SETTING THE SCENE
The field of antioxidants and free radicals is often perceived as focusing around the use of antioxidant supplements to prevent human disease. In fact, antioxidants/free radicals permeate the whole of life, creating the field of redox biology. Free radicals are not all bad, nor antioxidants all good. Life is a balance between the two: Antioxidants serve to keep down the levels of free radicals, permitting them to perform useful biological functions without too much damage (Halliwell and Gutteridge, 2006). This is especially true in plants, as the rest of this issue reveals. Yet some damage is inevitable, requiring repair systems to maintain cell viability. The purpose of this article is to take a broad view of the field, and highlight some of the fascinating differences between plants and other organisms.
HOW DID REDOX BIOLOGY BEGIN?
All animals need O2 for efficient production of energy in mitochondria. This need for O2 obscures the fact that it is a toxic mutagenic gas; aerobes survive only because they have evolved antioxidant defenses (Halliwell and Gutteridge, 2006).
Oxygen appeared in significant amounts in the Earth's atmosphere over 2.2 billion years ago, largely due to the evolution of photosynthesis by cyanobacteria. They evolved to use energy from the sun to split water. Thereby, they gained reducing power (hydrogen equivalents) to drive their metabolism, but the byproduct, tonnes of O2, was discarded into the atmosphere (Lane, 2002) as an early case of air pollution. Initially, most of this O2 was consumed by the formation of the metallic oxide deposits that exist in rocks and ores today. Only when this was largely complete did O2 build up in the atmosphere. The rise in atmospheric O2 was advantageous in at least two ways; it led to formation of the ozone (O3) layer in the stratosphere that protects living organisms from UV-C radiation (that may have helped organisms to leave the sea and colonize land), and it removed ferrous iron (Fe2+) from aqueous environments by forming insoluble ferric complexes. Most Fe2+ was precipitated from solution, leaving sea and river waters today containing only trace amounts of soluble iron (Lane, 2002).
What was the advantage of removing Fe2+? This species reacts rapidly with hydrogen peroxide (H2O2) to yield highly toxic hydroxyl radical (a superscript ⋅ denotes a free radical).
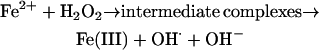 |
The above reaction is called the Fenton reaction, after its discoverer in 1876. Fenton chemistry occurs in vivo, but organisms carefully control it by limiting the availability of both Fe2+ and H2O2 (Halliwell and Gutteridge, 1984, 1990, 2006). It would have been difficult to evolve aerobic life in a world awash with Fe2+.
When living organisms first appeared on Earth, they did so under an atmosphere containing much N2 and CO2 but little O2, i.e. they were anaerobes (Kasting, 1993). Anaerobes still exist today, but usually their growth is inhibited or they are killed by exposure to 21% O2, the current atmospheric level. As atmospheric O2 rose, many anaerobes must have died out. Present-day ones are presumably the descendants of organisms that adapted to rising O2 by restricting themselves to anoxic microenvironments. Other organisms instead began to evolve antioxidant defenses (producing new ones and realigning ancient molecules to new functions) to protect against O2 toxicity. One of the earliest of these may have been to develop proteins that bind and detoxify iron to protect DNA against Fenton chemistry (Wiedenheft et al., 2005). In retrospect, this development of antioxidants was a fruitful path to follow. Organisms that tolerated O2 could further evolve to use it for metabolic transformations catalyzed by oxidase, oxygenase, and hydroxylase enzymes, such as the Pro and Lys hydroxylases needed for collagen biosynthesis. Indeed, large multicellular animals need collagen for making their support tissues (bone and cartilage). Best of all, O2 could facilitate efficient energy production, employing electron-transport chains with O2 as the terminal electron acceptor. This switch to aerobic metabolism increased the yield of ATP that could be made from food molecules such as Glc by over 15-fold.
Evolution of efficient energy production allowed the development of complex multicellular organisms, which then also needed systems to ensure that O2 can be distributed throughout their bodies (mechanisms varying from spiracles in insects to the human vascular system). A further advantage of evolving such systems is that delivery of O2 can be controlled: for example, most cells in the human body are never exposed to the full force of atmospheric O2 because blood pO2 is much lower than that of air. Similarly, insects control the opening of their spiracles to keep internal O2 low (Hetz and Bradley, 2005).
Photosynthesis: The Ultimate Paradox?
Since O2 is poisonous and photosynthetic organisms produce it, how could cyanobacteria evolve photosynthesis in a preantioxidant world? Even in present-day plants, which are full of antioxidants, much of the protein synthetic activity of chloroplasts is used to replace oxidatively damaged D1 and other proteins. Were some antioxidants in place already? Perhaps PSII evolved from a manganese-containing form of the enzyme catalase (Lane, 2002; Olson and Blankenship, 2004). Catalases, most of which are haem-containing proteins (Halliwell and Gutteridge, 2006), nowadays (although some bacteria still have manganese-containing ones [Horsburgh et al., 2002]) catalyze the rapid breakdown of H2O2.
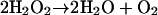 |
If this hypothesis is true, then catalase-like enzymes might have been present prior to a rise in atmospheric O2. How can this be? Was H2O2 present to drive its evolution?
Under an atmosphere mainly composed of N2 and CO2 with no O3 screen, UV radiation must have bombarded the face of the Earth. Even at the low O2 levels present 3.5 billion years ago (<0.1%), there could have been substantial H2O2 levels in rainwater generated by photochemical reactions with traces of O2 (Lane, 2002). Since Fe2+ was freely available, Fenton chemistry was a threat, so H2O2 must be eliminated. One suggestion is that the evolutionary precursors of PSII used H2O2 as a substrate (Olson and Blankenship, 2004) and only later evolved the increased chemical ferocity needed to split water.
Atmospheric O2 levels may have been higher at periods in the Earth's history, perhaps reaching 35% by the late Carboniferous era. As plant life flourished, CO2 levels fell drastically, and huge deposits of coal and oil formed (Graham et al., 1995; Lane, 2002). This increased O2 concentration may have permitted insects (whose O2 distribution system depends largely on diffusion) to become larger. For example, the giant Carboniferous dragonfly Meganeura monyi was bigger than any dragonfly that exists today (Lane, 2002). The plants and animals existing in Carboniferous times may have had enhanced antioxidant defenses, which would be fascinating to study if they could be resurrected. Even today, some of the plants that evolved at that time can resist damage by elevated O2 better than plants that evolved more recently (Beerling et al., 1998).
Oxygen Toxicity
All aerobes including plants, aerobic bacteria, and humans, suffer damage when exposed to O2 concentrations higher than normal (Gilbert, 1981; Balentine, 1982; Halliwell and Gutteridge, 2006), signifying that they have no excess of antioxidant defenses. Indeed, as we learned how to measure oxidative damage (for review, see Halliwell and Whiteman, 2004), it was found to occur in aerobes even at normal O2 levels. What causes this damage? Many scientists believe that O2 toxicity is due to excess formation of the superoxide radical O2⋅− (Fridovich, 1995), this is the superoxide theory of O2 toxicity. But let us step back for a moment and examine some basics.
