Abstract
The chloroplast NAD(P)H dehydrogenase (NDH) complex is involved in photosystem I cyclic electron transport and chlororespiration in higher plants. An Arabidopsis (Arabidopsis thaliana) chlororespiratory reduction 6 (crr6) mutant lacking NDH activity was identified by means of chlorophyll fluorescence imaging. Accumulation of the NDH complex was impaired in crr6. Physiological characterization of photosynthetic electron transport indicated the specific defect of the NDH complex in crr6. In contrast to the CRR7 protein that was recently identified as a potential novel subunit of the NDH complex by means of the same screening, the CRR6 protein was stable under the crr2 mutant background in which the NDH complex does not accumulate. The CRR6 gene (At2g47910) encodes a novel protein without any known motif. Although CRR6 does not have any transmembrane domains, it is localized in the thylakoid membrane fraction of the chloroplast. CRR6 is conserved in phototrophs, including cyanobacteria, from which the chloroplast NDH complex has evolutionally originated, but not in Chlamydomonas reinhardtii, in which the NDH complex is absent. We believe that CRR6 is a novel specific factor for the assembly or stabilization of the NDH complex.
The light reactions of photosynthesis involve electron transport through the thylakoid membrane in the chloroplast. Two photosystems, PSI and PSII, convert light energy into the chemical energy that drives electron transport. Coupled with this electron transport, the cytochrome (Cyt) b6f complex is involved in generating the proton gradient across the thylakoid membrane (ΔpH), which is utilized by H+-ATPase for ATP synthesis. In addition to this linear electron transport, PSI cyclic electron transport is solely driven by PSI and generates ΔpH without any net accumulation of NADPH (for review, see Munekage and Shikanai, 2005). In higher plants, PSI cyclic electron transport consists of two partially redundant pathways (Munekage et al., 2004). The main pathway requires a thylakoid protein, PROTON GRADIENT REGULATION 5 (PGR5), and is essential for photoprotection (Munekage et al., 2002). The chloroplast NAD(P)H dehydrogenase (NDH) complex functions in the minor pathway and is essential under certain stress conditions (Endo et al., 1999; Horváth et al., 2000; Munné-Bosch et al., 2005; Wang et al., 2006). Notably, the NDH complex is crucial under the mutant background of pgr5, strongly supporting the idea that the NDH complex protects the chloroplast from stromal overreduction (Munekage et al., 2004).
The machinery of photosynthetic electron transport is embedded in the thylakoid membrane and consists of multiple subunits encoded by both the nuclear and the chloroplast genomes in plants. Their functional assembly requires multiple steps that include gene expression in both the nuclear and the chloroplast genomes, which are developmentally and environmentally regulated (for review, see Barkan and Goldschmidt-Clermont, 2000), targeting and insertion of proteins into the thylakoid membrane (for review, see Bauer et al., 2001), and protein folding with cofactors to form an active complex (for review, see Pilon et al., 2006). Because the plastids have evolutionally originated from cyanobacteria, both prokaryotic and eukaryotic machinery are utilized in the biogenesis of the photosynthetic apparatus in the chloroplast. For example, the system of plastid gene expression is essentially similar to that of prokaryotes (for review, see Sugiura et al., 1998). In contrast, gene expression is posttranscriptionally regulated by numerous nuclear-encoded factors, including members of the pentatricopeptide repeat family that are specific to eukaryotes (Lurin et al., 2004). For another example, assembly of PSI requires a plastid-encoded gene, ycf3, which originated from cyanobacteria (Boudreau et al., 1997). On the other hand, a member of the ACCUMULATION OF PHOTOSYSTEM ONE1 (APO1) family that is specific to eukaryotes is involved in the cofactor assembly in PSI (Amann et al., 2004). To survey the genes involved in the biogenesis of the photosynthetic apparatus, the genetic approaches focusing on the activity of the specific complex have significantly contributed (Miles, 1980; Meurer et al., 1996; Shikanai et al., 1999; Hashimoto et al., 2003).
Activity of the chloroplast NDH complex is monitored as a transient increase in chlorophyll fluorescence after turning off actinic light (AL; Burrows et al., 1998; Kofer et al., 1998; Shikanai et al., 1998). This fluorescence change was monitored under a CCD camera to screen the Arabidopsis (Arabidopsis thaliana) chlororespiratory reduction (crr) mutant specifically defective in NDH activity (Hashimoto et al., 2003). Because 11 subunits of the NDH complex are encoded by the chloroplast genome (Matsubayashi et al., 1987), several crr mutants are defective in the expression of chloroplast ndh genes. CRR2 is essential for intergenic RNA cleavage between rps7 and ndhB (Hashimoto et al., 2003), whereas CRR4 is involved in the RNA editing that creates the translational initiation codon of ndhD (Kotera et al., 2005). Both genes encode members of the pentatricopeptide repeat family, which is specific to eukaryotes (Lurin et al., 2004).
