Abstract
Fruit development in higher plants normally requires pollination and fertilization to stimulate cell division of specific floral tissues. In some cases, parthenocarpic fruit development proceeds without either pollination or fertilization. Parthenocarpic fruit without seed has higher commercial value than seeded fruit. Several apple (Malus domestica) mutants (Rae Ime, Spencer Seedless and Wellington Bloomless) are known to produce only apetalous flowers that readily go on to develop into parthenocarpic fruit. Through genetics, a single recessive gene has been identified to control this trait in apple. Flower phenotypes of these apple mutants are strikingly similar to those of the Arabidopsis mutant pistillata (pi), which produces flowers where petals are transformed to sepals and stamens to carpels. In this study, we have cloned the apple PI homolog (MdPI) that shows 64% amino acid sequence identity and closely conserved intron positions and mRNA expression patterns to the Arabidopsis PI. We have identified that in the apetalous mutants MdPI has been mutated by a retrotransposon insertion in intron 4 in the case of Rae Ime and in intron 6 in the case of Spencer Seedless and Wellington Bloomless. The insertion apparently abolishes the normal expression of the MdPI gene. We conclude that the loss of function mutation in the MdPI MADS-box transcription factor confers parthenocarpic fruit development in these apple varieties and demonstrates another function for the MADS- box gene family. The knowledge generated here could be used to produce parthenocarpic fruit cultivars through genetic engineering.
Flowers of dicotyledonous plants consist of four basic floral organs: sepals, petals, stamens, and carpels. In Arabidopsis thaliana and Antirrhinum majus, floral homeotic mutants have been described lacking one or two floral organs but having increased numbers of other organs (1, 2). It has been found that the majority of these mutants are different mutational alleles within one or more MADS-box genes that encode putative transcription factors (1). The floral organ identity MADS-box genes have been grouped into three different classes by function, according to the proposed ABC model (1). In summary for Arabidopsis, the class A gene APETALA1 (AP1) specifies sepal formation; AP1 and two class B genes [APETALA3 (AP3) and PISTILLATA (PI)] together specify petal formation; AP3 and PI and the class C gene AGAMOUS (AG) together are required for stamen formation; and AG alone controls carpel formation (1). PI and AP3 proteins are functional partners and form a heterodimer. A loss of function mutation in either PI or AP3 produces flowers that contain two whorls of sepals and double numbers of carpels, but without stamens or petals (3, 4). In A. majus, similar mutant phenotypes are described, which are due to a mutation in the gene GLOBOSA (GLO) or DEFICIENS (DEF) (5, 6), homologs to PI and AP3, respectively.
In contrast to flowers, fruit structure varies significantly among dicotyledonous species depending on which tissues contribute to the development of the fruit after pollination and fertilization. In some cases, parthenocarpic fruit is produced without the need for pollination. Genetic control of parthenocarpy is poorly understood, although seedlessness is a highly desirable trait for many fruit cultivars.
Apple fruit is a pome derived from the floral tube, a fusion between the base of sepal, petal, and stamen tissues (7). This pome differs greatly from the Arabidopsis fruit (silique) that is derived from ovary tissue alone. A few apple mutants, including Spencer Seedless, Wellington Bloomless, and Rae Ime, are known to produce flowers without petals or stamens, but with two sets of sepals and two sets of carpels (7, 8). The phenotype of these apple flowers is conferred by a single recessive gene as confirmed by genetic studies (9) and is similar to Arabidopsis pi and ap3 mutants. In addition, the apple apetalous flowers can develop into seedless fruit in the absence of pollination and fertilization. However, seed is produced on hand pollination (7–9), indicating that the carpels are functional. We have chosen to clone and characterize the gene responsible for this trait to gain an insight into the control of parthenocarpy. This might lead to a novel approach to seedless fruit cultivar production.
We postulated that the Rae Ime phenotype is due to mutation in MdPI or MdAP3, the apple homologs of PI and AP3, respectively, because of similarities between apple and Arabidopsis mutant flower phenotypes. In a previous study (10), we isolated seven apple MADS-box genes, but none of these appeared to be a homolog of either PI or AP3. We therefore conducted studies to clone and characterize both MdPI and MdAP3 genes. This study shows that transposon insertion mutants of MdPI are responsible for the flower and fruiting phenotype in Rae Ime and related apple mutants.
