Abstract
Nitric oxide (NO) and hydrogen peroxide (H2O2) are regulatory molecules in various developmental processes and stress responses. Tobacco (Nicotiana tabacum) leaves exposed to moderate high light dramatically potentiated NO-mediated cell death in catalase-deficient (CAT1AS) but not in wild-type plants, providing genetic evidence for a partnership between NO and H2O2 during the induction of programmed cell death. With this experimental model system, the specific impact on gene expression was characterized by either NO or H2O2 alone or both molecules combined. By means of genome-wide cDNA-amplified fragment length polymorphism analysis, transcriptional changes were compared in high light-treated CAT1AS and wild-type leaves treated with or without the NO donor sodium nitroprusside. Differential gene expression was detected for 214 of the approximately 8,000 transcript fragments examined. For 108 fragments, sequence analysis revealed homology to genes with a role in signal transduction, defense response, hormone interplay, proteolysis, transport, and metabolism. Surprisingly, only 16 genes were specifically induced by the combined action of NO and H2O2, whereas the majority were regulated by either of them alone. At least seven transcription factors were mutually up-regulated, indicating significant overlap between NO and H2O2 signaling pathways. These results consolidate significant cross-talk between NO and H2O2, provide new insight into the early transcriptional response of plants to increased NO and H2O2 levels, and identify target genes of the combined action of NO and H2O2 during the induction of plant cell death.
Reactive oxygen and nitrogen species have wide-ranging effects on many biological systems (Durner and Klessig, 1999; Finkel and Holbrook, 2000). Nitric oxide (NO) and hydrogen peroxide (H2O2) function in the plant response to various environmental stresses, such as drought, temperature, UV, ozone, and biotic stresses (Mittler et al., 2004; Delledonne, 2005). In addition, both molecules are involved in various developmental processes, such as seed germination, gravitropism, cell wall lignification, and root development, indicating that they play a key role in the regulation of plant responses to a range of endogenous signals and stimuli, such as auxin and abscisic acid (Desikan et al., 2004). Both NO and H2O2 are capable, independently, of inducing expression of the pathogenesis-related protein PR-1 and Phe ammonia lyase, the first enzyme within the phenylpropanoid biosynthesis pathway (for review, see Neill et al., 2002). Both molecules also steer salicylic acid (SA) production (Chamnongpol et al., 1998; Durner et al., 1998) and, according to Phe ammonia lyase expression, are involved in phytoalexin biosynthesis (Dat et al., 2000; Romero-Puertas et al., 2004). Other defense-related genes induced independently by NO and H2O2 include the alternative oxidase, a glutathione S-transferase, and key enzymes of the jasmonic acid biosynthesis pathway (for review, see del Río et al., 2004; Foyer and Noctor, 2005). In contrast, antimicrobial flavonoids and peroxidases are up-regulated by NO (Huang et al., 2002; Modolo et al., 2002; Polverari et al., 2003) but not by H2O2 (Vanderauwera et al., 2005).
Although knowledge of their direct targets and their regulatory effect on gene expression remains scarce, some potential targets of NO and reactive oxygen species (ROS) are known in plants. For instance, NO targets include metal- and thiol-containing proteins, such as catalase and peroxidase (Clark et al., 2000), aconitase (Navarre et al., 2000), glutathione S-transferase, copper/zinc-superoxide dismutase, thioredoxin, and glutaredoxin, as well as five enzymes involved in glycolysis and S-adenosyl-l-Met synthetase (Lindermayr et al., 2005). ROS act on proteins directly or indirectly via ubiquitous redox-sensitive molecules, such as glutathione or thioredoxins, which control the cellular redox state (Vranová et al., 2002b). Mitogen-activated protein kinase (MAPK) cascades appear to operate downstream of both H2O2 and NO. H2O2 activates NPK1 in a MAPK cascade that translates extracellular stimuli into defense gene expression (Grant et al., 2000; Kovtun et al., 2000; Desikan et al., 2001a), whereas NO triggers activation of a SA-induced protein kinase in tobacco (Nicotiana tabacum; Kumar and Klessig, 2000) and an unknown MAPK in Arabidopsis (Arabidopsis thaliana; Clarke et al., 2000). Both oxidants rapidly activate MAPKs in shoots, even when applied to roots (Capone et al., 2004). Some common intermediates of both ROS and NO signaling pathways are Ca2+ and Ca2+-binding proteins, such as calmodulin (del Río et al., 2004; Mittler et al., 2004). In contrast, production of cyclic GMP seems to be exclusive of NO signaling, whereas activation of G proteins and phospholipid signaling, which results in the accumulation of phosphatidic acid, is now specifically attributed to ROS signaling (Desikan et al., 2004; Mittler et al., 2004).
In summary, despite hints for synergy, NO and H2O2 can also act independently in the same signaling pathways with similar downstream responses as a consequence. In addition to their individual roles, NO and H2O2 work in strong partnership during induction of the hypersensitive response (HR). Whereas uncontrolled NO production in animal cells may lead to either apoptotic or necrotic cell death, depending on the severity and the context of the damage (Murphy, 1999), in plant cells NO needs well-balanced H2O2 levels to be channeled into the cell death pathway (Delledonne et al., 2001; de Pinto et al., 2002). This particular cell death is characterized by dehydration of the infected leaf segments as a result of the complete destruction of all cell layers. In fact, the collapse of the infected cells delimits the infected zone and, hence, avoids multiplication and spread of the pathogen (Heath, 1998). On the contrary, cell death triggered by H2O2 exhibits a different phenotype. Indeed, high H2O2 levels trigger the bleaching of leaves, characterized by patches of gray, chlorotic areas in the interveinal area, indicative of the selective collapse of palisade parenchyma cells, whereas both the upper and lower epidermal cell layers and most of the spongy parenchyma cells remain undamaged (Dat et al., 2003; Montillet et al., 2005). In contrast to the triggering role of H2O2 during cell death, NO alone is not able to kill cells. Indeed, when high levels of NO were supplied to soybean (Glycine max) cell suspensions, cell viability remained unaltered unless NO was supplemented with high levels of ROS (Delledonne et al., 2001). Recently, several lines of evidence that indicated different cell death phenotypes, induced by either H2O2 alone or together with NO, have been correlated with the accumulation of various fatty acid hydroperoxides. Accordingly, cell death activated by sustained increased H2O2 levels was associated with an intense free radical-mediated lipid peroxidation of membranes, whereas moderate levels of NO and H2O2 triggered cell death without massive lipid peroxidation (Montillet et al., 2005).