In addition to the 11 chloroplast ndh genes, the nuclear genome also encodes several subunits of the NDH complex (Rumeau et al., 2005). Although 14 subunits have been identified for the chloroplast NDH complex so far, the subunit functioning in electron donor binding is still unclear (for review, see Shikanai and Endo, 2000). Most probably, the chloroplast NDH complex consists of more than 14 subunits. However, due to the fragile nature of the NDH complex during purification, it has been difficult to derive the entire subunit composition of the NDH complex in both the chloroplast and the cyanobacteria. The genetic approach focusing on the activity of the NDH complex complemented this biochemical approach. The Arabidopsis CRR7 protein was identified from the crr7 mutant and is a candidate for a subunit of the most fragile subcomplex in the chloroplast NDH complex (Munshi et al., 2005). Here we report characterization of the Arabidopsis crr6 mutant, which is distinctly different from crr7.
RESULTS
crr6 Is Defective in Accumulation of the Chloroplast NDH Complex
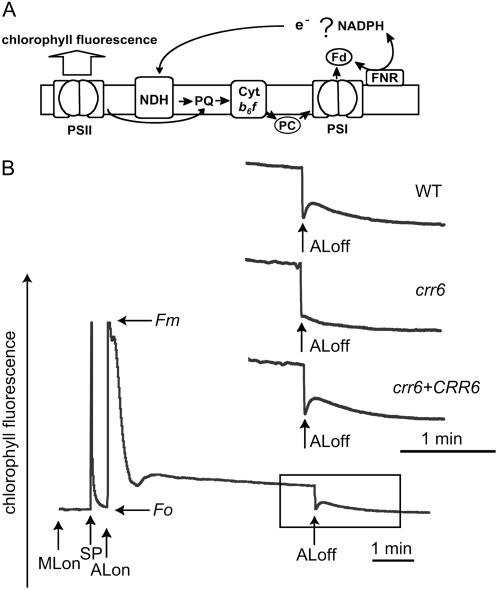
The NDH complex is involved in PSI cyclic electron flow in both the chloroplast and the cyanobacteria. Arabidopsis crr mutants, specifically defective in NDH activity, were identified based on monitoring of chlorophyll fluorescence levels under a CCD camera (Hashimoto et al., 2003). In the wild type, the chlorophyll fluorescence level transiently increased after turning off AL (Fig. 1, A and B). This change in chlorophyll fluorescence level is ascribed to the reduction of plastoquinone (PQ) via NDH activity (Burrows et al., 1998; Kofer et al., 1998; Shikanai et al., 1998) and was impaired in the crr6 mutant (Fig. 1B). Although PQ is also reduced via a non-NDH pathway after heat stress (Sazanov et al., 1998), the fluorescence change exclusively depends on NDH activity under the conditions used in this study (Hashimoto et al., 2003).
Figure 1.
Monitoring of NDH activity using chlorophyll fluorescence analysis. A, Schematic model of NDH function. The NDH complex functions in electron transport from an unidentified electron donor, possibly NAD(P)H or ferredoxin (Fd) to PQ. PQ reduction was monitored by chlorophyll fluorescence emitted from PSII. PQ reduction in the dark depends on NDH activity and can be monitored as a transient increase in chlorophyll fluorescence after AL illumination. PC, Plastocyanin; FNR, ferredoxin-NADP+ oxidoreductase. B, Analysis of the transient increase in chlorophyll fluorescence after turning off AL. The bottom curve indicates a typical trace of chlorophyll fluorescence in the wild type (WT). Leaves were exposed to AL (50 μmol photons m−2 s−1) for 5 min. AL was turned off and the subsequent change in chlorophyll fluorescence level was monitored. Insets are magnified traces from the boxed area. The fluorescence levels were normalized by Fm levels. ML, Measuring light; SP, saturating pulse of white light; crr6 + CRR6, crr6 complemented by introduction of the wild-type genomic CRR6.
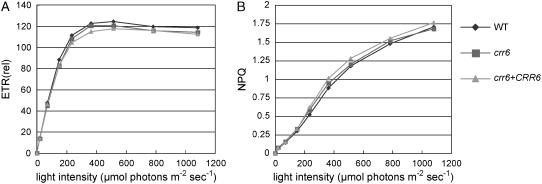
To characterize photosynthetic electron transport in crr6, the light intensity dependence of two chlorophyll fluorescence parameters was compared between the wild type and crr6 (Fig. 2). These parameters are influenced by even subtle defects in photosynthetic electron transport. The electron transport rate (ETR) reflects the relative rate of electron transport through PSII and is not affected in crr6 (Fig. 2A). Nonphotochemical quenching (NPQ) is mainly related to the size of energy dissipation as heat from PSII (thermal dissipation), which is a protective mechanism of PSII from oxidative damage. Thermal dissipation is triggered by acidification of the thylakoid lumen under excessive light conditions (Niyogi et al., 2005). Although PGR5-dependent PSI cyclic electron flow is essential for the induction of NPQ via ΔpH generation (Munekage et al., 2002), the NDH complex scarcely contributes to NPQ induction at least in the air (Shikanai et al., 1998; Munekage et al., 2004). In crr6, the light-intensity dependence of NPQ was not altered (Fig. 2B), unlike in other crr mutants (Hashimoto et al., 2003; Kotera et al., 2005; Munshi et al., 2005). We conclude that crr6 is specifically defective in the activity of the chloroplast NDH complex.