Materials and Methods
Plant Materials, DNA, and RNA Samples.
The trees of Rae Ime have been maintained in the germplasm collection plot at HortResearch in Havelock North, New Zealand. DNA samples of Spencer Seedless and Wellington Bloomless were kindly provided by M. Hemmat (Cornell University, New York). For cDNA synthesis, total RNA was isolated from flowers of the normal apple cultivar Granny Smith by using the method described by Chang et al. (11). Poly(A) mRNA was purified from the total RNA by using the mRNA purification kit (Amersham Pharmacia). cDNA was synthesized from the mRNA by using the ZAP cDNA synthesis kit (Stratagene).
Cloning PI Homolog by Using PCR Approaches.
DNA fragments were amplified from templates of cDNA by using two degenerative PCR primers P1 5′-CGGAATTCATGGGNMGNGGNAARRT and P2 5′-CGCTCGAGGATCCGGYTGNATNGGYTGNAC (n = ATGC, M = AC, R = AG, Y = CT). The primers were designed according to the conserved amino acid sequences MGRGKI in the MADS-box domain and VQPM/IQP in the C-terminal region in an alignment of PI GLOBOSA, FBP3, SLM2, and pMADS2 (GenBank accession nos. D30807, X68831, X71417, X80489, and X69947, respectively). The underlined EcoRI and BamHI sites were used for cloning PCR products. The PCR amplification conditions were as follows: initial denaturation at 94°C for 4 min; then 35 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 3 min, with a final extension of 5 min at 72°C. Several bands were detected from the PCR on agarose gels, and DNA in a band of the expected size (630 bp) was cloned into Bluescript SK (Stratagene) after EcoRI and BamHI digestion. Sequences of cloned fragments were determined. Two nested PCR primers, P3 (5′-CAATGACAGCATGCAAGTAGAG) and P4 (5′-TCAGGCATCTGAAGGGAGAGG), were designed from the sequences within the K-box region and used to amplify the 3′ region of MdPI cDNA together with a 3′ rapid amplification of cDNA ends primer 5′-GAGAGAGAACTAGTCTCGAG. The PCR conditions were the same as described above except that the annealing temperature was reduced to 50°C. The amplified fragments were cloned into pGEM-T EASY vector (Promega).
Genomic fragments of MdPI were amplified by using primers P5 (5′-GAGATCAAGAGGATTGAGAACTC), P6 (5′-CACAAACCGAGTTCATGCACGCC), and P3 and P7 (5′-GCCTCATAACGAAGCTAATTCC). PCR conditions were as follows: initial denaturation at 94°C for 2 min; then 10 cycles of 94°C for 15 s, 58°C for 30 s; and 20 cycles of 94°C for 15 s, 58°C for 30 s, and 68°C for 5 min plus cycle elongation of 20 s for each cycle; and with a final extension of 5 min at 68°C. The amplified fragments were cloned into pGEM-T EASY vector. Expand High Fidelity PCR System (Roche Diagnostics) was used in all PCR experiments.
DNA Sequence Determination.
Nucleotide sequences of MdPI clones were determined by using the automatic sequencer ABI PRISM model 377 (Perkin–Elmer) with universal forward and reverse primers. To obtain complete sequences, subclones were made, and gene-specific primers were designed and purchased from GIBCO/BRL.
Northern and Southern Analysis Using MdPI on Apple Tissues.
Total RNA was isolated as described by Chang et al. (11) from normal Granny Smith and Royal Gala and from mutant Rae Ime apple tissues. Northern blots were prepared as described by Dong et al. (12). The Northern blot contained RNA isolated from expanding leaves, unopened flowers, and fruit at 2 days and 1, 4, and 8 weeks after manual pollination. At 4 weeks after pollination, apple fruit is large enough to allow for easy separation into the three main tissue types, namely core, cortex, and skin.
DNA was isolated from leaf tissue of Granny Smith, Royal Gala, and Rae Ime by using the method of Rogers and Bendich (13). Southern blots were prepared by digesting apple DNA (approximately 20 μg per lane) with EcoRI or HindIII, separating DNA fragments on 0.7% agarose gel and transferring them to Hybond-N+ membrane (Amersham Pharmacia).