WHAT ARE FREE RADICALS?
A free radical is any species capable of independent existence (hence the term free) that contains one or more unpaired electrons (Halliwell and Gutteridge, 2006). An unpaired electron is one that occupies an atomic or molecular orbital by itself. The simplest free radical is atomic hydrogen. Since a hydrogen atom has only one electron, it must be unpaired. Many free radicals exist in living systems (some bad, some good, and some both), although most molecules in vivo are nonradicals. Radicals can be formed by several mechanisms, such as adding a single electron to a nonradical. They can form when a covalent bond is broken if one electron from the bonding pair remains on each atom (homolytic fission). Some bonds are hard to break, e.g. temperatures of 450°C to 600°C are often required to rupture C–C, C–H, or C–O bonds. Indeed, combustion of organic compounds proceeds by free radical mechanisms. Other covalent bonds fragment more easily: Just trimming your fingernails can cleave disulphide bonds in keratin to generate sulfur radicals (Symons, 1996).
Another example, the O–O bond in H2O2, is readily split by exposing it to UV light, generating OH⋅.
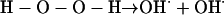 |
Homolytic fission of one of the O–H covalent bonds in water requires more energy (γ-rays or x-rays) and yields H⋅ and a hydroxyl radical (I write this as OH⋅, but some authors write it as ⋅OH, presumably to emphasize the location of the unpaired electron on the oxygen). Formation of OH⋅ accounts for much of the damage done to living organisms by ionizing radiation (von Sonntag, 1987).
The Oxygen Radicals and Related Species
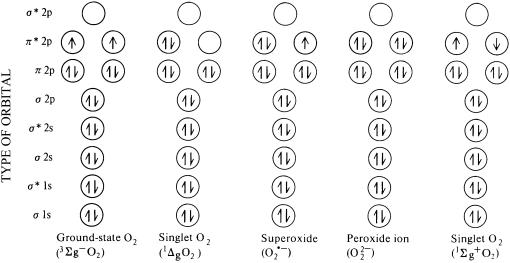
There are many types of free radicals in living systems, but I focus here on the oxygen radicals. In fact, the O2 molecule is a free radical (Gilbert, 1981; Halliwell and Gutteridge, 2006); it has two unpaired electrons. It should thus be written as O2⋅, but nobody ever does so I won't either. The two electrons in O2 have the same spin quantum number (or, as is often written, they have parallel spins). This is the most stable state, or ground state, of O2, and is the form that exists in the air around us. Oxygen is, thermodynamically, a potent oxidizing agent. However, if O2 tries to oxidize a nonradical by accepting a pair of electrons from it, both these electrons must have the same spin to fit into the vacant spaces in the π* orbitals (Fig. 1). A pair of electrons in an atomic or molecular orbital cannot meet this criterion, since they have opposite spins (+½ and −½). This spin restriction makes O2 prefer to accept its electrons one at a time, and helps explain why O2 reacts sluggishly with most nonradicals. By contrast, it often reacts outstandingly fast with other radicals by single electron transfers. Thus, if we heat human bodies or plants to a high enough temperature to cause some homolytic bond fission, O2 leaps onto the radicals formed and combustion begins (Gilbert, 1981; Halliwell and Gutteridge, 2006). Fortunately, most molecules in living organisms are nonradicals.
Figure 1.
A simplified version of bonding in the diatomic oxygen molecule and its derivatives. The oxygen atom has eight electrons, O2 has 16 electrons. Adapted from Halliwell and Gutteridge (2006) by courtesy of Oxford University Press.
Singlet Oxygens
More reactive forms of O2, the singlet oxygens, can be generated by an input of energy that rearranges the electrons (Foote et al., 1985). In both forms of singlet O2 the spin restriction is removed (Fig. 1) and the oxidizing ability greatly increased; singlet O2 can directly oxidize proteins, DNA, and lipids (Foote et al., 1985). The 1Σg+ state rapidly decays to the 1Δg state, so only the latter is usually encountered in biological systems. Note that 1ΔgO2 is not a free radical, it has no unpaired electrons (Fig. 1). Singlet O2 1Δg (written just as singlet O2 or 1O2 from now on) is the curse of the illuminated chloroplast; insufficient energy dissipation during photosynthesis can lead to formation of a chlorophyll triplet state that can transfer its excitation energy onto ground-state O2 to make 1O2 (Holt et al., 2005). This can oxidize chloroplast molecules and can trigger cell death (Wagner et al., 2004). The plant counters this by regulating energy distribution between the light harvesting complexes and by judicious use of certain carotenoids, which can quench both the triplet chlorophyll state and the 1O2 itself (Holt et al., 2005). Singlet O2 is also sometimes used as a signaling molecule.
Animals are not exempt from singlet O2 formation; it occurs in the skin and eye (Halliwell and Gutteridge, 2006). Photosensitizers of singlet O2 can be consumed in dietary plants (e.g. hypericin in St. John's Wort [Hypericum perforatum] and psoralens in celery [Apium graveolens]) or taken as drugs (e.g. the fluoroquinolone antibiotics), and subsequent sunbathing can cause skin damage (Morison, 2004; Halliwell and Gutteridge, 2006).
Superoxide Radical
If a single electron is supplied to O2, it enters one of the π* antibonding orbitals (Fig. 1). The product is superoxide radical, O2⋅−, full name superoxide radical anion. With only one unpaired electron, superoxide is less radical than O2, despite its super name (Fridovich, 1995).
Addition of another electron to O2⋅− will give O22−, the peroxide ion, a nonradical (no unpaired electrons left) with a weaker oxygen-oxygen bond. Addition of two more electrons to O22− eliminates the bond entirely, giving two O2− (oxide ions). In biology, the two-electron reduction product of O2 is H2O2, and the four-electron product, water.
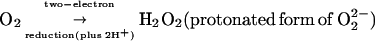 |
 |
Mitochondria take up O2 and reduce 95% or more of it to water, a process achieved by cytochrome oxidase. It removes one electron from each of four reduced (Fe2+-haem) cytochrome c molecules, oxidizing them to ferric cytochrome c, and adds the four electrons on to O2 as shown above. However, it is impossible to add four (not even two!) electrons to O2 at once. Cytochrome oxidase is a large and complex multiprotein assembly, both because it catalyzes several reduction steps and also because it must hold onto toxic partially reduced oxygen species until they can be fully reduced to water (Babcock, 1999).
Aspects of Terminology: The Reactive Oxygen Species
Many different terms are used in the literature to describe oxygen radicals and related (nonradical) species such as 1O2 and H2O2. I prefer the term reactive oxygen species (ROS), a collective descriptor that includes not only the oxygen radicals but also some nonradical derivatives of O2 (Table I). Hence, all oxygen radicals are ROS, but not all ROS are oxygen radicals (Halliwell and Gutteridge, 2006). Reactive is a relative term; O2⋅− and H2O2 are highly selective in their reactions with biological molecules (e.g. H2O2 can inactivate chloroplast Fru and sedoheptulose bisphosphatases, but not most other enzymes), whereas OH⋅ attacks everything around it. The term reactive species has been expanded to include reactive nitrogen, chlorine, and bromine species (Table I). Nitric oxide (NO⋅), an important signaling molecule in animals and plants, is a free radical, and its radical properties explain many of its biological actions (Halliwell et al., 1999).