We report that the NO donor sodium nitroprusside (SNP) provokes cell death in catalase-deficient tobacco leaves exposed to moderate high light (HL) stress, a condition synonymous with sublethal H2O2 accumulation, whereas a similar treatment is ineffective in inducing cell death in wild-type plants. This experimental system provides genetic evidence for a role of H2O2 in channeling NO through the cell death pathway. In addition, our results show that H2O2 and NO can act either individually or in partnership in regulating gene expression and, most interestingly, during the execution of hypersensitive cell death.
RESULTS AND DISCUSSION
The Balanced Action of H2O2 and NO Induces HR-Like Cell Death in Tobacco Leaves
Although the involvement of NO and H2O2 in plant cell death has already been established, the downstream signaling pathways and the gene products directly involved in the induction of cell death are not completely elucidated. To identify the key players in the signal transduction pathway leading to H2O2- and/or NO-dependent cell death, we used transgenic tobacco (CAT1AS) plants that retain only 10% of the wild-type catalase activity (Dat et al., 2001). When CAT1AS plants are exposed to HL, photorespiratory H2O2 accumulates, ultimately provoking cell death. However, modulation in either light intensity and/or duration of exposure allows control of production of photorespiratory H2O2 in CAT1AS leaves (Willekens et al., 1997; Dat et al., 2003). Likewise, variable NO concentrations can be achieved by infiltrating tobacco leaves with different concentrations of SNP.
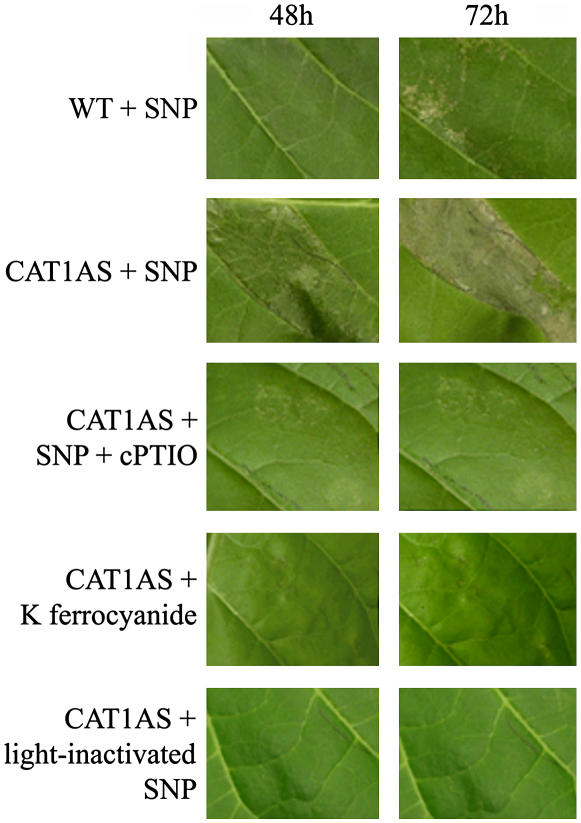
Wild-type and CAT1AS plants were infiltrated with 1 mm SNP and subsequently exposed to transient and moderate HL stress (500 μmol m−2 s−1 fluence rate) for 6 h. Only a few lesions appeared in SNP-treated wild-type plants, and no symptoms were observed in water-infiltrated wild-type and CAT1AS leaves (data not shown) or in SNP-infiltrated wild-type and CAT1AS plants kept under low light conditions (80 μmol m−2 s−1 fluence rate; data not shown). In contrast, exposing CAT1AS leaves infiltrated with the NO donor to the same moderate HL treatment led to the appearance of cell death lesions within 24 h. These lesions became more prominent after 48 h, leading to complete dehydration of the infiltrated leaf area within 72 h (Fig. 1). The simultaneous infiltration of 1 mm SNP and 1 mm 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO), a NO scavenger, abolished lesion formation (Fig. 1). These results, together with the lack of lesions in CAT1AS leaves exposed to moderate HL after infiltration with 1 mm potassium ferrocyanide, an analog of SNP that does not release NO, or with 1 mm SNP that has previously been exposed to light (1,000 μmol m−2 s−1 fluence rate for 24 h) to drive off NO (Fig. 1), eliminate the possibility that other by-products of SNP decomposition were responsible for the observed effect. The HR-like cell death phenology was similar to that induced by Pseudomonas sp. and cryptogein in tobacco (Montillet et al., 2005), but significantly differed from the chlorotic lesions observed in CAT1AS plants after prolonged exposure to HL (1,000 μmol m−2 s−1 fluence rate; Dat et al., 2003).
Figure 1.
Leaf symptoms induced by infiltration of SNP in catalase-deficient tobacco (CAT1AS) and wild-type plants exposed to a moderate HL treatment. Leaf of wild-type (WT) or catalase-deficient (CAT1AS) plant infiltrated with 1 mm of the NO donor SNP and exposed to a HL treatment (500 μmol m−2 s−1) for 6 h is shown. CAT1AS plants were also infiltrated with SNP plus the NO scavenger cPTIO, with potassium ferrocyanide, or with light-inactivated SNP. Pictures were taken after 48 and 72 h.
