Abstract
In this study, we analyzed double-strand break (DSB) repair in Arabidopsis (Arabidopsis thaliana) at various developmental stages. To analyze DSB repair, we used a homologous recombination (HR) and point mutation reversion assays based on nonfunctional β-glucuronidase reporter genes. Activation of the reporter gene through HR or point mutation reversion resulted in the appearance of blue sectors after histochemical staining. Scoring of these sectors at 3-d intervals from 2 to 31 d post germination (dpg) revealed that, although there was a 100-fold increase in the number of genomes per plant, the recombination frequency only increased 30-fold. This translates to a recombination rate at 31 dpg (2.77 × 10−8) being only 30% of the recombination rate at 2 dpg (9.14 × 10−8). Conversely, the mutation frequency increased nearly 180-fold, resulting in a 1.8-fold increase in mutation rate from 2 to 31 dpg. Additional analysis of DSBs over the early developmental stages revealed a substantial increase in the number of strand breaks per unit of DNA. Furthermore, RNA analysis of Ku70 and Rad51, two key enzymes in two different DSB repair pathways, and further protein analysis of Ku70 revealed an increase in Ku70 levels and a decrease of Rad51 levels in the developing plants. These data suggest that DSB repair mechanisms are developmentally regulated in Arabidopsis, whereby the proportion of breaks repaired via HR substantially decreases as the plants mature.
The genetic material of any organism is constantly fluctuating, with hundreds of mutations varying from silent-base substitutions to large deletions/insertions being introduced upon each genome replication (Tuteja et al., 2001; Kunz et al., 2005). The frequency with which these mistakes persist depends on several parameters, such as the competence of polymerase proofreading activity, the effectiveness of the proteins involved in early DNA damage recognition, the efficiency of chromatin modifiers, and the precision of the core DNA repair enzymes. In many cases, the same type of lesion can be repaired by several different DNA surveillance mechanisms. The balance between these mechanisms maintains the relative genome stability of a given organism.
Single- and double-strand breaks (SSBs and DSBs) are good examples of the lesions that are processed by the various repair pathways broadly grouped to nonhomologous end joining (NHEJ) and homologous recombination (HR; Sargent et al., 1997; Liang et al., 1998). These lesions can be extremely deleterious as even a single, unprocessed break may lead to cell death (Karanjawala et al., 2002).
NHEJ and HR have different repair fidelities. NHEJ is believed to result in various mutations, varying from single nucleotide substitutions to deletions/insertions of one to several thousand nucleotides (Roth and Wilson, 1986; Brennan and Schiestl, 1998; Jeggo, 1998; Ries et al., 2000; Ikeda et al., 2001; Pelczar et al., 2003; Kovalchuk et al., 2004). Conversely, HR, although generally believed to be free of repair mistakes, frequently results in large segmental duplications, gene duplication, gene loss, or gene inactivation. Presently, it is not clear which mechanism more significantly contributes to genome rearrangements and, therefore, to genome evolution (Gorbunova and Levy, 1997; Critchlow and Jackson, 1998; Kirik et al., 2000; Smith et al., 2001).
It has been well documented that the contribution of either NHEJ or HR to the repair of strand breaks varies from organism to organism (Cromie et al., 2001) and from tissue to tissue (Essers et al., 2000). For example, the frequency of HR-based repair was found to be different in various tissues of mammalian organisms, whereby embryonic stem cells displayed a higher frequency of HR when compared to other, differentiated cells (Essers et al., 2000). Moreover, various areas of the genome appear to have different rates of HR (Puchta et al., 1995; Filkowski et al., 2004) and, perhaps, NHEJ (Kovalchuk et al., 2000). However, the phenomenon of cell type-specific HR rates, as displayed in mammals, to the best of our knowledge, has not been studied in plants.
Given that the rates of HR and NHEJ differ according to the circumstance, the contribution of various DNA repair mechanisms to each specific lesion may also vary at different stages of organism development. Providing that the efficiency of any process depends on two major factors, cost and precision, the balance between reasonable costs and reasonable precision defines what is typical organism development. Therefore, as HR and NHEJ have different repair fidelities and different costs, their contribution to strand break repair at different developmental stages may also vary.
In this study we analyzed the HR events in Arabidopsis (Arabidopsis thaliana) at different developmental stages. We found that the rate of HR decreased with plant age, while the frequency of strand breaks and point mutation rates increased. These results were paralleled by a decrease in the abundance of Rad51 and an increase in the abundance of Ku70 transcripts. This phenomenon may reflect the existence of a mechanism that provides tight control over extensive recombination in polyploid plant cells.
RESULTS
Transgenic Lines
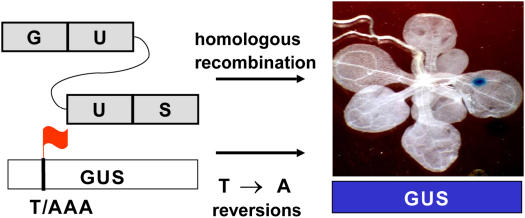
Genome stability was analyzed in transgenic Arabidopsis plants with a uidA HR (cultivar [cv] C24; line no. 11) or point mutation (cv Columbia; line no. 166_4) substrate. The recombination substrate consisted of two truncated, nonfunctional, overlapping copies of the uidA gene (Swoboda et al., 1994), whereas the mutation substrate consisted of a single copy of a stop codon-inactivated uidA gene (Kovalchuk et al., 2000; Fig. 1). In HR plants, any strand break generated in the region of homology between the truncated uidA genes could potentially be repaired via HR using the second copy as a template, thus possibly restoring reporter gene function. Similarly, in mutation plants, reversions to the original nucleotide could restore the reporter gene. In both cases, transgene activation could be visualized as sectors of blue via histochemical staining (Fig. 1).
