Abstract
Reactive oxygen species (ROS) are key players in the regulation of plant development, stress responses, and programmed cell death. Previous studies indicated that depending on the type of ROS (hydrogen peroxide, superoxide, or singlet oxygen) or its subcellular production site (plastidic, cytosolic, peroxisomal, or apoplastic), a different physiological, biochemical, and molecular response is provoked. We used transcriptome data generated from ROS-related microarray experiments to assess the specificity of ROS-driven transcript expression. Data sets obtained by exogenous application of oxidative stress-causing agents (methyl viologen, Alternaria alternata toxin, 3-aminotriazole, and ozone) and from a mutant (fluorescent) and transgenic plants, in which the activity of an individual antioxidant enzyme was perturbed (catalase, cytosolic ascorbate peroxidase, and copper/zinc superoxide dismutase), were compared. In total, the abundance of nearly 26,000 transcripts of Arabidopsis (Arabidopsis thaliana) was monitored in response to different ROS. Overall, 8,056, 5,312, and 3,925 transcripts showed at least a 3-, 4-, or 5-fold change in expression, respectively. In addition to marker transcripts that were specifically regulated by hydrogen peroxide, superoxide, or singlet oxygen, several transcripts were identified as general oxidative stress response markers because their steady-state levels were at least 5-fold elevated in most experiments. We also assessed the expression characteristics of all annotated transcription factors and inferred new candidate regulatory transcripts that could be responsible for orchestrating the specific transcriptomic signatures triggered by different ROS. Our analysis provides a framework that will assist future efforts to address the impact of ROS signals within environmental stress conditions and elucidate the molecular mechanisms of the oxidative stress response in plants.
Since the early identification of hydrogen peroxide (H2O2) as a key molecule orchestrating the hypersensitive response and inducing the expression of glutathione-S-transferase and glutathione peroxidase genes (Levine et al., 1994), numerous studies have supported the signaling role of reactive oxygen species (ROS) during different environmental responses and developmental processes, including biotic and abiotic stress responses, allelopathic plant-plant interactions, cell division and elongation, and programmed cell death (Pei et al., 2000; Bethke and Jones, 2001; Bais et al., 2003; Foreman et al., 2003; Apel and Hirt, 2004; Foyer and Noctor, 2005). Because ROS are constantly produced during normal cell metabolism, it is important that their basal levels are tightly controlled. This control is provided by a complex gene network that acts in all subcellular compartments and through elaborate feed-forward/feed-back loops between oxidants and antioxidants. In Arabidopsis (Arabidopsis thaliana), this ROS gene network comprises at least 152 genes (Mittler et al., 2004). While the importance of ROS for cellular signaling has been established, relatively little is known about how ROS stimuli are perceived, transduced, and finally result in specific cellular responses. It is necessary that ROS signals possess a certain degree of specificity and selectivity allowing them to act efficiently in a variety of developmental processes and environmental responses. The chemical nature of ROS and/or their subcellular site of production could be critical for the specificity and selectivity of ROS signals.
ROS with documented signaling functions include H2O2, singlet oxygen (1O2), hydroxyl radical (OH·), and superoxide anion radical (O2·−; Laloi et al., 2004). Chloroplasts, mitochondria, and peroxisomes are organelles with highly oxidizing metabolic activities or with intense rate of electron flow and are, hence, major sources of ROS production in plant cells. Therefore, it is not surprising that many of the ROS-scavenging enzymes, including superoxide dismutases (SODs), ascorbate peroxidases (APXs), catalases (CATs), glutathione peroxidases, and peroxiredoxins are localized within these subcellular compartments (Mittler et al., 2004). The main reasons why the proposed selective signaling by specific ROS remained difficult to tackle experimentally were because until recently, no accurate and quantifiable detection methods for different ROS were available, and there were no suitable experimental systems to modulate in planta the levels of a specific ROS at a given time. In recent years, the production of various transgenic Arabidopsis plants with compromised levels of specific antioxidant enzymes and the identification of the conditional fluorescent (flu) mutant provided the necessary tools to assess the specific effects of O2·−, H2O2, and 1O2 within a particular subcellular compartment (Meskauskiene et al., 2001; Pnueli et al., 2003; Rizhsky et al., 2003; Vandenabeele et al., 2004). Genome-wide microarrays provided the means to assess the specificity of ROS signaling by analyzing expression profiles that are influenced by increased ROS levels (op den Camp et al., 2003; Gechev et al., 2004, 2005; Vanderauwera et al., 2005). Transcriptomic footprints were produced for plants with compromised levels of individual ROS-scavenging enzymes, such as chloroplastic Cu/ZnSOD, cytosolic APX, peroxisomal CAT, and mitochondrial alternative oxidase (AOX; Rizhsky et al., 2003; Davletova et al., 2005; Umbach et al., 2005; Vanderauwera et al., 2005), as well as of plants treated with ROS-generating agents, such as methyl viologen (MV), 3-aminotriazole (AT), and the fungal Alternaria alternata f. sp. lycopersici (AAL) toxin (Gechev et al., 2004, 2005; http://www.uni-frankfurt.de/fb15/botanik/mcb/AFGN/AFGNHome.html). These genome-wide expression inventories have shed light on early response and downstream transcripts specifically altered in their expression by a particular type of ROS and hinted at transcripts or pathways that serve as integrative points of ROS-mediated plant responses. For example, cytosolic H2O2 was shown to control the expression of heat shock proteins during light stress (Pnueli et al., 2003), peroxisomal photorespiration-dependent H2O2 was shown to negatively impinge on the high light (HL) stress induction of transcripts within the anthocyanin biosynthesis pathway (Vanderauwera et al., 2005), and the production of 1O2 within plastids was shown to induce at least 70 nuclear transcripts that could not be activated by the exogenous application of the superoxide-generating MV (op den Camp et al., 2003). Collectively, these reports not only demonstrated that increased ROS levels lead to a clear reorientation of the plant's transcriptome, but also provided experimental evidence for the specific signaling capacities of different ROS and for the importance of the subcellular localization of the different antioxidant systems in the plant cell. Here, we present an integrative comparison of these and other oxidative stress-related genome-wide expression studies. Our analysis provides a general survey of oxidative stress-induced transcripts in Arabidopsis and defines common and specific responses toward different types of ROS signals.