The use of CAT1AS plants allowed in planta modulation of H2O2 levels, thus without the need of administering exogenous ROS donors. Hence, our results confirm previous observations obtained with NO and ROS donors in plant cell suspensions (Delledonne et al., 2001; de Pinto et al., 2002). SNP is the most widely used NO donor in planta, but its ability to produce NO depends on the presence of reducing agents, such as glutathione, or on nonlinear photochemical degradation (Feelisch, 1998). Therefore, our data clearly indicate that the light intensity-dependent cell death observed by several research groups using SNP infiltration is not related to variations in the kinetics of NO release, but is merely due to variations in the level of ROS, as demonstrated by the dramatic potentiation of cell death when SNP is applied to CAT1AS when compared to wild-type plants under the same light conditions.
cDNA-AFLP Profiling of Gene Expression Induced by NO-H2O2 Interplay during HR Cell Death
The experimental in planta model system allowed independent modulation of H2O2 and NO levels, thus enabling, under the same light conditions, a comparison between NO- and H2O2-dependent transcriptomic changes, as well as identifying genes whose expression is altered when H2O2 channels NO into the cell death pathway. Transcriptomes of two interveinal segments of the sixth leaf of both CAT1AS and wild-type tobacco plants that had been infiltrated with either 1 mm SNP or water and exposed to moderate HL stress (500 μmol m−2 s−1) were compared. Samples were harvested 0, 1, and 3 h after HL exposure and subjected to comprehensive cDNA-amplified fragment length polymorphism (AFLP) analysis.
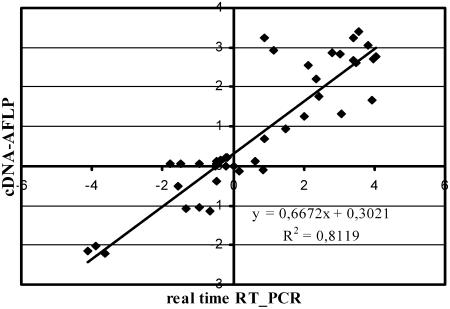
This approach delivers quantitative gene expression profiles and does not require prior sequence information or the availability of microarrays (Breyne et al., 2003). The expression kinetics of approximately 8,000 transcripts were monitored with 128 different BstYI + 1/MseI + 2 primer combinations for selective amplification. For each primer combination, 60 to 90 transcript fragments could be detected, varying from 50 to 600 bp in length. All detectable transcript fragments were quantified with the AFLP-QuantarPro software (Keygene N.V.), resulting in individual intensities per fragment for each time point. The raw data were processed with a tailor-made, in-house-developed arrayan cDNA-AFLP software package (Vandenabeele et al., 2003). Processed expression values were corrected for in-between lane differences with a total lane intensity correction, and a coefficient of variance (CV) was used as a selection criterion (see “Materials and Methods”). In total, 304 transcript fragments were identified as differentially expressed during the different treatments. These fragments were all excised from the gels and reamplified by PCR with their respective selective cDNA-AFLP primers (data not shown). Direct sequencing of the PCR products gave good-quality sequences for approximately 70% of the fragments (214). For the remaining 30%, no unique sequence could be attributed unambiguously, probably because of two or more comigrating cDNA-AFLP fragments. All 214 sequences were compared with those in available public databases. With the blast algorithm, 108 fragments were found to be similar to genes with a known function, 23 to sequences without any assigned function (unknowns), and 83 without homology to any other sequence in the public databases (see Supplemental Table I). To validate the reproducibility of the cDNA-AFLP data, the selective amplification reaction of 10 primer combinations was replicated twice and was >98% reproducible. A cDNA-AFLP analysis with 11 randomly chosen BstYI + 1/MseI + 2 primer combinations on a biological repeat experiment gave approximately 90% reproducibility. In addition, reproducibility of the cDNA-AFLP results was confirmed by real-time reverse transcription (RT)-PCR. The expression patterns of 10 candidate genes, belonging to five different clusters (see below), were examined at the 3-h time point. Although the absolute values for fold induction were not identical to those of the cDNA-AFLP analysis, a significant correlation (r2 = 0.812) was evident between the expression data generated by the two methods (Fig. 2).
Figure 2.
Correlation between cDNA-AFLP expression values and real time RT-PCR data. Log-transformed fold-change expression (base 2) for 10 differentially expressed transcripts in wild-type and CAT1AS leaves infiltrated with water or SNP and collected after 3 h is shown.
NO and H2O2 Steer Overlapping Signaling Pathways
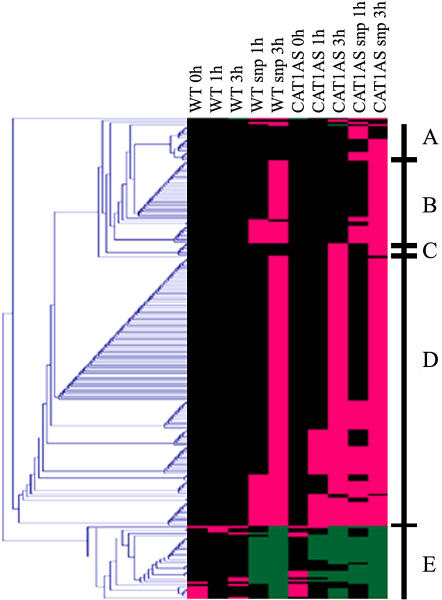
Expression values of the 214 successfully sequenced transcript fragments were variance normalized and clustered (see “Materials and Methods”). Figure 3 presents five prominent clusters. Cluster A represents 16 transcripts whose induction was dependent on the presence of both H2O2 and NO. Thirty-six genes in cluster B were classified as NO-dependent genes because they were only induced during HL in wild-type and CAT1AS leaves infiltrated with SNP. Cluster C contains 10 transcripts that were specifically induced by HL in CAT1AS leaves and are, hence, classified as H2O2-dependent genes. The largest set of genes (152) groups together clusters D (117) and E (35) because genes in these clusters are all independently induced or repressed by either NO or H2O2. This result indicates that, in addition to a specific response, a strong overlap exists in the signaling pathways triggered by either molecule.