Figure 2.
In vivo analysis of electron transport activity. A, Light-intensity dependence of ETR. ETR was depicted relative to ΦPSII × light intensity (μmol photons m−2 s−1). sds are <10% of values (n = 5). B, Light-intensity dependence of NPQ of chlorophyll fluorescence. sds are <20% of values (n = 5). crr6 + CRR6, crr6 transformed with the genomic wild-type CRR6.
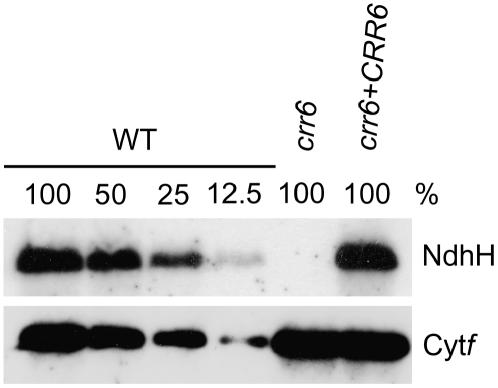
To assess the possibility that the level of the NDH complex is reduced in crr6, accumulation of NdhH, a subunit of the NDH complex, was analyzed in a protein blot (Fig. 3). The NdhH subunit is unstable without other Ndh subunits (Munekage et al., 2004; Munshi et al., 2005; Rumeau et al., 2005) and the protein blot can be used to monitor accumulation of the NDH complex. In crr6, the NdhH level was drastically reduced to an undetectable level (at least <12.5% of the wild type). In contrast, the level of Cyt f, a subunit of the Cyt b6f complex, was not affected in crr6. These results are consistent with those of chlorophyll fluorescence analyses, which indicated a specific loss of NDH activity in crr6 (Figs. 1 and 2). We conclude that the accumulation of the NDH complex is specifically impaired in crr6.
Figure 3.
Protein-blot analysis of the NDH complex. Immunodetection of an NDH subunit, NdhH, and a subunit of the Cyt b6f complex, Cyt f. Proteins were extracted from the thylakoid membrane fraction of the chloroplasts. Lanes were loaded with the protein samples corresponding to 0.2 μg chlorophyll for Cyt f and 5 μg chlorophyll for NdhH (100%) and the series of dilutions indicated. crr6 + CRR6, crr6 transformed by wild-type genomic CRR6.
CRR6 Encodes a Novel Protein Conserved in Phototrophs
The gene affected in crr6 was identified by map-based cloning. The crr6 mutant (Columbia gl1 background) was crossed to a polymorphic wild-type strain (Landsberg erecta), and the mutation was mapped to the bottom of chromosome 2. Fine mapping using 294 F2 plants identified a 124-kb region between the markers T30B22 and T9J23. The nucleotide sequences of candidate genes that encode the predicted plastid-targeting proteins were determined. Although TargetP (http://www.cbs.dtu.dk/services/TargetP) predicted that At2g47910 encodes a mitochondrial protein, this gene was included in the candidates because the homologs to At2g47910 are present in the cyanobacterial genomes. This is a criterion for candidates for novel subunits of the NDH complex because the chloroplast NDH complex is believed to have originated from the cyanobacterial complex. Finally, a sequence alteration was found in a single gene, At2g47910 in crr6.
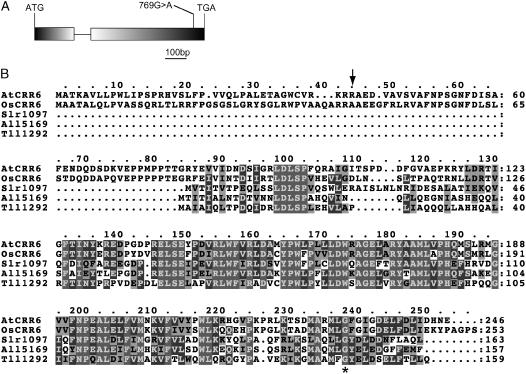
Direct sequencing of the reverse transcription-PCR product showed that At2g47910 consists of two exons and one intron (Fig. 4A). A single-nucleotide substitution in crr6 resulted in an amino acid alteration from well-conserved Gly to Arg in the C-terminal region of CRR6 (Fig. 4, A and B). To confirm that the crr6 phenotype is ascribed to the mutation in At2g47910, the wild-type genomic sequence of At2g47910 was introduced into crr6. This complementation fully restored the transient increase in chlorophyll fluorescence after turning off AL illumination (Fig. 1B). The NdhH level was also complemented by the transformation (Fig. 3). We conclude that the crr6 defect was due to the mutation in At2g47910 (CRR6).
Figure 4.