Northern and Southern blots were probed with 32P-dCTP-labeled MdPI cDNA fragments (450 bp) lacking the MADS-box sequence to significantly reduce cross hybridization to MADS-box DNA fragments. The blots were hybridized in 0.5 M NaPO4 buffer (pH 7.2) with 1 mM EDTA and 7% SDS at 65°C and washed with 0.4 × SSC and 0.2% SDS at 65°C. Hybridization signals were detected by using a Storm 840 PhosphorImager (Molecular Dynamics).
Results
Apple Pistillata-Like Flowers Develop into Parthenocarpic Fruit.
Normal apple flowers consist of four whorls of floral organs. There are usually five sepals, five petals, 9–20 stamens, and five carpels within an inferior ovary (ref. 7, Fig. 1A). In contrast, flowers of the mutant Rae Ime (Fig. 1B) have no petals or stamens but two whorls of five sepals and an increased number (up to 15) of styles and carpels.
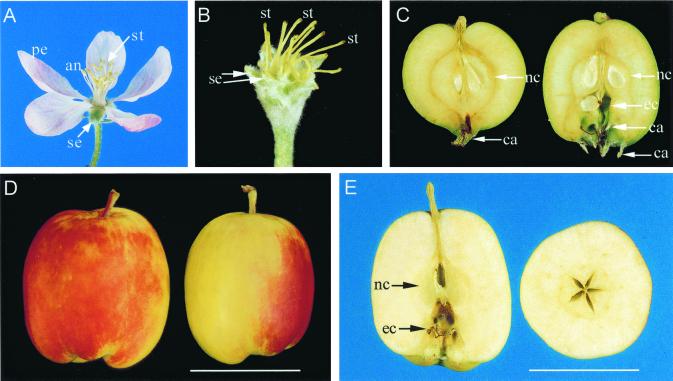
Figure 1.
Normal type and Rae Ime apple flowers and fruit. (A) A normal apple flower showing sepals (se), petals (pe), anthers (an), and styles (st). (B) A Rae Ime flower showing no petals or stamens but increased numbers of sepals and styles. (C) A young (5-week-old) fruit of normal apple (Left) contains normal carpels (nc) with seeds and one whorl of calyxes (ca). The same-age fruit of Rae Ime (Right) contains normal carpels, ectopic carpels (ec), and two whorls of calyxes. The seeds in the Rae Ime fruit were produced by manual pollination. (D and E) Mature parthenocarpic fruit of Rae Ime. (Bar = 5 cm.)
Fruit development on normal apple flowers requires pollination and seed development (Fig. 1C, Left). Normal apple fruit contains up to 10 seeds (7) depending on the level of pollination. Bees in an orchard do not normally pollinate the apetalous flowers of Rae Ime apple, as there is little or no nectar to attract the bees. On manual pollination, the apetalous flowers produced fruit-containing seeds in both the normal and ectopic carpels (Fig. 1C, Right). This indicates ovules in both whorls of carpels are functional. However, seed and embryos produced in the ectopic carpels are smaller. There are five normal carpels and 9–10 ectopic carpels. The fruit also have duplicated whorls of calyxes that are the remains of sepals, compared with one calyx whorl in a normal apple (Fig. 1C). Without manual pollination, these apetalous flowers readily bear parthenocarpic fruit without seeds (Fig. 1 D and E). The mature seedless fruit was close to normal apple fruit size, up to 7 cm long and 5 cm wide (Fig. 1 D and E).
Several other apple mutants, such as Spencer Seedless and Wellington Bloomless (8, 9), have been described with a very similar flower and fruit structure to that of Rae Ime. Anatomical studies of the vascular connections showed that the ectopic carpels have been transformed from the stamens and the second whorl of sepals from petals (8). These apple mutants also produce parthenocarpic fruit (9). In the Arabidopsis pi and ap3 mutants, flowers have no sepals or stamens but have double the number of sepals and carpels (3, 4). Therefore, homeotic changes in apple apetalous flowers closely resemble those in pi and ap3 mutants of Arabidopsis. In addition, apple apetalous flowers produce parthenocarpic fruit.
Transcription of MdPI in Rae Ime Mutant Is Abolished.