Figure 3.
Cluster analysis of genes modulated by NO and/or H2O2. Hierarchical clustering of cDNA-AFLP differentially expressed transcript tags in control and catalase-deficient tobacco plants after SNP or water infiltration during the HL time course (0, 1, and 3 h) is shown. A fold change of 0.5 was fixed to discriminate significant expression changes from the signal background. All expression differences <50% were set to 0. Those higher than the fixed threshold were set to 1 and −1 for positive and negative expression changes, respectively. Magenta and green correspond to up-regulation and down-regulation, respectively, and black indicates time points when no relevant change in transcript level was measured. Prominent clusters A to E are indicated in the figure. For details, see “Materials and Methods.”
Approximately one-half of the sequenced fragments share significant homology with available public sequences from plants or other organisms. Based on this homology, sequences were classified into seven major functional categories: signal transduction, defense response, metabolism, proteolysis, transport, hormone interplay, and mobile elements. The copious presence of these functional categories clearly reflects the involvement of both NO and H2O2 in stress responses. The same functional categories dominated the list of genes in other stress-related gene discovery experiments (Vranová et al., 2002a; Polverari et al., 2003; Vandenabeele et al., 2003). Based on the relative presence of the functional categories, apparently no specific category within the different clusters is overrepresented.
Homologous sequences identified in clusters B, C, and D have previously been described as responsive to NO or H2O2 in Arabidopsis (Desikan et al., 2001b; Huang et al., 2002; Polverari et al., 2003; Vandenabeele et al., 2003; Parani et al., 2004; Vanderauwera et al., 2005). For a complete overview of all sequences, see Supplemental Table I, which provides the 214 sequenced fragments and their expression characteristics, together with a comparative analysis of their expression (or that of homologs) in other NO- and H2O2-dependent transcriptome analyses.
In cluster A (Table I; Fig. 3), two genes are responsive within 1 h (GenBank accession no. DQ460115 shows similarity to a putative ADP ribosylation factor, homologous to TTN5 involved in the regulation of intracellular vesicle transport during seed development [McElver et al., 2000], and GenBank accession no. DQ460119 shows homology with S-adenosyl-l-Met synthetase, which catalyzes the conversion of ATP and l-Met into the ethylene precursor S-adenosyl-l-Met [Boerjan et al., 1994]). Accordingly, an ethylene-responsive element-binding protein-like transcription factor (GenBank accession no. DQ460050) is induced after 3 h, together with a late-embryogenesis abundant protein 5 (GenBank accession no. DQ460208), myoinositol-1-P synthase (MIPS), and two other genes involved in metabolism. MIPS catalyzes the first and rate-limiting step in the biosynthesis of all inositol-containing compounds (Majumder et al., 1997). Myoinositol, a 6-carbon sugar alcohol, is a precursor to compounds that function not only in phosphorus storage, but also in signal transduction, stress protection, hormonal homeostasis, and cell wall biosynthesis in plants (Loewus and Murthy, 2000). The other eight sequences in cluster A are not homologous with any sequence in public databases. Because these transcripts are already induced 1 or 3 h after the applied NO/H2O2 trigger, it is very unlikely that they represent downstream effects provoked by cell death lesions. Cell death becomes only macroscopically apparent approximately 24 h after the HL stress. Thus, we propose that these genes are controlled by a signaling pathway that is specifically induced or repressed by synergism between both molecules. Previously reported genes involved in plant cell death, such as genes encoding Ser hydroxymethyltransferase (SHMT1;Moreno et al., 2005), LSD1 and LOL1 (Dietrich et al., 1997; Epple et al., 2003), cyclic nucleotide-gated channel (HLM1/CNGC; Balagué et al., 2003), bax inhibitor (Kawai et al., 1999; Sanchez et al., 2000), Ser palmitoyltransferase (SPT; Perry et al., 2000), and MAPK (Yang et al., 2001) were not found in our analysis. At first sight, this suggests that these genes are not controlled by concerted H2O2 and NO action. Alternatively, and as likely, they are not covered by the 128 cDNA-AFLP primer combinations or are only induced at a later stage.
Table I.