RESULTS AND DISCUSSION
ROS-Induced Transcript Expression in Diverse Genome-Wide Transcriptome Data Sets
We performed a comparative analysis on various ROS-related data sets to obtain a global insight into the complexity of the transcriptomic response of plants to oxidative stress. To this end, we compiled the data from all relevant publicly available or in-house stored ROS-dependent microarray experiments. To allow a comprehensive comparison, only genome-wide profiles generated with Affymetrix ATH1 or Agilent Arabidopsis2 oligonucleotide arrays of high technical quality, performed on leaf material of at least 2-week-old Arabidopsis plants, were retained. Table I summarizes the nine selected experiments. Four data sets were obtained by exogenous application of oxidative stress-causing agents. Ozone (O3) is an atmospheric pollutant that breaks down in the apoplast, forming mainly O2·− and H2O2. O3 exposure triggers a hypersensitive response-like oxidative burst leading to the development of cell death lesions (Sandermann, 2004). A data set of O3-induced transcriptional changes generated by the group of A. Shirras (http://affymetrix.arabidopsis.info/narrays/experimentpage.pl?experimentid=26) was used. The herbicide AT is a potent inhibitor of CAT (Gechev et al., 2002). Application of AT to Arabidopsis plants dramatically reduces total CAT activity, increases peroxisomal H2O2 concentrations, and leads subsequently to cell death. Treatment of the toxin-sensitive Arabidopsis LAG one homolog 2 (LOH2) mutant with nanomolar quantities of the fungal AAL toxin perturbs the sphingolipid metabolism, followed by a drastic increase in H2O2 levels and subsequent cell death (Gechev et al., 2004). MV is a redox-cycling herbicide that generates O2·− in the chloroplasts and mitochondria. Most of this O2·− subsequently dismutates to H2O2, either spontaneously or enzymatically via SODs (Bowler et al., 1992). A very detailed and robust expression data set of Arabidopsis seedlings exposed to 10 μmol MV was generated by the group of D. Bartels (http://www.uni-frankfurt.de/fb15/botanik/mcb/AFGN/AFGNHome). One 1O2-related data set was obtained from the conditional flu mutant shifted from darkness to light. flu plants grown from seed under continuous light overaccumulate the potent photosensitizer protochlorophyllide when kept in the dark for 8 h. A dark-to-light switch leads to rapid production of 1O2 within the plastid compartment (op den Camp et al., 2003). The four remaining data sets describe transcriptomic changes in different mutants or transgenic plants in which the activity of a particular antioxidant enzyme was reduced or completely abolished, perturbing the ability to detoxify ROS at a specific subcellular location. A T-DNA knockout (KO) mutation of the key cytosolic H2O2-scavenging enzyme APX1 results in the accumulation of H2O2 in the cytosol, protein oxidation, and the collapse of the chloroplastic antioxidant system (Davletova et al., 2005). Arabidopsis plants with a T-DNA insertion in the promoter of the thylakoid-bound Cu/ZnSOD that partially reduces the levels of this enzyme had low chlorophyll levels and photosynthesis rate, and delayed flowering (Rizhsky et al., 2003). Peroxisomal CATs are key components of the ROS-scavenging network that function as a sink for photorespiratory H2O2 (Dat et al., 2003). The CAT2-deficient plants CAT2HP1 accumulate H2O2 and subsequently develop cell death within 8 h under conditions that promote photorespiration (Vanderauwera et al., 2005). The mitochondrial AOX diverts electrons from the ubiquinone pool to oxygen, thus reducing the risk of ROS formation. The basal oxidative state of the leaf tissue of an Arabidopsis AOX1a-AS line was not affected under nonlimiting growth conditions, but the silencing of this gene impacted the chloroplasts and several carbon metabolism pathways (Umbach et al., 2005).
Table I.
Overview of the nine ROS-generating systems included in the comprehensive analysis
For each experimental system, the identities of the major generated ROS and the intracellular sites of production are indicated. Sample and treatment descriptions together with other relevant experimental parameters are listed. Expression analysis of all experiments was performed using the Affymetrix ATH1 (ATH1) or Agilent Arabidopsis2 (Arab2) arrays. CAT2HP1, CAT-deficient plants; WT, wild type; KD, knockdown; AS, antisense; Chl, chloroplasts; Mit, mitochondria; Pl, plastids; Cyt, cytosol; Per, peroxisomes; Apo, apoplast. 1, op den Camp et al. (2003); 2, Rizhsky et al. (2003); 3, Umbach et al. (2005); 4, Davletova et al. (2003); 5, Vanderauwera et al. (2005); 6, Gechev et al. (2004); 7, Gechev et al. (2005); 8, D. Bartels (Bonn), AtGenExpress repository; 9, A. Shirras (Lancaster, UK), NASCArray repository.
