito (Stanford University, Stanford, CA). pKH3HA3-RhoA and RhoA mutants (L63 and N19), pKH3HA3-RacL61, and pKH3HA3-Cdc42L61 were kind gifts from Dr. D. Manor (Cornell University, Ithaca, NY). pEYFP-Flag-Smurf1 and the ubiquitinylation-deficient mutant Smurf1C699A were kind gifts from Dr. J. Wrana (University of Toronto, Toronto, Canada). For real-time quantitative PCR, RNA was treated with DNase I and was reverse-transcribed using random priming and Superscript II reverse transcriptase according to the manufacturer's instructions (Invitrogen). Quantitative PCR was performed by monitoring in real time the increase in fluorescence of SYBR Green on an ABI PRISM 7000 sequence detector system (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. Gene-specific primers were designed using Primer Express software (Applied Biosystems). SB34 was used for normalization. Sequences for Smurf1 were 5′-CAACAGTCCAGGGCCAAGTT and 5′-CTTGGTATCCTGGGGTCGT; for SB34, they were 5′-TGCATCAGTACCCCATTCTATCAT and 5′-AAGGTGTAATCCGTCTCCACAGA.
Immunofluorescence and Time-Lapse Imaging
For immunofluorescence experiments, cells were fixed in 4% paraformaldehyde (Sigma-Aldrich) and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich). Actin cytoskeleton was visualized using 1 μg/ml fluorescein isothiocyanate-conjugated phalloidin (Sigma-Aldrich). Fluorescent signals were analyzed with a TCS-SP confocal microscope (Leica, Heidelberg, Germany) using a 63× magnification lens. Each picture represents the projection of four serial confocal optical sections. For time-lapse imaging experiments, cells were filmed for 6 h with or without 10 nM CNF1 in constant conditions of 5% CO2 and 37°C. Cells were analyzed using a 20× phase-contrast optic lens on an Axiovert 200 microscope, under shutter-controlled illumination (Carl Zeiss, Göttingen, Germany), and the image was captured by a cooled, digital charge-coupled device camera (Roper Scientific, Trenton, NJ). Images were recorded at 1 frame/30 s and processed using MetaMorph 2.0 image analysis software (Molecular Devices, Sunnyvale, CA) and QuickTime pro5 software (Apple Computer, Cupertino, CA).
Metal-Affinity Precipitation and Pull-Down of Activated Rho GTPases
For metal-affinity precipitation, cells other than MEF cells were electroporated with pRBG4-6HisUb alone or in combination with pKH3HA3-RacL61, RhoL63, or Cdc42L61 (15 μg of each plasmid for 5 × 106 cells) (Lin et al., 1999). In histidine-tagged Ub (His-Ub) control experiments, pCDNA3 empty-vector (15 μg), instead of pRBG4-6HisUb, was added to pKH3HA3-Rho protein expression vectors. MEF cells were transfected using the MEF Nucleofector kit 1 (Amaxa Biosystems, Gaithersburg, MD). Briefly, for each condition twice 2 × 106 cells were electroporated with 2 μg of the indicated plasmids, using the T-20 program. Cells were transfected 8 h for RhoL63 expression and 16 h for either RacL61 or Cdc42L61 expression. Cells were lysed at room temperature in the ULB buffer (8 M urea, 20 mM Tris-HCl, pH 7.5, 200 mM NaCl, 10 mM imidazole, and 0.1% Triton X-100). Quantities of exogenous hemagglutinin (HA)-Rho proteins present in the clarified lysates were first determined on immunoblots to use equal quantities of HA-Rho proteins in the assay. Normalized HA-Rho proteins lysates were purified for His-Ub on 30 μl of cobalt chelated resin (Clontech), preincubated in 0.4% bovine serum albumin (radioimmunoassay grade; Sigma-Aldrich). After 1 h of lysate incubation at room temperature, beads were washed five times in ULB and resuspended in 1 volume of Laemmli buffer. Proteins were resolved on a 12% SDS-PAGE, and polyubiquitylated Rho proteins were visualized by anti-HA immunoblotting assay. Pull-down assays were performed as described previously (Manser et al., 1998; Ren et al., 1999).
