Abstract
Arabidopsis (Arabidopsis thaliana) was transformed with a redox-sensing green fluorescent protein (reduction-oxidation-sensitive green fluorescent protein [roGFP]), with expression targeted to either the cytoplasm or to the mitochondria. Both the mitochondrial and cytosolic forms are oxidation-reduction sensitive, as indicated by a change in the ratio of 510 nm light (green light) emitted following alternating illumination with 410 and 474 nm light. The 410/474 fluorescence ratio is related to the redox potential (in millivolts) of the organelle, cell, or tissue. Both forms of roGFP can be reduced with dithiothreitol and oxidized with hydrogen peroxide. The average resting redox potentials for roots are −318 mV for the cytoplasm and −362 mV for the mitochondria. The elongation zone of the Arabidopsis root has a more oxidized redox status than either the root cap or meristem. Mitochondria are much better than the cytoplasm, as a whole, at buffering changes in redox. The data show that roGFP is redox sensitive in plant cells and that this sensor makes it possible to monitor, in real time, dynamic changes in redox in vivo.
Cellular redox status influences many processes in plants, including apoptosis (Cai and Jones, 1999), oxidative defense mechanisms (Foyer and Noctor, 2005), senescence (Groten et al., 2005), allosteric control of enzyme activities, transcription and translation (Apel and Hirt, 2004), and a variety of signal transduction pathways (Dröge, 2002; Ermak and Davies, 2002; Neill et al., 2002). Yet, as central as is redox status to these processes, the redox potentials (oxidation-reduction potential) of living plant cells have rarely been measured during the occurrence of these activities (Renew et al., 2005). Rather, most often plant tissues are homogenized and the homogenates subsequently assayed, either with redox-sensing electrodes, or, by measuring the ratios of the reduced and oxidized forms of glutathione and ascorbate, the two principal redox regulators in living systems (Foyer and Noctor, 2003). Recently the redox state of plant tissues has also been assessed using the dyes 5- (and 6-) carboxy-2′, 7′-dichlorodihydrofluorescein diacetate (C-400; Jiang et al., 2003) and dihydrofluorescein diacetate (N. Smirnoff, personal communication). While such approaches allow one to sum the oxidized and reduced species, and thereby to infer the overall redox status of a tissue, it is not possible with these approaches to obtain a measure of redox potential at the time the events of interest are occurring. Moreover, whole tissue homogenization does not allow one to more finely resolve redox status within the various compartments and organelles comprising a typical plant cell, nor does this approach allow for an assessment of the redox status of the cell wall. As well, homogenizing a tissue precludes the possibility of monitoring dynamic changes of redox status, including reversibility. As a consequence, plant biologists lack knowledge of the rapidity of redox changes in plant cells.
Recently Hanson et al. (2004) described a redox-sensitive green fluorescent protein (GFP) that allows for nondestructive, real-time measurement of redox potential in HeLa cells. For this work GFP was modified (reduction-oxidation-sensitive GFP [roGFP]) to have two fluorescence excitation maxima (at approximately 400 and 475–490 nm) that show rapid and reversible ratiometric changes in their fluorescence in relation to ambient redox status. Depending on the targeting sequence of the GFP, redox potentials could be measured either in the cytoplasm or in the mitochondrial matrix. With this probe Dooley et al. (2004) were able to image dynamic changes in redox status in mammalian cells and demonstrate that both the cytosol and mitochondrial matrix have a relatively reducing redox status. Prior to the development of the roGFP probe, redox status in living cells could be measured by continuous wave electron paramagnetic resonance imaging that uses free radical contrast agents that are converted to a nonmagnetic species by intracellular redox processes (Fagan et al., 2001; Kuppusamy and Krishna, 2002). However, this approach is technically complex and requires an electron paramagnetic resonance imaging apparatus.
Here we report the development of a roGFP for plants. For this effort the roGFP1 originally designed for expression in HeLa cells (Hanson et al., 2004) has been modified for expression in the cytoplasm and mitochondrial matrix of Arabidopsis (Arabidopsis thaliana). We document that the roGFP1 in Arabidopsis shows similar spectra as in animal cells, and that it responds ratiometrically and with similar sensitivity to changes in redox.