| Experiment | Major ROS | Localization | Plant Age | Sample Description | Treatment/Growth Conditions | Time Points | Replicates | Microarray Platform | Source |
|---|---|---|---|---|---|---|---|---|---|
| weeks | h after Treatment | ||||||||
| flu mutant | 1O2 | Pl | 3 | flu mutant | Dark-to-light shift (80–100 μmol m−2 s−1) | 0.5, 1, 2 | 1 | ATH1 | 1 |
| KD-SOD | O2·− | Chl | 3 | Cu/Zn-SOD-KD plants | Standard | – | 3 | ATH1 | 2 |
| AOX1a-AS | O2·−, H2O2 | Mit | 3 | AOX1a-AS plants | Standard | – | 2 | ATH1 | 3 |
| KO-Apx1 + HL | H2O2 | Cyt | 3 | KO-Apx1 plants | HL shift: 25 to 250 μmol m−2 s−1 | 0, 0.25, 0.5, 1.5, 3, 6, 24 | 2 | ATH1 | 4 |
| CAT2HP1 + HL | H2O2 | Per | 6 | CAT2HP1 plants | HL shift: 140 to 1,800 μmol m−2 s−1 | 0, 3, 8 | 2 | ATH1 | 5 |
| LOH2 + AAL toxin | H2O2 | – | 4 | LOH2 plants | Water/AAL toxin injection, 200 nm | 7, 24, 48, 72 | 1 | Arab2 | 6 |
| AT spray | H2O2 | Per | 4 | WT plants | AT spray, 20 mm | 7 | 1 | Arab2 | 7 |
| MV | O2·− | Mit/Chl | 2.5 | WT plants | MV, 10 μmol | 0.5, 1, 3, 6, 12, 24 | 2 | ATH1 | 8 |
| O3 fumigation | O3, O2·−, H2O2 | Apo | 2 | WT plants | O3, 500 μg L−1 | 6 | 3 | ATH1 | 9 |
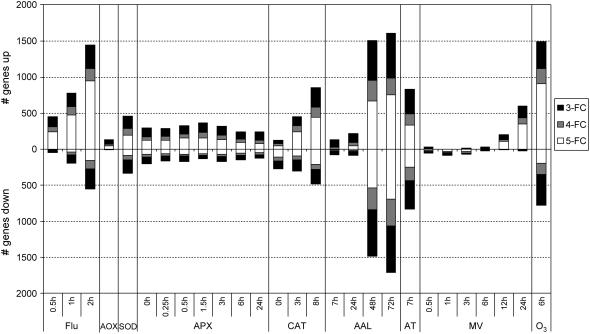
The collection of data sets and the preprocessing and processing of data are described in “Materials and Methods.” Five of the nine experiments were time series. Therefore, we assessed the dynamics of the transcriptional response in these five independent experiments by a hierarchical average linkage clustering analysis of their individual transcriptional profile. Supplemental Figure 1 presents the major trends in the transcriptomic behavior for these time course experiments. Whereas in some of these experiments, such as flu, a rapid response is provoked, in others, such as MV, the response is clearly delayed. These differences can be attributed to the intrinsic properties of the various experimental systems. 1O2 is generated almost immediately within chloroplasts of the flu mutant, while the exogenously applied MV requires more time to penetrate the leaves and perform its action. These differences in the kinetics have been taken into account in the comparative analysis described below. Following data processing, fold-change values were used as a selection criterion for differential transcript levels. Due to the heterogeneity of the experimental setups of the different experiments, no robust statistical analysis could be performed. Therefore, the use of relative expression data seemed opportune, although one should be aware of possible false positives/negatives associated with this approach. Fold-changes were calculated for all time points of all nine individual experiments, using the expression values of the mutant or treated samples relative to the wild-type or control (mock-treated) samples. Thresholds for positive response were set at 3-, 4-, and 5-fold change in expression. Figure 1 graphically presents the intensity of the transcriptomic response within the individual experiments. The strongest response was observed in the flu, AAL toxin, and O3 experiments. Whereas most treatments provoke a predominant inductive response, both AAL toxin and AT treatments have an equivalent number of repressed and induced transcripts. As noticed before (Supplemental Fig. 1), differentially expressed transcripts in KO-Apx1 plants were not further influenced by the exposure to a 10-fold higher light intensity.
Figure 1.
Transcript level changes within the individual ROS experiments. The number of transcripts with a 3-, 4-, or 5-fold increase or decrease in expression in transgenic or treated plants compared to wild-type or control plants at different time points are represented by black, gray, and white bars, respectively. Data were processed as described in “Materials and Methods.” FC, Fold change.