Table I.
Some reactive species
ROS is a collective term that includes both oxygen radicals and certain nonradicals that are oxidizing agents and/or are easily converted into radicals (HOCl, HOBr, O3, ONOO−, 1O2, and H2O2). All oxygen radicals are ROS, but not all ROS are oxygen radicals. Reactive nitrogen species is a similar collective term that includes NO⋅ and NO2⋅, as well as nonradicals such as HNO2 and N2O4. Reactive is not always an appropriate term: H2O2, NO⋅, and O2⋅− react fast with few molecules, whereas OH⋅ reacts fast with almost everything. Species such as RO2⋅, NO3⋅, RO⋅, HOCl, HOBr, CO3⋅−, CO2⋅−, NO2⋅, ONOO−, NO2+, and O3 have intermediate reactivities.
| Free Radicals | Nonradicals |
|---|---|
| ROS | ROS |
| Superoxide, O2⋅− | H2O2 |
| Hydroxyl, OH⋅ | Hypobromous acid, HOBra |
| Hydroperoxyl, HO2⋅ (protonated superoxide) | Hypochlorous acid, HOClb |
| Carbonate, CO3⋅− | Ozone, O3c |
| Peroxyl, RO2⋅ | Singlet oxygen (O21Δg) |
| Alkoxyl, RO⋅ | Organic peroxides, ROOH |
| Carbon dioxide radical, CO2⋅− | Peroxynitrite, ONOO−d |
| Singlet O21Σg + | Peroxynitrate, O2NOO−d |
| Peroxynitrous acid, ONOOHd | |
| Peroxomonocarbonate, HOOCO2− | |
| Reactive chlorine species | Reactive chlorine species |
| Atomic chlorine, Cl⋅ | Hypochlorous acid, HOClb |
| Nitryl chloride, NO2Cle | |
| Chloramines | |
| Chlorine gas (Cl2) | |
| Bromine chloride (BrCl)a | |
| Chlorine dioxide (ClO2) | |
| Reactive bromine species | Reactive bromine species |
| Atomic bromine, Br⋅ | Hypobromous acid (HOBr) |
| Bromine gas (Br2) | |
| Bromine chloride (BrCl)a | |
| Reactive nitrogen species | Reactive nitrogen species |
| Nitric oxide, NO⋅ | Nitrous acid, HNO2 |
| Nitrogen dioxide, NO2⋅c | Nitrosyl cation, NO+ |
| Nitrate radical, NO3⋅c,f | Nitroxyl anion, NO− |
| Dinitrogen tetroxide, N2O4 | |
| Dinitrogen trioxide, N2O3 | |
| Peroxynitrite, ONOO−d | |
| Peroxynitrate, O2NOO−d | |
| Peroxynitrous acid, ONOOHd | |
| Nitronium cation, NO2+ | |
| Alkyl peroxynitrites, ROONO | |
| Alkyl peroxynitrates, RO2ONO | |
| Nitryl chloride, NO2Cl | |
| Peroxyacetyl nitrate, CH3C(O)OONO2c |
HOBr and BrCl could also be regarded as reactive bromine species.
HOCl and HOBr are often included as ROS.
Oxidizing species formed in polluted air that are toxic to plants and animals.
ONOO−, ONOOH, and O2NOO− are often included as ROS.
NO2Cl can also be regarded as a reactive nitrogen species.
This species may cause formation of allergenic nitrated proteins in pollens.
WHAT DAMAGE CAN FREE RADICALS AND OTHER ROS DO?
If two free radicals meet, they can join their unpaired electrons to form a covalent bond. Thus, NO⋅ and O2⋅− react fast to form a nonradical product, peroxynitrite (Beckman and Koppenol, 1996).
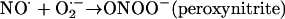 |
At physiological pH, ONOO− rapidly protonates to peroxynitrous acid, ONOOH. This powerful oxidizing and nitrating agent can directly damage proteins, lipids, and DNA. It can also cause damage by undergoing homolytic fission to give noxious products.
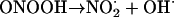 |
Peroxynitrite also reacts with CO2:
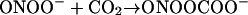 |
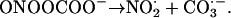 |
Both NO2⋅ (nitrogen dioxide) and CO3⋅− (carbonate radical) are powerful oxidizing agents: not as bad as OH⋅ but not nice either. Thus, any system producing NO⋅ and O2⋅− can cause biological damage, and this occurs in many human diseases (Beckman and Koppenol, 1996; Halliwell et al., 1999; Greenacre and Ischiropoulos, 2001; Alvarez and Radi, 2003). It may happen in plants as well, since they can make both O2⋅− and NO⋅ (Vanin et al., 2004), but few data are available.
However, most biological molecules are nonradicals. When a free radical reacts with a nonradical, a new radical results, and chain reactions may occur. There are several types of reactions (Halliwell and Gutteridge, 2006).
(1) A radical may add to another molecule; the adduct still has an unpaired electron. For example, OH⋅ adds to position 8 in the ring structure of guanine in DNA; the initial product is an 8-hydroxy-2′-deoxyguanosine radical. Among other fates, this can undergo oxidation to the mutagenic lesion 8-hydroxy-2′-deoxyguanosine, 8OHdG (Evans et al., 2004). Hydroxyl radical also attacks other bases and the deoxy-Rib sugar in DNA, producing massive damage (Evans et al., 2004).
(2) A radical may be a reducing agent, donating a single electron. Thus the toxicity of paraquat (PQ2+) to plants, animals, and bacteria involves its reduction to paraquat radical cation (PQ⋅+) by cellular enzymes.
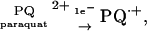 |
followed by PQ⋅+ reducing O2 to O2⋅−:
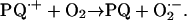 |
Thus, paraquat catalyzes O2⋅− formation (Halliwell and Gutteridge, 2006).
(3) A radical may be an oxidizing agent, taking a single electron from a nonradical to leave an unpaired electron behind. For example, OH⋅ oxidizes carbonate to carbonate radical.
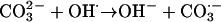 |
(4) A reactive radical (e.g. OH⋅ or NO2⋅) may abstract a hydrogen atom from a C–H bond, e.g. from a hydrocarbon side chain of a polyunsaturated fatty acid (PUFA) residue in a membrane. As H⋅ has only one electron, an unpaired electron remains on the carbon.
Carbon-centered radicals react fast with O2 (like many radicals do) to generate peroxyl radicals
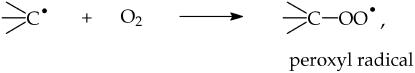 |
which are reactive enough to both oxidize membrane proteins and attack adjacent PUFA side chains, propagating a chain reaction.
A new C⋅ radical is formed to continue the chain, and a lipid hydroperoxide forms. If the PUFA chain is long enough, the peroxyl radicals can whip around and abstract H⋅ from the same PUFA, giving cyclic peroxides (Fam and Morrow, 2003).