GST-RhoA Pull-Down of Smurf1
Cells were electroporated with pEYFP-Flag-Smurf1C699A in combination of pLef, pLef-RhoA, or RhoAL63 or RhoAN19, respectively (15 μg of each plasmid for 5 × 106 cells). Transgene expression was allowed for 8 h. Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 1% Triton X-100, 10 mM MgCl2, 0.5% sodium deoxycholate, and 0.1% SDS). Relative quantities of yellow fluorescent protein (YFP)-Flag-Smurf1C699A proteins present in clarified lysates were determined by anti-FLAG immunoblot assay. Glutathione S-transferase (GST)-tagged Rho proteins were purified using 30 μl of glutathione-agarose beads (Sigma-Aldrich) from 1 mg of cell lysate. After incubation at 4°C for 1 h, beads were washed three times in lysis buffer and resuspended in 1 volume of Laemmli buffer. Samples were resolved by 12% SDS-PAGE followed by immunoblotting anti-GST and by immunoblotting anti-FLAG (YFP-Flag-Smurf1C699A detection).
RESULTS
CNF1-induced Proteasomal Degradation of RhoA Is Impaired in Smurf1-deficient Cells
Smurf1 is a HECT domain-containing protein that bears an Ub-ligase activity on wild-type RhoA both in vitro and in vivo (Wang et al., 2003; Ozdamar et al., 2005). A key question raised by these findings is whether Smurf1 ubiquitylates the active form of RhoA. To clarify this question, we have further investigated the molecular mechanism of CNF1 toxin. This toxin catalyzes the permanent activation of Rho proteins, which then become sensitive to Ub-mediated proteasomal degradation. We reasoned that if Smurf1 was entirely responsible for activated RhoA ubiquitylation, its cellular loss would impair the CNF1-induced ubiquitylation and proteasomal degradation of RhoA. We first addressed this question by using a system of RNAi-mediated Smurf1 knockdown described previously (Wang et al., 2003). The activity of Smurf1-RNAi expression vector was first verified. Cotransfection of increasing amounts of Smurf1-RNAi expression vector was found to efficiently block the ectopic expression of Smurf1 (Figure 1A). The interference of Smurf1-RNAi on the activity of CNF1 toward RhoA was next assessed. We observed a marked reduction of the depletion of RhoA induced by CNF1 upon Smurf1 RNAi-mediated knockdown (Figure 1B). We next took advantage of recently established Smurf1-deficient mice to ascertain the involvement of Smurf1 in the CNF1-induced proteasomal degradation of RhoA (Yamashita et al., 2005). We observed that, in contrast to wild-type MEF cells, intoxication of Smurf1−/− MEF cells by CNF1 failed to deplete RhoA (Figure 1, C and D). That CNF1 produced the depletion of Rac in both cell types was indicative of the specificity of the Ub-ligase activity of Smurf1 on RhoA, as reported previously (Wang et al., 2003). Finally, we observed an accumulation of activated RhoA upon CNF1-treatment of Smurf1−/−, in contrast to wild-type, MEF cells (Figure 1, E and F). Hence, we observed an increase of activated RhoA ubiquitylation in Smurf1-deficient MEF cells upon Smurf1 ectopic expression (Supplemental Figure 1). In conclusion, both approaches concurred to show that Smurf1 is a rate-limiting factor involved in the CNF1-induced cellular depletion of activated RhoA.
Relationship between Smurf1 and the CNF1-induced Reorganization of the Actin Cytoskeleton
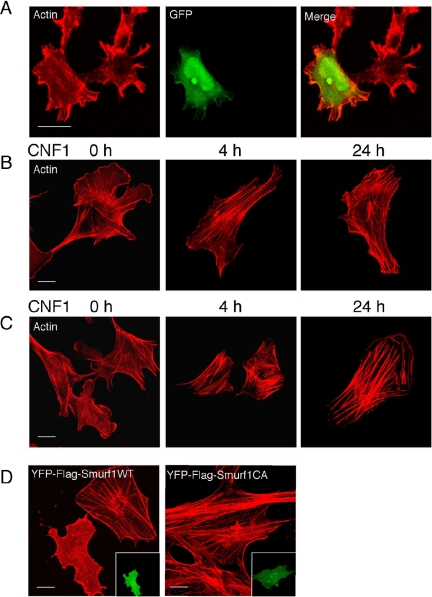
We noticed that the intoxication of HEK293 cells by CNF1 resulted in a higher content of actin cables upon Smurf1-RNAi knockdown (Figure 2A). Consistent with this, we also observed in CNF1-intoxicated cells a marked increase of the thickness of actin cables in Smurf1−/−, compared with wild-type MEF cells (Figure 2, B and C). Finally, we observed that the ectopic expression of Smurf1 in Smurf1-deficient cells abolished the formation of actin cables induced by CNF1, in contrast to the expression of the catalytic inactive mutant Smurf1-C699A (Figure 2D). Collectively, these results established a direct relationship between the expression of Smurf1, the cellular depletion of activated RhoA, and the extent of actin cables formation produced by CNF1.