RESULTS
roGFP1 Is Expressed in Arabidopsis
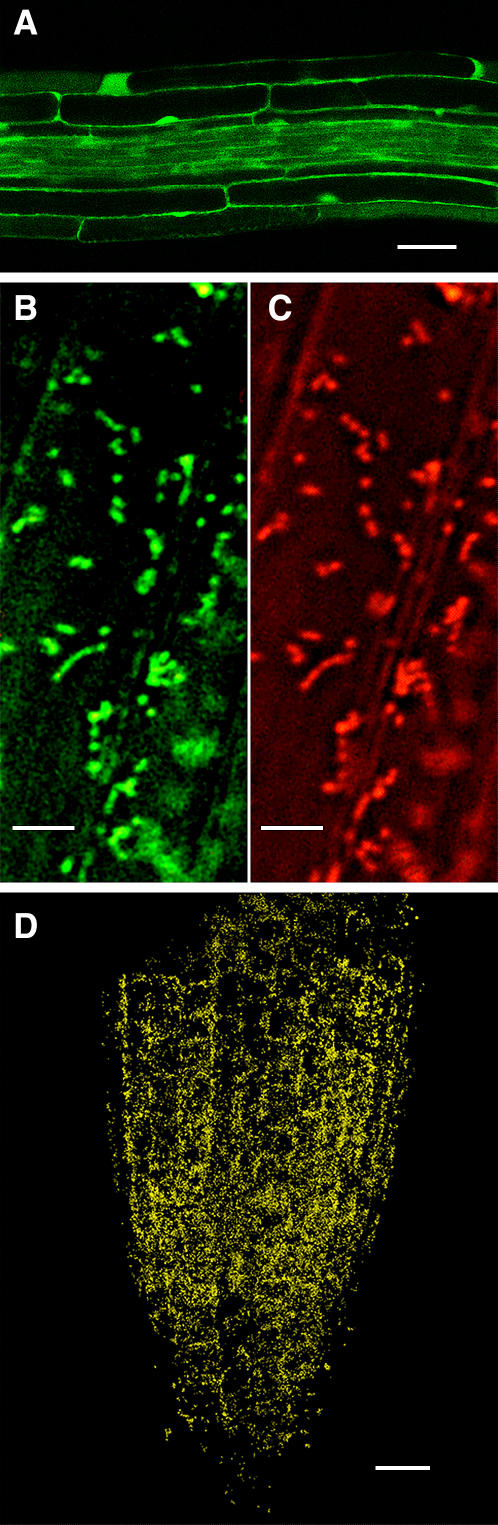
We placed the roGFP1 gene under the control of the cauliflower mosaic virus-35S promoter and also targeted expression to mitochondria, using a mitochondria-targeting sequence from tobacco (Nicotiana tabacum; Logan and Leaver, 2000). roGFP1-transformed Arabidopsis (roGFP1) expresses both the mitochondrial- and cytosolic-targeted forms of roGFP1, designated mt-roGFP1 and c-roGFP1, respectively (Fig. 1, A and B). Plants expressing either form of roGFP1 are morphologically normal, flower, and set seed. The presumed mitochondrial-targeted expression (Logan and Leaver, 2000) was reconfirmed by showing that a mitochondria-specific dye (MitoTracker Orange; Fig. 1C) and the putative mitochondria roGFP1 colocalize in the mitochondria (Fig. 1D).
Figure 1.
Expression of roGFP1 in roots of Arabidopsis. A, Confocal microscopy scan of the c-roGFP1. Bar = 20 μm. B and C, High power view of an Arabidopsis cortical cell from a plant transformed with mt-roGFP1 (B) and stained with MitoTracker Orange (C). Bar = 5 μm. D, A low power view of the entire Arabidopsis root tip showing the colocalization (yellow) of mt-roGFP1 and MitoTracker Orange. Bar = 20 μm.