Positional cloning of crr6. A, Structure of CRR6. Exons (boxes) and an intron (horizontal thin line) were determined by direct sequencing of the reverse transcription-PCR products. The position of the crr6 mutation is indicated. B, Alignment of CRR6 homolog sequences. The predicted cleavage site of the target signal (TargetP) is indicated by a vertical arrow. AtCRR6, Arabidopsis; OsCRR6, rice; Slr1097, Synechocystis sp. PCC 6803; All5169, Anabaena sp. PCC 7120; Tll1292, Thermocynechococcus elongatus BP-1. The position of the crr6 mutation is indicated by an asterisk.
The CRR6 protein consists of 246 amino acids and does not contain any known motifs (Fig. 4B). In addition to higher plants, CRR6 is conserved in cyanobacteria, but not in nonphototrophs. In higher plants, CRR6 has a short N-terminal extension that is absent in the cyanobacterial version (Fig. 4B). Interestingly, CRR6 is not conserved in Chlamydomonas reinhardtii in which the chloroplast NDH complex is absent (http://genome.jgi-psf.org/chlre2/chlre2.home.html). This fact suggests that the function of CRR6 is specific to the NDH complex and includes the possibility that CRR6 is a novel subunit of the complex.
CRR6 Localizes to the Chloroplast
Although TargetP and Predotar (http://urgi.infobiogen.fr/predotar/predotar.html) predict mitochondrial localization of CRR6, the crr6 phenotype strongly suggests that CRR6 is a chloroplast protein (Figs. 1–3). Predotar does, in fact, predict chloroplast localization for rice (Oryza sativa) CRR6. To assess this possibility, we determined the subcellular localization of CRR6 using antibodies. We constructed a chimeric gene in which the C terminus of CRR6 was fused with the influenza hemaglutinin protein (HA) epitope (CRR6-HA) and transcribed it under the control of the CRR6 promoter. The construct was then introduced into crr6. The transformation complemented the activity of the NDH complex, which was monitored as a transient increase in chlorophyll fluorescence after turning off AL (data not shown). The level of NdhH was also restored to that in the wild type (Fig. 5A). This result indicates that the fusion protein was functional and localized to the proper position.
Figure 5.
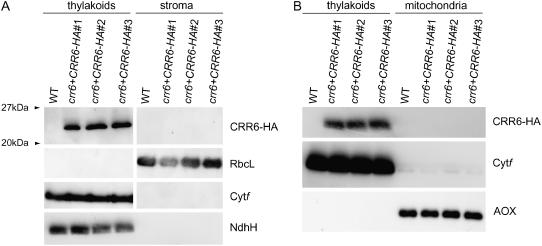
Protein-blot analysis of HA-tagged CRR6. A, Immunodetection of CRR6 protein using a monoclonal antibody against the HA tag. Chloroplast preparations were further fractionated to obtain a thylakoid membrane fraction and a stromal fraction. Large subunit of Rubisco (RbcL) and Cyt f were detected as the control for the fractionation. B, CRR6 protein was absent in the mitochondrial fraction. The lanes were loaded with protein samples corresponding to 5 μg (CRR6-HA), 5 μg (NdhH), 0.2 μg (Cyt f), and 0.01 μg (RbcL) chlorophyll. The crude mitochondrial protein was loaded so that it corresponded to 0.2 μg chlorophyll of the thylakoid fraction. Alternative oxidase was detected as a marker of the mitochondrial fraction. crr6 + CRR6-HA, crr6 transformed by genomic CRR6 fused to the HA epitope tag. Three independent lines (nos. 1–3) were analyzed.
From leaves of crr6 transformed with CRR6-HA, chloroplasts were isolated and further fractionated into the stromal fraction and the thylakoid membrane fraction that possibly also contains the chloroplast envelopes. The monoclonal antibody against the HA tag detected a protein in the thylakoid membrane fraction isolated from the transgenic lines. The size of the protein is approximately consistent with that of CRR6 without the putative transit peptide (23.5 kD) plus the linker and the HA tag (1.6 kD). However, the protein was absent in both fractions isolated from the wild type that do not carry the transgene. Although CRR6 is unlikely to have any transmembrane domains (http://sosui.proteome.bio.tuat.ac.jp/sosuimenu0.html), it localized to the thylakoid membrane fraction of the chloroplast. We speculate that CRR6 interacts with other membrane proteins.
Because TargetP predicted the mitochondrial localization of CRR6, we assessed the possibility of whether CRR6 colocalizes in both the chloroplast and the mitochondria. The crude mitochondrial fraction containing a marker protein, alternative oxidase, did not include CRR6-HA protein (Fig. 5B). We conclude that CRR6 exclusively localizes to the chloroplast.