Figure 1.
Detection of HR and point mutations in plants. The HR substrate consisted of 5′ end (UG) and 3′ end (US) of the uidA transgene. The 5′ end of uidA gene was cloned in an inverted orientation (Swoboda et al., 1994). A recombination event between two regions of homology (U) would result in the restoration of the active uidA gene. The mutation substrate consisted of a stop codon-inactivated uidA gene, whereby the AAA triplet was changed to a TAA stop codon. The T/A reversion mutation restored the active uidA gene. HR or point mutation events were visualized as sectors of blue in the stained Arabidopsis leaves.
To confirm the results found with uidA-based recombination lines, we used another Arabidopsis recombination reporter line based on the luciferase (LUC) substrate. These plants also carry a single copy of two truncated nonfunctional marker genes cloned in direct orientation (Kovalchuk et al., 2003). HR events restore transgene activity, which can be monitored as fluorescing spots under a CCD camera (see “Materials and Methods”).
Recombination Rates at Different Stages of Development
To understand at what point during plant development HR events were most prevalent, the following experiment was performed.
Twelve groups of line number 11 plants were germinated on soil and harvested for histochemical staining at 2, 3, 5, 7, 10, 13, 16, 19, 22, 25, 28, and 31 d post germination (dpg). It has been previously shown that the DNA content in Arabidopsis leaves increases linearly until 20 to 25 d after germination (Draper and Hays, 2000); therefore, DNA content from all twelve groups was analyzed. Sixty plants of ages 2, 3, and 5 dpg, and four to 20 plants of ages 7 to 31 dpg were harvested for DNA content.
Calculating the number of genomes present per plant revealed that there was an approximately 100-fold increase in genomes from 2 to 31 dpg (5.47 × 105 and 5.42 × 107, respectively). Counting the recombination events (sectors of blue) in populations of 200 to 500 plants per group, and relating these events to the total number of screened plants, revealed a linear increase in HR frequencies. HR frequencies increased by a factor of 30.0, from 0.05/plant at 2 dpg to 1.5/plant by 31 dpg (Fig. 2; Supplemental Table I). The actual recombination rate (RR), the ratio of HR frequency to number of genomes present, dropped significantly and at 31 dpg was 30% of the RR at 2 dpg. There was, however, a transient increase in RR from day 5 to day 19 (Fig. 2; Supplemental Table I). This experiment suggested that plants older than 19 d use HR less frequently when compared to younger plants.
Figure 2.
HR frequency and RRs at different plant ages. Plants of line number 11 were harvested at different times post germination. Each group consisted of 200 to 500 plants. The data is presented as logarithmic scale of fold induction of HR frequency (HRF), number of genomes (Genomes), and RR in plants of different ages (3–31 dpg) as compared to 2 dpg.
The finding that HR rates decrease with plant age is important. To be sure that these findings were not an artifact from one particular marker gene or one particular transgenic line, we performed a similar experiment with another Arabidopsis transgenic line. Line 15D8 carries in its genome a single copy of a different recombination substrate that is based on the LUC reporter gene. The advantage of using this line is that it allows one to evaluate the recombination frequency without harvesting the plant. In this case, multiple evaluations of the recombination frequency can be done in the same plants throughout their entire life cycle (Kovalchuk et al., 2003). This allowed us to carry the experiment until 56 dpg. Calculation of HR frequency and RR was done similarly to that of line number 11 (see “Materials and Methods”). In correspondence to line number 11, line 15D8 showed a nearly 2-fold decrease of RR between 2 and 31 dpg (Fig. 3A; Supplemental Table II). Moreover, the RR continued to decrease until 56 dpg, where it was but 27% of the rate at 2 dpg. This experiment confirmed that older plants use HR less frequently than younger plants.
Figure 3.
HR frequency, genome number, and HR rates at different plant ages as calculated from the 15D8 line. HR frequency was calculated from 20 to 200 plants (depending on plant age). A, Shows the fold increase in HR frequency, number of genomes, and HR rate in plants of different ages (2–56 dpg, x axis) as compared to 2 dpg, expressed in logarithmic scale (y axis). B, Shows the same for plants grown in the presence of 4 μm RB.
Exposure to Rose Bengal Results in Increased HR Rate in All Plant Age Groups
One remaining question was whether there is a particular limit in the application of HR repair. That is, whether there is a limiting number of times a plant cell can utilize HR machinery for repair. If this were the case, plants exposed to high levels of stress would not be able to increase the number of times they utilized the HR pathway past the limiting point.
To examine whether the HR rate changed upon exposure to a mutagen, we grew plants in the presence of rose Bengal (RB), an oxidative stress-generating compound. Our previous work showed that this chemical substantially increases the HR frequency; likely from the induction of breaks from the oxygen radicals it produces (Filkowski et al., 2004). When RB was applied in the current experiment, we found a 2.8-fold increase in HR rate in plants from 2 to 28 dpg. Furthermore, the RR remained higher than that of 2 dpg until the last day of measurement (Figs. 3B and 4; Supplemental Table II). This experiment showed that plants exposed to this stress were able to utilize HR repair machinery at a higher frequency, thus indicating that the number of times HR is used in the cell does not limit the potential for the stress-induced HR repair.
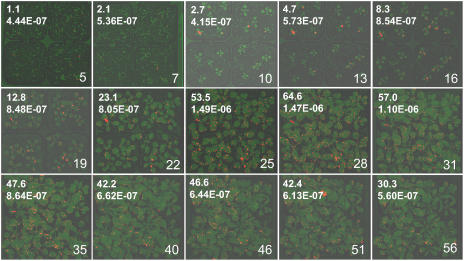
Figure 4.