A single initiation event thus has the potential to generate multiple peroxide molecules by a chain reaction. The initial H⋅ abstraction from a PUFA can occur at different points on the carbon chain, giving complex mixtures of peroxides. Singlet O2 can react directly with PUFAs to give peroxides, e.g. it converts linoleic acid to 9-, 10-, 12-, and 13-hydroperoxides (Foote et al., 1985). The lifetime of singlet O2 in the hydrophobic interior of membranes is greater than in aqueous solution. Hence, illumination of lipids in the presence of sensitizers of 1O2 formation induces rapid peroxide formation. Such reactions are important in the eye and in plants.
The overall effects of lipid peroxidation are to decrease membrane fluidity, make it easier for phospholipids to exchange between the two halves of the bilayer, increase the leakiness of the membrane to substances that do not normally cross it other than through specific channels (e.g. K+ and Ca2+), and damage membrane proteins, inactivating receptors, enzymes, and ion channels. Iron and copper irons accelerate lipid peroxidation by two mechanisms (Halliwell and Gutteridge, 1984). First, they convert H2O2 to OH⋅ by splitting the O–O bond (the Fenton reaction, in the case of Fe2+). In an analogous reaction, they can split lipid hydroperoxides,
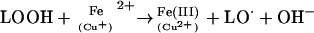 |
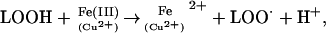 |
giving alkoxyl (LO⋅) and more peroxyl (LOO⋅) radicals to keep the chain reaction going.
Continued oxidation of fatty acid side chains and their fragmentation to produce aldehydes and hydrocarbons (see below) will eventually lead to loss of membrane integrity, e.g. rupture of lysosomal or central vacuolar membranes. In addition, some end products of lipid peroxidation have direct damaging effects. These include the isoprostanes (IPs), cyclic peroxides formed from PUFAs with at least three double bonds, including linolenic acid, arachidonic acid (F2 IPs), eicosapentaenoic acid (F3 IPs), and docosahexaenoic acid (F4 IPs, sometimes called neuroprostanes). Increased formation of IPs is observed in many human diseases and after exposure of animals to a range of toxins (Fam and Morrow, 2003). Some act as vasoconstrictors and can damage tissues. Linoleate-derived IPs (phytoprostanes) are abundant in plants but their roles have not been studied as much as the animal IPs (Mueller, 2004).
Decomposition of lipid peroxides accelerated by iron and copper ions or by heating (e.g. the use of oxidized cooking oils) generates a complex mixture of toxic products including epoxides, saturated aldehydes, unsaturated aldehydes, ketones, and hydrocarbons such as ethane and pentane (Esterbauer et al., 1991). Particularly toxic aldehydes include malondialdehyde (formed from peroxidation of linolenic, arachidonic, or docosahexaenoic acids), and 4-hydroxynonenal, formed from linolenic and arachidonic acid peroxides. Both bind avidly to membrane proteins, inactivating enzymes and receptors. Both can attack DNA, forming mutagenic lesions (Esterbauer et al., 1991).
ANTIOXIDANT DEFENSES
Making Less ROS
How do organisms deal with O2 toxicity? One strategy is to minimize the levels of O2, or deter its conversion to ROS (Halliwell and Gutteridge, 2006). Some mobile organisms avoid O2 toxicity by swimming away from high O2 regions, and Caenorhabditis elegans can cluster together to regulate the group O2 level to the value they like (Gray et al., 2004). The most important source of O2⋅− in vivo in many (perhaps all) aerobic animal cells is probably the mitochondrial electron-transport chain (in plants, chloroplasts make a lot of O2⋅− as well, of course). Whereas cytochrome oxidase releases no ROS (a beautifully evolved enzyme complex), some earlier components of the mitochondrial electron-transport chain can leak electrons directly to O2, although passing the bulk of them onto the next component in the chain (Turrens, 2003). This leakage generates O2⋅−. Whereas mammalian cytochrome oxidase is saturated at low O2 tensions, the rate of electron leakage (and hence O2⋅− production) by mitochondria is, in general, increased by raising O2 levels (although it is not quite so simple; leakage is also favored by high levels of reduced carriers, which can drop when O2 is high).
Aerobes may have evolved to pack the redox constituents of the mitochondrial and other electron-transport chains together in a way that makes escape of electrons to O2 to form O2⋅− less likely. Plants may additionally use alternative oxidase systems to minimize mitochondrial O2⋅− formation (Moller, 2001), and both plants and animals have uncoupling proteins in their inner mitochondrial membranes. These may act as antioxidants, e.g. by allowing more proton leak and preventing a back up of electrons in the chain to escape to O2. Formation of 4-hydroxynonenal or O2⋅− within mitochondria seems to activate uncoupling proteins, which should then decrease the membrane potential and limit O2⋅− formation (Brand et al., 2004). Proteins that bind metal ions (such as transferrin, ferritins, and metallothioneins) and so hinder both Fenton chemistry and the acceleration of lipid peroxide ion by iron and copper, are other mechanisms deterring ROS formation (Halliwell and Gutteridge, 1990).
Scavenging ROS
The superoxide dismutase enzymes (SODs) remove O2⋅− by catalyzing its dismutation, one O2⋅− being reduced to H2O2 and another oxidized to O2.
 |
Animals have SODs containing active-site manganese (MnSOD) in the mitochondrial matrix, plus SODs with copper and zinc (CuZnSOD) in the mitochondrial intermembrane space and in the rest of the cell (Fridovich, 1995). Plants have more or less the same, but some have iron-containing SODs (FeSOD) in the chloroplast (in addition to CuZnSOD; Alscher et al., 2002). Bacteria often have CuZnSOD plus MnSOD and/or FeSOD; a few even have nickel-containing SOD (Halliwell and Gutteridge, 2006). Whatever the metal, all SODs catalyze the above reaction (Fridovich, 1995; Halliwell and Gutteridge, 2006). Some anaerobic bacteria cope with O2⋅− (generated if they are transiently exposed to O2) in a different way: they use superoxide reductase (Niviere and Fontecave, 2004) proteins, which catalyze the overall reaction.
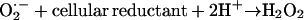 |
Unlike SOD, no O2 is produced, an obvious advantage for an anaerobe, although the H2O2 has to be dealt with.
Superoxide dismutases and reductases must work together with enzymes that remove H2O2. Catalases are not the most important in this context since there is little or no catalase in mitochondria and chloroplasts, where much O2⋅− is generated (Halliwell and Gutteridge, 2006). Most or all catalase in plants and animals is in peroxisomes, to deal with H2O2 produced by oxidase enzymes acting on such substrates as glycollate, urate, and D amino acids (Schrader and Fahimi, 2004). Plants are rich in peroxidases, enzymes that remove H2O2 by using it to oxidize a cosubstrate. Many plant peroxidases are nonspecific, using multiple cosubstrates (horseradish [Armoracia lapathifolia] peroxidase is perhaps the most-studied example); ascorbate peroxidases in plant chloroplast and cytosol can remove H2O2 by using vitamin C as a cosubstrate, oxidizing it to a (poorly reactive) ascorbyl free radical (Mano et al., 2001).