The NDH Complex Is Not Essential for Stabilizing CRR6
CRR6 is essential for the accumulation of the NDH complex (Figs. 1 and 3) and localizes to the thylakoid membrane fraction (Fig. 5). CRR6 is conserved in phototrophs, but not in C. reinhardtii in which the chloroplast NDH complex is absent (Fig. 4). All the characters are identical to those of CRR7 (Munshi et al., 2005), NdhN, and NdhO (Rumeau et al., 2005), suggesting that CRR6 is a novel subunit of the NDH complex. To assess this possibility, we analyzed the stability of CRR6 under the mutant background lacking the NDH complex. If CRR6 were a subunit of the NDH complex, CRR6 would be unstable without the accumulation of the other subunits.
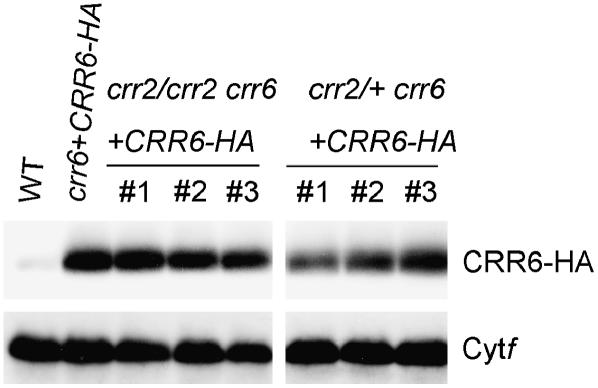
In the Arabidopsis mutant crr2-2, expression of the chloroplast ndhB gene encoding a subunit of the NDH complex is impaired due to lack of intergenic RNA cleavage between rps7 and ndhB (Hashimoto et al., 2003). NdhB is essential for stabilizing other subunits (Hashimoto et al., 2003; Munshi et al., 2005). To assess the stability of CRR6, the CRR6-HA construct was introduced into the double mutant crr2-2 crr6. In the double mutant, CRR6 function was complemented by the transgene (Fig. 5). Unexpectedly, the CRR6-HA protein was detected in the double-mutant lines (Fig. 6). To quantitatively assess the effect of the crr2 mutant background on CRR6 accumulation, the double mutants were crossed with the single mutant of crr6. In the resulting F1 plants, the crr6 locus is homozygous for the mutant allele, whereas the crr2 locus is heterozygous, indicating that ndhB expression had been restored. Seedlings carrying the transgene were used for chloroplast isolation and the CRR6 level was evaluated by protein-blot analysis (Fig. 6). The level of CRR6 was identical between seedlings carrying the homozygous crr2/crr2 and the heterozygous crr2/+. This result indicates that the NDH complex is not essential for stabilizing CRR6 and does not influence the level of CRR6.
Figure 6.
Immunodetection of CRR6-HA protein under the crr2-2 mutant background. The lanes were loaded with the thylakoid membrane protein samples corresponding to 0.2 μg chlorophyll for Cyt f and 5 μg chlorophyll for CRR6-HA. crr2-2 crr6 + CRR6-HA, crr2-2 crr6 transformed by the chimeric gene encoding CRR6-HA. Three independent lines (nos. 1–3) were analyzed and then used for crosses with crr6 to generate crr2-2/+ crr6 + CRR6-HA (heterozygous for crr2-2, homozygous for crr6, and containing the transgene).
DISCUSSION
In Synechocystis sp. PCC 6803, four distinct protein complexes contain ndh gene products (Zhang et al., 2004). The largest NDH-1L complex includes at least 15 subunits and is essential for heterotrophic growth (Ohkawa et al., 2000; Zhang et al., 2004). In addition to 11 subunits, NdhA to NdhK, which are encoded by the chloroplast genome in higher plants, four subunits, NdhL to NdhO, have recently been identified (Ogawa, 1992; Prommeenate et al., 2004; Battchikova et al., 2005). At least for NdhM, NdhN, and NdhO, the orthologs were discovered in the nuclear genome of higher plants (Rumeau et al., 2005). Although the NDH-1M complex lacks NdhD1 and NdhF1 subunits, it interacts with two versions of the NDH-1S complex and functions in CO2 uptake (Zhang et al., 2004, 2005). Whereas the NDH-1S1 complex consists of NdhD3, NdhF3, CupA, and Sll1735, forming the inducible high-affinity CO2 transporter, the NDH-1S2 complex consists of NdhD4, NdhF4, and CupB, forming the constitutive low-affinity CO2 transporter (Ohkawa et al., 2000; Shibata et al., 2001; Zhang et al., 2004). In higher plants, ndhD and ndhF are single-copy genes and homologous to ndhD1/D2 and ndhF1 of Synechocystis sp. PCC 6803, respectively. This fact suggests that the chloroplast NDH complex is similar to the cyanobacterial NDH-1L complex. This is consistent with the results indicating that the chloroplast NDH complex is involved in PSI cyclic electron flow in the light and also in chlororespiration in the dark (Burrows et al., 1998; Shikanai et al., 1998). Despite extensive biochemical studies, the entire subunit composition of the NDH complex has remained unclear in both the cyanobacteria and the chloroplast.