HR events as observed in plants of the 15D8 line. HR events were observed in several groups of 20 to 200 plants (depending on age). Plant age is marked in the bottom right corner. The HR frequency (top) and the HR rate (bottom) is shown in the top left corner.
Strand Break Levels during Development
Our data showed that the contribution of HR to DNA repair decreases dramatically with maturity in Arabidopsis. As HR is a strand break repair mechanism, two alternate scenarios could explain this phenomenon. Either the level of strand breaks decreases with age, or the cell utilizes other repair mechanisms as the plant matures. The latter explanation seems counterintuitive, as the quantity of DNA per cell increases as a plant cell ages. As such, it seems more likely that the amount of strand breaks would increase under constant conditions purely through the increase in DNA per cell.
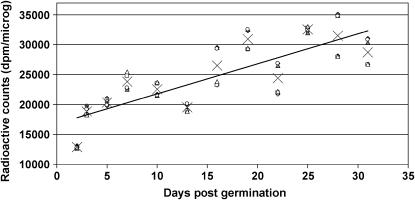
To test this hypothesis, we analyzed the level of strand breaks in developing plants at the aforementioned days post germination. To measure the level of strand breaks, we used the random oligonucleotide primed synthesis (ROPS) assay. This assay is based on Klenow polymerase-aided incorporation of [3H]-dCTP into newly synthesized DNA at the break points (see “Materials and Methods” for details). The average data from three independent experiments (with two independent measurements per each data point) showed that the level of strand breaks (shown per microgram of DNA) increased gradually through development and was 220% of the 2 dpg value at 31 dpg (Fig. 5).
Figure 5.
Strand breaks at different plant ages. Strand breaks were analyzed in plants of different age groups (2–31 dpg, x axis). The y axis shows the radioactive incorporation (dpm/μg) into the DNA breaks. This figure shows the individual data points for three independent experiments, with independent measurements per each experiment. X indicates the average of the data points. The trend line shows the steady linear increase of the strand break number.
This experiment showed that the decrease in HR rate was not due to a decrease in the level of breaks. Thus, another strand break repair mechanism must have been employed to compensate for the additional breaks and decrease in HR activity.
Point Mutation Rates at Different Developmental Stages
The previous experiments showed that the RR decreases with plant age, and that this decrease is not due to a decrease in the level of strand breaks. Given that we observed an increase in the number of breaks, and these breaks cannot persist in the genome unrepaired, another mechanism must be used to deal with this increased damage. As mentioned, NHEJ is known as an error-prone repair mechanism that creates point mutations and deletions/insertions.
To analyze the contribution of NHEJ to strand break repair, we measured the level of point mutations (Fig. 1) in plants of different ages. The data revealed a near constant level of point mutations in plants of all groups. From 2 to 31 dpg, the mutation frequency increased by a factor of 180, whereas number of genomes increased by a factor of 100 (Fig. 6). This resulted in a slightly elevated, but statistically insignificant (except for 16 and 31 dpg), increase in mutation rate (Supplemental Table III). This experiment showed us that NHEJ repair activity either increases or remains constant as plants mature.
Figure 6.
Mutation frequency, number of genomes, and mutation rate at different plant ages. Plants of line number 166.1 were used. The y axis shows fold difference in mutation frequency (MF), genome number (Genomes), and mutation rate (MR) in different plant ages as compared to 2 dpg. The x axis shows days post germination. Detailed data on MF, genome number, and MR is presented in Supplemental Table II.
The Activity of Ku70 and Rad51, Major HR and NHEJ Repair Genes
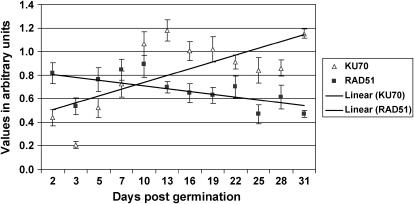
The previous experiments showed that while RR decreases with plant age, the mutation rate increases. It was important to support these findings with an analysis of the expression of key proteins involved in both repair pathways. We analyzed the steady-state mRNA levels for Ku70 and Rad51 genes involved in NHEJ and HR repair, respectively. Real-time PCR (RT-PCR) data revealed a 1.6-fold decrease in the steady-state mRNA level of Rad51 and a 3.0-fold increase in the steady-state mRNA level of Ku70 between 2 and 31 dpg (Fig. 7). Three independent experiments revealed this statistically significant trend of changing expression patterns in response to aging of the plant (Fig. 7). To confirm the mRNA expression values with protein data, we performed western-blot analyses using anti-Ku70 antibodies (Fig. 8). Three independent western blots carried out with tissue from plants of the aforementioned ages (5–50 plants per age group) showed a substantial increase in Ku70 protein levels as the plants aged (Fig. 8B). Plotting the data from the RT-PCR and western-blot analyses of Ku70 together with the strand break levels at different plant ages revealed nearly identical trend lines (Supplemental Fig. 1). These experiments confirmed that the decrease in RR and the increase in mutation rate are at least in part due to the change in the expression of SSB and DSB repair genes.
Figure 7.
Ku70 and Rad51 steady-state mRNA levels. The data for RT-PCR analysis of Ku70 and Rad51 was standardized to the actin activity and expressed in arbitrary units (y axis). The x axis shows the plant age (days post germination). Bars represent se as calculated from three independent experiments. The trend lines show the increase in the amount of Ku70 and the decrease in the amount of Rad51 RNA.
Figure 8.
Western-blot analysis of Ku70 protein. A, The top section shows the representative gel. The bottom section shows the loading control. B, The average band intensity (in arbitrary units) was calculated from three independent western blots. Bars show the se.