Because Rae Ime shows a similar flower phenotype to Arabidopsis pi mutant, we chose to determine whether the PI homolog in Rae Ime is mutated. As the first step, we cloned the apple PI homolog MdPI by using a PCR approach. DNA fragments of 630 bp were amplified from Granny Smith apple flower cDNA by using two degenerate PCR primers (P1 and P2, Fig. 2) against conserved sequences in the MADS-box and in the C-terminal region of PI and its homologs. After these DNA fragments were cloned, six randomly chosen clones were sequenced and found to contain the same sequence. The cloned cDNA sequences started from the first presumed ATG start codon, contained MADS-box, K-box, and most of the C-terminal region, and had high homology to PI. The C-terminal and the 3′ untranslated regions were further amplified by using two nested PCR primers (P3 and P4, Fig. 2) within the K-box and a 3′ rapid amplification of cDNA ends primer. Six clones containing the 3′ fragments were sequenced, and all were found to contain the same DNA sequence overlapping with those in the 5′ clone. Sequences from the 5′ and 3′ clone were assembled together (Fig. 2). The assembled sequences show highest homology to PI, with 64% amino acid sequence identity over the whole gene. This putative apple PI homolog was named as MdPI.
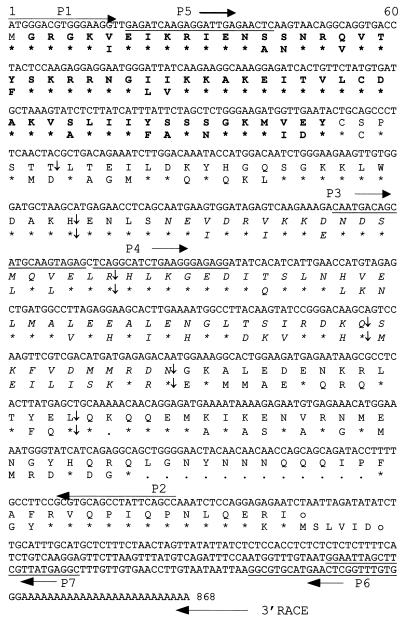
Figure 2.
Sequence information of MdPI. The cDNA sequences and deduced amino acid sequences of MdPI isolated from Granny Smith apple and Arabidopsis PI protein are shown in the top, middle, and bottom lines, respectively. Asterisks indicate amino acid identity. Gene-specific PCR primers are underlined. Primer directions are indicated with horizontal arrows. Intron positions are indicated with vertical arrows.
MdPI was found to be highly expressed in petals and stamens as determined by Northern analysis. Expression in young leaves, sepals, ovaries, flower peduncles, and 1-week-old and 4-week-old fruit was not detected (Fig. 3). This expression pattern is essentially the same as that shown for the Arabidopsis PI gene (3). Genomic sequences of MdPI were amplified by using the PCR primers P5 within the MADS-box and P6 within the 3′ nontranslated region. Two clones containing the MdPI genomic DNA were sequenced and found to contain the same sequences having six easily identifiable introns. The relative positions of intron 2 to intron 6 were strictly conserved compared with the positions of the five introns in the PI gene (Fig. 2). We conclude that MdPI is the PI homolog based on its sequence identity, conserved intron positions, and mRNA expression patterns.
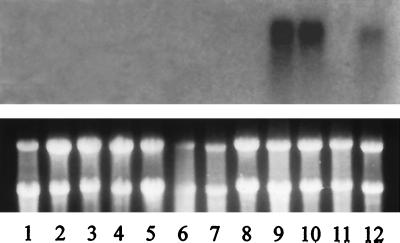
Figure 3.
Northern blot analysis of apple RNA sample using MdPI cDNA as a probe. All of the lanes are for Granny Smith tissue except for lane 11, which is for Rae Ime. RNA samples were prepared from ovaries (1), sepals (2), young leaves (3), skin (4), cortex (5), and core (6) tissue of 4-week-old fruit, 1-week-old fruit (7), flower peduncles (8), stamens (9), petals (10), flower buds (11), and flower buds (12). (Top) Hybridization signal with MdPI cDNA probe. (Bottom) Stained gel showing RNA loading.
To examine whether there is a mutation in MdPI of Rae Ime, the transcript level of MdPI in flower buds was determined. Transcription of MdPI was not detected in Rae Ime flower buds but readily detected in normal flower buds of the Granny Smith (Fig. 3) and Royal Gala (data not shown). PI transcription is usually abolished in flower buds in Arabidopsis pi mutants (3). Our expression data suggested that MdPI be mutated in Rae Ime.