Our genetic approach aims to complement the biochemical approach in which the fragility of the NDH complex makes it difficult to isolate the entire complex. To select the candidate genes for the novel subunits of the NDH complex from the crr mutant pool, our criteria are as follows: (1) the protein is essential for stabilizing the NDH complex; (2) conversely, the NDH complex is required for stabilizing this protein; and (3) the gene is specifically conserved in phototrophs, including cyanobacteria, but not in C. reinhardtii. CRR7 fits all these criteria and is therefore a candidate for biochemical confirmation such as by use of an ultraviolet cross-linking technique. Although CRR6 was also applicable to criteria 1 and 3, it was stable under the crr2 mutant background. CRR6 is classified into a new group of CRR proteins.
The most straightforward interpretation of the results is that CRR6 is a nonsubunit factor involved in the accumulation of the NDH complex in the chloroplast. CRR6 may be a chaperone-like protein for assembly of the NDH complex. We cannot completely exclude the possibility that CRR6 is involved in chloroplast gene expression as CRR2 and CRR4, although the sequence information does not suggest this. In any case, characterization of the crr6 phenotype (Figs. 1–3) indicates that the function of CRR6 is specific to the NDH complex. A defect in the more general machinery involved in the protein complex assembly is likely to result in a more severe phenotype. An Arabidopsis CE26-48 mutant was isolated based on its defective NDH activity and was shown to have an amino acid alteration in a putative α-subunit of the Cpn60 chaperone in the chloroplast (At2g28000). In contrast to typical crr mutants, CE26-48 also showed the pale-green leaf phenotype and a drastic reduction in electron transport activity, even though gene function was partially impaired in the mutant (T. Shikanai, unpublished data). The knockout of this gene results in a defect in embryogenesis (Apuya et al., 2001).
Our results do not suggest that CRR6 is a subunit of the chloroplast NDH complex, which is similar to the cyanobacterial NDH-1L complex. No subunits have thus far been shown to be stable under the mutant background of other subunits. However, it is still possible that CRR6 is a component of another protein complex that interacts with the chloroplast NDH complex. The cyanobacterial NDH-1S complexes are stable even under the mutant background of M55, in which both NDH-1L and NDH-1M complexes are absent (Zhang et al., 2004). Although the NDH-1L complex does not interact with the NDH-1S complexes in cyanobacteria, and higher plants are unlikely to contain the NDH-1S-like complex, the chloroplast NDH complex may be associated with an unknown peripheral complex that is essential for stabilizing the chloroplast NDH complex. Although CRR6 does not have any transmembrane domains, it localizes to the thylakoid membrane fraction of the chloroplast (Fig. 5), suggesting that CRR6 anchors to a membrane protein. Our genetic approach has identified two new pieces in the puzzle of the NDH complex: CRR6 and CRR7. It is essential to fit them into place and continue the survey of the entire complex using them as tags.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seedlings were grown in soil under growth chamber conditions (50 μmol photons m−2 s−1) for 3 to 4 weeks. crr6 was mutagenized by ethyl methanesulfonate (Hashimoto et al., 2003).
Chlorophyll Fluorescence Analysis
Chlorophyll fluorescence was measured using a minipulse-amplitude modulation portable chlorophyll fluorometer (Walz). Minimal fluorescence at open PSII centers in the dark-adapted state (F0) was excited by a weak measuring light (650 nm) at a light intensity of 0.05 to 0.1 μmol photons m−2 s−1. A saturating pulse of white light (800 ms, 3,000 μmol photons m−2 s−1) was applied to determine the maximal fluorescence at closed PSII centers in the dark-adapted state (Fm) and during AL illumination (Fm′). The steady-state fluorescence level (Fs) was recorded during AL illumination (15–1,000 μmol photons m−2 s−1). NPQ was calculated as (Fm − Fm′)/Fm′. The quantum yield of PSII (ΦPSII) was calculated as (Fm′ − Fs)/Fm′ (Genty et al., 1989). ETR was calculated as ΦPSII × light intensity (μmol photons m−2 s−1). The transient increase in chlorophyll fluorescence after turning off AL was monitored as described previously (Shikanai et al., 1998).
Map-Based Cloning
The crr6 mutation was mapped with molecular markers based on a cleaved amplified polymorphic sequence (Konieczny and Ausubel, 1993). Primer sequences and the restriction enzymes are 5′-CATAGACTTGAAGAACCGTGC-3′ and 5′-TGGCTCACTCTGTTTCATCTG-3′, BamHI for T30B22 and 5′-CCTTTTTCTGCCACATTCCC-3′ and 5′-CATACTGAGCGGAATATTCTG-3′, RsaI for T9J23. Genomic DNA was isolated from F2 plants derived from a cross between crr6 and the wild type (Landsberg erecta). The genomic sequences of the candidate genes were amplified by PCR using ExTaq DNA polymerase (Takara). The resulting PCR products were directly sequenced using a dye terminator cycle sequencing kit and an ABI PRISM 3100 sequencer (Applied Biosystems).
For complementation of the crr6 mutation, the 2.1-kb wild-type genomic sequence surrounded by 5′-TCTAGAAATGTTGGAACTTC-3′ and 5′-TTTGGGTATTTGATACACAC-3′ was cloned in pBIN19 and introduced into crr6 via Agrobacterium tumefaciens MP90.