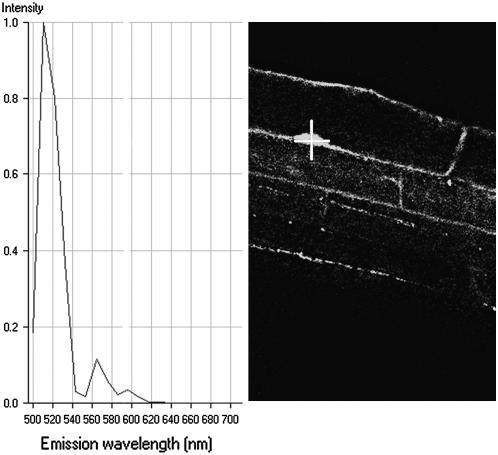
c-roGFP1/mt-roGFP1 in Vivo Emission Spectra
We characterized in vivo the emission spectrum of this distinctive GFP and observed the major emission peak at 510 nm, which is characteristic for roGFP1 (Fig. 2; Hanson et al., 2004).
Figure 2.
In vivo emission spectrum of c-roGFP1. The cursor (+) is placed at the point from which emission wavelengths are recorded (left section).
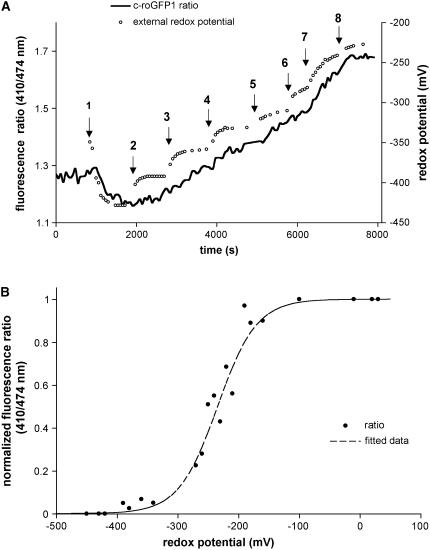
Redox Titration of roGFP1
Treating roots expressing either c- or mt-roGFP1 with buffers of differing redox status demonstrated that the ratio of light emitted following 410/474 nm irradiation (defined as the fluorescence ratio) changed as the external redox potential was altered using hydrogen peroxide (H2O2) and dithiothreitol (DTT; Fig. 3A). For these experiments a stable baseline was established and the root was then perfused with solutions containing differing ratios of the membrane-permeant, redox-active species DTT and H2O2. As the redox potential of the solution became increasingly more negative (more reducing) as measured by a redox electrode (Fig. 3A, small circles), we observed a decrease in the 410/474 fluorescence ratio (Fig. 3A, solid line). At a 410/474 fluorescence ratio of about 1.15, the ratio curve flattened, indicating that the c-roGFP1 had reached the lower limit of its sensitivity to solutions of increasingly more negative redox potentials. After reaching this limit the root was then perfused with new solutions containing varying amounts and ratios of DTT and H2O2. Significantly, the 410/474 fluorescence ratio paralleled the changes (increases) in the external redox potential, thereby indicating little lag time between the addition of new solutions of differing redox potentials and the response of the roGFP1. A summary of experiments like those in Figure 3A shows the dependence of the fluorescence ratio on redox status (Fig. 3B), thereby demonstrating that redox-sensing roGFP1 functions similarly in Arabidopsis as does the recombinant roGFP1 used by Hanson et al. (2004).
Figure 3.
c-roGFP1 responds to changes in redox. A, Increasing the H2O2:DTT ratio changes the 410/474 fluorescence ratio of c-roGFP1 in the Arabidopsis root. Small circles denote external redox potential (in millivolts) as measured with a redox-sensing electrode; solid line indicates the c-roGFP1 fluorescence ratio. Arrows indicate the times when changes were made with solutions with the following redox potentials (in millivolts): 1 = −430; 2 = −390; 3 = −360; 4 = −330; 5 = −310; 6 = −280; 7 = −240; and 8 = −220. B, Normalized c-roGFP1 fluorescence ratios (black circles) recorded from eight different roots by titration with DTT: H2O2 solutions as shown in A. Ratio values were recorded after reaching a steady state (5–10 min) in each solution. Increase in oxidation of the extracellular solution was associated with increase in the 410/474 c-roGFP fluorescence ratio. Dashed line shows all data fitted to nonlinear regression.