Insertion Mutation Detected in MdPI of Three Apetalous and Parthenocarpic Apples.
Evidence for mutation in Rae Ime MdPI came from experiments comparing restriction fragment-length polymorphism (RFLP) patterns for Rae Ime with normal apple cultivars by using the MdPI cDNA as a probe. Southern hybridization showed different RFLP patterns between Rae Ime and Granny Smith with both EcoRI and HindIII digestion, although Granny Smith RFLP pattern was conserved in another apple cultivar Royal Gala (data not shown). The RFLP data also indicated that the MdPI gene in Rae Ime has been mutated. As both enzyme digestions revealed RFLP differences, the mutation was likely to be a gross change in gene structure rather than a point mutation.
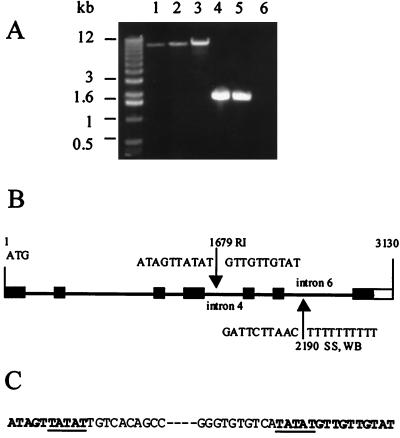
Furthermore, genomic DNA fragments were amplified from three apetalous apple mutants (Spencer Seedless, Wellington Bloomless, and Rae Ime) and two normal flower cultivars (Granny Smith and Royal Gala) by using two PCR primers (P3 and P7, Fig. 2) designed with MdPI cDNA sequence. The amplified DNA fragments were 10–11 kb for the three apetalous mutants in contrast to a 2-kb amplified product for the two normal flower cultivars (Fig. 4). All these fragments hybridized to MdPI cDNA probe in a Southern blot analysis (data not shown). The 11-kb fragment for Rae Ime was cloned and fully sequenced. The two ends of the fragments showed identical sequence to the Granny Smith MdPI, but an insertion in intron 4 of the MdPI gene in Rae Ime was detected. Clones containing the PCR fragment for Spencer Seedless and Wellington Bloomless were partially sequenced from the 5′ ends. The sequence for these two mutants was identical but contained an insert in intron 6 when compared with Granny Smith MdPI (Fig. 4). This result confirmed that there was an insertion mutation in the MdPI gene in the case of Rae Ime, as well as in Spencer Seedless and Wellington Bloomless.
Figure 4.
Identification of an insertion in MdPI of Rae Ime. (A) Genomic DNA fragments were amplified from Spencer Seedless (1), Wellington Bloomless (2), Rae Ime (3), Granny Smith (4), and Royal Gala (5) by using primers P3 and P7. No DNA was amplified from the control reaction without apple DNA as template (6). (B) Arrows indicate insertion sites in MdPI for Rae Ime (RI), Spencer Seedless (SS), and Wellington Bloomless (WB). The numbers indicate nucleotide positions starting from the ATG codon in the wild-type MdPI of Granny Smith. Dark boxes are coding regions, and the white box is the 3′ noncoding region. Lines between the boxes represent introns. (C) Five-base pair duplication (underlined) of MdPI sequence was found in the insertion site for Rae Ime.
Inserts in MdPI Are Retrotransposons.