Until recently, it was thought that the most important H2O2-removing enzymes in animals are glutathione peroxidases, a family (Brigelius-Flohe, 1999) of selenium-containing enzymes that remove H2O2 by coupling its reduction to water with oxidation of reduced glutathione (GSH), a thiol-containing tripeptide (glu-cys-gly).
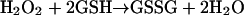 |
The product, oxidized glutathione (GSSG), consists of two GSH linked by a disulphide bridge, and can be converted back to GSH by glutathione reductase enzymes.
At least four types of GPx exist. One is the classical enzyme, often now called GPx1. Mammalian body fluids contain low levels of a different form, GPx3. Another type, GPx2, is found in the cells lining the gastrointestinal tract and may help to metabolize peroxides in ingested food lipids. The fourth member of the family is phospholipid hydroperoxide glutathione peroxidase (PHGPx or GPx4), with the ability to reduce not only H2O2 but also fatty acid hydroperoxides (to alcohols) that are still esterified in lipids of membranes or lipoproteins. In all four types of GPx, the selenium is essential for catalysis (Brigelius-Flohe, 1999). In marked contrast, selenium plays little role in plants. The chloroplast appears to lack selenoprotein GPx activity (indeed selenoprotein GPx enzymes are rare in plants) and ascorbate peroxidase takes over some or all of the job of H2O2 removal (Mano et al., 2001). GPx-like activity has been identified in chloroplasts and cytoplasm in some plant species, and genes similar to those encoding GPxs in animals (most commonly resembling PHGPx genes) have been identified in several plant genomes. Plant GPxs have Cys rather than seleno Cys at the active sites, which decreases their catalytic activity as compared with selenoprotein GPx enzymes. Indeed, at least some of the plant enzymes may prefer thioredoxin to GSH as a substrate (Herbette et al., 2002; Rodriguez Milla et al., 2003).
It has now been realized that peroxiredoxins may be the most important H2O2-removal systems in animals, bacteria, and possibly plants (Rhee et al., 2005). They are homodimers and contain no prosthetic groups: The redox reactions are dependent on Cys at the active sites. There are at least three classes: the typical 2-cys (the most common), the atypical 2-cys, and the 1-cys peroxiredoxins. In all cases, H2O2 oxidizes an -SH group on the peroxiredoxin to a sulfenic acid, cys-SOH. In the 2-cys enzymes, this reacts with another -SH on the protein to give a disulphide that is then reduced by thioredoxin. In 1-cys peroxiredoxins, another cellular reductant (not yet identified) regenerates the -SH group.
Peroxiredoxins are slower at catalyzing H2O2 removal than GPx, although the large amounts present (up to 0.8% of total soluble protein in some animal cells) and their low Km for H2O2 (<20 μm) can compensate for this, as can their presence in all subcellular organelles and in the cytosol. Peroxiredoxins are readily inactivated by H2O2, the eukaryotic ones being more susceptible to this than bacterial ones (Georgiou and Masip, 2003). Cells have various mechanisms for reactivating oxidized peroxiredoxins (Rhee et al., 2005). In animals, selenium is essential for the activity of thioredoxin reductase, the enzyme that keeps thioredoxins in the reduced state for peroxiredoxins (and for the many other metabolic activities of thioredoxins). Thus lack of selenium in animals impairs the peroxiredoxin system as well as GPx (Su et al., 2005).
Finally, there are sacrificial antioxidants, agents that are preferentially oxidized by reactive species to preserve more important biomolecules. For example, ascorbate can scavenge most ROS, including O2⋅−, OH⋅, RO2⋅, and ONOOH, as can GSH. Tocopherols (Tocs) are good scavengers of peroxyl radicals and help to protect membranes against lipid peroxidation by interrupting the chain reaction. Reaction is with the phenolic -OH group of the Toc structure (Halliwell and Gutteridge, 2006).
The essence of the antioxidant actions of ascorbate and Toc is that the radicals they form are poorly reactive (Smirnoff, 2001; Halliwell and Gutteridge, 2006). Plants can make all the ascorbate and Tocs they want, we poor humans need to eat the plants to get them! Most animals can still make ascorbate, but the terminal step in its synthesis generates H2O2 (Puskas et al., 1998).
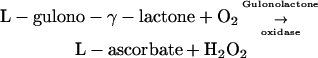 |
Is this why humans, evolving on a plant-rich diet, lost this enzyme? It is an interesting speculation. Also interesting, ascorbate synthesis in plants uses a different pathway that does not make H2O2 (Smirnoff, 2001).
Other examples of sacrificial antioxidants include carotenoids, urate, plasma albumin, and GSH.
WHAT'S SO BAD ABOUT SUPEROXIDE?
Transgenic animal experiments show that SODs are important enzymes in animals. Even bacteria and yeasts (Saccharomyces cerevisiae) lacking SODs are pretty sick (for review, see Halliwell and Gutteridge, 2006) and plants have problems as well (e.g. Rizhsky et al., 2003). In one study (Lebovitz et al., 1996), most MnSOD-knockout mice died within 10 d after birth, with cardiac abnormalities, fat accumulation in liver and skeletal muscle, metabolic acidosis, and severe mitochondrial damage in heart and, to a lesser extent, in other tissues. Those mice that manage to survive longer than 10 d soon succumb to a variety of pathologies, including anemia, retinal defects, and neurodegeneration (Melov et al., 1998). Even MnSOD (+/−) heterozygous mice, which at first seem normal, show increased mitochondrial oxidative damage as they age, as well as increased nuclear oxidative DNA damage and elevated cancer rates (Van Remmen et al., 2003).
Mice lacking CuZnSOD have also been obtained. When young, they appear normal (although they are more sensitive to ROS-generating toxins) but as they age, neurological damage, hearing loss, and cancers (especially liver cancer) can develop at an accelerated rate. They have reproductive problems and show impaired vascular function (Elchuri et al., 2005).
It follows that O2⋅− can cause severe damage. But why? Superoxide in aqueous solution does not react with many biomolecules. However, those few that it does attack are very important. First, its reaction with NO⋅ to give ONOO− is fast. A second clue came from studies with bacteria (Imlay, 2003). Superoxide inactivates several enzymes important in energy production and amino acid metabolism. These enzymes have iron-sulfur clusters; inactivation is caused by oxidation of the cluster, leading to release of iron, followed by Fenton chemistry. The oxidized enzymes can be repaired in vivo by reassembling the iron clusters. Indeed, even the low basal levels of O2⋅− production in Escherichia coli damage these enzymes, and activity is maintained by constantly repairing them (Imlay, 2003). If O2⋅− levels rise (e.g. at elevated O2), inactivation rates accelerate, repair cannot keep up, and metabolic pathways are inhibited. Similar damage to Fe-S cluster enzymes occurs in yeast and animals. Superoxide can release additional iron from ferritins, and degradation of haem proteins by H2O2 can also release iron. Peroxynitrite also displaces iron from Fe-S proteins (Halliwell and Gutteridge, 2006).