Figure 2.
Relationship between Smurf1 and actin reorganization induced by CNF1. (A–D) Cells were fixed and processed for actin cytoskeleton labeling using tetramethylrhodamine B isothiocyanate-phalloidin. Bars, 10 μm. (A) Effect of Smurf1-RNAi on actin cytoskeleton reorganization induced by CNF1 in HEK293 cells. Cells were cotranfected with Smurf1-RNAi expression plasmid together with pEGFP-C2. Twenty-four hours after transfection cells were intoxicated for another 24 h with 10−9 M CNF1. This picture shows a transfected cell, which displays actin cables in contrast to nontransfected cells. (B and C) Reorganization of the actin cytoskeleton in either wild-type (B) or Smurf1-deficient (C) MEF cells intoxicated 4 or 24 h by 10−9 M CNF1. (D) Ectopic expression of Smurf1, at the difference with Smurf1-C699A, impairs the formation of actin cables induced by 24 h of intoxication of Smurf1-deficient MEF cells by 10−9 M CNF1. Smurf1-deficient MEF cells were transfected with pEYFP-Flag-Smurf1 or pEYFP-Flag-Smurf1C699A expression plasmids and intoxicated 16 h after transfection. Inset pictures show the transfected cells.
Specificity of Actin Reorganization in Cell Lines Intoxicated by CNF1
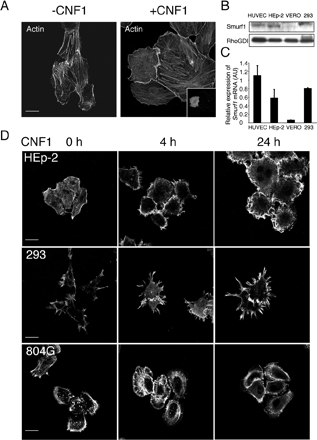
The relationship observed between CNF1-activated RhoA, Smurf1, and the actin cytoskeleton phenotype produced by CNF1 led us to reinvestigate the effect of CNF1 in Vero cells. In these cells, CNF1 treatment as well as injection of purified recombinant permanently activated RhoA (RhoAQ63E) produces a massive formation of thick actin cables (Flatau et al., 1997). Interestingly, expression of Smurf1 in Vero cells intoxicated by CNF1 prevents the formation of actin cables (Figure 3A). These results suggest that Vero cells may be deficient in Smurf1 activity, thus producing a phenotype similar to that produced by CNF1 in Smurf1−/− MEF cells. Consistently, we observed a lower expression level of Smurf1 in Vero cells, which correlated with the lower level of Smurf1 mRNA expression (Figure 3, B and C). In subsequent analyses of the effects of CNF1 on different cell lines, we made a complementary observation that CNF1 produced a massive formation of actin-containing membrane ruffles in HEp-2 cells, whereas the toxin produced large filopodia in HEK293 cells (Figure 3D). In contrast, CNF1 did not produce significant actin phenotypes after 24 h of intoxication in 804G cells (Figure 3D). The effects of CNF1 on these different cell lines were also documented by time-lapse videoimaging. Shortly after addition of the toxin, we observed formation of massive membrane ruffles in HEp-2 cells (Supplemental Videos 1 and 2). CNF1 was found to induce the spreading of Vero cells together with limited membrane ruffles (Supplemental Videos 3 and 4). In correlation with the actin phenotype of CNF1-intoxicated HEK293 cells, we observed the formation of large filopodia protrusions (Supplemental Videos 5 and 6). Collectively, these observations show that CNF1 induces a reorganization of actin cytoskeleton specific of activated RhoA, Rac1, or Cdc42 in Vero, HEp-2, and HEK293 cells, respectively.
Figure 3.
Specificity of actin cytoskeleton reorganization induced by CNF1 in cell lines. (A and D) Cells were fixed and processed for actin cytoskeleton labeling using TRITC-phalloidin. Bars, 10 μm. (A) Ectopic expression of Smurf1 impairs the formation of actin cables induced by 24 h of Vero cell intoxication by 10−9 M CNF1. Vero cells were transfected with pEYFP-Flag-Smurf1 expression plasmid and intoxicated 16 h after transfection. Inset picture shows the transfected cell. (B) Immunoblot anti-Smurf1. Cell protein extracts (100 μg) were resolved on 7% SDS-PAGE before anti-Smurf1 immunoblotting (H-60 used at 1/400). (C) Histogram showing the relative expression level of Smurf1 mRNA in cell lines and HUVECs determined by real-time quantitative PCR (mean values of 2 independent experiments). Total RNA was prepared from 106 cells using RNeasy kit (QIAGEN). (D) HEp-2, HEK293, and 804G cells were intoxicated with 10−9 M CNF1 for the indicated times.