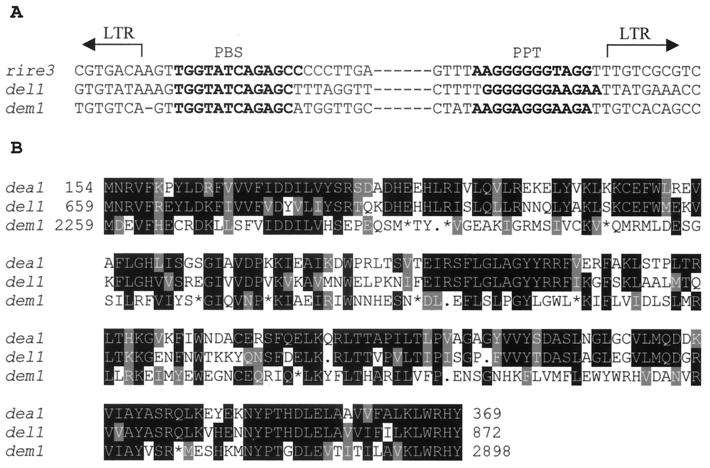
The insert in MdPI of Rae Ime was fully sequenced and found to be 9,332 bp. It contains long-terminal repeats (LTRs) of 2,773 bp (left) and 2,769 bp (right), both in the same orientation. The length difference between the two repeats is due to a shorter microsatellite (TA) repeat in the right LTR. There was no significant homology identified between the LTR and sequences in GenBank. In the internal sequences, a primer-binding site complementary to 3′ of tRNAMet and a polypurine tract were identified (Fig. 5A). Both primer-binding site and polypurine tract are essential cis elements for cDNA synthesis in retrotransposons (14, 15). The primer-binding site is the initiation site of cDNA synthesis. Homology was identified between part of the internal sequences and reverse transcriptase in retrotransposon del1 of lily (14) and dea1 of pineapple (16) (Fig. 5B). Five-base pair duplication of the target sequence was found at the insertion site (Fig. 4C). These data indicate that the insert in MdPI in Rae Ime is an LTR-type retrotransposon. Due to the highest homology to del1 and dea1, the apple retrotransposon is named as dem1. The occurrence of numerous stop codons in reading frames (Fig. 5B) and frameshifts indicates that the cloned element of dem1 is no longer an active element.
Figure 5.
Sequence homology between the apple transposon and other plant transposons. (A) The apple transposon (dem1) has a primer-binding site and polypurine tract similar to that of transposon in rice (rire3) and Lilium henryi (del1). (B) dem1 shears significant homology to the amino acid sequences in the reverse transcriptase domain of retrotransposons of pineapple (dea1) and L. henryi (del1). Eight stop codons were detected (shown as asterisks) in the apple transposon. The numbers indicate amino acid positions for dea1 (GenBank accession no. Y12432) and del1 (accession no. X13886) as previously published and nucleotide positions for dem1 as the sequence submitted to GenBank (accession no. AJ291492).
The insert in MdPI of Spencer Seedless and Wellington Bloomless was partially sequenced from the left LTR. The sequences of the inserts in these two mutants appeared to be identical. Because the insertion sites in these two mutants are also the same, we concluded that Spencer Seedless and Wellington Bloomless carry an identical mutation in MdPI. The insert in Spencer Seedless was further sequenced from the left LTR into the internal region for 4 kb. This 4-kb sequence showed approximately 90% homology to the insert in Rae Ime. This result indicates that the insert in Spencer Seedless and Wellington Bloomless is also an LTR retrotransposon that is related to that inserted into MdPI of Rae Ime.
Discussion
We have shown that MdPI has functions similar to those of PI gene in regulation of flower development. In Arabidopsis, expression of PI is required for development of sepals and stamens. In mutants lacking PI expression, the petals are transformed to sepals and stamens to carpels (3). In apple MdPI mutants containing a transposon insertion, transcript of MdPI in flower buds was not detected, and the floral organ transformation is the same as that in Arabidopsis pi mutants. Functional homologs of floral homeotic genes in tree species have usually been identified by expressing tree genes in Arabidopsis, as it is difficult to generate tree mutants (17–19). We report here the use of a naturally occurring apple mutant for analyzing the function of a MADS-box transcription factor. We have demonstrated that there is remarkable functional similarity between MdPI in a perennial woody tree species and PI in an annual weed in control of floral organ development. In addition, MdPI is a key regulator of apple fruit development.
A difference between MdPI mutant apple and pi mutant Arabidopsis is that the former produces parthenocarpic fruit but the latter does not. Genetic and molecular evidence show that both apetaly and parthenocarpy in apple are due to mutations in MdPI. Genetic analysis has been performed by using two apetalous-parthenocarpic mutants, Spencer Seedless and Wellington Bloomless. Crossing pollen from the cultivar Wijcik McIntosh with normal flowers to Wellington Bloomless generates hybrids that all produce normal flowers (9). Crossing the pollen from these hybrids to Spencer Seedless generates plants, half of which produce normal flowers and half apetalous flowers and parthenocarpic fruit (9). This result indicates that a single recessive gene controls apetalous flower development and subsequently parthenocarpic fruit formation.
