Abstract
Shoots can be regenerated from Arabidopsis (Arabidopsis thaliana) root explants in tissue culture through a two-step process requiring preincubation on an auxin-rich callus induction medium. Regenerating tissues can be directed along different developmental pathways leading to the formation of shoots, new roots, or callus by transferring to the appropriate organ induction medium. Using gene-profiling methods, we identified groups of genes that serve as molecular signatures of the different developmental processes, i.e. genes that were specifically up- or down-regulated on one developmental pathway, but not on others. One transcription factor gene that was up-regulated during early shoot development was RAP2.6L (At5g13330), a member of the ERF (ethylene response factor) subfamily B-4 of the ERF/APETALA2 transcription factor gene family. RAP2.6L functions in shoot regeneration because T-DNA knockdown mutations in the gene reduced the efficiency of shoot formation in tissue culture, but not normal embryo or seedling development. RAP2.6L promoter:β-glucuronidase fusions demonstrated that the up-regulation of the gene during shoot regeneration was, at least in part, transcriptionally controlled. The promoter:β-glucuronidase fusions also demonstrated that RAP2.6L expression was localized to the shoot and emerging leaves, but expression declined in the leaf lamina as leaves expanded. T-DNA knockdown mutations in RAP2.6L reduced the expression of many genes that are normally up-regulated during shoot development including CUP-SHAPED COTYLEDON2 that is involved in shoot meristem specification. Thus, RAP2.6L appears to be part of a network involved in regulating the expression of many other genes in shoot regeneration.
Nearly a half century ago, Skoog and Miller (1957) showed that the developmental fate of regenerating tobacco (Nicotiana tabacum) pith tissue in culture could be directed by the plant hormones cytokinin and auxin. Shoots were produced at high concentrations of cytokinin relative to auxin, while roots were formed when the ratios were reversed. Undifferentiated tissue or callus formed at hormone concentrations that were optimal for callus growth, but not for shoot or root formation. The ability to direct the course of development by two simple plant hormones has intrigued plant biologists for years.
Much has been learned in the past few years about cytokinin and auxin signaling, but less is known about the developmental events downstream. Cytokinin signal transduction involves a multicomponent phosphorelay signaling system (Imamura et al., 1999; Hutchison and Kieber, 2002; Hwang et al., 2002; Oka et al., 2002; Sheen, 2002) in which sensory His kinases (HKs) such as AHK2, AHK3, and CRE1/AHK4 in Arabidopsis (Arabidopsis thaliana) serve as cytokinin receptors (Inoue et al., 2001; Schmulling, 2001; Suzuki et al., 2001; Yamada et al., 2001; Oka et al., 2002). The cytokinin signal is transduced to the nucleus via His phosphotransfer proteins (HPts or AHPs), which belong to a family of six genes in Arabidopsis (Hwang and Sheen, 2001; Hwang et al., 2002; Suzuki et al., 2002). The signal is relayed to two types of gene expression regulators, A- and B-type response regulators (ARRs; Imamura et al., 1999; Hutchison and Kieber, 2002; Hwang et al., 2002). B-type ARRs have DNA binding and transcriptional activator domains, while A types do not (Sakai et al., 1998). Cytokinin signaling is thought to activate gene expression through the action of B-type ARRs, which constitute a family of 11 members in Arabidopsis (Sakai et al., 2000; Hwang and Sheen, 2001; Hutchison and Kieber, 2002; Hwang et al., 2002). Members of the B-1 subfamily of B-type ARRs activate some of the genes encoding A-type ARRs (Hwang and Sheen, 2001; Sakai et al., 2001). There are 10 to 12 genes encoding A-type ARRs in Arabidopsis (Sakai et al., 2000; Hwang and Sheen, 2001; Hutchison and Kieber, 2002; Hwang et al., 2002; Mason et al., 2004) and some are negative feedback regulators of cytokinin responses (To et al., 2004; Leibfried et al., 2005). Other targets of B-type ARR action are being sought and some may include genes that are rapidly up-regulated by cytokinin (Rashotte et al., 2003; Brenner et al., 2005).