DISCUSSION
The repair of DBSs is an important cell task that is performed by two competing mechanisms, HR and NHEJ. In this study, we showed that there appears to be a developmental control over the rate of involvement of these mechanisms. Specifically, we showed that between the ages of 2 and 31 dpg (1) RR decreased 3-fold, (2) mutation rate slightly increased, (3) strand breaks increased by 220%, and (4) Rad51 activity dropped, whereas Ku70 activity increased. Our data suggest that there exists a developmental control over the involvement of HR and NHEJ in DSB repair, whereby the more mature the plant, the lower the contribution of HR.
The question remained of whether recombination was occurring randomly over the growth period of Arabidopsis, and whether it was directly proportional to cell division or genome replication. The data reported in this study shows that the occurrence of HR is nonrandom and does not directly depend on the number of genomes present in a plant. Recombination events were more frequent in early developmental stages and became less frequent as the number of genomes present in the plant increased.
After a certain developmental stage, cell divisions decrease drastically while genome duplications may continue to occur. This endoreduplication can continue throughout the entire life cycle of the plant (Galbraith et al., 1991), greatly increasing the number of genomes per cell while cell and nuclear sizes persist. This increase in ploidy directly results in an increase of gene copies; therefore, one would expect a proportional increase in the number of recombination events reflecting the proportional increase in DNA breaks. For example, if mutation rates were held constant, it would be expected that the number of mutation events would double with an increase in ploidy from two to four genomes. This would inevitably increase the number of gene rearrangements, duplications, and reshuffling events resulting from DNA repair malfunction. Although it is unlikely that any highly endoreduplicated cells will give rise to a new generation, such an increase in gene rearrangements seems unnecessary and, perhaps, deleterious. In this respect, it is sensible that plant cells possess a specialized mechanism for controlling recombination in highly endoreduplicated tissue. The question of whether genome fidelity functions as efficiently during endoreduplication as during DNA replication in dividing cells is now answered (Hays, 2002).
SSB and DSB Levels
The decrease in RR we experienced could primarily be due to a lower level of breaks that occurs in older plants. In other words, the 3-fold reduction in RR between 2 and 31 dpg could be explained by a similar decrease in the number of strand breaks per DNA unit. However, such a scenario would be highly unlikely. In most cases, DNA damage is a random process and, thus, should remain approximately constant when related to a haploid genome. That is, the more copies of the genome in the nucleus, the greater the amount of DNA damage. Furthermore, the rate of break occurrence (frequency of occurrence related to the number of replication events) should also remain more or less constant. Another contribution to the increase in strand breaks in older cells could be the increasing contribution of apoptosis and DNA fragmentation-related breaks in aging cells (Yoshida, 2003; Huang et al., 2005). Measurement of breaks in plants of different age groups supported our prediction, as the rate of breaks did not decrease with plant maturity (Fig. 5). In actuality, we experienced a 220% increase in SSBs and DSBs from 2 to 31 dpg, indicating that there must be another factor contributing to the decrease in RR.
The method we used for strand break analysis is based on the detection of strand breaks with retained 3′OHs (Basnakian and James, 1996; James et al., 2003). The number of lesions, including direct breaks of the phosphodiester backbone and converted excision repair intermediates, as well as those associated with bypass repair at the replication fork, could contribute to the formation of a strand break. Importantly, all of the aforementioned lesions could potentially be repaired by either NHEJ or HR.
It has been suggested that an efficient method for detection of strand breaks is the Comet assay (Rundell et al., 2003; Collins, 2004). This method is based on the detection of gel shifts between damaged and nondamaged DNA due to a difference in supercoiling and relies on software-based measurements of the tail length of relaxed DNA molecules (Rundell et al., 2003; Collins, 2004). This method is very sensitive when used under alkaline conditions and detects SSBs, DSBs, and other lesions that relax DNA (Cotelle and Ferard, 1999; Collins, 2004). To be specific to DSBs, this method should be used under a neutral pH, where the sensitivity of this method drops significantly (Rundell et al., 2003; Collins, 2004). However, several papers suggest the existence of false-positive results in apoptotic cells, potentially confounding the results of an experiment such as ours that focuses on the developmental stages of plants (Choucroun et al., 2001; Czene et al., 2002). Since the method that we used is based on the direct incorporation of radionucleotides into 3′OH ends (see “Materials and Methods”), we warranted it more appropriate than the Comet assay for our experiments (Rundell et al., 2003; Collins, 2004).
The Role of Ku70 and Rad51
According to Ray and Langer (2002), NHEJ and HR compete for available DNA ends generated at break sites. This competition is mirrored at the molecular level by an equilibrium between Rad52 (HR) and the Ku70-Ku80 dimer (NHEJ) in animals. If Rad52 is not available, a Ku70-Ku80 complex binds to the DNA ends at the break site and triggers a mitotic checkpoint arrest by signaling through a DNA-dependent protein kinase. Binding of the Ku70-Ku80 complex allows the recruitment of DNA ligase IV (Dn14) and its accessory factor, Lif1 (XRCC4). In this instance, the DNA ends are joined via NHEJ (Ray and Langer, 2002). To date, no Rad52 homolog has been found in plants, which could be one of the explanations for low levels of HR in flowering plants. However, an Arabidopsis homolog of Rad51, AtRad51, has been identified (Doutriaux et al., 1998).
It remains unclear what, if any, are the key regulatory elements that direct which mechanism will be used for break repair. It is possible that the availability of key proteins, such as Ku70 and Rad51, at the time of DBS repair is one of these mechanisms. Interestingly, the level of the Ku70-Ku80 complex is much lower in meiotic mice cells when compared to somatic cells (Goedecke et al., 1999). This implies that HR acts preferentially when the levels of NHEJ proteins are low. Reduced levels of Lif1 expression in meiosis-competent yeast (Saccharomyces cerevisiae) cells suggests that NHEJ is low when HR is high (Valencia et al., 2001).