Depletion of Rho GTPases Induced by CNF1 in Cell Lines and Primary Cell Types
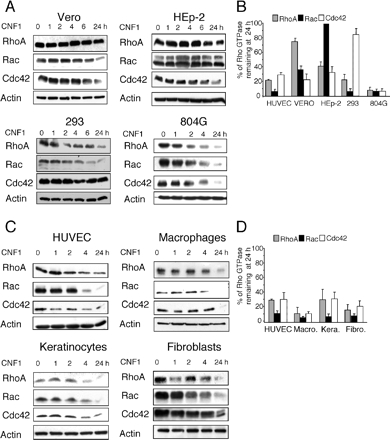
Our findings prompted us to investigate the kinetics of Rho protein depletion induced by CNF1 in different cell types. We observed a specific absence of cellular depletion of Rho in Vero cells, Cdc42 in HEK293 cells, and Rac in Hep-2 cells (Figure 4, A and B). We next addressed the question of whether these differences might reflect cell line or cell-type specificity. We thus analyzed the kinetics of Rho, Rac, and Cdc42 depletion produced by CNF1 in four different human cell types. All primary cell types tested showed an efficient depletion of Rho, Rac, and Cdc42 after 24 h of treatment with the CNF1 toxin (Figure 4, C and D). Consequently, the specific absence of depletion of Rho proteins observed after CNF1 intoxication of cell lines seemed to be a characteristic of these cell lines, rather than a consequence of their differentiation status.
Figure 4.
Specificity of CNF1-induced Rho protein depletion in cell lines and primary cell types. (A) Immunoblots showing the kinetics of CNF1-induced Rho, Rac, and Cdc42 depletion in HEp-2, Vero, HEK293, and 804G cell lines. Cells were treated for different times with 10−9 M CNF1. Cells were lysed and immunoblots were performed on 40 μg of the total protein lysate. Immunoblots anti-actin were used to evaluate equal loading and normalize Rho GTPase signals. (B) Quantification of the CNF1-induced depletion of Rho proteins. Cellular depletion of Rho proteins was measured at 24 h of cell intoxication with 10−9 M CNF1 (mean values of 2 independent experiments). (C) Immunoblots showing the kinetics of CNF1-induced Rho, Rac, and Cdc42 depletion in primary HUVECs, macrophages, keratinocytes, and fibroblasts. Cells were treated for different times with 10−9 M CNF1. Cells were lysed and immunoblots were performed using 40 μg of the total protein lysate. Immunoblots anti-actin were used to evaluate equal loading and normalize Rho GTPases signals. (D) Quantification of the CNF1-induced depletion of Rho proteins. Cellular depletion of Rho proteins was measured at 24 h of cell intoxication with 10−9 M CNF1 (mean values of 2 independent experiments).
Relationship between CNF1-induced Depletion and Activation of Rho Proteins
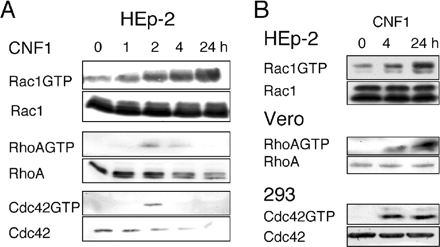
In 804G cells, the CNF1-induced cellular depletion of Rho proteins results in a transient activation of all three Rho proteins (Doye et al., 2002). In contrast, the absence of cellular depletion of activated Rac in HEp-2 cells results in its sustained activation (Figure 5A). Consistent with our observation on HEp-2 cells, we observed a sustained activation of Rho in Vero cells and a sustained activation of Cdc42 in HEK293 cells (Figure 5B). Collectively these results demonstrated the direct correlation that exists between the CNF1-induced depletion of Rho proteins and the corresponding reorganization of the actin cytoskeleton induced by the toxin. These observations were suggestive of a specific lack of Ub-mediated proteasomal degradation of permanently activated Rho proteins in cell lines. Consistently, CNF1 produces a sustained activation of RhoA in Smurf1-deficient cells (Figure 1E).
Figure 5.
Kinetics of CNF1-induced Rho protein activation. (A) Immunoblots showing the kinetics of activation and degradation of Rho proteins in HEp-2 cells after different times of 10−9 M CNF1 exposure. RhoGTP, RacGTP, and Cdc42GTP indicate the extent of Rho protein activation measured using GST pull-down assays. Bottom, corresponding kinetics of CNF1-induced Rho proteins depletion. (B) Immunoblots showing the CNF1-induced Rho, Rac, and Cdc42 sustained activation in, respectively, HEp-2, Vero, and 293 cells. Activation of Rho proteins was measured using GST pull-down assays.