In this report, we used gene expression profiling to highlight genes that are specifically up- or down-regulated during the regeneration of shoots, roots, or calli from root explants with the goal of identifying molecular signatures for these developmental processes. In Arabidopsis, shoots are typically regenerated from root and/or hypocotyl explants by indirect organogenesis, which involves a period of callus formation prior to shoot induction (Valvekens et al., 1988). Explants are preincubated on an auxin-rich callus induction medium (CIM) and then are transferred to a cytokinin-rich shoot induction medium (SIM) for shoot formation. During CIM preincubation, root explants acquire competence to respond to shoot induction signals during subsequent incubation on SIM. What acquisition of competence is in cellular or molecular terms is not known. It is generally thought that preincubation on CIM is required to permit the dedifferentiation of tissues that will ultimately redifferentiate into organs (Gautheret, 1966; Hicks, 1980).
When CIM-preincubated root explants are transferred to a cytokinin-rich SIM, they first become committed to form shoots (will form shoots if transferred to basal medium) and then shoots emerge (Cary et al., 2002). Earlier gene expression profiling studies in Arabidopsis revealed progressive waves of gene expression changes involving hundreds of genes during shoot regeneration (Che et al., 2002). Such large-scale changes in the transcriptome during these developmental processes must involve the deployment of many transcription factors. The Arabidopsis genome encodes over 1,500 transcription factors (Riechmann et al., 2000). One of the large and diverse families of transcription factors in Arabidopsis is the APETALA2 (AP2)/ERBP family involved in many different developmental processes and environmental response events (Riechmann and Meyerowitz, 1998). The family is composed of 144 members in Arabidopsis and has been divided into five subfamilies: the AP-2 subfamily, RAV subfamily, DREB (A) subfamily, ERF (ethylene response factor; B) subfamily, and others (Sakuma et al., 2002). Some AP-2 subfamily members have been shown to affect shoot regeneration such as ENHANCER OF SHOOT REGENERATION1 (Banno et al., 2001).
In this study, we conducted a global analysis of gene expression during the acquisition of competence and during the regeneration of shoots, roots, and callus. In addition, we focused on the role of ERF/AP2 transcription factor RAP2.6L (a B-4 subfamily member), encoded by a gene that was specifically up-regulated during shoot regeneration. T-DNA knockdown mutations in RAP2.6L reduced the efficiency of shoot development and impacted the expression of shoot meristem-specifying genes. This analysis, therefore, allowed us to link an early responder in cytokinin signaling to events in shoot development.
RESULTS
Gene Expression Programs during CIM Preincubation
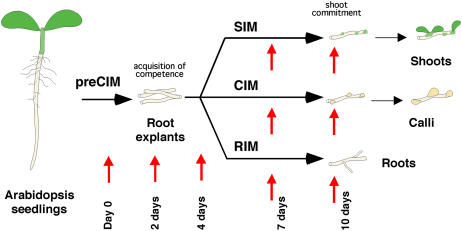
Shoots, callus, or roots can be regenerated from root explants in Arabidopsis tissue culture (Fig. 1). Shoots are regenerated in a two-step process whereby root explants are preincubated for a few days on an auxin-rich CIM (we refer to the preincubation period on CIM as CIM preincubation or preCIM to distinguish it from later incubation on CIM; Valvekens et al., 1988). During CIM preincubation, root explants acquire competence to respond to shoot induction signals when transferred to a cytokinin-rich SIM (Cary et al., 2002).
Figure 1.
Arabidopsis root explants regenerating in tissue culture form shoots, calli, or roots depending on culture conditions. Illustration shows that explants were preincubated on CIM for 4 d and then transferred to cytokinin-rich SIM, fresh CIM, or auxin-rich RIM. Red arrows show times during development when RNA samples were taken.
To gain a better understanding of the molecular events surrounding the acquisition of competence and the early developmental events in shoot, callus, and root development, gene expression patterns were profiled during CIM preincubation and SIM, CIM, and root induction medium (RIM) incubation (Fig. 1). Affymetrix Arabidopsis gene chips (ATH1) were used to profile gene expression in a randomized complete block design with two independent replications. In each replicated experiment, root explant samples were randomly collected for RNA extraction at each of nine time points: day 0, two time points during the preincubation period on CIM, and at two time points during incubation on SIM, RIM, or on further incubation on fresh CIM (Fig. 1). A standard ANOVA conducted for each gene indicated that thousands of genes exhibited some evidence of differential expression across the nine time points. Using the method of Storey and Tibshirani (2003), nearly half (10,700 out of 22,810) of the probe sets exhibited nonconstant expression profiles when controlling the false discovery rate (FDR) at the 0.01 level.