Redox Buffering Capacity of the Cytoplasm and Mitochondria
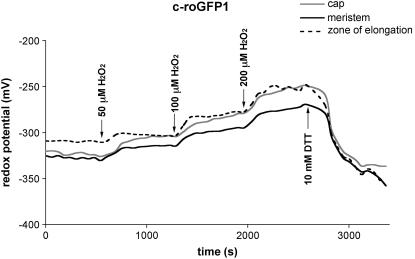
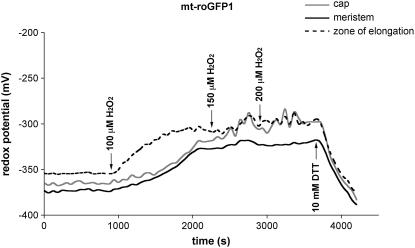
Using roGFP1 we measured the resting redox potentials of the cytoplasm and mitochondria under control conditions. In addition, by titrating roots with 50 to 200 μm H2O2, we investigated the buffering capacities of these two compartments and also measured the resulting changes in redox potential. On average, the cytoplasm from all three root zones (root cap, zone of elongation, and meristem) had a significantly more oxidized resting redox status (−318 ± 13 mV; Fig. 4; Table I) than the mitochondria (−362 ± 10 mV; Fig. 5; Table II), agreeing with similar observations reported by Hanson et al. (2004) of a more reduced status for mitochondria in mammalian cells. By titrating with increasing amounts of H2O2 it appeared that mitochondria were better able to buffer (ameliorate) increases in oxidized species than was the cytoplasm. At 200 μm H2O2, mitochondria showed a change of only −35 mV (−362 to −327 mV), whereas the same concentration of H2O2 produced about a −66 mV change in the redox status of the cytoplasm (−318 to −252 mV).
Figure 4.
Changes over time in cytosolic redox potential in a representative root, beginning with the addition of H2O2, and followed by DTT; from three zones of the Arabidopsis root. Arrows indicate the time of addition and concentration of H2O2 or DTT.
Table I.
Buffering capacity of c-roGFP1
Redox potentials in the cytosol in various zones of the root following equilibration with bathing solutions of varying H2O2 concentration (± sd; n = 6).
| H2O2 Concentration | Redox Potential
|
|||
|---|---|---|---|---|
| Cap | Meristem | Zone of Elongation | Whole Root Averagea | |
| μm | mV | |||
| 0 | −319 ± 9 | −325 ± 16 | −309 ± 8.4 | −318 ± 13 |
| 100 | −292 ± 19 | −289 ± 22 | −281 ± 10 | −287 ± 17 |
| 150 | −290 ± 14 | −286 ± 7 | −252 ± 47 | −276 ± 30 |
| 200 | −255 ± 28 | −255 ± 25 | −246 ± 22 | −252 ± 24 |
Averaging cap, meristem, and zone of elongation.
Figure 5.
Changes over time in mitochondrial redox potential in a representative root, beginning with the addition of H2O2, and followed by DTT; from three zones of the Arabidopsis root. Arrows indicate the time of addition and concentration of H2O2 or DTT.
Table II.
Buffering capacity of mt-roGFP1
Redox potentials of mitochondria in various zones of the root following equilibration with bathing solutions of varying H2O2 concentration (± sd; n = 8).
| H2O2 Concentration | Redox Potential
|
|||
|---|---|---|---|---|
| Cap | Meristem | Zone of Elongation | Whole Root Averagea | |
| μm | mV | |||
| 0 | −363 ± 6 | −367 ± 6 | −354 ± 13 | −362 ± 10 |
| 100 | −351 ± 14 | −352 ± 12 | −334 ± 19 | −346 ± 17 |
| 150 | −340 ± 20 | −342 ± 17 | −327 ± 20 | −336 ± 19 |
| 200 | −329 ± 24 | −336 ± 15 | −317 ± 20 | −327 ± 20 |
Averaging cap, meristem, and zone of elongation.
Although the shapes of the roGFP1 titration curves were similar for both the cytoplasm and mitochondria (in both the root cap and meristem), the zone of elongation responded faster (a steeper slope) to oxidative challenge (Figs. 4 and 5), suggesting that the externally supplied oxidant may be less well buffered in the zone of elongation or that the oxidant entered the cells more rapidly in the zone of elongation, compared to either the root cap or meristem region. We also showed that the redox state of both the cytoplasm and mitochondria can be returned to their starting levels by adding reductant (DTT) after the curves began to flatten (Figs. 4 and 5). Indeed, if sufficient reductant (e.g. 10 mm) was added, the mitochondria and cytosol became even more reduced than the starting (resting) redox levels (Figs. 4 and 5).