ROS-Related Transcriptomic Signatures
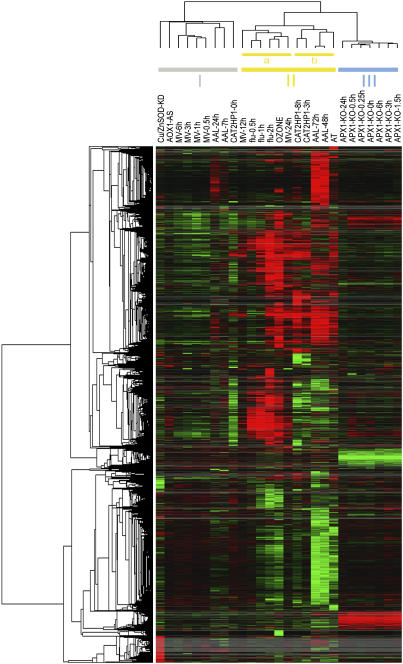
Overall, 8,056, 5,312, and 3,925 different transcripts responded with at least a 3-, 4-, or 5-fold difference in expression, respectively, in at least one time point across all integrated experiments. To assess the most prominent differences in transcript levels between the selection of ROS-dependent expression profiles, we focused on the transcripts that matched a stringent selection criterion of a 5-fold change within at least one time point across all experiments. A total of 3,925 probe sets matched this criterion, and their expression values in all selected experiments were hierarchically clustered in two dimensions: transcripts and experiments (Eisen et al., 1998). Figure 2 shows that the experiments clustered into three main branches. Cluster I comprises the earliest time points of the MV and AAL toxin treated plants, the unchallenged CAT2HP1, AOX1a-AS, and KD-SOD transgenics. In this cluster, transcriptional profiles are not highly dynamic or differentiating, except for a group of transcripts that is exclusively up-regulated in the KD-SOD plants. Cluster II encompasses the O3 and AT experiments, the later time points of the CAT2HP1 plants, MV, and AAL toxin treated plants together with the complete set of time points of the flu experiment. In this cluster, two subclusters can be distinguished in which the flu, O3, and MV experiments (IIa) separated from the AAL toxin, AT, and the CAT2HP1 time points (IIb). Although there seem to be some similarities between the 1O2- and O2·−-dependent experiments, it is not surprising that the transcriptomic footprints of the AT, and AAL toxin-treated and CAT2HP1 plants, cluster together. All three experimental systems are known to provoke an increase in peroxisomal H2O2. A molecular link between sphingolipid (AAL toxin) and H2O2 signaling during programmed cell death has recently been supported by the isolation of mutants that are more tolerant to both ROS and fungal toxins (Danon et al., 2005; Gechev and Hille, 2005; T. Gechev, M.A. Ferwerda, L. Bernier, and J. Hille, unpublished data). A third cluster (III) groups exclusively the complete HL-stress response of the KO-Apx1 time course. Two distinct groups of transcripts were recognized in this cluster: transcripts with enhanced and suppressed expression in the KO-Apx1 plants. The levels of these transcripts are not significantly responsive to the HL exposure.
Figure 2.
Hierarchical clustering of the 3,925 transcripts across the different ROS-generating conditions. Transcripts with at least a 5-fold difference in expression within one time point across all experiments were subjected to hierarchical average linkage clustering. Each horizontal line displays the expression data for one gene. Red and green indicate up- and down-regulation in transgenic or treated plants compared to wild-type or untreated plants, respectively. Intensity of the colors is proportional to the absolute value of the fold difference. Gray corresponds to missing expression values for transcripts that are not represented on either the Affymetrix ATH1 or Agilent Arabidopsis2 microarrays. The horizontal dendrogram (top) indicates the relationship among the experiments across all transcripts included in the cluster analysis. Three main clusters (I, II, and III) are indicated.
Besides the visualization of these general ROS-related transcriptomic footprints, we also identified transcripts that are differentially expressed after the generation of a particular ROS signal. To facilitate this effort, we selected a representative individual time point within each time course experiment, based on both the expression dynamics of each time course experiment (Supplemental Fig. 1) and the abundance of transcripts induced at a specific time point (Fig. 1). As a result, data points for 8-h HL stress of CAT2HP1 plants, 2-h light exposure of the flu mutant, 1.5-h HL stress of the KO-Apx1 plants, 48 h of AAL toxin treatment, and 24 h of MV treatment were retained together with the single time points of the KD-SOD, AT, and O3 experiments. The AOX1a-AS experiment was omitted from further analysis because of the minor variability in transcript levels observed in these transgenic plants. Starting from this smaller data set, a total of 3,293 transcripts, with at least a 5-fold difference in at least one experiment, was analyzed. To verify the validity of this selection, we hierarchically clustered the expression values in two dimensions: vertical and horizontal, encompassing the transcripts and the experiments, respectively. As in the complete data set (Fig. 2), three main horizontal clusters stood out: cluster I with the MV, flu, and O3 experiments; subcluster II containing the KD-SOD and KO-Apx1 transgenics; and subcluster III comprising the AAL toxin, AT, and CAT2HP1 experiments (Supplemental Fig. 2).
Marker Transcripts for ROS-Induced Gene Expression
Five transcripts were up-regulated more than 5-fold in at least seven of the eight experiments (Supplemental Table IA). These transcripts could be considered as hallmarks for the general oxidative stress response, regardless of the type of ROS or its subcellular production site. It is very likely that these transcripts are responding to a signal that is provoked by oxidative cellular damage. One of these transcripts, encoding an 89-amino acid defensin-like protein (At2g43510), was at least 5-fold induced in all analyzed experiments. Defensins form a diverse group of evolutionarily conserved Cys-rich peptides involved in pathogen defense and reproduction (Silverstein et al., 2005). Ten of the 317 defensin-like genes in the Arabidopsis genome were at least 5-fold induced within our composite data set. Increased ROS levels and namely H2O2 have been shown to increase the expression of defensins in plants (Hu et al., 2003). One transcript, encoding a protein of unknown function (At2g21640), was at least 5-fold elevated in all experiments, except in the KO-Apx1 plants, while three transcripts were induced in all experiments, except in the KD-SOD plants. The latter represent two proteins of unknown function (At1g19020 and At1g05340) and a Toll-Interleukin-1(TIR)-class disease resistance protein (At1g57630). Whereas the expression of the two transcripts with unknown function was far below the imposed threshold of 5-fold difference, the TIR-class disease resistance protein transcript showed a 4.6-fold increase in expression in the KD-SOD plants (Supplemental Table IA). TIR proteins are key players in pathogen recognition and defense response, both closely linked to oxidative stress and ROS signaling (Ausubel, 2005). When both KD-SOD and KO-Apx1 data were omitted from the comparison, 27 transcripts had at least a 5-fold increase in expression in all remaining experiments: MV, O3, flu, AAL, AT, and CAT2HP1 (Supplemental Table IB). In addition to several transcripts that encoded proteins of unknown function, two WRKY transcription factors (AtWRKY6 and AtWRKY75), a bHLH transcription factor, four glutathione-S-transferases, three glycosyl transferases, two Cyt P450, and various oxidoreductases could be recognized. One of these oxidoreductases, din11 (At3g49620), was induced 44-fold in the flu mutant, 65-fold in AAL toxin and AT experiments, and more than 100-fold in the CAT2HP1, MV, and O3 experiments (Supplemental Table IB). Transcript levels of din11 rapidly increased also during dark-induced leaf senescence (Fujiki et al., 2001). Glutathione-S-transferases and some of the other transcripts have already been reported to be responsive to H2O2 (Kovtun et al., 2000; Gechev et al., 2005), but we show that their induction is common for diverse oxidative stress-promoting conditions. Six transcripts were up-regulated at least 5-fold, specifically in the photorespiratory H2O2-related experiments CAT2HP1, AAL toxin, and AT (Supplemental Table IC). They encode an L-Asp oxidase family protein (At5g14760), xyloglucan endotransglycosylase 4 (XTR4, At1g32170), a Pro-rich extensin family protein (EXTENSIN3, At1g21310), a 2-oxoglutarate-dependant dioxygenase (At3g13610), and two unknown proteins (At3g26440 and At3g10320). The oxoglutarate-dependent oxidoreductase has previously been implicated in H2O2-induced cell death in Arabidopsis, and KO plants are impaired in AT-induced cell death (Gechev et al., 2005). A group of 66 transcripts is common between and exclusive for the flu, MV, and O3 experiments (Supplemental Table ID). This large number of transcripts could be explained by the presence of cell death, leading to secondary effects that could affect gene expression.