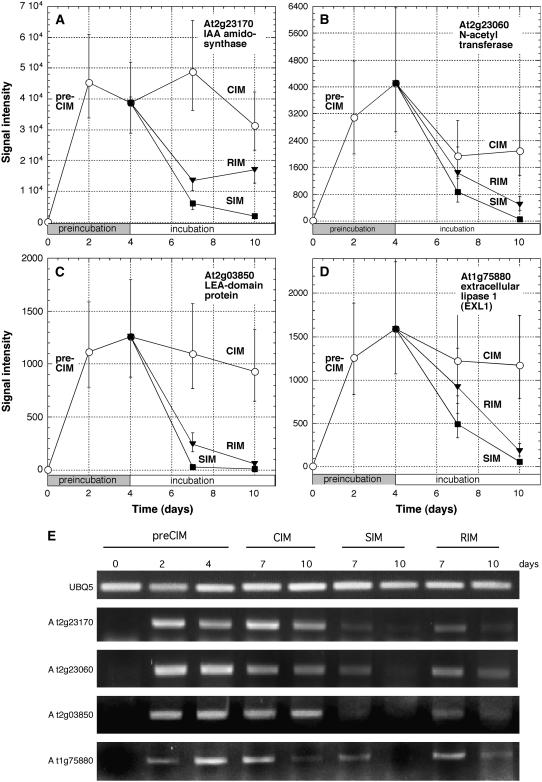
To identify genes that were up-regulated during CIM preincubation, we required that the estimated mean level of expression at 4 d preCIM be significantly greater than the estimated mean prior to preincubation (0 d preCIM) when controlling the FDR at the level of 0.02 using the method of Storey and Tibshirani (2003). Of the genes that met these criteria, we rank ordered them by fold change (FC) in expression at 4 d preCIM versus 0 time (Supplemental Table I). The genes topping this list (Table I) ranged from >200-fold to approximately 50-fold up-regulated at 4 d CIM, and the most highly up-regulated gene (At2g23170; Fig. 2A) and one further down the list (At2g14960) encode 3-indoleacetic acid (IAA)-amido synthases. The medium on which root segments are preincubated (CIM) is an auxin-rich medium, and IAA-amido synthases function in auxin homeostasis to congujate Asp and other amino acids to auxin (Staswick et al., 2005). The next most highly up-regulated gene encodes a GCN5-related N-acetyltransferase (GNAT; At2g23060; Fig. 2B). Some members of this gene family encode enzymes involved in histone acetylation, a chromatin modification that is thought to be critical to reprogram cells for different developmental functions (Jenuwein and Allis, 2001; Loidl, 2004). Others high on the list encode a late-embryogenesis abundant-domain protein (At2g03850; Fig. 2C) and an extracellular lipase (At1g75880; Fig. 2D). The expression patterns of these four genes were also confirmed by semiquantitative reverse transcriptase (RT)-PCR analysis (Fig. 2E). Two hundred and thirty genes were up-regulated more than 10-fold at 4 d preCIM, and when they were functionally categorized, it was found that genes encoding transcription factor activity were overrepresented by 2.3-fold in comparison to their frequency in the total genome (Table II). This suggests that considerable reprogramming of gene expression occurs during CIM preincubation.
Table I.