DISCUSSION
Redox-Sensing GFP Functions in Plants
We describe the transformation of Arabidopsis with a redox-sensing GFP and show that this redox reporter behaves in vivo in a manner similar to that reported for animal cells and for the recombinant protein. The titration of c-roGFP1 in Arabidopsis using DTT:H2O2 (Fig. 3A) showed a fluorescence ratio/redox potential dependency (Fig. 3B) very similar to that generated by Hanson et al. (2004) for the recombinant roGFP1 titrated with DTTred:DTTox solutions, with redox potentials subsequently calculated according to the Nernst equation (figure 1C in Hanson et al., 2004). Moreover, because both the c-roGFP1 and mt-roGFP1 probes in plants responded in a manner similar to that reported for the recombinant roGFP1 in vitro (Hanson et al., 2004), we used the in vitro-generated calibration curves (figure 1C in Hanson et al., 2004) to convert roGFP1 fluorescence ratios in Arabidopsis to millivolts (Tables I and II). Using this titration curve as a template and based on the assumption that the mitochondrial pH in Arabidopsis was similar to that reported for other plant mitochondria (pH 7.8 in isolated mitochondria from mung bean [Vigna radiata]; Moore et al., 1978), we corrected mitochondrial redox potentials in Arabidopsis for a pH more alkaline than pH 7. The calibration of the recombinant roGFP1 using DTTred:DTTox solutions at pH 7, and the Nernst equation, includes a pH dependency of 60.1 mV per pH unit (Hanson et al., 2004). If we assume that the mitochondrial pH is similar in Arabidopsis and mung bean, we can correct for this difference of 0.8 pH units between the mitochondrial matrix pH of 7.8 and the in vitro calibration of roGFP1 at pH 7.0 (Hanson et al., 2004) by subtracting 48.08 mV (0.8 × 60.1 mV = 48.08 mV). After performing this correction, the average resting redox potential in Arabidopsis mitochondria was found to be −362 mV (Table II), nearly identical to that reported for mitochondria in HeLa cells (−360 mV; Hanson et al., 2004). However, if in the future the pH of the Arabidopsis mitochondrial matrix is found to differ from 7.8, redox values would need to be corrected. Paralleling the similarities of mitochondrial redox potentials in Arabidopsis and HeLa cells, we also find that the average resting redox potential for the cytoplasm in Arabidopsis root cells (−318 mV; Table I) is almost identical to that reported for HeLa cells (−315 mV; Dooley et al., 2004). Thus, based on this initial application of a redox reporter in plants, we would conclude that the resting redox status of plant and animal cells is quite similar.
Redox Status in Different Regions of the Arabidopsis Root
We measured the redox status in three regions of the root; the root cap, the meristem (zone of rapid cell division), and the zone of elongation. Both forms of roGFP1 (cytosolic and mitochondrial) in the zone of elongation showed a more oxidized resting redox status than the meristem (P = 0.0045 for c-roGFP1 [n = 6]; P = 0.008 for mt-roGFP1 [n = 8]). Moreover, of the three regions of the root, the zone of elongation responded most rapidly (a steeper slope) to changing (increasing) concentrations of H2O2 (for five of six roots for c-roGFP1 and for six of eight roots for mt-roGFP1; Figs. 4 and 5; Tables I and II), suggesting that the elongation zone was less able to buffer changes in redox, compared to either the cap or meristem. Alternatively, the zone of elongation may be more permeable to the H2O2 in the external buffer than are the other two regions of the root. This slightly more oxidized status of the zone of elongation may also be related to factors that loosen the cell wall and permit plant cells to expand. Joo et al. (2001) showed that relatively high levels of reactive oxygen species correlated with the greatest rates of cell elongation in graviresponding roots and Sánchez-Fernández et al. (1997) reported that a highly reducing environment inhibits cell (trichoblast) elongation. Thus, as Schopfer (2001) noted, there appears to be a role for reactive oxygen species in cell wall loosening and in the control of cell elongation.