The fact that we have found lower RRs in mature plants with an increasing number of strand breaks suggests that another mechanism, perhaps NHEJ, took over DSB repair. Here we showed that the steady-state mRNA level of a key NHEJ repair protein, Ku70, tripled between 2 and 31 dpg (Fig. 7). These changes in steady-state mRNA expression were echoed by changes observed on the protein level using anti-Ku70 antibodies, whereby a steady increase in the amount of protein was observed (Fig. 8). Concurrently, the mRNA level of the HR repair protein, Rad51, decreased by 1.6-fold (Fig. 7). These results suggest that NHEJ is indeed compensating for the decrease in HR.
Increased Mutation Rate
To support the aforementioned results, we had to show that there was an increase in the outcome of NHEJ repair. It is known that NHEJ repair is an error-prone mechanism that frequently results in various types of point mutations, deletions, and insertions (Pfeiffer, 1998; Kirik et al., 2000; Kovalchuk et al., 2004). Previously, we generated a transgenic Arabidopsis line that could monitor the level of point mutations in the plant genome (Kovalchuk et al., 2000). This line was successfully used in a number of mutagenesis assays, as well as in the analysis of the contribution of DSB repair to the generation of point mutations (Kovalchuk et al., 2004; Ilnytskyy et al., 2005). The experiments in this study showed that point mutation rates increased slightly, although insignificantly, throughout the period between 2 and 31 dpg. To support our findings, it was sufficient to show that the point mutation rate did not change; nevertheless, it seems unusual that the mutation rate did not increase dramatically. However, as we measured only one of the possible NHEJ repair mistakes, it was difficult to estimate the real contribution of NHEJ, as deletions and insertions of various sizes could account for a number of errors. Moreover, NHEJ is not the only mechanism that could have potentially contributed to the reversion events, as mistakes in mismatch repair, replication bypass, base- and nucleotide-excision repair can also contribute to the appearance of point mutations.
Regulation of HR/NHEJ in Animal Cells
Information about the contribution of either HR or NHEJ to the strand break repair in different developmental stages in plants is scarce. In contrast, a substantial body of information on DSB repair in mammalian cells has accumulated. Pierce et al. (2001) showed that the loss of most HR factors leads to early or mid-embryonic lethality in mice, suggesting an essential role for HR in development. Loss of NHEJ factors, however, results in late embryonic death for only particular factors, XRCC4 and DNA ligase IV, but not for others, Ku80, Ku70, and DNA-PKcs (summarized in Couëdel et al., 2004). This suggests that NHEJ does not play as important a role in development as does HR, and that the factors critical to embryo development in NHEJ may have alternative developmental functions.
Several studies have shown that the number of DSBs increase in the tissues of old mice (Singh et al., 2001; Sedelnikova et al., 2004). This can directly lead to the increased accumulation of various mutations. For example, an analysis of the mutation spectra in mice of different ages found that genomic rearrangements may occur as a result of NHEJ (Dolle et al., 1997; Vijg and Dolle, 2002). These results can be interpreted as the NHEJ repair pathway being responsible for age-related genomic instability. However, regions of extended homology were not found at these breakpoints, suggesting that rearrangements may have resulted from mistakes in NHEJ repair (Dolle et al., 1997; Vijg and Dolle, 2002). These results suggest that as postmitotic and senescent cells accumulate, the mode of repair will shift from HR toward NHEJ (Gorbunova and Seluanov, 2005). In agreement with this hypothesis is the observation that HR is more efficient in embryonic than in adult cells (Arbones et al., 1994).
CONCLUSION
HR is a complex and versatile process of DNA repair. This research contributes to the understanding of how HR mechanisms are regulated during plant maturity. The finding that HR is suppressed in highly endoreduplicated cells is the most intriguing, as it demonstrates the developmental regulation of processes involved in DNA repair and gene rearrangements. Speculation as to why HR is down-regulated with plant maturity could lead one to believe that HR in mature cells with increased ploidy have a deleterious effect; and/or HR in mature cells is not an efficient mechanism for dealing with strand breaks, as recombination events are less likely to be passed on to the next generation.
MATERIALS AND METHODS
Plant Growth and Sampling
Plants of two transgenic lines (cv C24, line no. 11 and cv Columbia, line no. 166_4) were germinated and grown on soil at 22°C with a 16/8 d/night light regime, and illumination at 100 μm m−2 s−1. The recombination substrate consisted of two truncated, nonfunctional, overlapping copies of the uidA gene (Swoboda et al., 1994), whereas the mutation substrate consisted of a single copy of a stop codon-inactivated uidA gene (Kovalchuk et al., 2000; Fig. 1). Twelve groups of plants of each line were germinated on soil and sampled for histochemical staining and DNA extraction at 2, 3, 5, 7, 10, 13, 16, 19, 22, 25, 28, and 31 dpg.
The Generation of Plants Carrying the LUC Recombination Substrate
The construction of the LUC recombination substrate was described previously (Gorbunova et al., 2000). It consisted of two nonfunctional, overlapping copies of the LUC transgene cloned in an inverted orientation. The recombination events can be visualized as bright sectors on a dark background with the aid of a LUC CCD camera. The number 15D8 LUC line used in the experiments carried a single-transgene copy.
Histochemical Staining Procedure
Histochemical staining, as described by Jefferson (1987), was done with plants at the aforementioned developmental stages. For destructive staining, plants were vacuum infiltrated for 10 min in a sterile staining buffer containing 100 mg of 5-bromo-4-chloro-3-indolyl glucuronide substrate (Jersey Labs) in 300 mL 100 mm phosphate buffer (pH 7.0), 0.05% NaN3, 0.05% Tween 80, and 1 mL dimethylformamide. Plants were then incubated at 37°C for 48 h and subsequently bleached with ethanol (Fig. 1).