Besides the mutually expressed transcripts, a vast majority of transcripts were up- or down-regulated in only one experiment. These transcripts can be considered as hallmarks for a specific oxidative stress characterized by the chemical identity of the produced ROS and/or the subcellular site of its production. Supplemental Tables II, III, and IV list the transcripts that are specifically responsive to 1O2 (flu experiment), O2·− (KD-SOD and MV experiments), and H2O2 (KO-Apx1, CAT2HP1, and the overlapping genes between CAT2HP1, AAL toxin, and AT experiments), respectively.
A group of 146 transcripts was specifically induced in the unchallenged KD-SOD plants (Fig. 2; Supplemental Table III). Within this group, plastome transcripts encoding the major constituents of the photosynthetic apparatus are overrepresented. Of the 77 probes for chloroplast-encoded genes present on the ATH1 Affymetrix Genome array, 61 were represented in this cluster. Induction of chloroplastic transcripts has already been reported (Rizhsky et al., 2003), but here we show that decreasing the Cu/ZnSOD activity at the thylakoid membranes specifically leads to the up-regulation of chloroplastic transcripts. This observation suggests a highly specific O2·− sensor mechanism in the thylakoids. Even though photosynthesis is also negatively affected in most of the other experiments, they fail to influence the expression of these photosynthesis-related transcripts. Also a clear antagonistic expression pattern could be observed. Anthocyanin biosynthesis transcripts were up-regulated in the KD-SOD plants, while their induction during HL exposure was compromised in CAT2HP1 plants. The differential accumulation of anthocyanins in these transgenic lines has already been described previously (Rizhsky et al., 2003; Vanderauwera et al., 2005). From our analysis, we infer further consolidation that transcripts involved in the production of anthocyanins are transcriptionally coregulated in response to O2·− and that H2O2 negatively impinges on the expression of these transcripts. The inverse correlation between the effect of O2·− and H2O2 on anthocyanin accumulation clearly demonstrates specific signaling capacities of particular ROS. In the KO-Apx1 plants, 74 and 50 transcripts were exclusively induced and suppressed, respectively (Supplemental Table IV). Remarkably, their expression is hardly influenced by the shift from low light to HL. Within these 74 up-regulated transcripts, 10 transcripts encode disease resistance proteins. In addition, three transcripts encoding a bHLH and a MADS-box transcription factor and ICE1, which is known to regulate the transcription of CBF genes during cold, were present (Chinnusamy et al., 2003). Taken together, this result and the observation of Pnueli et al. (2003) demonstrate that cytosolic H2O2 could play an important role in signaling during abiotic stress and acclimation to low or high temperatures, as well as in response to pathogen infection.
The largest group of transcripts specifically regulated in only one experiment is found in the flu mutant: 289 and 29 transcripts were up- or down-regulated, respectively, at least 5-fold specifically after the release of 1O2, and were not affected by stress conditions generating O2·− or H2O2 (Supplemental Table II). Three of the 20 highest specifically induced transcripts in the flu mutant are ethylene-responsive element-binding proteins that are indicative of ethylene signaling (Supplemental Table II). Hence, the previously reported tight interplay between ethylene and 1O2 is nicely reflected in the ROS-dependent transcriptomic signatures and reiterates the predictive power of such an analysis (Danon et al., 2005). In addition, these results confirm and extend previous observations that demonstrated the importance of the chemical identity of a given ROS in determining its signaling specificity (op den Camp et al., 2003).