In comparing the response of the mitochondria and cytosol to increasing concentrations of H2O2 in the external bathing buffer, it appears that the mitochondria were better able to maintain a relatively reduced state following equilibration of roots with 200 μm H2O2. Roots treated with a high H2O2 concentration exhibited, on average, a change in mitochondrial-redox potential of −35 mV (a change from −362 to −327 mV), compared to a change in cytoplasmic-redox potential of about −66 mV (a change from −318 to −252 mV; Tables I and II). This difference may reflect dissimilarities in the capacity of the mitochondria and cytoplasm to buffer (ameliorate) redox changes, though these results may also be explained by the fact that the outside cytoplasm, or even the cell wall, buffers some of the incoming H2O2 before it reaches the mitochondria.
In summary, we have developed and characterized reporters (c-roGFP1 and mt-roGFP1) for measuring in vivo redox status in plants. The availability of these reporters now provides plant biologists with the opportunity to measure and quantify, in real time, the redox state of a cell or tissue under defined experimental conditions. The cytosolic form of this redox reporter should be particularly useful in understanding the roles of oxidative stress in a variety of plant processes, while the mitochondrial-targeted form may have special application in studies of pathogenesis and apoptosis, processes that have been linked to changes induced in mitochondria as a consequence of oxidative stress.
MATERIALS AND METHODS
Design and Construction of the Plasmids
All presequence cDNAs and the roGFP1 gene were subcloned into the NcoI and BstEII restriction sites in the plant binary vector pCAMBIA-1304, downstream of the cauliflower mosaic virus-35S promoter, replacing the sequence encoding the mGFP5-GUSA fusion protein. For cytosolic-expressed redox probes, roGFP1 was PCR amplified from pEFGP-N1roGFP (Dooley et al., 2004) using the forward primer CGACGGTACCGCGGGCCCGGG (roGFP-NcoI) and the reverse primer GGAAGGTGACCTTACTTGTACAGCTCGTCCATG (roGFP-BstEII). For mitochondrial localization, the 261 bp of the signal sequence, which corresponds to the first 87 amino acids of the tobacco (Nicotiana tabacum) β-ATPase (GeneBank X02868), was PCR amplified from pBINmgfp5-atpase (a kind gift from Dr. D.C. Logan [Logan and Leaver, 2000]). The forward primer used for this amplification is CATGCCATGGCTTCTCGGAGGCT (atpase-NcoI) and the reverse primer is GACTAGTACCAGCGCCGGTGAACTCATC (atpase-SpeI). The roGFP1 fragment for mitochondrial expression was amplified using GACTAGTAAGGGCGAGGAGCTGTTCACC (roGFP-SpeI) as the forward primer and roGFP-BstEII as the reverse primer. The original start codon for the roGFP1 was excised. The plasmid clones were sequenced to confirm that they were in frame before transformation with Agrobacterium.
Plant Materials, Growth Conditions, and Transformation
Arabidopsis (Arabidopsis thaliana; ecotype Columbia) was used as the wild type for transformation employing the standard Agrobacterium-mediated floral dip method (Clough and Bent, 1998), and seed later harvested. Putative transformants were selected by plating seed on Murashige and Skoog basal medium supplied with 15 mg mL−1 of hygromycin. Hygromycin-resistant plants were examined microscopically under UV light using a Leica DM microscope. Seedling plants showing GFP fluorescence were transplanted to soil, allowed to self, and the seed (T1 generation) collected, replated, and screened again, as above. Seedlings showing GFP fluorescence were transferred to the greenhouse and allowed to set seed (T2 generation).
Spectroscopy and Redox Titration
In Vivo Characterization and Localization of roGFP
To characterize roGFP1 expression in Arabidopsis and to demonstrate that this expression is ratiometrically redox sensitive, various types of spectroscopy were performed. Roots expressing both c- and mt-roGFP1 were examined with a Zeiss LSM 510 meta laser scanning confocal microscope equipped with an argon-ion laser, using single wavelength excitation at 488 nm and a dichroic reflector at HFT488 nm. Emission was characterized from 494 to 719 nm using the Zeiss meta detector (10.7 nm steps).