Six apple mutants have been recorded to produce apetalous flowers and parthenocarpic fruit in different countries. Many of these records can be traced back to several centuries ago (8, 9). The wide geographic origin of these apple mutants indicates the possibility of independent events of mutation. We have analyzed MdPI genomic DNA sequences of three mutants and found insertions in all three. Spencer Seedless and Wellington Bloomless have the same insertion site in intron 6, which is different from that in Rae Ime. Independent mutant alleles at the same locus are good evidence that a single gene is involved in the development of apetalous flower and parthenocarpic fruit in these apple mutants.
The difference in fruit development between Rae Ime apple and Arabidopsis pi mutant may be explained in two different ways. First, MdPI may have a different function compared with PI in influencing ovary and fruit development. Functional differences have been shown for homologs of floral homeotic genes in different plant species. These differences are particularly clear for the C-function genes, although the differences between B-function genes (including PI) are less clear (20). Second, the most likely explanation is that apple fruit develops from different floral tissues compared with Arabidopsis fruit (silique).
Apple differs from tomato and Arabidopsis, two model systems often used in studies of fruit development, where the fruit or silique develops from ovary tissue alone (1, 21). Apple has an inferior ovary that only contributes tissues to form the fruit core. The major part of apple fruit, the fleshy cortex, develops from the floral tube. According to the appendicular hypothesis, the floral tube consists of the fused bases of the sepals, petals, and stamens (7). However, recent studies on apple MADS-box genes by others and us have produced evidence that apple fruit flesh derives from sepal tissue and contains little contribution from petal and stamen tissues. The apple MADS-box gene MdAP1 is exclusively expressed in sepals during flower development (22) and in the flesh cortex during fruit development (10). This indicates that base of sepals contributes to apple flesh cortex formation. We have shown that MdPI is strongly expressed in petals and stamens but not in developing fruit (Fig. 3). This indicates that petals and stamens contribute no or little tissue for fruit development. A loss of function mutation in MdPI removes a repressor from MdAP1 and enhances sepal development. This enhancement may trigger apple cortex development without the need for pollination and fertilization.
Some parthenocarpic fruit cultivars have considerable advantages over seeded fruit cultivars. This is clearly demonstrated with several types of fruits, such as banana and seedless forms of watermelon, grape, and orange. These fruits are more convenient than seeded fruit to consumers. Parthenocarpic apple trees can be cropped without pollination, which reduces dependence on bees, pollinator varieties, and warm weather at flowering. For Spencer Seedless, the weight of parthenocarpic fruit is approximately 70% of seeded fruit (23). Seedless apple cultivars avoid or reduce biennial bearing tendencies that have been attributed to the inhibition of flower bud formation by developing seed (23). Finally, seedless apple fruit is much less susceptible to codling moth, a major pest on apple trees, compared with seeded fruit (24).
Fruit quality of the current seedless apple mutants is poor. Attempts are being made to breed this seedlessness trait into commercial apple cultivars. However, breeding for seedlessness in woody fruit trees is a slow process when conventional hybridization methods are used because of the long juvenile period and many generations required (9). With identification of MdPI sequence, genetic transformation could be used to generate high quality seedless apple cultivars by down-regulation of MdPI expression by using antisense or cosuppression techniques (25) together with Agrobacterium-mediated transformation systems developed for apple cultivars (26). No pollen production on these transgenic trees would alleviate environmental concerns regarding the transfer of transgenes to nontransgenics by cross-pollination. The MdPI clone may also be used as a marker for selection of the seedless trait in apple breeding. Knowledge gained from this study on apple may be applied to other fruit species, particularly those (like pear) with a similar fruit structure as apple.
Acknowledgments
We thank Dr. Minou Hemmat for kindly providing the genomic DNA of Spencer Seedless and Wellington Bloomless, Allan White and Mike Malone for Rae Ime plant material, Kent Tobutt for a discussion on his breeding data with Spencer Seedless and Wellington Bloomless, and Drs. Andrew Gleave and Dan Cohen for critical reading of this manuscript. This research was supported by New Zealand Foundation for Research, Science, and Technology contract number C06411.