Genes most highly up- or down-regulated during CIM preincubation
Genes listed showed an estimated mean level of expression (from two independent replications) at 4 d CIM that was greater or less than the estimated mean prior to preincubation (0 d CIM) when controlling the FDR at the level of 0.02. The genes that met these criteria were rank ordered by FC in the comparison of expression at 4 d CIM versus 0 time, and the top 25 most highly up- and down-regulated genes are shown.
| Pub Locus ID | FCa | P Valuesa | q Valuesa | Descriptions |
|---|---|---|---|---|
| Up-regulated genes | ||||
| At2g23170 | 215.3 | 1.240E-06 | 2.320E-05 | IAA-amido synthase |
| At2g23060 | 215.0 | 2.300E-05 | 1.368E-04 | GNAT |
| At2g03850 | 188.5 | 6.380E-06 | 6.170E-05 | Late-embryogenesis abundant domain-containing protein |
| At1g75880 | 156.0 | 1.880E-05 | 1.198E-04 | Family II extracellular lipase 1 |
| At1g08430 | 139.7 | 1.040E-05 | 8.270E-05 | Expressed protein |
| At2g18660 | 117.3 | 2.380E-05 | 1.392E-04 | Expansin family protein |
| At3g60420 | 108.4 | 8.950E-06 | 7.600E-05 | Expressed protein |
| At5g40645 | 106.2 | 1.500E-06 | 2.570E-05 | Expressed protein |
| At2g38340 | 92.5 | 5.290E-05 | 2.442E-04 | ERF/AP2 transcription factor subfamily A-2 |
| At4g04490 | 87.8 | 1.320E-05 | 9.650E-05 | Putative receptor-like protein kinase |
| At3g22360 | 83.0 | 8.790E-06 | 7.560E-05 | Alternative oxidase 1b precursor |
| At1g59860 | 80.0 | 1.754E-04 | 5.674E-04 | 17.6-kD class I heat shock protein (HSP17.6A-CI) |
| At1g74110 | 76.0 | 3.980E-05 | 1.996E-04 | Cytochrome P-450 |
| At5g65510 | 72.4 | 2.670E-07 | 9.420E-06 | Similar to AP2/EREBP transcription factor BABY BOOM1 |
| At3g52780 | 69.4 | 8.580E-06 | 7.470E-05 | Protein Ser/Thr phosphatase |
| At2g14960 | 68.9 | 1.993E-04 | 6.205E-04 | IAA-amido synthase |
| At1g09310 | 67.7 | 2.596E-04 | 7.464E-04 | Expressed protein |
| At4g37770 | 63.8 | 3.234E-04 | 8.783E-04 | 1-Aminocyclopropane-1-carboxylate synthase-like protein |
| At2g38540 | 59.8 | 4.410E-09 | 1.510E-06 | Nonspecific lipid transfer protein |
| At2g29940 | 59.2 | 8.745E-04 | 1.847E-03 | Putative ABC transporter |
| At4g36260 | 56.0 | 1.110E-06 | 2.190E-05 | Zinc finger protein, similar to lateral root primordium 1 |
| At5g52390 | 54.2 | 7.730E-07 | 1.780E-05 | Photoassimilate-responsive protein |
| At3g60140 | 50.1 | 2.820E-05 | 1.570E-04 | β-Glucosidase-like protein |
| At1g74670 | 49.8 | 1.370E-05 | 9.860E-05 | GA-regulated protein 4 precursor |
| At3g10870 | 49.7 | 6.410E-05 | 2.780E-04 | Putative α-hydroxynitrile lyase |
| Down-regulated genes | ||||
| At5g67400 | 0.001 | 1.07E-05 | 8.39E-05 | Peroxidase 73 |
| At3g49960 | 0.001 | 2.51E-06 | 3.39E-05 | Peroxidase ATP21a |
| At3g01190 | 0.002 | 2.37E-06 | 3.27E-05 | Peroxidase 27 |
| At4g30170 | 0.002 | 8.27E-06 | 7.32E-05 | Peroxidase ATP8a |
| At5g49080 | 0.003 | 1.67E-04 | 5.45E-04 | Pro-rich extensin-like family protein |
| At3g53980 | 0.003 | 1.32E-05 | 9.65E-05 | Protease inhibitor/seed storage/lipid transfer protein |
| At3g62680 | 0.003 | 3.04E-07 | 9.98E-06 | Pro-rich family protein |