What about H2O2? Knockout of catalase or GPx1 in animals does not cause many problems, but removal of both GPx1 and GPx2 predisposed mice to inflammation and cancer of the intestines, and knockout of GPx4 is embryonic lethal (Chu et al., 2004; Halliwell and Gutteridge, 2006). Knockouts of peroxiredoxins in mice also cause problems (anemia, cancer, etc.) later in life (Lee et al., 2003).
ROS: NOT AS BAD AS THEY LOOK
Antioxidant defenses are not 100% effective, since oxidative damage to DNA, proteins, and lipids is demonstrable in all aerobes under ambient O2. Indeed, this damage may contribute to the age-related development of cancer in animals (Ames, 1983; Halliwell, 2002) and perhaps even to ageing itself. Hence, even 21% O2 is toxic. So why are all the ROS not eliminated? Probably because ROS perform important roles, so the challenge was to evolve antioxidant defenses that allow such roles while minimizing damage. Mechanisms to cope with oxidative damage are required, e.g. to repair oxidized DNA, reassemble [Fe-S] clusters in enzymes, repair oxidized Met residues on proteins (using Met sulfoxide reductases, some of which are selenium requiring in animals, but not in plants; Moskovitz and Stadtman, 2003), and destroy oxidized lipids and proteins.
In what ways could ROS be beneficial? Plants provide lots of examples. ROS production in animals by phagocytes and by other cells in the gastrointestinal and respiratory tracts is a defense against microorganisms (Fang, 2004; Donko et al., 2005). Everyone is familiar with regulation of cellular processes by phosphorylation and dephosphorylation of enzymes and transcription factors, but we have realized in the past decade that regulation by oxidation and reduction (redox regulation) is equally important. Not only that, the two systems cross talk, i.e. the redox state of the cell influences phosphorylation, and vice versa. For example, binding of ligands to growth factor receptors on animal cells activates protein kinases that then phosphorylate and activate subsequent proteins in the signal cascade. Often at the same time, cellular ROS levels increase and facilitate the signaling. Although some kinases can be directly affected by ROS (e.g. some isoforms of protein kinase C), in general ROS tend not to stimulate phosphorylation directly. Instead, they increase net phosphorylation by inhibiting protein dephosphorylation (Rhee et al., 2005). Protein phosphatase enzymes are constantly active in cells, but can be attacked and inactivated by ROS.
Thus, the ligand binding increases kinase activity, and the ROS assist by transiently inactivating phosphatases. Where do the ROS come from? Sometimes the ligand increases O2⋅− production, e.g. by activating O2⋅−-producing NADPH oxidase enzymes. These were originally described in phagocytes (Fang, 2004), but are now known to be widespread in animal (and plant, see the rest of this issue) cells. In addition, when cells are exposed to extra H2O2 (e.g. at a site of injury or inflammation or when NADPH oxidase enzymes are activated), the peroxiredoxins are partially inactivated to allow signaling, neatly explaining why the animal enzymes are more sensitive to inactivation. The cell then rapidly makes more peroxiredoxin, and reactivates the inactive form, so that the extra H2O2 can be removed after it has done its job (Georgiou and Masip, 2003; Rhee et al., 2005).
Mitochondrial ROS production is often thought of as a nuisance, an unavoidable consequence of electron leakage under O2, a view consistent with the severe phenotype of MnSOD-knockout mice. However, another view is that variations in mitochondrial H2O2 production are a signal that advises the cytoplasm and nucleus what the mitochondria are doing, leading to changes in nuclear gene transcription via redox regulation and phosphorylation of transcription factors. Mitochondrially targeted antioxidants are proving useful in attempts to study the physiological roles of mitochondrial ROS (Sheu et al., 2006). Chloroplast ROS may perform similar roles.
OXIDATIVE STRESS AND DAMAGE
What happens if the balance between ROS and antioxidants is upset? Having too many ROS in relation to the available antioxidants is said to be a state of oxidative stress. Sies (1991) defined this term as a disturbance in the prooxidant-antioxidant balance in favor of the former, leading to potential damage. Such damage is often called oxidative damage, which has been defined as the biomolecular damage caused by attack of reactive species upon the constituents of living organisms (Halliwell and Whiteman, 2004). Increased oxidative damage can result not only from more oxidative stress, but also from failure to repair or replace damaged biomolecules. Oxidative stress can result from decreases in antioxidant levels, e.g. mutations decreasing the levels of MnSOD. Depletions of dietary antioxidants and other essential dietary constituents (e.g. copper, iron, zinc, and magnesium) can also cause it. For example, children with the protein deficiency disease kwashiorkor suffer oxidative stress, involving low GSH levels (lack of sulfur-containing amino acids in the diet) and iron overload (inability to make enough transferrin; Fuchs, 2005). Oxidative stress can also be due to increased ROS production, e.g. by exposure to elevated O2, the presence of toxins that produce ROS (e.g. paraquat), or excessive activation of natural systems producing ROS, e.g. inappropriate activation of phagocytes (Halliwell and Gutteridge, 2006).
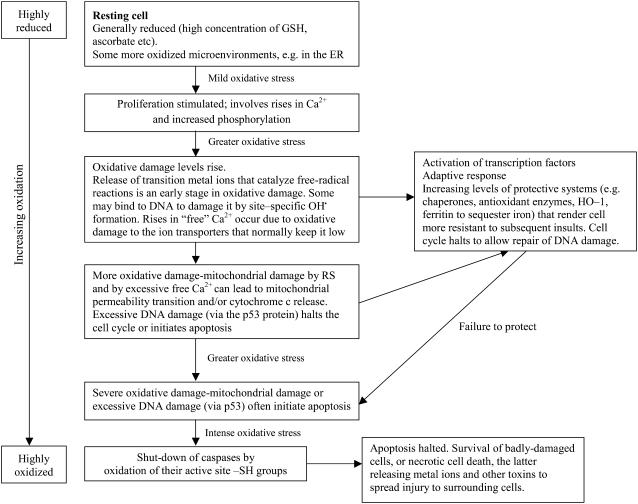
What do cells do when under oxidative stress? It depends on the cell and the level of stress applied (Fig. 2). Usually intracellular free Ca2+ levels rise; so do levels of iron catalytic for free radical reactions (Fig. 2). Several cell types respond to mild oxidative stress by proliferating, which can be good in wound healing but bad if it leads to tissue fibrosis (Cave et al., 2005). Cells may adapt to the stress by up-regulation of defense and/or repair systems. This may completely protect against damage, protect against damage to some extent but not completely, or sometimes overprotect; the cells are then resistant to higher levels of oxidative stress imposed subsequently. Adaptation need not always involve increases in antioxidants: there can be decreases in ROS-producing systems, increases in other protective mechanisms (such as chaperones), or changes in oxidative damage targets (e.g. E. coli under oxidative stress can replace a fumarase enzyme sensitive to inactivation by O2⋅− with one that resists O2⋅− [Liochev and Fridovich, 1993]). Moderate oxidative stress usually halts the cell cycle, or can drive cells into senescence; the cell survives but can no longer divide. Severe oxidative damage, especially to DNA, may trigger death by apoptosis, necrosis, or mechanisms with features of both. Indeed, ROS act as triggers of apoptosis, and as participants in apoptosis induced by other mechanisms, in both plants and animals.