Transcript fragments, accession numbers, homology, functional category, and nucleotide identities of transcripts specifically induced by the combined action of NO and H2O2 (cluster A)
| Name | Accession No. | Description | Induction Time | Functional Class | E Value |
|---|---|---|---|---|---|
| BT4M34-408 | DQ460208 | MIPS | 3 h | Defense response | 2.00E-06 |
| BC3M41-3 | DQ460136 | Late-embryogenesis abundant protein 5 | 3 h | Defense response | 5.00E-12 |
| BC3M24-1 | DQ460119 | S-adenosyl-l-Met synthetase | 1 h | Hormone interplay | 4.00E-25 |
| BC3M41-4 | DQ460137 | Ribulose bisphosphate carboxylase | 3 h | Metabolism | 6.00E-35 |
| BT4M34-134 | DQ460206 | Iron-sulfur cluster assembly complex protein | 3 h | Metabolism | 1,00E-57 |
| BC1M11-154 | DQ460050 | Transcription factor EREBP-like protein | 3 h | Signal transduction | 2.00E-10 |
| BC3M22-144 | DQ460115 | Putative ADP ribosylation factor | 1 h | Transport | 2.00E-10 |
The transcripts in cluster B are specifically induced by NO regardless of the presence or absence of H2O2 and are therefore good candidates for target genes specifically modulated by NO-dependent pathways. Identification of this specific cluster also illustrates the strength of our approach. In previous transcriptome efforts on NO-mediated gene expression with SNP, these genes could not be distinguished from others up-regulated by ROS signals. Within our experimental setup, we can clearly discriminate between the more general gene expression response triggered by both H2O2 and NO (clusters D and E) from target genes exclusively induced by either of these signals. The transcripts in cluster C are specifically induced by H2O2 regardless of the presence or absence of NO. Surprisingly, only a minor fraction of the transcripts is under specific control of H2O2, again indicative of mutual pathways in the regulation of gene expression (Supplemental Table I). Among the genes in this cluster, there is a tobacco transcription factor (NtWRKY1) and a member of a class of heat shock proteins (HSP17.4) that has been shown to be tightly regulated by photorespiratory H2O2 in Arabidopsis (Vanderauwera et al., 2005). Of the total differentially expressed genes, 71% belong to clusters D and E. These clusters include genes that are induced or repressed within 1 or 3 h in both wild-type and CAT1AS plants infiltrated with SNP or with either water or SNP (all exposed to HL stress), respectively. Among the genes that are up-regulated (cluster D), 13 are potentially involved in signal transduction: seven are transcription factors and six are kinases, of which two are receptor protein kinases and one is a homolog of an Arabidopsis transcription factor involved in phytochrome A signaling (Bolle et al., 2000). In cluster E, one Zip gene, one kinase, two phosphatases, and one proteinase are present. The fact that the most dominant cluster contains genes that are modulated by both NO and H2O2 independently is a strong indication that some signal transduction pathways activated by both molecules converge and, hence, share common components or act on the same transcription factors. These genes probably are not candidates involved in the induction or execution of cell death because their induction is not directly related to lesion development, but more probably involved in other non-cell death-related processes in which NO and H2O2 intervene. However, because they are also induced in leaves that develop lesions (CAT1AS infiltrated with SNP), we cannot exclude the possibility that they are somehow necessary for an appropriate induction of the cell death program in concert with other genes in this cluster. Multiple transcripts belong to the defense-response functional category. Besides genes involved in oxylipin metabolism, a number of genes encoding putative components of disease resistance pathways, such as glucanase-like proteins, the FIERG1 protein, a putative Cys proteinase inhibitor, R3a, and ELI3, are induced by both NO and H2O2. The presence of these genes in this cluster is in agreement with their signaling role during plant-pathogen interactions (Neill et al., 2002).
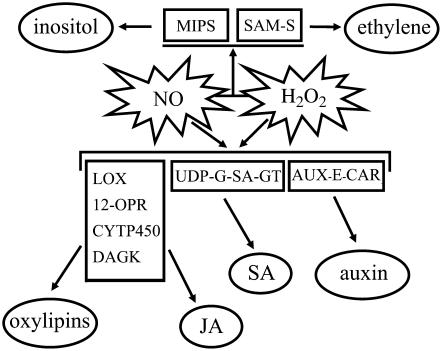
As noted before, this experimental approach allowed us to gain further knowledge of NO- and H2O2-dependent gene expression in plants. Most surprisingly, both signals individually appear to drive mutual signaling pathways, which might be attributed to the strong interplay of both molecules with other phytohormones (Pastori and Foyer, 2002; Desikan et al., 2004; Wendehenne et al., 2004), as evidenced by the high number of transcripts related to their biosynthesis and signaling. Accordingly, previous studies on CAT1AS plants have found an increase in SA and ethylene levels in these plants as early as 3 h after HL treatment (Takahashi et al., 1997; Chamnongpol et al., 1998). Transcripts encoding lipoxygenase (GenBank accession no. DQ460188), cytochrome P450-dependent fatty acid hydroxylase (GenBank accession no. DQ460205), diacylglycerol kinase (GenBank accession no. DQ460183), lipase (GenBank accession no. DQ460127), and 12-oxophytodienoate reductase (GenBank accession no. DQ460062), all involved in the biosynthesis of oxylipins and jasmonic acid, are up-regulated by NO and H2O2. Similarly, a UDP-Glc:SA glucosyltransferase (GenBank accession no. AJ538414) that converts SA to SA-glucoside, a conjugated and stable form of SA, is also induced by both molecules (Fig. 4), illustrating not only the additional complexity of the interaction scheme between NO and H2O2 in and outside cell death programs, but also a favorable hypothesis to explain the overlapping transcriptomic signatures written by NO and H2O2.
Figure 4.
Model of the NO and H2O2 signaling pathway. SAM-S, S-adenosyl-l-Met synthetase; LOX, lipoxygenase; 12-OPR, 12-oxophytodienoate reductase; CYTP450, cytochrome P450-dependent fatty acid hydroxylase; DAGK, diacylglycerol kinase; UDP-G-SA-GT, UDP-Glc:SA glucosyltransferase; AUX-E-CAR, auxin efflux carrier family protein.
MATERIALS AND METHODS
Plant Growth Conditions and Treatments
CAT1AS and wild-type SR1 tobacco (Nicotiana tabacum) plants were grown under a normal light regime (14-h-light/10-h-dark photoperiod at 100 μmol m−2 s−1) at 25°C and 70% relative humidity. Treatments were performed on the sixth leaf of 6- to 8-week-old plants. Leaves were infiltrated with 1 mm SNP by means of a 1-mL syringe without a needle. As a negative control, water, 1 mm potassium ferrocyanide, 1 mm light-inactivated SNP, or 1 mm SNP plus 1 mm cPTIO was used. To allow infiltration of the solution and to ensure that wet leaves were not exposed to HL, plants were exposed 15 min after infiltration to moderate HL treatment (500 μmol m−2 s−1, 6 h light/18 h dark) for 6 h in a Phytotron chamber PLC SCG 970 (Sanyo Gallenkamp). Phenotypes were observed in five independent experiments. Each treatment consisted of infiltration in two interveinal segments from two plants per sample. All experiments gave similar results. Infiltrated leaves were collected 1 and 3 h after HL treatment and immediately frozen in liquid nitrogen for the subsequent cDNA-AFLP analysis. RNA was extracted from pools of two interveinal segments of the same leaf. Each treatment was done on different plants and the cDNA-AFLP analysis was undertaken on two biological repeats.