| At4g26010 | 0.003 | 2.14E-06 | 3.08E-05 | Peroxidase ATP13a |
| At5g57530 | 0.004 | 4.58E-06 | 4.93E-05 | Xyloglucan:xyloglucosyl transferase |
| At4g13770 | 0.004 | 2.16E-05 | 1.31E-04 | Cytochrome P450 |
| At5g60660 | 0.004 | 3.14E-06 | 3.92E-05 | Membrane intrinsic protein family protein |
| At5g53250 | 0.004 | 3.58E-08 | 3.66E-06 | Arabinogalactan protein |
| At4g28850 | 0.004 | 1.66E-04 | 5.44E-04 | Xyloglucan:xyloglucosyl transferase |
| At2g01520 | 0.005 | 1.55E-04 | 5.17E-04 | Major latex protein |
| At4g34580 | 0.005 | 3.69E-05 | 1.89E-04 | SEC14/phosphatidylinositol transfer-like protein IV |
| At1g05250 | 0.005 | 1.20E-06 | 2.27E-05 | Peroxidase ATP11a |
| At3g18170 | 0.005 | 1.21E-04 | 4.30E-04 | Expressed protein |
| At3g19710 | 0.005 | 6.17E-07 | 1.53E-05 | Branched-chain amino acid aminotransferase |
| At4g11290 | 0.006 | 3.55E-07 | 1.07E-05 | Peroxidase ATP19a |
| At5g35190 | 0.006 | 3.56E-04 | 9.44E-04 | Pro-rich extensin-like protein |
| At1g32450 | 0.006 | 4.38E-07 | 1.25E-05 | Proton-dependent oligopeptide transport protein |
| At3g45710 | 0.006 | 2.13E-06 | 3.08E-05 | Proton-dependent oligopeptide transport protein |
| At4g22080 | 0.007 | 6.30E-04 | 1.43E-03 | Pectate lyase |
| At5g38550 | 0.007 | 1.60E-04 | 5.28E-04 | Jacalin lectin family protein |
| At5g38930 | 0.007 | 3.55E-04 | 9.43E-04 | Germin-like protein |
For the comparison between 4 d CIM and 0 time.
Figure 2.
Expression profiles for genes highly up-regulated during preCIM. Explants were preincubated on CIM for 4 d and then transferred to SIM, RIM, or to fresh CIM. RNA was extracted at various times and subjected to Affymetrix DNA chip analysis. A to D, Expression profiles are shown of genes most highly up-regulated (greatest FC) on 4 d CIM versus 0 time. Data are from Supplemental Table I and Table I. E, Semiquantitative RT-PCR analysis of the expression profiles for the genes shown in A to D. UBIQUITIN5 (UBQ5) was used as a control.
Table II.
Functional categories of up- and down-regulated genes
Genes up- or down-regulated 10-fold or more during CIM preincubation were functionally categorized according to GO at The Arabidopsis Information Resource (fold up- or down-regulation was calculated from the estimated mean levels of expression at 4 d CIM compared to 0 d when controlling for a FDR at the level of 0.02). Genes specifically up- or down-regulated during shoot-, root-, or callus development on SIM, RIM, and CIM, respectively, by the criteria indicated in Supplemental Table I, were also assigned to functional categories. Fisher's exact test (Fisher, 1934) was used to identify categories which were significantly (P < 0.05) over- or underepresented among the identified genes relative to counts expected under simple random sampling from the entire genome. Bold text in the table corresponds to overrepresented categories while italic text corresponds to underrepresented categories. Although Fisher's exact test is commonly used for these types of analyses, see Allison et al. (2006) for criticisms of this approach and Barry et al. (2005) for an alternative strategy.