Figure 2.
How cells respond to oxidative stress. Adapted from Halliwell and Gutteridge (2006) by courtesy of Oxford University Press. Stimulation of proliferation by low levels of reactive species is associated with increased net phosphorylation of multiple proteins. The cell is generally a reducing environment, especially the mitochondria (GSH/GSSG > 100) and cytosol (GSH/GSSG > 100), but less so in the endoplasmic reticulum (ER) lumen (GSH/GSSG = approximately 3), since a more-oxidizing environment is required for optimal protein folding and disulphide bridge formation. HO-1, Haem oxygenase 1; RS, reactive species.
SUMMARY
ROS are all over the place in plants, animals, and aerobic bacteria. We cannot live without them or we will probably die from infections. Yet, they often kill us in the end; over the long human lifespan, the continual damage by ROS, if not properly repaired (and repair efficiency tends to drop in the aged) can contribute to the age-related development of cancer, neurodegenerative diseases, and many other disorders (Ames, 1983; Halliwell and Gutteridge, 2006). This may be a by product of evolution; ROS are essential for defense against infection and signaling, keeping you alive until your reproduction has finished and children have grown up. Who cares if they kill you in the later postreproductive years? Evolution doesn't. So why does taking antioxidant supplements not make us live healthily for ever? Simply because the human body regulates the ROS/antioxidant balance so carefully that feeding antioxidants does not disturb it much, and oxidative damage does not decrease (Halliwell, 1999, 2000).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Barry Halliwell (bchbh@nus.edu.sg).
References
- Alscher RG, Erturk N, Heath LS (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53: 1331–1341 [PubMed] [Google Scholar]
- Alvarez B, Radi R (2003) Peroxynitrite reactivity with amino acids and proteins. Amino Acids 25: 295–311 [DOI] [PubMed] [Google Scholar]
- Ames BN (1983) Dietary carcinogens and anticarcinogens: oxygen radicals and degenerative diseases. Science 221: 1256–1264 [DOI] [PubMed] [Google Scholar]
- Babcock GT (1999) How oxygen is activated and reduced in respiration. Proc Natl Acad Sci USA 96: 13114–13117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balentine JD (1982) Pathology of Oxygen Toxicity. Academic Press, New York
- Beckman JS, Koppenol WH (1996) Nitric oxide, superoxide and peroxynitrite: the good, the bad, and ugly. Am J Physiol 271: C1424–C1437 [DOI] [PubMed] [Google Scholar]
- Beerling DJ, Woodward FI, Lomas MR, Wills MA, Quick WP, Valdes PJ (1998) The influence of carboniferous palaeoatmospheres on plant function: an experimental and modelling assessment. Philos Trans R Soc B Biol Sci 353: 131–139 [Google Scholar]
- Brand MD, Affourtit C, Esteves TC, Green K, Lambert AJ, Miwa S, Pakay JL, Parker N (2004) Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radic Biol Med 37: 755–767 [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R (1999) Tissue-specific functions of individual glutathione peroxidases. Free Radic Biol Med 27: 951–965 [DOI] [PubMed] [Google Scholar]
- Cave A, Grieve D, Johar S, Zhang M, Shah AM (2005) NADPH oxidase-derived reactive oxygen species in cardiac pathophysiology. Philos Trans R Soc Lond B Biol Sci 360: 2327–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu FF, Esworthy RS, Chu PG, Longmate JA, Huycke MM, Wilczynski S, Doroshow JH (2004) Bacteria-induced intestinal cancer in mice with disrupted Gpx1 and Gpx2 genes. Cancer Res 64: 962–968 [DOI] [PubMed] [Google Scholar]
- Donko A, Peterfi Z, Sum A, Leto T, Geiszt M (2005) Dual oxidases. Philos Trans R Soc Lond B Biol Sci 360: 2301–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elchuri S, Oberley TD, Qi WB, Eisenstein RS, Roberts LJ, Van Remmen H, Epstein CJ, Huang TT (2005) CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene 24: 367–380 [DOI] [PubMed] [Google Scholar]
- Esterbauer H, Schaur RJ, Zollner H (1991) Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med 11: 81–128 [DOI] [PubMed] [Google Scholar]
- Evans MD, Dizdaroglu M, Cooke MS (2004) Oxidative DNA damage and disease: induction, repair and significance. Mutat Res 567: 1–61 [DOI] [PubMed] [Google Scholar]
- Fam SS, Morrow JD (2003) The isoprostanes: unique products of arachidonic acid oxidation - a review. Curr Med Chem 10: 1723–1740 [DOI] [PubMed] [Google Scholar]
- Fang FC (2004) Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol 2: 820–832 [DOI] [PubMed] [Google Scholar]
- Foote CS, Valentine JS, Greenberg A, Liebman JF, editors (1985) Active Oxygen in Chemistry. Chapman and Hall, New York
- Fridovich I (1995) Superoxide radical and SODs. Annu Rev Biochem 64: 97–112 [DOI] [PubMed] [Google Scholar]
- Fuchs GJ (2005) Antioxidants for children with kwashiorkor. BMJ 330: 1095–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiou G, Masip L (2003) Biochemistry: an overoxidation journey with a return ticket. Science 300: 592–594 [DOI] [PubMed] [Google Scholar]
- Gilbert DL, editor (1981) Oxygen and Living Processes: An Inter-Disciplinary Approach. Springer, New York
- Graham JB, Aguilar NM, Dudley R, Gans C (1995) Implications of the late Palaeozoic oxygen pulse for physiology and evolution. Nature 375: 117–120 [Google Scholar]
- Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta MA, Bargmann CI (2004) Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430: 317–322 [DOI] [PubMed] [Google Scholar]
- Greenacre SA, Ischiropoulos H (2001) Tyrosine nitration: localisation, quantification, consequences for protein function and signal transduction. Free Radic Res 34: 541–581 [DOI] [PubMed] [Google Scholar]
- Halliwell B (1999) Establishing the significance and optimal intake of dietary antioxidants: the biomarker concept. Nutr Rev 57: 104–113 [DOI] [PubMed] [Google Scholar]
- Halliwell B (2000) The antioxidant paradox. Lancet 355: 1179–1180 [DOI] [PubMed] [Google Scholar]
- Halliwell B (2002) Effect of diet on cancer development: is oxidative DNA damage a biomarker? Free Radic Biol Med 32: 968–974 [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC (1984) Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J 219: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC (1990) The antioxidants of human extracellular fluids. Arch Biochem Biophys 280: 1–8 [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC (2006) Free Radicals in Biology and Medicine, Ed 4. Clarendon Press, Oxford