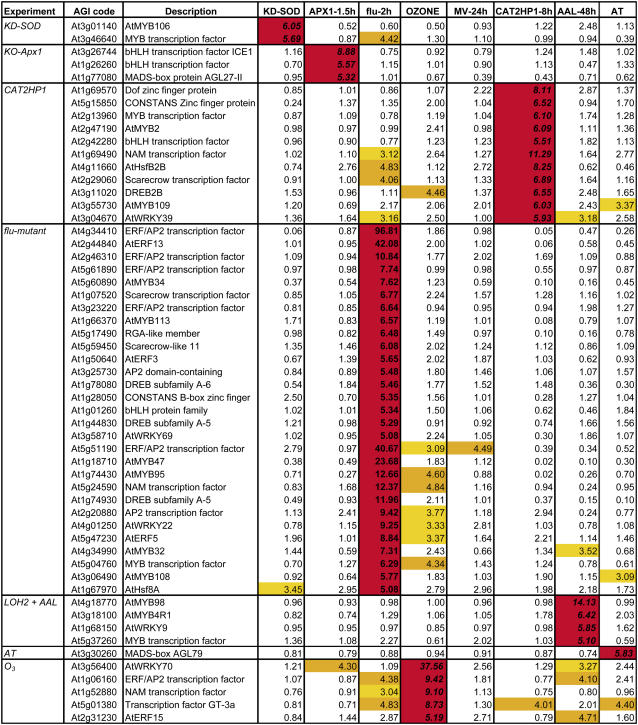
To correlate ROS-dependent transcriptomic signatures with the expression of transcripts encoding putative transcription factors, we assessed the expression characteristics of all annotated transcription factors in Arabidopsis in our microarray data sets (Riechmann et al., 2000). Of the approximately 1,500 genes, 552, 406, and 316 were at least 3-, 4-, or 5-fold differentially expressed in at least one of the selected experiments, respectively. The fact that one-third of all known transcription factors are 3-fold or more up- or down-regulated indicates the great impact of increased ROS levels on the cell. We screened for those transcription factors that were particularly modulated within the individual experiments. Table II provides a detailed overview of the transcription factors that are specifically induced within a certain experiment. In addition, Supplemental Table V gives a selection of all transcription factors with an increase in expression of at least 5-fold that are common among different experiments. Noteworthy is the presence of two WRKY family transcription factors, AtWRKY6 and AtWRKY75, which together with a bHLH transcription factor were induced more than 5-fold in seven of the nine experiments. WRKY transcription factors bind to the W-box within the promoter of many pathogenesis-related genes. According to previous studies, WRKY genes are induced under pathogen attack and oxidative stress conditions and regulate various aspects of plant development and cell death connected with ROS signaling events (Ülker and Somssich, 2004; Davletova et al., 2005; Guo and Gan, 2005).
Table II.
Transcription factors specifically up-regulated within one ROS experiment
A selection of transcription factors showing at least a 5-fold increase in expression within a single oxidative stress-related experiment is presented. Yellow, orange, and red indicate transcript levels between 3- and 4-fold, 4- and 5-fold, and above 5-fold, respectively.
Expression of ROS-Responsive Transcripts during Abiotic Stress Conditions
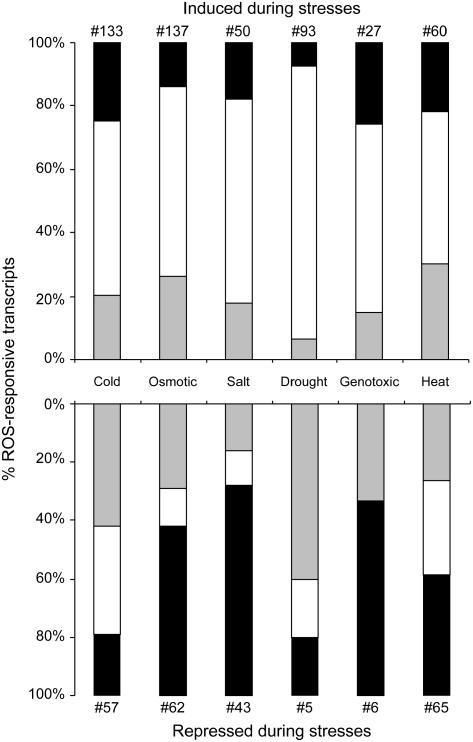
During abiotic stress conditions, ROS accumulation could represent toxic by-products of stress, as well as important signal transduction molecules responsible for the activation of defense mechanisms (Mittler, 2002). As a first step toward determining the involvement of the identified ROS-responsive transcripts in basic biological functions of plants, we analyzed the expression of ROS-induced transcripts in microarray data sets obtained from plants exposed to different abiotic stress conditions. Figure 3 summarizes the relative abundance of O2·−, 1O2, and H2O2 positively responsive transcripts in plant tissues subjected to different abiotic stresses. For each stress, the total numbers of up-regulated ROS-responsive transcripts (listed in Supplemental Tables II–IV) were determined and the relative contribution of each specific ROS-response cluster was calculated from this value. As shown in Figure 3, a complex pattern of different ROS-responsive transcripts could be found in all stress conditions, suggesting that different ROS are generated in cells during abiotic stresses. Transcripts responsive to 1O2 represent the largest fraction of ROS-related genes induced during these stresses. In contrast, H2O2- and O2·−-responsive transcripts were mainly repressed during the abiotic stresses. These findings suggest that different abiotic stress conditions provoke the production of different ROS, and that transcriptome profiling analysis could predict the degree of involvement of a specific ROS during a specific stress condition. With the availability of new transcriptome analysis data that include more time points and additional stresses, as well as data obtained from ROS-response mutants subjected to abiotic stresses, the analysis shown in Figure 3 could be refined and diverse stresses could be linked with specific ROS produced at specific subcellular compartments.
Figure 3.
Relative abundance of specific ROS in abiotic stresses. The relative contribution of specific ROS-induced transcripts that respond positively or negatively to different abiotic stresses is presented. For each abiotic stress, the total numbers of ROS-responsive transcripts are indicated. Black, gray, and white bars represent O2·−-, H2O2-, and 1O2-responsive transcripts, respectively. Data were processed as described in “Materials and Methods.”