| Functional Category | PreCIM
|
SIM (Shoot)
|
RIM (Root)
|
CIM (Callus)
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Upa | Up | Downb | Down | Upb | Up | Downb | Down | Upc | Up | Downc | Down | Upd | Up | Downd | Down | Genomee | |
| % | Pvalues | % | P values | % | P values | % | P values | % | P values | % | P values | % | P values | % | P values | % | |
| Keyword category: Cellular component | |||||||||||||||||
| Cell wall | 3.9 | 6.50E-06 | 2.5 | 1.28E-05 | 1.0 | 1.93E-01 | 1.2 | 9.94E-02 | 0.6 | 6.44E-01 | 2.8 | 6.34E-07 | 1.1 | 1.51E-01 | 0.9 | 2.04E-01 | 0.7 |
| Cellular component unknown | 25.8 | 3.26E-01 | 20.1 | 1.10E-05 | 9.9 | 8.52E-24 | 24.1 | 4.02E-02 | 21.3 | 6.02E-05 | 18.5 | 1.71E-08 | 25.0 | 2.17E-01 | 25.8 | 2.16E-01 | 28.7 |
| Chloroplast | 6.0 | 1.61E-03 | 6.6 | 1.15E-05 | 28.3 | 1.32E-20 | 12.3 | 9.04E-01 | 8.8 | 4.71E-03 | 13.7 | 3.64E-01 | 9.2 | 1.15E-01 | 11.0 | 3.78E-01 | 12.6 |
| Cytosol | 0.9 | 6.77E-01 | 0.7 | 9.29E-01 | 0.8 | 8.39E-01 | 0.7 | 9.55E-01 | 0.8 | 8.53E-01 | 1.9 | 1.22E-02 | 1.8 | 1.29E-01 | 1.4 | 2.36E-01 | 0.9 |
| Endoplasmic reticulum | 0.9 | 1.16E-01 | 0.5 | 2.51E-01 | 0.0 | 3.18E-01 | 0.7 | 1.33E-01 | 0.5 | 3.43E-01 | 0.6 | 3.63E-01 | 0.0 | 7.94E-01 | 0.2 | 8.36E-01 | 0.4 |
| Extracellular | 3.0 | 3.31E-05 | 1.6 | 1.49E-03 | 0.6 | 3.98E-01 | 0.9 | 8.92E-02 | 0.5 | 5.90E-01 | 1.0 | 4.55E-02 | 0.4 | 6.49E-01 | 0.2 | 9.18E-01 | 0.5 |
| Golgi apparatus | 0.0 | 9.93E-01 | 0.0 | 4.32E-01 | 0.0 | 4.68E-01 | 0.5 | 2.39E-01 | 0.9 | 1.45E-02 | 0.0 | 3.38E-01 | 0.0 | 9.63E-01 | 0.0 | 6.42E-01 | 0.3 |
| Mitochondria | 6.0 | 2.05E-02 | 6.5 | 9.47E-04 | 5.7 | 1.45E-04 | 6.7 | 8.59E-03 | 9.2 | 3.10E-01 | 10.0 | 6.17E-01 | 12.3 | 3.22E-01 | 7.3 | 2.79E-02 | 10.7 |
| Nucleus | 13.3 | 9.33E-05 | 4.5 | 4.89E-02 | 3.4 | 3.00E-03 | 13.9 | 1.14E-07 | 8.5 | 5.90E-02 | 8.1 | 1.15E-01 | 6.7 | 8.16E-01 | 5.9 | 7.01E-01 | 6.5 |
| Other cellular components | 3.0 | 1.58E-02 | 6.1 | 5.60E-01 | 8.5 | 1.32E-01 | 6.9 | 8.57E-01 | 7.1 | 8.43E-01 | 8.7 | 7.23E-02 | 9.5 | 9.42E-02 | 5.6 | 3.66E-01 | 6.9 |
| Other cytoplasmic components | 3.4 | 2.33E-01 | 2.5 | 7.05E-01 | 2.4 | 9.89E-01 | 3.2 | 2.35E-01 | 4.2 | 7.71E-03 | 3.7 | 6.16E-02 | 3.5 | 2.80E-01 | 2.6 | 6.25E-01 | 2.5 |