- Halliwell B, Whiteman M (2004) Measuring reactive species and oxidative damage in vivo and in cell culture: how should you do it and what do the results mean? Br J Pharmacol 142: 231–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Zhao K, Whiteman M (1999) Nitric oxide and peroxynitrite: the ugly, the uglier and the not so good: a personal view of recent controversies. Free Radic Res 31: 651–669 [DOI] [PubMed] [Google Scholar]
- Herbette S, Lenne C, Leblanc N, Julien JL, Drevet JR, Roeckel-Drevet P (2002) Two GPX-like proteins from Lycopersicon esculentum and Helianthus annuus are antioxidant enzymes with phospholipid hydroperoxide glutathione peroxidase and thioredoxin peroxidase activities. Eur J Biochem 269: 2414–2420 [DOI] [PubMed] [Google Scholar]
- Hetz SK, Bradley TJ (2005) Insects breathe discontinuously to avoid oxygen toxicity. Nature 433: 516–519 [DOI] [PubMed] [Google Scholar]
- Holt NE, Zigmantas D, Valkunas L, Li XP, Niyogi KK, Fleming GR (2005) Carotenoid cation formation and the regulation of photosynthetic light harvesting. Science 307: 433–436 [DOI] [PubMed] [Google Scholar]
- Horsburgh MJ, Wharton SJ, Karavolos M, Foster SJ (2002) Manganese: elemental defence for a life with oxygen. Trends Microbiol 10: 496–501 [DOI] [PubMed] [Google Scholar]
- Imlay JA (2003) Pathways of oxidative damage. Annu Rev Microbiol 57: 395–418 [DOI] [PubMed] [Google Scholar]
- Kasting JF (1993) Earth's early atmosphere. Science 259: 920–926 [DOI] [PubMed] [Google Scholar]
- Lane N (2002) Oxygen, the Molecule That Made the World. Oxford University Press, Oxford
- Lebovitz RM, Zhang H, Vogel H, Cartwright J Jr, Dionne L, Lu N, Huang S, Matzuk MM (1996) Neurodegeneration, myocardial injury, and perinatal death in mitochondrial superoxide dismutase-deficient mice. Proc Natl Acad Sci USA 93: 9782–9787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Kim SU, Yu SL, Kim SH, Park DS, Moon HB, Dho SH, Kwon KS, Kwon HJ, Han YH, et al (2003) Peroxiredoxin II is essential for sustaining life span of erythrocytes in mice. Blood 101: 5033–5038 [DOI] [PubMed] [Google Scholar]
- Liochev SI, Fridovich I (1993) Modulation of the fumarases of E. coli in response to oxidative stress. Arch Biochem Biophys 301: 379–384 [DOI] [PubMed] [Google Scholar]
- Mano J, Ohno C, Domae Y, Asada K (2001) Chloroplastic ascorbate peroxidase is the primary target of methyl viologen-induced photooxidative stress in spinach leaves: its relevance to monodehydroascorbate radical detected with in vivo ESR. Biochim Biophys Acta 504: 275–287 [DOI] [PubMed] [Google Scholar]
- Melov S, Schneider JA, Day BJ, Hinerfeld D, Coskun P, Mirra SS, Crapo JD, Wallace DC (1998) A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nat Genet 18: 159–163 [DOI] [PubMed] [Google Scholar]
- Moller IM (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52: 561–591 [DOI] [PubMed] [Google Scholar]
- Morison WL (2004) Clinical practice: photosensitivity. N Engl J Med 350: 1111–1117 [DOI] [PubMed] [Google Scholar]
- Moskovitz J, Stadtman ER (2003) Selenium-deficient diet enhances protein oxidation and affects methionine sulfoxide reductase (MsrB) protein level in certain mouse tissues. Proc Natl Acad Sci USA 100: 7486–7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller MJ (2004) Archetype signals in plants: the phytoprostanes. Curr Opin Plant Biol 7: 441–448 [DOI] [PubMed] [Google Scholar]
- Niviere V, Fontecave M (2004) Discovery of superoxide reductase: an historical perspective. J Biol Inorg Chem 9: 119–123 [DOI] [PubMed] [Google Scholar]
- Olson JM, Blankenship RE (2004) Thinking about the evolution of photosynthesis. Photosynth Res 80: 373–386 [DOI] [PubMed] [Google Scholar]
- Puskas F, Braun L, Csala M, Kardon T, Marcolongo P, Benedetti A, Mandl J, Banhegyi G (1998) Gulonolactone oxidase activity-dependent intravesicular glutathione oxidation in rat liver microsomes. FEBS Lett 430: 293–296 [DOI] [PubMed] [Google Scholar]
- Rhee SG, Chae HZ, Kim K (2005) Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med 38: 1543–1552 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R (2003) The water-water cycle is essential for chloroplast protection in the absence of stress. J Biol Chem 278: 38921–38925 [DOI] [PubMed] [Google Scholar]
- Rodriguez Milla MA, Maurer A, Huete AR, Gustafson JP (2003) Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J 36: 602–615 [DOI] [PubMed] [Google Scholar]
- Schrader M, Fahimi HD (2004) Mammalian peroxisomes and reactive oxygen species. Histochem Cell Biol 122: 383–393 [DOI] [PubMed] [Google Scholar]
- Sheu SS, Nauduri D, Anders MW (2006) Targeting antioxidants to mitochondria: a new therapeutic direction. Biochim Biophys Acta 1762: 256–265 [DOI] [PubMed] [Google Scholar]
- Sies H (1991) Oxidative Stress. Oxidants and Antioxidants. Academic Press, New York [DOI] [PubMed]
- Smirnoff N (2001) L-ascorbic acid biosynthesis. Vitam Horm 61: 241–266 [DOI] [PubMed] [Google Scholar]
- Su D, Novoselov SV, Sun QA, Moustafa ME, Zhou Y, Oko R, Hatfield DL, Gladyshev VN (2005) Mammalian selenoprotein thioredoxin-glutathione reductase: roles in disulfide bond formation and sperm maturation. J Biol Chem 280: 26491–26498 [DOI] [PubMed] [Google Scholar]
- Symons MCR (1996) Radicals generated by bone cutting and fracture. Free Radic Biol Med 20: 831–835 [DOI] [PubMed] [Google Scholar]
- Turrens JF (2003) Mitochondrial formation of reactive oxygen species. J Physiol 552: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, Huang TT, et al (2003) Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics 16: 29–37 [DOI] [PubMed] [Google Scholar]
- Vanin AF, Svistunenko DA, Mikoyan VD, Serezhenkov VA, Fryer MJ, Baker NR, Cooper CR (2004) Endogenous superoxide production and the nitrite/nitrate ratio control the concentration of bioavailable free nitric oxide in leaves. J Biol Chem 279: 24100–24107 [DOI] [PubMed] [Google Scholar]
- von Sonntag C (1987) The Chemical Basis of Radiation Biology. Taylor & Francis, London
- Wagner D, Przybyla D, Op den Camp R, Kim C, Landgraf F, Lee KP, Wursch M, Laloi C, Nater M, Hideg E, et al (2004) The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306: 1183–1185 [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Mosolf J, Willits D, Yeager M, Dryden KA, Young M, Douglas T (2005) An archaeal antioxidant: characterization of a Dps-like protein from Sulfolobus solfataricus. Proc Natl Acad Sci USA 102: 10551–10556 [DOI] [PMC free article] [PubMed] [Google Scholar]