Comparative Efficiency of Ubiquitylation of Permanently Activated Rho Proteins
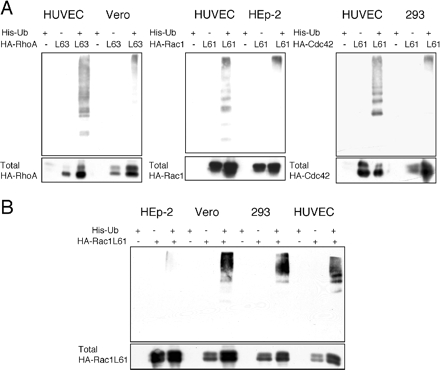
We then addressed the question of whether the absence of cellular depletion of Rho proteins observed in these different cell lines could be attributed to a specific loss of Rho protein ubiquitylation. This question was directly addressed by comparing the levels of ubiquitylation of permanently activated Rho proteins in cell lines and HUVECs. Consistent with our hypothesis, we observed a lower efficiency of ubiquitylation of activated RhoA in Vero, of activated Rac1 in HEp-2, and of activated Cdc42 in HEK293 cells (Figure 6A). In addition, we observed that in contrast to HEp-2 cells, other cell lines were all equally competent for permanently activated Rac ubiquitylation (Figure 6B). In conclusion, we demonstrate that the absence of CNF1-induced depletion of Rho proteins observed in HEp-2, Vero, and 293 cells correlates with the lower level of ubiquitylation of specific Rho proteins in these cell lines.
Figure 6.
Specificity of permanently activated Rho protein ubiquitylation. (A) Comparison of permanently activated Rho protein cellular ubiquitylation efficiencies. Cells expressing permanently activated HA-tagged Rho proteins and His-Ub were processed for histidine-tag purification after HA-Rho protein levels normalization. His-Ub cross-linked forms of HA-Rho proteins were visualized by immunoblotting anti-HA. An anti-HA immunoblot was performed in parallel on 2% of the total lysates to compare the amounts of total HA-Rho proteins used in the His-Ub purification (labeled total HA-Rac, HA-Rho, and HA-Cdc42). (B) Comparison of permanently activated Rac ubiquitylation efficiencies in four cell lines. Cells expressing permanently activated HA-tagged Rac and His-Ub were processed for histidine-tag purification after HA-Rho protein levels normalization. His-Ub cross-linked forms of HA-Rac were visualized by immunoblotting anti-HA. An anti-HA immunoblot was performed in parallel on 2% of the total lysates to compare the amounts of total HA-Rac used in the His-Ub purification (labeled total HA-Rac).
Expression of Smurf1 in Vero Cells Restores the Ubiquitylation of Activated RhoA
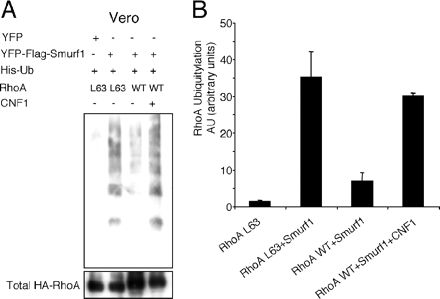
We showed that some cell lines have a reduced ubiquitylation efficiency of specific Rho proteins. Based on our model, we hypothesized that the reduced level of RhoA ubiquitylation in Vero cells could be a consequence of decreased Smurf1 activity. We thus investigated the effect of Smurf1 ectopic expression in Vero cells on activated RhoA ubiquitylation. We observed that Smurf1 expression in Vero cells resulted in an increase of ubiquitylation of permanently activated RhoA (Figure 7, A and B). In addition, we observed that Smurf1 expression in Vero cells restored the ubiquitylation of wild-type RhoA after cell intoxication by CNF1 (Figure 7, A and B). Collectively, these results demonstrate that Smurf1 ubiquitylates the activate form of RhoA, lending further support to the role of Smurf1 in the ubiquitylation of the active form of RhoA induced by CNF1.
Figure 7.