| Other intracellular components | 3.4 | 1.99E-01 | 3.9 | 1.32E-01 | 9.2 | 7.82E-04 | 5.8 | 7.59E-01 | 5.8 | 7.13E-01 | 6.2 | 4.41E-01 | 6.3 | 5.35E-01 | 5.6 | 8.47E-01 | 5.6 |
| Other membranes | 29.6 | 1.30E-03 | 42.8 | 1.16E-29 | 19.4 | 5.31E-01 | 21.5 | 7.54E-01 | 30.1 | 9.09E-08 | 21.0 | 8.58E-01 | 21.1 | 8.05E-01 | 30.8 | 3.45E-06 | 20.8 |
| Plasma membrane | 0.9 | 5.54E-01 | 1.3 | 2.12E-01 | 0.7 | 9.51E-01 | 0.9 | 4.24E-01 | 1.4 | 7.59E-02 | 1.6 | 3.59E-02 | 0.7 | 5.97E-01 | 0.5 | 8.57E-01 | 0.8 |
| Plastid | 0.0 | 1.90E-01 | 0.4 | 2.29E-01 | 6.4 | 4.27E-17 | 0.5 | 4.57E-01 | 0.3 | 1.42E-01 | 0.9 | 9.27E-01 | 1.1 | 4.54E-01 | 2.1 | 2.82E-02 | 1.0 |
| Ribosome | 0.0 | 1.02E-01 | 0.0 | 2.93E-03 | 3.6 | 9.11E-05 | 0.2 | 7.06E-02 | 0.0 | 1.23E-03 | 1.5 | 4.22E-01 | 1.4 | 7.38E-01 | 0.0 | 1.58E-02 | 1.3 |
| Total | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | ||||||||
| Keyword category: Molecular function | |||||||||||||||||
| DNA or RNA binding | 7.5 | 1.50E-01 | 3.4 | 4.39E-02 | 4.8 | 6.59E-01 | 8.5 | 7.05E-03 | 4.7 | 5.40E-01 | 5.8 | 5.80E-01 | 3.9 | 4.78E-01 | 3.6 | 1.12E-01 | 5.4 |
| Hydrolase activity | 10.5 | 1.30E-01 | 10.3 | 3.65E-02 | 6.5 | 2.93E-01 | 8.7 | 4.50E-01 | 8.7 | 3.87E-01 | 10.5 | 2.13E-02 | 11.2 | 5.16E-02 | 8.7 | 5.36E-01 | 7.9 |
| Kinase activity | 4.1 | 6.27E-01 | 3.6 | 7.52E-01 | 3.2 | 4.13E-01 | 6.3 | 2.06E-02 | 4.2 | 6.86E-01 | 4.6 | 3.67E-01 | 5.6 | 2.08E-01 | 4.5 | 4.67E-01 | 4.0 |
| Molecular function unknown | 22.2 | 2.31E-02 | 23.0 | 3.38E-03 | 15.9 | 2.99E-11 | 19.0 | 3.10E-06 | 21.2 | 1.55E-05 | 17.3 | 4.51E-11 | 18.8 | 2.90E-04 | 23.0 | 1.19E-02 | 29.0 |
| Nucleic acid binding | 1.5 | 3.34E-01 | 0.7 | 2.40E-03 | 1.8 | 4.78E-01 | 1.9 | 6.49E-01 | 1.1 | 4.73E-02 | 2.6 | 7.97E-01 | 2.6 | 8.07E-01 | 0.9 | 3.00E-02 | 2.5 |
| Nucleotide binding | 3.8 | 2.94E-01 | 3.8 | 6.40E-02 | 3.8 | 7.04E-02 | 6.5 | 4.17E-01 | 4.2 | 1.30E-01 | 9.9 | 3.26E-05 | 6.9 | 3.00E-01 | 3.1 | 3.63E-02 | 5.7 |
| Other binding | 7.5 | 4.65E-01 | 9.0 | 2.10E-02 | 8.0 | 1.52E-01 | 9.1 | 3.50E-02 | 9.2 | 9.97E-03 | 6.5 | 8.79E-01 | 9.2 | 7.95E-02 | 7.6 | 3.41E-01 | 6.5 |
| Other enzyme activity | 12.0 | 2.49E-01 | 13.2 | 2.09E-02 | 18.6 | 6.68E-09 | 9.3 | 6.53E-01 | 13.3 | 1.01E-02 | 12.2 | 8.68E-02 | 10.9 | 6.26E-01 | 15.6 | 6.40E-04 | 10.1 |
| Other molecular functions | 3.0 | 1.39E-01 | 6.2 | 3.49E-01 | 5.0 | 8.57E-01 | 2.8 | 1.72E-02 | 4.4 | 3.42E-01 | 4.0 | 1.37E-01 | 2.6 | 4.74E-02 | 5.8 | 5.38E-01 | 5.4 |