RNA Extraction and cDNA-AFLP Analysis
Total RNA was extracted from samples using TRIzol reagent (Invitrogen). Starting from 2.5 μg of total RNA, the cDNA-AFLP-based transcript-profiling procedure was performed as described by Breyne et al. (2003). The restriction enzymes used were BstYI and MseI (New England Biolabs). For preamplification, a MseI primer without a selective nucleotide was combined with a BstYI primer containing a T or a C as a selective nucleotide at the 3′ end. Of a 600-fold dilution of the preamplified samples, 5 μL were used for the final selective amplifications with a BstT/C primer with one selective nucleotide and an MseI primer with two selective nucleotides. All 128 possible primer combinations were performed. Selective [γ-33P]ATP-labeled amplification products were separated on 4.5% polyacrylamide gels with the Sequigel system (Bio-Rad). Dried gels were scanned with a phosphor imager (GE Healthcare) and then exposed to Biomax films (Kodak).
Quantitative Measurement of Expression Profiles and Data Analysis
Gel images were quantitatively analyzed with AFLP-QuantarPro image analysis software (Keygene N.V.), by which all visible cDNA-AFLP fragments were scored and individual band intensities were measured in each lane. Using a tailor-made, in-house-developed arrayan cDNA-AFLP software package (Vandenabeele et al., 2003), the raw data were corrected for differences by using a total lane intensity correction: The intensity of each band was divided by a correction factor calculated by dividing the sum of the intensities of all individual bands within one lane by the average of all sums within the respective primer combination. A CV was calculated as the ratio of sd on all time course values and the average expression over the time course. The CV was used as a selection criterion for differential expression. Expression values of genes with a CV > 0.80 were taken for further analysis. Subsequently, expression values of selected genes were log transformed and variance normalized according to Tavazoie et al. (1999). A fold change of 0.5 was fixed to discriminate significant expression changes from the signal background. Thus, all expression differences <50% were set to 0 to prevent them from negatively influencing the clustering process. Moreover, as we were not interested in absolute expression variation, all expression differences higher than the fixed threshold were set to 1 and −1 for positive and negative expression changes, respectively. All data processing was custom made with a Python script. Hierarchical clustering was performed by using Euclidean distance and single-linkage clustering. Clustering and generation of dendrograms were done with the Multiexperiment viewer from the Institute of Genome Research microarray software suite (http://www.tm4.org).
Sequence Analysis of cDNA-AFLP Fragments
Bands corresponding to differentially expressed genes were excised from the gels and the eluted DNA was reamplified under the same conditions as those for selective amplification. Sequence information was obtained by direct sequencing of the reamplified product with the BstYI primer and compared to nucleotide and protein sequences in the available public databases by blast sequence alignments (Altschul et al., 1997). Some of the differentially expressed transcripts were not sequenced again because they matched perfectly with fragments in the cDNA-AFLP gels generated in previously published work on H2O2-induced gene expression in tobacco (Vandenabeele et al., 2003). These fragments are flagged in Supplemental Table I.
Real-Time RT-PCR Analysis
Real-time RT-PCR was carried out on the same RNA used in the cDNA-AFLP analysis. The samples analyzed were wild type (3 h), wild-type SNP (3 h), CAT1AS (0 h), CAT1AS (3 h), and CAT1AS SNP (3 h). Total RNA was treated with RNase-free DNase (Sigma-Aldrich), according to the manufacturer's instructions. Of DNase-treated total RNA, 3 μg was used for the RT reaction, utilizing Ready-To-Go you-prime first-strand beads (GE Healthcare). For real-time PCR analysis, 5 μL of 1:10 diluted cDNA samples was used in 25-μL reactions containing 0.4 μm gene-specific primers and 12.5-μL platinum SYBR Green qPCR SuperMix with ROX (Invitrogen). Triplicate quantitative assays were performed with a 7000 sequence detection system, according to the manufacturer's protocol (Applied Biosystems). Fold change in RNA expression was estimated using threshold cycles. The amplicon of tobacco ACTIN (forward, 5′-AGTGCTCAGTGGTGGCTCAAC-3′ and reverse, 5′-CTGGAGCAACAACCTTAATCTTC-3′) was used as an internal control to normalize all data (Burger et al., 2003). A control experiment without cDNA was included for each PCR mix.
The following gene-specific primers were used: BC3M24-1 (forward, 5′-TCCAGGTTTCTTATGCAATTGGTG-3′; reverse, 5′-AGTCAAAGTTCTCCTTGATCAGAG-3′); BC3M41-3 (forward, 5′-GTTTGTGGTTCAAGAGCATC-3′; reverse, 5′-CAAGCAAGAAGACAACATCATG-3′); BT2M14-236 (forward, 5′-CTCAGAGGTCAAGAAGATGAATG-3′; reverse, 5′-ACTCCATTGATCTCCAACAATTC-3′); BC2M33-455 (forward, 5′-GTCCCTCATTTCCTTCCATGTC-3′; reverse, 5′-CTGGAACATTCAACGGAAACTTC-3′); BC2M22-432 (forward, 5′-TTGCCAACGCTAGGATTGATTG-3′; reverse, 5′-CAGCACTTTGCCTTCTTCCAC-3′); BT4M31-256 (forward, 5′-CCGCAATGTGTGTCCAGGAAC-3′; reverse, 5′-GGAGAGGAGGATCAGTTTCAG-3′); BC2M21-465 (forward, 5′-CTCTCATCGTTCTTTCGCTTC-3′; reverse, 5′-GCTTGAGATTTCCTTCGGTTG-3′); BT3M13-430 (forward, 5′-TCATATGGGTGGCTTCTGCTC-3′; reverse, 5′-TGCCCGGCTCAGGCATGAATC-3′); BC2M14-230 (forward, 5′-TGAAACCCATTGAGTCAGGTTC-3′; reverse, 5′-CACCTCTATGTGAAAAGAGCTG-3′); and BC4M23-428 (forward, 5′-GGAGGCCAGCTCTCCTTCTC-3′; reverse, 5′-GTACTGTTCTCGGAATCAGCAG-3′).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ460050 to DQ460212.