Detection of LUC Recombination Events
The recombination events were visualized as bright sectors on a dark background in the LUC CCD camera (Fig. 1C; Gloor Instruments AG) 1 to 2 h after the cleavage substrate luciferine was applied.
Calculation of Number of Genomes
Total DNA of the respective transgenic lines was isolated from whole plants at the full rosette stage or at the different development stages using a Nucleon phytopure plant DNA extraction kit (Amersham Life Science). The yield of total DNA (micrograms/plant) was compared with the DNA content (0.16 pg) of an Arabidopsis (Arabidopsis thaliana) cell, to give an estimate of the number of genomes present (Swoboda et al., 1993). The DNA was prepared from 12 groups of plants sampled at different ages (4–60 plants per group). The average DNA content was used to estimate the number of genomes present.
To find out whether the DNA extraction method had a significant influence on the yield, we prepared DNA using another protocol (Boyko et al., 2005). Although the DNA yield was somewhat different (about 50% higher than from Nucleon Phytopure kit), the ratio between the amounts of DNA in plants of different ages was the same.
RT-PCR Analysis of Gene Expression
For real-time expression, all plant lines were grown as previously mentioned. These plants were harvested and frozen in liquid nitrogen at different developmental stages. Two independent RNA samples per each treatment group (20 plants per sample on average) were prepared using Trizol reagent from Invitrogen. Reverse transcriptase PCR (You-Prime-First-Strand, ready to go PCR beads, Amersham) was carried out on all samples providing a transcriptome copy for each of the mutant lines. RT-PCR was performed in a total volume of 25 μL using 1 μL of the first-strand cDNA synthesis mixture as a template, 300 nm forward primer, 300 nm reverse primer, and 12.5 μL of 2xSYBRGreen PCR Master Mix (Applied Biosystems). The duplicate reactions were carried out with the 1:3 and 1:15 dilutions of the first-strand cDNA synthesis mixture. A SmartCycler (Cepheid) was used to perform the PCR cycles and fluorescence was quantified against standards. The cDNAs were amplified under the following conditions: (1) 95°C for 5 min for one cycle; (2) 94°C for 30 s, 57°C to 62°C (depending on the primers used) for 30 s, 72°C for 1 min for 30 cycles; and (3) 72°C for 10 min for one cycle. The melting temperatures were estimated for every gene product. The standards for the expression of each gene were amplified from the cDNA of following dilutions: 1 μL, 1:4, 1:20, and 1:100. For the RT-PCR analysis, the following primers were used: Ku70 forward 5′-AGACCTAATTCCTCAGCAACC-3′, reverse 5′-TATCAAATATAGGGAACTCTGC-3′; Rad51 forward 5′-TTGTGTTGTGACGACAAGC-3′, reverse 5′-ATCAATCTGCTCAAGAACACC-3′; and AtActin-1 (internal control) forward 5′-TGGACAAGTCATAACCATCGGAGC-3′, reverse 5′-TGTGAACAATCGATGGACCTGAC-3′. An average of four reactions (two dilutions per each of two RNA preparations) was obtained and the fold induction was calculated. The experiment was repeated three times, and the statistical significance of the experiment was confirmed by performing the Student's t test (two-tailed paired or nonpaired).
Western Immunoblotting
Western immunoblotting for AtKu70 was conducted using plants (5–50 plants per sample, on average) of different ages ground in 0.4 to 0.6 mL of ice-cold protein extraction buffer (100 mm NaHPO4 pH 8.0, 0.1% TritonX-100, 20% glycerol) supplemented with Complete protease inhibitor (Roche). Homogenates were spun for 1 h at 1,600g at 4°C. Supernatant was spun the second time at the aforementioned conditions, and collected again, aliquoted, and stored at −80°C. Extracts were boiled for 3 min in 0.6 mL of hot 1% SDS. Small aliquots (10 μL) of homogenate were reserved for protein determination using protein assay reagents from Bio-Rad. Equal amounts of proteins (20 μg) were separated by SDS-PAGE in slab gels of 12% polyacrylamide, made in duplicates, and transferred to polyvinylidene difluoride membranes. Membranes were incubated with AtKu70 antibodies (1:500, Santa Cruz Biotechnology). Antibody binding was revealed by incubation with anti-goat secondary antibodies (1:5,000, Santa Cruz Biotechnology) and the ECL Plus immunoblotting detection system (Amersham). Chemiluminescence was detected by Biomax MR films (Kodak). Polyvinylidene difluoride membranes were stained with Coomassie Blue (Bio-Rad), scanned, and the intensity of the Mr-40,000 protein band was assessed as a loading control. Signals were quantified using NIH Image 1.43 software and normalized to the Mr-40,000 protein. The experiment was repeated three times.
Strand Break Measurement (ROPS Assay)
Quantification of 3′OH DNA breaks was performed using the ROPS assay (Basnakian and James, 1996). This assay is based on the ability of the Klenow fragment polymerase (New England Biolabs) to initiate ROPS from the reannealed 3′OH ends of single-stranded DNA. After a denaturation-reassociation step, the ssDNA serves as its own primer by randomly reassociating itself to other ssDNA molecules. Under strictly defined reaction conditions, the incorporation of [3H]-dCTP into newly synthesized DNA will be proportional to the initial number of 3′OH ends (breaks). A 1-μg aliquot of plant DNA just prior to ROPS reaction was denatured at 100°C for 5 min and then cooled on ice. The reaction mixture for one sample contained 1 μg heat denatured DNA, 2.5 μL 0.5 mm 3dNTPs (dGTP, dATP, and dTTP mix), 2.5 μL 10× Klenow fragment buffer, 0.45 μL 33 μm dCTP, 5 units Klenow enzyme, and 0.5 μL [3H]-dCTP. Reaction volume was adjusted to 25 μL with distilled sterile water. After incubation at 25°C for 30 min, the reaction was stopped by the addition of an equal volume of 25 mm EDTA pH 8.0. Following this, the 50 μL reaction volume of each sample was aliquoted to three 25-mm DE-81 ion-exchanging filter papers (Whatman), washed with 500 mm sodium phosphate buffer (pH 7.0) for 10 min, and repeated three times. Subsequently, filters were thoroughly dried and transferred to a vial containing 5 mL of scintillation cocktail. Radiation levels (3H decays per minute) were detected in a scintillation counter (Beckman LS 5000CE).