Isolation of Chloroplasts and Mitochondria
Leaves of 4- to 5-week-old plants were homogenized in a medium containing 330 mm sorbitol, 20 mm Tricine/NaOH, pH 7.6, 5 mm EGTA, 5 mm EDTA, 10 mm NaCO3, 0.1% (w/v) bovine serum albumin, and 1.87 mm ascorbate. After centrifugation for 5 min at 2,000g, the pellet was resuspended in 300 mm sorbitol, 20 mm HEPES/KOH, pH 7.6, 5 mm MgCl2, and 2.5 mm EDTA. Intact chloroplasts were purified by passing through 40% Percoll. The purified chloroplasts were suspended in 20 mm HEPES/KOH, pH 7.6, 5 mm MgCl2, and 2.5 mm EDTA. The insoluble fraction containing the thylakoids and the envelopes was separated from the stroma fraction by centrifugation for 5 min at 15,000g. The first low-speed centrifugation was followed by centrifugation for 10 min at 25,000g to pellet the crude mitochondrial fraction.
Protein Analysis
The genomic sequence used for the complementation was modified to carry the sequence encoding the HA tag (YPYDVPDYAG) so that the C terminus of CRR6 fused with the HA tag via a linker (PRGG). The fusion gene was cloned between the CRR6 promoter and the Nos terminator in pBIN19 and introduced into crr6 via A. tumefaciens MP90. The CRR6-HA protein was detected using a monoclonal antibody against the HA tag (Santa Cruz Biotechnology). Proteins were separated by 12.5% SDS-PAGE and the protein blot was analyzed as previously described (Munekage et al., 2002).
Acknowledgments
We thank Momoko Miyata and Etsuko Habe for their excellent technical assistance. We are grateful to Gilles Peltier, Tsuyoshi Endo, Amane Makino, Akiho Yokota, and Ko Noguchi for their gifts of antibodies.
This work was supported by a grant-in-aid for Scientific Research on Priority Areas (grant no. 16085206) and for Creative Scientific Research (grant no. 17GS0316) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Toshiharu Shikanai (shikanai@agr.kyushu-u.ac.jp).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.080267.
References
- Amann K, Lezhneva L, Wanner G, Herrmann RG, Meurer J (2004) ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe-4S] cluster-containing chloroplast complexes and antenna proteins. Plant Cell 16: 3084–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apuya NR, Yadegari R, Fischer RL, Harada JJ, Zimmerman JL, Goldberg RB (2001) The Arabidopsis embryo mutant schlepperless has a defect in the chaperonin-60α gene. Plant Physiol 126: 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Goldschmidt-Clermont M (2000) Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559–572 [DOI] [PubMed] [Google Scholar]
- Battchikova N, Zhang P, Rudd S, Ogawa T, Aro E-M (2005) Identification of NdhL and Ssl1690 (NdhO) in NDH-1L and NDH-1M complexes of Synechocystis sp. PCC 6803. J Biol Chem 280: 2587–2595 [DOI] [PubMed] [Google Scholar]
- Bauer J, Hiltbrunner A, Kessler F (2001) Molecular biology of chloroplast biogenesis: gene expression, protein import and intraorganellar sorting. Cell Mol Life Sci 58: 420–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau E, Takahashi Y, Lemieux C, Turmel M, Rochaix J-D (1997) The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J 16: 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows PA, Sazanov LA, Svab Z, Maliga P, Nixon PJ (1998) Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J 17: 868–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo T, Shikanai T, Takabayashi A, Asada K, Sato F (1999) The role of chloroplastic NAD(P)H dehydrogenase in photoprotection. FEBS Lett 457: 5–8 [DOI] [PubMed] [Google Scholar]
- Genty B, Briantais J-M, Baker NR (1989) The relationship between quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta 990: 87–92 [Google Scholar]
- Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T (2003) A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. Plant J 36: 541–549 [DOI] [PubMed] [Google Scholar]
- Horváth EM, Peter SO, Joët T, Rumeau D, Cournac L, Horváth GV, Kavanagh TA, Schäfer C, Peltier G, Medgyesy P (2000) Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol 123: 1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofer W, Koop H-U, Wanner G, Steinmüller K (1998) Mutagenesis of the genes encoding subunits A, C, H, I, J and K of the plastid NAD(P)H-plastoquinone-oxidoreductase in tobacco by polyethylene glycol-mediated plastome transformation. Mol Gen Genet 258: 166–173 [DOI] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433: 326–330 [DOI] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi T, Wakasugi T, Shinozaki K, Yamaguchi-Shinozaki K, Zaita N, Hidaka T, Meng BY, Ohto C, Tanaka M, Kato A, et al(1987) Six chloroplast genes (ndhA-F) homologous to human mitochondrial genes encoding components of the respiratory chain NADH dehydrogenase are actively expressed: determination of the splice sites in ndhA and ndhB pre-mRNAs. Mol Gen Genet 210: 385–393 [DOI] [PubMed] [Google Scholar]