Effect of Smurf1 on permanently activated RhoA ubiquitylation in Vero cells. (A) Immunoblots anti-HA showing RhoA ubiquitylation efficiencies. Vero cells were cotransfected 8 h with expression plasmids of His-Ub, HA-RhoAL63, or wild-type HA-RhoA and pYFP-Flag-Smurf1, as indicated. Cells were processed for histidine-tag purification directly or after 6 h of intoxication with 10−9 M CNF1, as indicated. Equal levels of HA-Rho proteins were engaged in the His-Ub purification assays. His-Ub cross-linked forms of HA-RhoA were visualized by immunoblotting anti-HA. In parallel, an anti-HA immunoblot was performed on 2% of the total lysates to compare the amounts of total HA-RhoA used in the His-Ub purification (labeled total HA-RhoA). (B) Two independent experiments were quantified for RhoA ubiquitylation levels, compared with total RhoA in 2% of the cell lysates.
DISCUSSION
Our study sheds light on the molecular mechanism of ubiquitylation of Rho proteins. Recent findings have raised the question of whether Smurf1 participates in the Ub-mediated proteasomal degradation of RhoA produced by CNF1, and consequently whether Smurf1 ubiquitylates the active form of RhoA (Jaffe and Hall, 2003; Wang et al., 2003). We report that Smurf1 is responsible for the ubiquitylation of activated RhoA. In addition, we show that some cell lines, in contrast to primary cells, display a lack of ubiquitylation of RhoA, Rac1, or Cdc42. The lack of ubiquitylation of specific activated Rho proteins in cell lines results in their sustained activation and remodeling of actin cytoskeleton, after CNF1 intoxication. Collectively, our findings suggest the existence of three distinct pathways for ubiquitylation of RhoA, Rac1, or Cdc42 and establish the need that Rho protein ubiquitylation should be analyzed in specific cell lines or preferentially in primary cells. Hence, our study demonstrates that Smurf1-deficient mice represent a good model to study the importance of RhoA ubiquitylation.
A question addressed by previous findings is whether RhoA is ubiquitylated before or after being activated by GEF factors (Wang et al., 2003). CNF1 is a deamidase that catalyzes the deamidation of glutamine 63 of RhoA, converting it into a glutamic acid (Flatau et al., 1997; Schmidt et al., 1997). This posttranslational modification of the Switch-II domain of RhoA impairs the intrinsic GTPase activity of RhoA. As a consequence, the deamidated form of RhoA behaves as a classical permanently activated mutant of RhoA, which becomes insensitive to regulation by GAPs. Consistent with previous observations, made in vitro, we have observed that in cells, wild-type and activated RhoA binds to Smurf1, although with a lower affinity than with dominant-negative form of RhoA (Supplemental Figure 2). In complement, we show that Smurf1 expression in Smurf1-deficient Vero cells promotes the ubiquitylation of RhoA, either activated by point mutation or by CNF1. Hence, CNF1 produces a transient activation of RhoA also reinforcing the idea that RhoA is degraded after being activated. Collectively, our findings favor a model in which Smurf1 binds to RhoA, during its GTP loading, but ubiquitylates RhoA once activated.
According to the findings that CNF1 induced the Ub-mediated proteasomal depletion of RhoA, Rac, and Cdc42 in primary endothelial HUVECs (Munro et al., 2004; Doye et al., 2006), we show that CNF1 induces a depletion of all three isoforms of Rho proteins in three other primary human cell types. This implies that the Ub-mediated proteasomal degradation of permanently activated RhoA, Rac1, and Cdc42 most likely corresponds to the bona fide molecular mechanism of cell intoxication by CNF1. In contrast, several studies have reported an absence of cellular depletion of some Rho proteins in cell lines intoxicated by CNF1 (Doye et al., 2002; Lerm et al., 2002; Hopkins et al., 2003). We have further screened cell lines for CNF1-induced actin cytoskeleton reorganization into membrane ruffles, filopodia, or actin cables (Supplemental Figure 3). Together, four of these cell lines, intoxicated by CNF1, showed an absence of RhoA, Rac1, or Cdc42 cellular depletion. Further analysis of these aspects with Vero cells, as a model, demonstrated that ectopic expression of Smurf1 in these cells rescued the ubiquitylation of activated RhoA. Collectively, our results not only demonstrate that ubiquitylation of Rho proteins can be specifically impaired in cell lines but also point to the existence of three independent pathways of either RhoA, Rac1, and Cdc42 ubiquitylation. Ub-ligases of Rac and Cdc42 thus remained to be identified.