Ratiometric Measurements
Seeds from the T1 generation of three transgenic lines of Arabidopsis plants expressing c- and mt-roGFP1 were sterilized, plated on 1× Murashige and Skoog medium containing 0.8% agar, pH 5.7, and exposed to cold for 3 d. Subsequently the plates were moved to long-day conditions (16 h of light, 8 h of darkness) at a temperature of 22°C, and reoriented vertically. Three- to 5-d-old seedlings were carefully removed from the plates and affixed to circular coverslips (25 mm) by embedding the seedlings in a 1% low temperature gelling agarose (at 30°C). Immobilizing the seedlings was necessary so that measurements could be obtained microscopically over long periods from the same cell or group of cells. Measurements of redox status were performed using previously described methodology (Hanson et al., 2004). Briefly, a Nikon Diaphot inverted microscope was used with a 20× lens. A CCD camera collected emission (510 ± 15 nm) images during alternate excitation at 410 nm and 474 nm using a filter wheel (Lambda-10, Sutter Instruments). Emission data were collected over time. Axon Imaging Workbench 4.0 (Axon Instruments) controlled both filters and the collection of data. Fluorescence ratios were obtained by dividing the intensities obtained at 410 nm and 474 nm (i.e. 410/474). Each image was corrected for background by subtracting the intensity of an adjacent cell-free region. We normalized the 410/474 fluorescence ratios using maximal reduced and oxidized values obtained after adding 10 mm H2O2 or DTT; maximal ratio under maximally oxidized conditions was set equal to 1.0 and minimum ratio measured during maximally reduced conditions was set to 0. These normalized fluorescence ratios were then converted to redox potentials using the calibration curve generated by Hanson et al. (2004) for recombinant roGFP in a cytosol-like solution with pH 7.0 (their figure 1C, p. 13047). Their experimentally determined data for fluorescence ratio values obtained from isolated recombinant roGFP1 titrated with DTTred to DTTox solutions with calculated redox potentials were fit using a nonlinear regression equation [f = y0 + a/(1 + exp(−(x − x0)/b); SigmaPlot 8.0] and the resulting curve (y0 = 0.01, a = 0.99, x0 = −288, and b = 13) was then used to calibrate these experiments. Redox potentials obtained from mt-roGFP1 were corrected for an alkaline pH of 7.8 by subtracting 48.08 mV from the values obtained from this calibration following Hanson et al. (2004).
In general, the experimental procedure was to begin each experiment by incubating the immobilized seedlings in 1× Murashige and Skoog buffer, pH 5.7, while allowing the 410/474 fluorescence ratio to stabilize for 5 to 10 min before titration with H2O2 or DTT. Following each addition (change in redox buffer) the curve was allowed to flatten (5–10 min) before adding the next solution, usually containing increasing amounts (50–200 μm) of H2O2. After attaining the maximal (oxidative) response 10 mm DTT was added to demonstrate reversibility of the sensor. When the buffer was changed the current (old) buffer was completely removed prior to adding the new buffer. Thus, addition of the new buffer was not cumulative.
Redox potentials (in millivolts) of solutions containing different amounts of the DTT/H2O2 mixture (X mm DTT + X mm H2O2 = 10 mm) were measured in a buffer containing 10 mm HEPES, 10 nm NaCl, 2.5 mm KCl, 2.0 mm CaCl2, and 2.0 mm MgSO4, pH 7.0, to generate solutions with defined redox potentials as measured by a redox-sensitive platinum electrode (Inlab 501 Pt redox electrode; Mettler). These solutions were applied to the seedlings and used to determine c-roGFP1 and mt-roGFP1 fluorescence properties in vivo and to compare the fluorescence ratios to those reported for the recombinant roGFP1 described by Hanson et al. (2004).
To determine if the differences between the meristem and the elongation zone are statistically significant (α = 0.05), data were analyzed in Excel (Microsoft) using the Student's two-tailed t test for paired samples.