Supplementary Material
Acknowledgments
We thank Dr. Martine De Cock for help in preparing the manuscript.
This work was supported by grants from the European Union-Human Potential Program (HPMT–CT–2000–00088), the Research Fund of the Ghent University (Geconcerteerde Onderzoeksacties no. 12051403), the EMBO Young Investigator Program, the Research Fund of the University of Verona, the Vlaamse Gemeenschap (Tournesol T2005.18), and the Ministère des Affaires Etrangères (Programme d'Actions Intégrées Tournesol 08993ZL).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Frank Van Breusegem (frank.vanbreusegem@psb.ugent.be).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.078444.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagué C, Lin B, Alcon C, Flottes G, Malmström S, Köhler C, Neuhaus G, Pelletier G, Gaymard F, Roby D (2003) HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide-gated channel ion channel family. Plant Cell 15: 365–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Bauw G, Van Montagu M, Inzé D (1994) Distinct phenotypes generated by overexpression and suppression of S-adenosyl-l-methionine synthetase reveal developmental patterns of gene silencing in tobacco. Plant Cell 6: 1401–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua N-H (2000) PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev 14: 1269–1278 [PMC free article] [PubMed] [Google Scholar]
- Breyne P, Dreesen R, Cannoot B, Rombaut D, Vandepoele K, Rombauts S, Vanderhaeghen R, Inzé D, Zabeau M (2003) Quantitative cDNA-AFLP analysis for genome-wide expression studies. Mol Genet Genomics 269: 173–179 [DOI] [PubMed] [Google Scholar]
- Burger C, Rondet S, Benveniste P, Schaller H (2003) Virus-induced silencing of sterol biosynthetic genes: identification of a Nicotiana tabacum L. obtusifoliol-14α-demethylase (CYP51) by genetic manipulation of the sterol biosynthetic pathway in Nicotiana benthamiana L. J Exp Bot 54: 1675–1683 [DOI] [PubMed] [Google Scholar]
- Capone R, Tiwari BS, Levine A (2004) Rapid transmission of oxidative and nitrosative stress signals from roots to shoots in Arabidopsis. Plant Physiol Biochem 42: 425–428 [DOI] [PubMed] [Google Scholar]
- Chamnongpol S, Willekens H, Moeder W, Langebartels C, Sandermann H Jr, Van Montagu M, Inzé D, Van Camp W (1998) Defense activation and enhanced pathogen tolerance induced by H2O2 in transgenic plants. Proc Natl Acad Sci USA 95: 5818–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D, Durner J, Navarre DA, Klessig DF (2000) Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol Plant-Microbe Interact 13: 1380–1384 [DOI] [PubMed] [Google Scholar]
- Clarke A, Desikan R, Hurst RD, Hancock JT, Neill SJ (2000) NO way back: nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J 24: 667–677 [DOI] [PubMed] [Google Scholar]
- Dat J, Vandenabeele S, Vranová E, Van Montagu M, Inzé D, Van Breusegem F (2000) Dual action of the active oxygen species during plant stress responses. Cell Mol Life Sci 57: 779–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dat JF, Inzé D, Van Breusegem F (2001) Catalase-deficient tobacco plants: tools for in planta studies on the role of hydrogen peroxide. Redox Rep 6: 37–42 [DOI] [PubMed] [Google Scholar]
- Dat JF, Pellinen R, Beeckman T, Van De Cotte B, Langebartels C, Kangasjärvi J, Inzé D, Van Breusegem F (2003) Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J 33: 621–632 [DOI] [PubMed] [Google Scholar]
- de Pinto MC, Tommasi F, De Gara L (2002) Changes in the antioxidant systems as part of the signaling pathway responsible for the programmed cell death activated by nitric oxide and reactive oxygen species in tobacco Bright-Yellow 2 cells. Plant Physiol 130: 698–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río LA, Corpas FJ, Barroso JB (2004) Nitric oxide and nitric oxide synthase activity in plants. Phytochemistry 65: 783–792 [DOI] [PubMed] [Google Scholar]
- Delledonne M (2005) NO news is good news for plants. Curr Opin Plant Biol 8: 390–396 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Zeier J, Marocco A, Lamb C (2001) Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc Natl Acad Sci USA 98: 13454–13459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Cheung M-K, Bright J, Henson D, Hancock JT, Neill SJ (2004) ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J Exp Bot 55: 205–212 [DOI] [PubMed] [Google Scholar]
- Desikan R, Hancock JT, Ichimura K, Shinozaki K, Neill SJ (2001. a) Harpin induces activation of the Arabidopsis mitogen-activated protein kinases AtMPK4 and AtMPK6. Plant Physiol 126: 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Mackerness SA-H, Hancock JT, Neill SJ (2001. b) Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol 127: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL (1997) A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88: 685–694 [DOI] [PubMed] [Google Scholar]
- Durner J, Klessig DF (1999) Nitric oxide as a signal in plants. Curr Opin Plant Biol 2: 369–374 [DOI] [PubMed] [Google Scholar]
- Durner J, Wendehenne D, Klessig DF (1998) Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc Natl Acad Sci USA 95: 10328–10333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple P, Mack AA, Morris VRF, Dangl JL (2003) Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc Natl Acad Sci USA 100: 6831–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feelisch M (1998) The use of nitric oxide donors in pharmacological studies. Naunyn Schmiedebergs Arch Pharmacol 358: 113–122 [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ (2000) Oxidants, oxidative stress and the biology of ageing. Nature 408: 239–247 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2005) Oxidant and antioxidant signalling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ 28: 1056–1071 [Google Scholar]