Here, we show that Ub-mediated proteasomal degradation of activated RhoA is impaired in Smurf1-deficient MEF cells. It is therefore likely that ubiquitylation of RhoA is also impaired in Smurf1−/− mice (Yamashita et al., 2005). High levels of activated RhoA suppresses epithelial to mesenchymal transition at the primitive streak during mouse gastrulation (Fuse et al., 2004). Considering that Smurf1−/− mice are born viable, this implies that during development the regulation of the level of activated RhoA is achieved mainly by factors other than Smurf1 or that compensatory mechanisms exist in Smurf1-deficient mice. Consistent with this we also did not observe differences in the levels of both total and activated RhoA between wild-type and Smurf1−/− MEFs. Smurf1-deficient mice exhibit an age-dependent increase of bone mass because of the increase of osteoblast activity, a phenomenon caused by the impaired Ub-mediated proteasomal degradation of MEKK2 by Smurf1 (Yamashita et al., 2005). Nevertheless, it cannot be excluded that ubiquitylation of RhoA fine-tunes other biological phenomenon, such as host responses against pathogens. Thus, our findings that activated RhoA is not ubiquitylated in Smurf1-deficient cells indicate that these mice represent a remarkable model for investigating the key question of whether the ubiquitylation of RhoA confers to this GTPase unsuspected signaling properties (Jaffe and Hall, 2003).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to D. Manor for pKH3-Rho proteins expression vectors and to A. A. Chassot for technical assistance with culture of primary human keratinocytes and fibroblasts. We thank Y. Le Marchand-Brustel and the Bettencourt-Schueller's foundation for videomicroscopy facilities. This work was supported by institutional funding from Institut National de la Santé et de la Recherche Médicale, by Grant ANR A05135AS from the Agence Nationale de la Recherche, by Grant ARC 3337 from the Association pour la Recherche sur le Cancer (to E. L.), and a fellowship from the Ligue Nationale Contre le Cancer (to L. B.). M. Y. and Y.E.Z. were supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-09-0876) on March 15, 2006.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Atfi A., Djelloul S., Chastre E., Davis R., Gespach C. Evidence for a role of Rho-like GTPases and stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) in transforming growth factor beta-mediated signaling. J. Biol. Chem. 1997;272:1429–1432. doi: 10.1074/jbc.272.3.1429. [DOI] [PubMed] [Google Scholar]
- Barbieri J. T., Riese M. J., Aktories K. Bacterial toxins that modify the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 2002;18:315–344. doi: 10.1146/annurev.cellbio.18.012502.134748. [DOI] [PubMed] [Google Scholar]
- Boettner B., Van Aelst L. The role of Rho GTPases in disease development. Gene. 2002;286:155–174. doi: 10.1016/s0378-1119(02)00426-2. [DOI] [PubMed] [Google Scholar]
- Boquet P., Lemichez E. Bacterial virulence factors targeting Rho GTPases: parasitism or symbiosis? Trends Cell Biol. 2003;13:238–246. doi: 10.1016/s0962-8924(03)00037-0. [DOI] [PubMed] [Google Scholar]
- Burridge K., Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–179. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- Capo C., Meconi S., Sanguedolce M. V., Bardin N., Flatau G., Boquet P., Mege J. L. Effect of cytotoxic necrotizing factor-1 on actin cytoskeleton in human monocytes: role in the regulation of integrin-dependent phagocytosis. J. Immunol. 1998;161:4301–4308. [PubMed] [Google Scholar]
- Del Pozo M. A., Kiosses W. B., Alderson N. B., Meller N., Hahn K. M., Schwartz M. A. Integrins regulate GTP-Rac localized effector interactions through dissociation of Rho-GDI. Nat. Cell Biol. 2002;4:232–239. doi: 10.1038/ncb759. [DOI] [PubMed] [Google Scholar]
- Doye A., Boyer L., Mettouchi A., Lemichez E. Ubiquitin-mediated proteasomal degradation of Rho proteins by the CNF1 toxin. Methods Enzymol. 2006;406:447–456. doi: 10.1016/S0076-6879(06)06033-2. [DOI] [PubMed] [Google Scholar]
- Doye A., Mettouchi A., Bossis G., Clement R., Buisson-Touati C., Flatau G., Gagnoux L., Piechaczyk M., Boquet P., Lemichez E. CNF1 exploits the ubiquitin-proteasome machinery to restrict Rho GTPase activation for bacterial host cell invasion. Cell. 2002;111:553–564. doi: 10.1016/s0092-8674(02)01132-7. [DOI] [PubMed] [Google Scholar]