Abbreviations
- RFLP
restriction fragment-length polymorphism
- LTR
long-terminal repeat
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AJ291490, AJ291491, and AJ291492).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031502498.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031502498
References
- 1.Weigel D, Meyerowitz E M. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 2.Schwarz-Sommer Z, Huijser P, Nacken W, Saedler H, Sommer H. Science. 1990;50:931–936. doi: 10.1126/science.250.4983.931. [DOI] [PubMed] [Google Scholar]
- 3.Goto K, Meyerowitz E. Genes Dev. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- 4.Jack T, Brockman L, Meyerowitz E M. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- 5.Sommer H, Beltran J, Huijser P, Pape H, Lonning W, Saedler H, Schwarz-Sommer Z. EMBO J. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trobner W, Ramirez L, Motte P, Hue I, Huijser P, Lonning W, Saedler H, Sommer H, Schwarz-Sommer Z. EMBO J. 1992;11:4693–4704. doi: 10.1002/j.1460-2075.1992.tb05574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pratt C. Hortic Rev. 1988;10:273–308. [Google Scholar]
- 8.Brase K D. M. S. thesis. New York: Cornell University; 1937. [Google Scholar]
- 9.Tobutt K R. Euphytica. 1994;77:51–54. [Google Scholar]
- 10.Yao J-L, Dong Y-H, Kvarnheden A, Morris B A M. J Am Soc Hortic Sci. 1999;124:8–13. [Google Scholar]
- 11.Chang S, Puryear J, Cairney J. Plant Mol Biol Rep. 1993;11:113–116. [Google Scholar]
- 12.Dong Y-H, Janssen B, Bieleski L, Atkinson R, Morris B, Gardner R. J Am Soc Hortic Sci. 1997;122:752–757. [Google Scholar]
- 13.Rogers S O, Bendich A J. In: Plant Molecular Biology Manual. Gelvin S, Schilperoort R, editors. Dordrecht, The Netherlands: Kluwer; 1988. p. A6. :1–13. [Google Scholar]
- 14.Smyth D R, Kalistsis P, Joseph J L, Sentry J W. Proc Natl Acad Sci USA. 1989;86:5015–5019. doi: 10.1073/pnas.86.13.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumekawa N, Ohtsubo H, Horiuchi T, Ohtsubo E. Mol Gen Genet. 1999;260:593–602. doi: 10.1007/s004380050933. [DOI] [PubMed] [Google Scholar]
- 16.Thomson K G, Thomas J E, Dietzgen R G. Plant Mol Biol. 1998;38:461–465. doi: 10.1023/a:1006083200299. [DOI] [PubMed] [Google Scholar]
- 17.Kyozuka J, Harcourt R, Peacock W J, Dennis E S. Plant Mol Biol. 1997;35:573–584. doi: 10.1023/a:1005885808652. [DOI] [PubMed] [Google Scholar]
- 18.Rutledge R, Rean S, Nicolas O, Fobert P, Cote C, Bosnich W, Kauffeldt C, Sunohara G, Seguin A, Stewart D. Plant J. 1998;15:625–634. doi: 10.1046/j.1365-313x.1998.00250.x. [DOI] [PubMed] [Google Scholar]
- 19.Tandre K, Svenson M, Svensson M, Engstrom P. Plant J. 1998;15:615–623. doi: 10.1046/j.1365-313x.1998.00236.x. [DOI] [PubMed] [Google Scholar]
- 20.Causier B, Weir I, Davies B. In: Sex Determination in Plants. Ainsworth C C, editor. Oxford: BIOS Scientific; 1999. pp. 1–23. [Google Scholar]
- 21.Gillaspy G, Ben-David H, Gruissem W. Plant Cell. 1993;5:1439–1451. doi: 10.1105/tpc.5.10.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotoda N, Wada M, Komori S, Kidou S, Abe K, Masuda T, Soejima J. J Am Soc Hortic Sci. 2000;125:398–403. [Google Scholar]
- 23.Chan B G, Cain J G. Proc Am Soc Hortic Sci. 1967;91:63–68. [Google Scholar]
- 24.Goonewardene H F, Kwolek W F, Hayden R A. J Econ Entomol. 1984;77:1427–1431. [Google Scholar]
- 25.Montgomery M K, Fire A. Trends Genet. 1998;14:255–258. doi: 10.1016/s0168-9525(98)01510-8. [DOI] [PubMed] [Google Scholar]
- 26.Yao J-L, Cohen D, Atkinson R, Morris B. In: Biotechnology in Agriculture and Forestry: Transgenic Trees. Bajaj Y P S, editor. Vol. 44. Berlin: Springer; 1999. pp. 153–170. [Google Scholar]