- Grant JJ, Yun B-W, Loake GJ (2000) Oxidative burst and cognate redox signalling reported by luciferase imaging: identification of a signal network that functions independently of ethylene, SA and Me-JA but is dependent on MAPKK activity. Plant J 24: 569–582 [DOI] [PubMed] [Google Scholar]
- Heath MC (1998) Apoptosis, programmed cell death and the hypersensitive response. Eur J Plant Pathol 104: 117–124 [Google Scholar]
- Huang X, von Rad U, Durner J (2002) Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215: 914–923 [DOI] [PubMed] [Google Scholar]
- Kawai M, Pan L, Reed JC, Uchimiya H (1999) Evolutionally conserved plant homologue of the Bax inhibitor-1 (BI-1) gene capable of suppressing Bax-induced cell death in yeast. FEBS Lett 464: 143–147 [DOI] [PubMed] [Google Scholar]
- Kovtun Y, Chiu W-L, Tena G, Sheen J (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97: 2940–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Klessig DF (2000) Differential induction of tobacco MAP kinases by the defense signals nitric oxide, salicylic acid, ethylene, and jasmonic acid. Mol Plant-Microbe Interact 13: 347–351 [DOI] [PubMed] [Google Scholar]
- Lindermayr C, Saalbach G, Durner J (2005) Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol 137: 921–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus FA, Murthy PPN (2000) myo-Inositol metabolism in plants. Plant Sci 150: 1–19 [Google Scholar]
- Majumder AL, Johnson MD, Henry SA (1997) 1l-myo-inositol-1-phosphate synthase. Biochim Biophys Acta 1348: 245–256 [DOI] [PubMed] [Google Scholar]
- McElver J, Patton D, Rumbaugh M, Liu C-m, Yang LJ, Meinke D (2000) The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell 12: 1379–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) The reactive oxygen gene network in plants. Trends Plant Sci 9: 490–498 [DOI] [PubMed] [Google Scholar]
- Modolo LV, Cunha FQ, Braga MR, Salgado I (2002) Nitric oxide synthase-mediated phytoalexin accumulation in soybean cotyledons in response to the Diaporthe phaseolorum f. sp. meridionalis elicitor. Plant Physiol 130: 1288–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montillet J-L, Chamnongpol S, Rustérucci C, Dat J, Van de Cotte B, Agnel J-P, Battesti C, Inzé D, Van Breusegem F, Triantaphylidès C (2005) Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol 138: 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JI, Martin R, Castresana C (2005) Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J 41: 451–463 [DOI] [PubMed] [Google Scholar]
- Murphy MP (1999) Nitric oxide and cell death. Biochim Biophys Acta 1411: 401–414 [DOI] [PubMed] [Google Scholar]
- Navarre DA, Wendehenne D, Durner J, Noad R, Klessig DF (2000) Nitric oxide modulates the activity of tobacco aconitase. Plant Physiol 122: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hurst RD, Hancock JT (2002) Hydrogen peroxide and nitric oxide as signalling molecules in plants. J Exp Bot 53: 1237–1247 [PubMed] [Google Scholar]
- Parani M, Rudrabhatla S, Myers R, Weirich H, Smith B, Leaman DW, Goldman SL (2004) Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnol J 2: 359–366 [DOI] [PubMed] [Google Scholar]
- Pastori GM, Foyer CH (2002) Common components, networks, and pathways of cross-tolerance to stress. The central role of “redox” and abscisic acid-mediated controls. Plant Physiol 129: 460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry DK, Carton J, Shah AK, Meredith F, Uhlinger DJ, Hannun YA (2000) Serine palmitoyltransferase regulates de novo ceramide generation during etoposide-induced apoptosis. J Biol Chem 275: 9078–9084 [DOI] [PubMed] [Google Scholar]
- Polverari A, Molesini B, Pezzotti M, Buonaurio R, Marte M, Delledonne M (2003) Nitric oxide-mediated transcriptional changes in Arabidopsis thaliana. Mol Plant-Microbe Interact 16: 1094–1105 [DOI] [PubMed] [Google Scholar]
- Romero-Puertas MC, Perazzolli M, Zago ED, Delledonne M (2004) Nitric oxide signalling functions in plant-pathogen interactions. Cell Microbiol 6: 795–803 [DOI] [PubMed] [Google Scholar]
- Sanchez P, Zabala MD, Grant M (2000) AtBI-1, a plant homologue of Bax Inhibitor-1, suppresses Bax-induced cell death in yeast and is rapidly up regulated during wounding and pathogen challenge. Plant J 21: 393–399 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Chen Z, Du H, Liu Y, Klessig D (1997) Development of necrosis and activation of disease resistance in transgenic tobacco plants with severely reduced catalase levels. Plant J 11: 993–1005 [DOI] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM (1999) Systematic determination of genetic network architecture. Nat Genet 22: 281–285 [DOI] [PubMed] [Google Scholar]
- Vandenabeele S, Van Der Kelen K, Dat J, Gadjev I, Boonefaes T, Morsa S, Rottiers P, Slooten L, Van Montagu M, Zabeau M, et al (2003) A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc Natl Acad Sci USA 100: 16113–16118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inzé D, Van Breusegem F (2005) Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol 139: 806–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranová E, Atichartpongkul S, Villarroel R, Van Montagu M, Inzé D, Van Camp W (2002. a) Comprehensive analysis of gene expression in Nicotiana tabacum leaves acclimated to oxidative stress. Proc Natl Acad Sci USA 99: 10870–10875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranová E, Inzé D, Van Breusegem F (2002. b) Signal transduction during oxidative stress. J Exp Bot 53: 1227–1236 [PubMed] [Google Scholar]
- Wendehenne D, Durner J, Klessig DF (2004) Nitric oxide: a new player in plant signalling and defence responses. Curr Opin Plant Biol 7: 449–455 [DOI] [PubMed] [Google Scholar]
- Willekens H, Chamnongpol S, Davey M, Schraudner M, Langebartels C, Van Montagu M, Inzé D, Van Camp W (1997) Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J 16: 4806–4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K-Y, Liu Y, Zhang S (2001) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc Natl Acad Sci USA 98: 741–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.