Supplementary Material
Acknowledgments
We would like to thank Chris Picken for critical reading of the manuscript. We acknowledge the Natural Sciences and Engineering Research Council of Canada and Alberta Heritage for Science and Engineering grants for I.K.
This work was supported by the Natural Sciences and Engineering Research Council of Canada (Establishment Grant to I.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Igor Kovalchuk (igor.kovalchuk@uleth.ca).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.074658.
References
- Arbones ML, Austin HA, Capon DJ, Greenburg G (1994) Gene targeting in normal somatic cells: inactivation of the interferon-gamma receptor in myoblasts. Nat Genet 6: 90–97 [DOI] [PubMed] [Google Scholar]
- Basnakian AG, James SJ (1996) Quantification of 3'OH DNA breaks by random oligonucleotide-primed synthesis (ROPS) assay. DNA Cell Biol 15: 255–262 [DOI] [PubMed] [Google Scholar]
- Boyko A, Filkowski J, Kovalchuk I (2005) Homologous recombination in plants is temperature and day length dependent. Mutat Res 572: 73–83 [DOI] [PubMed] [Google Scholar]
- Brennan RJ, Schiestl R (1998) Free radicals generated in yeast by the Salmonella test-negative carcinogens benzene, urethane, thiourea and auramine O. Mutat Res 403: 65–73 [DOI] [PubMed] [Google Scholar]
- Choucroun O, Gillet D, Dorange G, Sawicki B, Dewitte JD (2001) Comet assay and early apoptosis. Mutat Res 478: 89–96 [DOI] [PubMed] [Google Scholar]
- Collins AR (2004) The comet assay for DNA damage and repair: principles, applications, and limitations. Mol Biotechnol 26: 249–261 [DOI] [PubMed] [Google Scholar]
- Cotelle S, Ferard JF (1999) Comet assay in genetic ecotoxicology: a review. Environ Mol Mutagen 34: 246–255 [PubMed] [Google Scholar]
- Couëdel C, Mills KD, Barchi M, Shen L, Olshen A, Johnson RD, Nussenzweig A, Essers J, Kanaar R, Li GC, et al (2004) Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev 18: 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchlow S, Jackson S (1998) DNA end-joining: from yeast to man. Trends Biochem Sci 23: 394–402 [DOI] [PubMed] [Google Scholar]
- Cromie GA, Connelly JC, Leach DR (2001) Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell 8: 1163–1174 [DOI] [PubMed] [Google Scholar]
- Czene S, Testa E, Nygren J, Belyaev I, Harms-Ringdahl M (2002) DNA fragmentation and morphological changes in apoptotic human lymphocytes. Biochem Biophys Res Commun 294: 872–878 [DOI] [PubMed] [Google Scholar]
- Dolle ME, Giese H, Hopkins CL, Martus HJ, Hausdorff JM, Vijg J (1997) Rapid accumulation of genome rearrangements in liver but not in brain of old mice. Nat Genet 17: 431–434 [DOI] [PubMed] [Google Scholar]
- Doutriaux M, Couteau F, Bergounioux C, White C (1998) Isolation and characterization of the Rad51 and DMC1 homologs from Arabidopsis thaliana. Mol Gen Genet 257: 283–291 [DOI] [PubMed] [Google Scholar]
- Draper CK, Hays JB (2000) Replication of chloroplast, mitochondrial, and nuclear DNA during growth of unirradiated and UVB-irradiated Arabidopsis leaves. Plant J 23: 255–265 [DOI] [PubMed] [Google Scholar]
- Essers J, van Steeg H, de Wit J, Swagemakers S, Vermeij M, Hoeijmakers J, Kanaar R (2000) Homologous and nonhomologous recombination differentially affect DNA damage repair in mice. EMBO J 19: 1703–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filkowski J, Kovalchuk O, Kovalchuk I (2004) Genome stability of vtc1, tt4 and tt5 Arabidopsis thaliana mutants impaired in protection against oxidative stress. Plant J 38: 60–69 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Knapp S (1991) Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol 96: 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedecke W, Eijpe M, Offenberg M, van Aalderen M, Heyting C (1999) Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet 23: 194–198 [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Avivi-Ragolski N, Shalev G, Kovalchuk I, Abbo S, Hohn B, Levy A (2000) A new hyperrecombinogenic mutant of Nicotiana tabacum. Plant J 24: 601–611 [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Levy AA (1997) Non-homologous DNA end-joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res 25: 4650–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova V, Seluanov A (2005) Making ends meet in old age: DSB repair and aging. Mech Ageing Dev 126: 621–628 [DOI] [PubMed] [Google Scholar]
- Hays JB (2002) Arabidopsis thaliana, a versatile model system for study of eukaryotic genome-maintenance functions. DNA Repair (Amst) 1: 579–600 [DOI] [PubMed] [Google Scholar]
- Huang X, Halicka HD, Traganos F, Tanaka T, Kurose A, Darzynkiewicz Z (2005) Cytometric assessment of DNA damage in relation to cell cycle phase and apoptosis. Cell Prolif 38: 223–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Takagi M, Tosa Y, Mayama S (2001) Heat shock, copper sulfate and oxidative stress activate the retrotransposon MAGGY resident in the plant pathogenic fungus Magnaporthe grisea. Mol Genet Genomics 266: 318–325 [DOI] [PubMed] [Google Scholar]