Mitochondrial-Specific Dyes
To reconfirm that the putative mitochondrial-targeted GFP indeed was expressed in the mitochondria, we treated transformed Arabidopsis (mt-roGFP1) with a mitochondria-specific dye (MitoTracker Orange [Invitrogen catalogue no. M7510]; Jiang et al., 2006). Arabidopsis roots were incubated for 1 to 2 h in the dark in 0.1 to 0.25 nm MitoTracker Orange dissolved in 10 mm K-P buffer, pH 5.7. Following incubation with the dye the roots were washed several times in plain buffer and then observed with 543 nm illumination using a Leica DM microscope. The mt-roGFP1 expression was superimposed on the MitoTracker Orange using color software by Imaris.
Acknowledgments
We thank Dr. David C. Logan for kindly providing the pBINmgfp5-atpase.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Lewis Feldman (feldman@nature.berkeley.edu).
References
- Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Cai J, Jones DP (1999) Mitochondrial redox signaling during apoptosis. J. Bioenerg Biomembr 31: 327–334 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY (2004) Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem 279: 22284–22293 [DOI] [PubMed] [Google Scholar]
- Dröge W (2002) Free radicals in the physiological control of cell function. Physiol Rev 82: 47–95 [DOI] [PubMed] [Google Scholar]
- Ermak G, Davies KJ (2002) Calcium and oxidative stress: from cell signaling to cell death. Mol Immunol 38: 713–721 [DOI] [PubMed] [Google Scholar]
- Fagan AJ, Davies GR, Hutchison JMS, Glasser FP, Lurie DJ (2001) Free radical imaging. Br J Radiol 74: 782–784 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2003) Redox sensing and signaling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol Plant 119: 355–364 [Google Scholar]
- Foyer CH, Noctor G (2005) Oxidant and antioxidant signaling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ 28: 1056–1071 [Google Scholar]
- Groten K, Vanacker H, Dutilleul C, Bastian F, Bernard S, Carzaniga R, Foyer C (2005) The roles of redox processes in pea nodule development and senescence. Plant Cell Environ 28: 1293–1304 [Google Scholar]
- Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ (2004) Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein. J Biol Chem 279: 13044–13053 [DOI] [PubMed] [Google Scholar]
- Jiang K, Ballinger T, Li D, Zhang S, Feldman LJ (2006) A role for mitochondria in the establishment and maintenance of the maize root quiescent center. Plant Physiol 140: 1118–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Meng YL, Feldman LJ (2003) Quiescent center formation in maize roots is associated with an auxin-regulated oxidizing environment. Development 130: 1429–1438 [DOI] [PubMed] [Google Scholar]
- Joo JH, Bae YS, Lee JS (2001) Role of auxin-induced reactive oxygen species in root gravitropism. Plant Physiol 126: 1055–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy P, Krishna MC (2002) EPR imaging of tissue redox status. Current Topics in Biophysics 26: 29–34 [Google Scholar]
- Logan DC, Leaver CJ (2000) Mitochondrial-targeted GFP highlights the heterogeneity of mitochondrial shape, size and movement within living plant cells. J Exp Bot 51: 865–871 [PubMed] [Google Scholar]
- Moore AL, Bonner WD Jr, Rich PR (1978) The determination of the proton-motive force during cyanide-insensitive respiration in plant mitochondria. Arch Biochem Biophys 186: 298–306 [DOI] [PubMed] [Google Scholar]
- Neill S, Desikan R, Hancock J (2002) Hydrogen peroxide signaling. Curr Opin Plant Biol 5: 388–395 [DOI] [PubMed] [Google Scholar]
- Renew S, Heyno E, Schopfer P, Liszkay A (2005) Sensitive detection and localization of hydroxyl radical production in cucumber roots and Arabidopsis seedlings by spin trapping electron paramagnetic resonance spectroscopy. Plant J 44: 342–347 [DOI] [PubMed] [Google Scholar]
- Sánchez-Fernández R, Fricker M, Corben LB, White NS, Sheard N, Leaver CJ, Van Montagu M, Inze D, May MJ (1997) Cell proliferation and hair tip growth in the Arabidopsis root are under mechanistically different forms of redox control. Proc Natl Acad Sci USA 94: 2745–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopfer P (2001) Hydroxyl radical-induced cell-wall loosening in vitro and in vivo: implications for control of elongation growth. Plant J 28: 679–688 [DOI] [PubMed] [Google Scholar]