- Meurer J, Meierhoff K, Westhoff P (1996) Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterisation by spectroscopy, immunoblotting and northern hybridisation. Planta 198: 385–396 [DOI] [PubMed] [Google Scholar]
- Miles D (1980) Mutants of higher plants: maize. Methods Enzymol 69: 3–23 [Google Scholar]
- Munekage Y, Hashimoto M, Miyake C, Tomizawa K, Endo T, Tasaka M, Shikanai T (2004) Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429: 579–582 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T (2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110: 361–371 [DOI] [PubMed] [Google Scholar]
- Munekage Y, Shikanai T (2005) Cyclic electron transport through photosystem I. Plant Biotech 22: 361–369 [Google Scholar]
- Munné-Bosch S, Shikanai T, Asada K (2005) Enhanced ferredoxin-dependent cyclic electron flow around photosystem I and α-tocopherol quinone accumulation in water-stressed ndhB-inactivated tobacco mutants. Planta 222: 502–511 [DOI] [PubMed] [Google Scholar]
- Munshi MK, Kobayashi H, Shikanai T (2005) Identification of a novel protein, CRR7, required for the stabilization of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant J 44: 1036–1044 [DOI] [PubMed] [Google Scholar]
- Niyogi KK, Li X-P, Rosenberg V, Jung H-S (2005) Is PsbS the site of non-photochemical quenching in photosynthesis? J Exp Bot 56: 375–382 [DOI] [PubMed] [Google Scholar]
- Ogawa T (1992) Identification and characterization of the ictA/ndhL gene product essential to inorganic carbon transport of Synechocystis PCC6803. Plant Physiol 99: 1604–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa H, Pakrasi HB, Ogawa T (2000) Two types of functionally distinct NAD(P)H dehydrogenases in Synechocystis sp. strain PCC6803. J Biol Chem 275: 31630–31634 [DOI] [PubMed] [Google Scholar]
- Pilon M, Abdel-Ghany SE, Van Hoewyk D, Ye H, Pilon-Smits EA (2006) Biogenesis of iron-sulfur cluster proteins in plastids. Genet Eng 27: 101–117 [DOI] [PubMed] [Google Scholar]
- Prommeenate P, Lennon AM, Markert C, Hippler M, Nixon PJ (2004) Subunit composition of NDH-1 complexes of Synechocystis sp. PCC 6803: identification of two new ndh gene products with nuclear-encoded homologues in the chloroplast Ndh complex. J Biol Chem 279: 28165–28173 [DOI] [PubMed] [Google Scholar]
- Rumeau D, Bécuwe-Linka N, Beyly A, Louwagie M, Garin J, Peltier G (2005) New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell 7: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazanov LA, Burrows PA, Nixon PJ (1998) The chloroplast Ndh complex mediates the dark reduction of the plastoquinone pool in response to heat stress in tobacco leaves. FEBS Lett 429: 115–118 [DOI] [PubMed] [Google Scholar]
- Shibata M, Ohkawa H, Kaneko T, Fukuzawa H, Tabata S, Kaplan A, Ogawa T (2001) Distinct constitutive and low-CO2-induced CO2 uptake systems in cyanobacteria: genes involved and their phylogenetic relationship with homologous genes in other organisms. Proc Natl Acad Sci USA 98: 11789–11794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, Endo T (2000) Physiological function of a respiratory complex, NAD(P)H dehydrogenase in chloroplasts: dissection by chloroplast reverse genetics. Plant Biotechnol 17: 79–86 [Google Scholar]
- Shikanai T, Endo T, Hashimoto T, Yamada Y, Asada K, Yokota A (1998) Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc Natl Acad Sci USA 95: 9705–9709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, Munekage Y, Shimizu K, Endo T, Hashimoto T (1999) Identification and characterization of Arabidopsis mutants with reduced quenching of chlorophyll fluorescence. Plant Cell Physiol 40: 1134–1142 [DOI] [PubMed] [Google Scholar]
- Sugiura M, Hirose T, Sugita M (1998) Evolution and mechanism of translation in chloroplasts. Annu Rev Genet 32: 437–459 [DOI] [PubMed] [Google Scholar]
- Wang P, Duan W, Takabayashi A, Endo T, Shikanai T, Ye J-Y, Mi H (2006) Chloroplastic NAD(P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol 141: 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Battchikova N, Jansen T, Appel J, Ogawa T, Aro E-M (2004) Expression and functional roles of the two distinct NDH-1 complexes and the carbon acquisition complex NdhD3/NdhF3/CupA/Sll1735 in Synechocystis sp. PCC 6803. Plant Cell 16: 3326–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Battchikova N, Paakkarinen V, Katoh H, Iwai M, Ikeuchi M, Pakrasi HB, Ogawa T, Aro E-M (2005) Isolation, subunit composition and interaction of the NDH-1 complexes from Thermosynechococcus elongatus BP-1. Biochem J 390: 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]