- Ilnytskyy Y, Yao Y, Kovalchuk I (2005) Double-strand break repair machinery is sensitive to UV radiation. J Mol Biol 345: 707–715 [DOI] [PubMed] [Google Scholar]
- James SJ, Pogribny IP, Pogribna M, Miller BJ, Jernigan S, Melnyk S (2003) Mechanisms of DNA damage, DNA hypomethylation, and tumor progression in the folate/methyl-deficient rat model of hepatocarcinogenesis. J Nutr 133: 3740S–3747S [DOI] [PubMed] [Google Scholar]
- Jefferson R (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Report 5: 387–405 [Google Scholar]
- Jeggo PA (1998) DNA breakage and repair. Adv Genet 38: 185–218 [DOI] [PubMed] [Google Scholar]
- Karanjawala ZE, Murphy N, Hinton DR, Hsieh CL, Lieber MR (2002) Oxygen metabolism causes chromosome breaks and is associated with the neuronal apoptosis observed in DNA double-strand break repair mutants. Curr Biol 12: 397–402 [DOI] [PubMed] [Google Scholar]
- Kirik A, Salomon S, Puchta H (2000) Species-specific double-strand break repair and genome evolution in plants. EMBO J 19: 5562–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk I, Kovalchuk O, Hohn B (2000) Genome-wide variation of the somatic mutation frequency in transgenic plants. EMBO J 19: 4431–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk I, Kovalchuk O, Kalck V, Boyko V, Filkowski J, Heinlein M, Hohn B (2003) Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 423: 760–762 [DOI] [PubMed] [Google Scholar]
- Kovalchuk I, Pelczar P, Kovalchuk O (2004) High frequency of nucleotide misincorporations upon the processing of the double-strand breaks. DNA Repair (Amst) 3: 217–223 [DOI] [PubMed] [Google Scholar]
- Kunz BA, Anderson HJ, Osmond MJ, Vonarx EJ (2005) Components of nucleotide excision repair and DNA damage tolerance in Arabidopsis thaliana. Environ Mol Mutagen 45: 115–127 [DOI] [PubMed] [Google Scholar]
- Liang F, Han M, Romanienko P, Jasin M (1998) Homology-directed repair is major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA 95: 5172–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelczar P, Kalck V, Kovalchuk I (2003) Different genome maintenance strategies in human and tobacco cells. J Mol Biol 331: 771–779 [DOI] [PubMed] [Google Scholar]
- Pfeiffer P (1998) The mutagenic potential of DNA double-strand break repair. Toxicol Lett 96–97: 119–129 [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M, Jasin M (2001) Double-strand breaks and tumorigenesis. Trends Cell Biol 11: S52–S59 [DOI] [PubMed] [Google Scholar]
- Puchta H, Swoboda P, Hohn B (1995) Induction of homologous DNA recombination in whole plants. Plant J 7: 203–210 [Google Scholar]
- Ray A, Langer M (2002) Homologous recombination: ends as the means. Trends Plant Sci 7: 435–440 [DOI] [PubMed] [Google Scholar]
- Ries G, Heller W, Puchta H, Sandermann H, Seidlitz HK, Hohn B (2000) Elevated UV-B radiation reduces genome stability in plants. Nature 406: 98–101 [DOI] [PubMed] [Google Scholar]
- Roth D, Wilson J (1986) Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol 6: 4295–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundell MS, Wagner ED, Plewa MJ (2003) The Comer assay: genotoxic damage or nuclear fragmentation? Environ Mol Mutagen 42: 61–67 [DOI] [PubMed] [Google Scholar]
- Sargent R, Brenneman M, Wilson J (1997) Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol 17: 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC (2004) Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol 6: 168–170 [DOI] [PubMed] [Google Scholar]
- Singh NP, Ogburn CE, Wolf NS, van Belle G, Martin GM (2001) DNA double-strand breaks in mouse kidney cells with age. Biogerontology 2: 261–270 [DOI] [PubMed] [Google Scholar]
- Smith J, Baldeyron C, De Oliveira I, Sala-Trepat M, Papadopoulo D (2001) The influence of DNA double-strand break structure on end-joining in human cells. Nucleic Acids Res 29: 4783–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda P, Gal S, Hohn B, Puchta H (1994) Intrachromosomal homologous recombination in whole plants. EMBO J 13: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda P, Hohn B, Gal S (1993) Somatic homologous recombination in planta: the recombination frequency is dependent on the allelic state of recombining sequences and may be influenced by genomic position effects. Mol Gen Genet 237: 33–40 [DOI] [PubMed] [Google Scholar]
- Tuteja N, Singh MB, Misra MK, Bhalla PL, Tuteja R (2001) Molecular mechanisms of DNA damage and repair: progress in plants. Crit Rev Biochem Mol Biol 36: 337–397 [DOI] [PubMed] [Google Scholar]
- Valencia M, Bentele M, Vaze M, Herrmann G, Kraus E, Lee S, Schar P, Haber J (2001) NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 414: 666–669 [DOI] [PubMed] [Google Scholar]
- Vijg J, Dolle ME (2002) Large genome rearrangements as a primary cause of aging. Mech Ageing Dev 123: 907–915 [DOI] [PubMed] [Google Scholar]
- Yoshida S (2003) Molecular regulation of leaf senescence. Curr Opin Plant Biol 6: 79–84 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.