CONCLUSION
With this analysis, we provide a framework of oxidative stress-dependent transcript expression in plants. It depicts specificity in the general characteristics of transcripts whose expression is influenced by elevated ROS and provides a transcriptomic footprint of the specific signaling capacities of different ROS in plants. The identification of ROS sensors and signaling components that are responsible for this remarkable selectivity and specificity of ROS signaling within the cell remains a major challenge. We think that our analysis will be instrumental in future wet-lab efforts to address the impact of specific ROS signals within environmental stress conditions and to elucidate the molecular mechanisms of the oxidative stress response in plants.
MATERIALS AND METHODS
Microarray Data
Microarray data of the published experiments (KD-SOD, KO-Apx1, CAT2HP1, AOX1a-AS, flu, AAL toxin, and AT) were downloaded from the journal Web resources or provided by the authors. For more information on the experimental details, we refer to the original publications (see Table I). MV treatment and O3 fumigation data were retrieved from the international AtGenExpress repository (the Arabidopsis Functional Genomics Network, http://www.uni-frankfurt.de/fb15/botanik/mcb/AFGN/AFGNHome.html) and from the European Arabidopsis Stock Center international transcriptomics service (NASCArrays; Craigon et al., 2004; http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl; ref. no. NASCARRAYS-26), respectively. The MV treatment and the O3 fumigation experiments were performed by the groups of Dr. D. Bartels (Bonn) and Dr. A. Shirras (Lancaster, UK), respectively. For more information on the exact conditions of both experiments, see http://web.uni-frankfurt.de/fb15/botanik/mcb/AFGN/atgenextable2.htm and http://affymetrix.arabidopsis.info/narrays/experimentpage.pl?experimentid=26, respectively.
Multiple time points (time course) were available for the KO-Apx1, CAT2HP1, flu, AAL toxin, and MV experiments, whereas KD-SOD, AOX1a-AS, AT, and O3 were single time point experiments.
Data Processing, Cross Platform Comparison, and Data Analysis
For Affymetrix ATH1 data sets, the raw data (CEL files) were collectively processed by Robust Multiarray Average (gcRMA; Bolstad et al., 2003; Irizarry et al., 2003a, 2003b) using the affy package of R/Bioconductor (Gautier et al., 2004). Resulting gcRMA expression values were inverse log transformed and, if possible, averaged over the biological replicates of the experiments. For Agilent Arabidopsis2 data sets, the processed data were extracted from the features gProcessedSignal and rProcessedSignal that were provided in the text output file, as supplied by the service provider (Service XS). Because sample handling, processing, and scanning can lead to certain biases, absolute probe signal intensity levels from samples processed in different laboratories should be compared with caution. Moreover, due to the heterogeneity of the experimental setup of the different integrated experiments (such as sufficient numbers of replicates), no careful statistical analysis could be performed. Therefore, relative expression data were used, although the possible generation of false positives/negatives by this approach should be kept in mind. Fold changes were calculated for all time points of all experiments, using the average expression values of the mutant or treatment samples relative to the wild-type or control samples. For the comparative analysis, a cross-platform comparison of the Affymetrix ATH1 and Agilent Arabidopsis2 arrays was performed. These microarrays carry 22,746 and 21,500 Arabidopsis (Arabidopsis thaliana) probes, respectively (Redman et al., 2004; Allemeersch et al., 2005). A previous cross-platform comparison of ATH1 and Agilent arrays revealed a high correlation coefficient and a fair agreement of signal intensities (Allemeersch et al., 2005). Based on the Arabidopsis Genome Initiative codes, 18,767 genes have corresponding probes on both arrays, whereas 3,979 and 2,918 genes are exclusively present on the Affymetrix and Agilent arrays, respectively. Probes for mitochondrion- and chloroplast-encoded genes are found only on the Affymetrix array. In total, the expression of nearly 26,000 Arabidopsis transcripts in response to different ROS was monitored. Thresholds for positive response were set at 3-, 4-, or 5-fold difference in expression within at least one time point across all experiments. Resulting data sets were log transformed, median centered across each gene, and subjected to hierarchical average linkage clustering (Euclidian distance) of both transcripts and experiments, with Cluster/Treeview (Eisen et al., 1998). Annotation was based on the The Arabidopsis Information Resource (Rhee et al., 2003; http://www.arabidopsis.org).
Specific ROS within Different Abiotic Stresses
Within the different tools of GENEVESTIGATOR, the Response Viewer was initially used to get an insight into the response profiles of individual ROS-regulated transcripts to different stimuli (Zimmermann et al., 2004, 2005). Out of the different conditions annotated, we selected the abiotic stress time series, cold, heat, salt, drought, osmotic, and genotoxic stress (Kudla's Laboratory, Germany) to reveal the response profiles of all ROS-induced transcripts presented in Supplemental Tables II to IV. For more information on the exact conditions of these different stress treatments, see http://web.uni-frankfurt.de/fb15/botanik/mcb/AFGN/atgenextable2.htm. Using the RMA-processed relative expression values of each abiotic stress experiment, the total numbers of ROS-specific transcripts (with at least 2-fold difference in expression) were determined. From the most responsive time point, relative contributions of each specific ROS-response cluster were calculated and graphically presented.
Supplementary Material
Acknowledgments
The authors thank Tineke Casneuf for help in normalizing the microarray data; Dr. Philip Zimmermann for providing the RMA-normalized data of the different abiotic stress experiments; Stéphane Rombauts, Dr. Fabio Fiorani, Dr. Philip Zimmermann, Dr. Marnik Vuylsteke, and Dr. Gerrit Beemster for useful contribution and discussions; Dr. Martine De Cock for assistance in preparing the manuscript; and Karel Spruyt for artwork.