- Engel M. E., Datta P. K., Moses H. L. RhoB is stabilized by transforming growth factor beta and antagonizes transcriptional activation. J. Biol. Chem. 1998;273:9921–9926. doi: 10.1074/jbc.273.16.9921. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Flatau G., Lemichez E., Gauthier M., Chardin P., Paris S., Fiorentini C., Boquet P. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- Fuse T., Kanai Y., Kanai-Azuma M., Suzuki M., Nakamura K., Mori H., Hayashi Y., Mishina M. Conditional activation of RhoA suppresses the epithelial to mesenchymal transition at the primitive streak during mouse gastrulation. Biochem. Biophys. Res. Commun. 2004;318:665–672. doi: 10.1016/j.bbrc.2004.04.076. [DOI] [PubMed] [Google Scholar]
- Hopkins A. M., Walsh S. V., Verkade P., Boquet P., Nusrat A. Constitutive activation of Rho proteins by CNF-1 influences tight junction structure and epithelial barrier function. J. Cell Sci. 2003;116:725–742. doi: 10.1242/jcs.00300. [DOI] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A. Cell biology. Smurfing at the leading edge. Science. 2003;302:1690–1691. doi: 10.1126/science.1092874. [DOI] [PubMed] [Google Scholar]
- Jaffe A. B., Hall A. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Lang P., Gesbert F., Thiberge J. M., Troalen F., Dutartre H., Chavrier P., Bertoglio J. Characterization of a monoclonal antibody specific for the Ras-related GTP-binding protein Rho A. Biochem. Biophys. Res. Commun. 1993;196:1522–1528. doi: 10.1006/bbrc.1993.2424. [DOI] [PubMed] [Google Scholar]
- Lemichez E., Flatau G., Bruzzone M., Boquet P., Gauthier M. Molecular localization of the Escherichia coli cytotoxic necrotizing factor CNF1 cell-binding and catalytic domains. Mol. Microbiol. 1997;24:1061–1070. doi: 10.1046/j.1365-2958.1997.4151781.x. [DOI] [PubMed] [Google Scholar]
- Lerm M., Pop M., Fritz G., Aktories K., Schmidt G. Proteasomal degradation of cytotoxic necrotizing factor 1-activated Rac. Infect Immun. 2002;70:4053–4058. doi: 10.1128/IAI.70.8.4053-4058.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerm M., Selzer J., Hoffmeyer A., Rapp U. R., Aktories K., Schmidt G. Deamidation of Cdc42 and Rac by Escherichia coli cytotoxic necrotizing factor 1, activation of c-Jun N-terminal kinase in HeLa cells. Infect. Immun. 1999;67:496–503. doi: 10.1128/iai.67.2.496-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Cerione R. A., Manor D. Specific contributions of the small GTPases Rho, Rac, and Cdc42 to Dbl transformation. J. Biol. Chem. 1999;274:23633–23641. doi: 10.1074/jbc.274.33.23633. [DOI] [PubMed] [Google Scholar]
- Manser E., Loo T. H., Koh C. G., Zhao Z. S., Chen X. Q., Tan L., Tan I., Leung T., Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol. Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- Moon S. Y., Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- Munro P., Flatau G., Doye A., Boyer L., Oregioni O., Mege J. L., Landraud L., Lemichez E. Activation and proteasomal degradation of Rho GTPases by cytotoxic necrotizing factor-1 elicit a controlled inflammatory response. J. Biol. Chem. 2004;279:35849–35857. doi: 10.1074/jbc.M401580200. [DOI] [PubMed] [Google Scholar]
- Ozdamar B., Bose R., Barrios-Rodiles M., Wang H. R., Zhang Y., Wrana J. L. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science. 2005;307:1603–1609. doi: 10.1126/science.1105718. [DOI] [PubMed] [Google Scholar]
- Ren X. D., Kiosses W. B., Schwartz M. A. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudert F., Visser E., Gradl G., Grandison P., Shemshedini L., Wang Y., Grierson A., Watson J. pLEF, a novel vector for expression of glutathione S-transferase fusion proteins in mammalian cells. Gene. 1996;169:281–282. doi: 10.1016/0378-1119(95)00820-9. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Schmidt G., Sehr P., Wilm M., Selzer J., Mann M., Aktories K. Gln 63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- Shi Y., Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Takai Y., Sasaki T., Matozaki T. Small GTP-binding proteins. Physiol. Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Turchi L., Chassot A. A., Bourget I., Baldescchi C., Ortonne J. P., Meneguzzi G., Lemichez E., Ponzio G. Cross-talk between RhoGTPases and stress activated kinases for matrix metalloproteinase-9 induction in response to keratinocytes injury. J. Investig. Dermatol. 2003;121:1291–1300. doi: 10.1111/j.1523-1747.2003.12627.x. [DOI] [PubMed] [Google Scholar]
- Wang H. R., Zhang Y., Ozdamar B., Ogunjimi A. A., Alexandrova E., Thomsen G. H., Wrana J. L. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]
- Yamashita M., Ying S. X., Zhang G. M., Li C., Cheng S. Y., Deng C. X., Zhang Y. E. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.