This work was supported by the Research Fund of the Ghent University (Geconcerteerde Onderzoeksacties no. 12051403), by the Institute for the Promotion of Innovation by Science and Technology in Flanders (grant no. 040134), by the Scientific Co-operation between Eastern Europe and Switzerland program and the International Atomic Energy Agency (predoctoral fellowship to I.G.), the North Atlantic Treaty Organization (postdoctoral fellowship [RIG 981271] to T.S.G.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Frank Van Breusegem (frank.vanbreusegem@psb.ugent.be).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.078717.
References
- Allemeersch J, Durinck S, Vanderhaeghen R, Alard P, Maes R, Seeuws K, Bogaert T, Coddens K, Deschouwer K, Van Hummelen P, et al (2005) Benchmarking the CATMA microarray: a novel tool for Arabidopsis transcriptome analysis. Plant Physiol 137: 588–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Ausubel FM (2005) Are innate immune signaling pathways in plants and animals conserved? Nat Immunol 6: 973–979 [DOI] [PubMed] [Google Scholar]
- Bais HP, Vepachedu R, Gilroy S, Callaway RM, Vivanco JM (2003) Allelopathy and exotic plant invasion: from molecules and genes to species interactions. Science 301: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Bethke PC, Jones RL (2001) Cell death of barley aleurone protoplasts is mediated by reactive oxygen species. Plant J 25: 19–29 [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Åstrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Bowler C, Van Montagu M, Inzé D (1992) Superoxide dismutase and stress tolerance. Annu Rev Plant Physiol Plant Mol Biol 43: 83–116 [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee B-h, Hong X, Agarwal M, Zhu J-K (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craigon DJ, James N, Okyere J, Higgins J, Jotham J, May S (2004) NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res 32: D575–D577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon A, Miersch O, Felix G, op den Camp RGL, Apel K (2005) Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. Plant J 41: 68–80 [DOI] [PubMed] [Google Scholar]
- Dat JF, Pellinen R, Beeckman T, Kangasjärvi J, Langebartels C, Inzé D, Van Breusegem F (2003) Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J 33: 621–632 [DOI] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang H, Zhong S, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R (2005) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. Plant Cell 17: 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JHF, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JDG, et al (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2005) Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ 28: 1056–1071 [Google Scholar]
- Fujiki Y, Yoshikawa Y, Sato T, Inada N, Ito M, Nishida I, Watanabe A (2001) Dark-inducible genes from Arabidopsis thaliana are associated with leaf senescence and repressed by genes. Physiol Plant 111: 345–352 [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Gechev T, Gadjev I, Van Breusegem F, Inzé D, Dukiandjiev S, Toneva V, Minkov I (2002) Hydrogen peroxide protects tobacco from oxidative stress by inducing a set of antioxidant enzymes. Cell Mol Life Sci 59: 708–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Gadjev IZ, Hille J (2004) An extensive microarray analysis of AAL-toxin-induced cell death in Arabidopsis thaliana brings new insights into the complexity of programmed cell death in plants. Cell Mol Life Sci 61: 1185–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Hille J (2005) Hydrogen peroxide as a signal controlling plant programmed cell death. J Cell Biol 168: 17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Minkov IN, Hille J (2005) Hydrogen peroxide-induced cell death in Arabidopsis: transcriptional and mutant analysis reveals a role of an oxoglutarate-dependent dioxygenase gene in the cell death process. IUBMB Life 57: 181–188 [DOI] [PubMed] [Google Scholar]
- Guo Y, Gan S (2005) Leaf senescence: signals, execution, and regulation. Curr Top Dev Biol 71: 83–112 [DOI] [PubMed] [Google Scholar]
- Hu X, Bidney DL, Yalpani N, Duvick JP, Crasta O, Folkerts O, Lu G (2003) Overexpression of a gene encoding hydrogen peroxide-generating oxalate oxidase evokes defense responses in sunflower. Plant Physiol 133: 170–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP (2003. a) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15 [DOI] [PMC free article] [PubMed]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003. b) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu W-L, Tena G, Sheen J (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97: 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C, Apel K, Danon A (2004) Reactive oxygen signalling: the latest news. Curr Opin Plant Biol 7: 323–328 [DOI] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79: 583–593 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc Natl Acad Sci USA 98: 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) The reactive oxygen gene network in plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- op den Camp RGL, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Göbel C, Feussner I, et al (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z-M, Murata Y, Benning G, Thomine S, Klüsener B, Allen GJ, Grill E, Schroeder JI (2000) Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731–734 [DOI] [PubMed] [Google Scholar]
- Pnueli L, Liang H, Rozenberg M, Mittler R (2003) Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. Plant J 34: 187–203 [DOI] [PubMed] [Google Scholar]
- Redman JC, Haas BJ, Tanimoto G, Town CD (2004) Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J 38: 545–561 [DOI] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al (2003) The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res 31: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C-Z, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R (2003) The water-water cycle is essential for chloroplast protection in the absence of stress. J Biol Chem 278: 38921–38925 [DOI] [PubMed] [Google Scholar]
- Sandermann H Jr (2004) Molecular ecotoxicology of plants. Trends Plant Sci 9: 406–413 [DOI] [PubMed] [Google Scholar]
- Silverstein KAT, Graham MA, Paape TD, VandenBosch KA (2005) Genome organization of more than 300 defensin-like genes in Arabidopsis. Plant Physiol 138: 600–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ülker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7: 491–498 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Fiorani F, Siedow JN (2005) Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol 139: 1806–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S, Vanderauwera S, Vuylsteke M, Rombauts S, Langebartels C, Seidlitz HK, Zabeau M, Van Montagu M, Inzé D, Van Breusegem F (2004) Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J 39: 45–58 [DOI] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inzé D, Van Breusegem F (2005) Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol 139: 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hennig L, Gruissem W (2005) Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci 10: 407–409 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.