| Protein binding | 3.0 | 2.83E-01 | 2.6 | 1.98E-02 | 3.0 | 6.52E-02 | 4.6 | 9.46E-01 | 4.7 | 9.05E-01 | 5.2 | 4.47E-01 | 6.3 | 2.22E-01 | 3.3 | 1.96E-01 | 4.7 |
| Receptor binding or activity | 0.8 | 4.25E-01 | 0.7 | 9.12E-01 | 0.7 | 8.35E-01 | 0.4 | 9.56E-01 | 0.4 | 4.86E-01 | 0.8 | 4.35E-01 | 1.0 | 4.65E-01 | 0.0 | 1.49E-01 | 0.7 |
| Structural molecule activity | 0.4 | 2.58E-01 | 2.8 | 2.23E-02 | 5.0 | 2.17E-07 | 0.2 | 2.89E-02 | 0.3 | 1.36E-02 | 2.1 | 2.35E-01 | 2.0 | 3.35E-01 | 0.4 | 1.46E-01 | 1.5 |
| Transcription factor activity | 11.3 | 7.96E-05 | 3.8 | 2.30E-01 | 4.8 | 9.44E-01 | 10.1 | 2.58E-05 | 7.0 | 2.86E-02 | 4.7 | 7.31E-01 | 3.6 | 4.44E-01 | 6.9 | 7.42E-02 | 5.0 |
| Transferase activity | 7.5 | 6.58E-01 | 9.2 | 4.22E-02 | 9.0 | 6.20E-02 | 9.5 | 3.76E-02 | 8.0 | 2.42E-01 | 8.1 | 1.88E-01 | 10.2 | 3.30E-02 | 6.3 | 6.59E-01 | 6.9 |
| Transporter activity | 4.9 | 7.33E-01 | 7.8 | 1.03E-03 | 9.7 | 1.46E-06 | 3.2 | 2.39E-01 | 8.5 | 4.03E-05 | 5.7 | 1.86E-01 | 5.3 | 4.38E-01 | 10.3 | 2.97E-06 | 4.6 |
| Total | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | ||||||||
| Keyword category: Biological process | |||||||||||||||||
| Biological process unknown | 10.6 | 1.26E-03 | 14.3 | 1.85E-02 | 9.9 | 3.04E-07 | 12.4 | 1.39E-03 | 12.6 | 1.82E-04 | 12.8 | 3.96E-04 | 13.9 | 9.53E-02 | 13.8 | 2.05E-02 | 18.4 |
| Cell organization and biogenesis | 1.5 | 9.20E-01 | 3.5 | 4.39E-02 | 1.9 | 8.87E-01 | 2.2 | 6.70E-01 | 1.9 | 9.12E-01 | 3.3 | 4.87E-02 | 2.3 | 8.07E-01 | 0.9 | 8.71E-02 | 2.1 |
| Developmental processes | 4.3 | 7.90E-03 | 1.5 | 8.24E-01 | 1.4 | 6.80E-01 | 3.0 | 7.02E-02 | 1.6 | 7.82E-01 | 2.4 | 2.53E-01 | 2.8 | 1.62E-01 | 1.8 | 7.67E-01 | 1.9 |
| DNA or RNA metabolism | 0.6 | 9.46E-01 | 0.0 | 5.93E-02 | 0.0 | 7.13E-02 | 0.5 | 9.63E-01 | 0.2 | 1.89E-01 | 1.2 | 9.87E-02 | 1.5 | 5.23E-02 | 0.0 | 1.50E-01 | 0.7 |
| Electron transport or energy pathways | 1.5 | 7.55E-01 | 2.8 | 6.48E-02 | 6.3 | 8.53E-10 | 1.1 | 6.03E-01 | 2.6 | 8.61E-02 | 1.2 | 3.93E-01 | 1.7 | 8.31E-01 | 3.0 | 6.30E-02 | 1.8 |
| Other biological processes | 11.2 | 3.33E-04 | 4.5 | 4.27E-01 | 5.0 | 7.60E-01 | 8.3 | 1.41E-02 | 6.5 | 2.37E-01 | 6.2 | 3.97E-01 | 6.2 | 5.03E-01 | 6.0 | 6.06E-01 | 5.5 |
| Other cellular processes | 15.1 | 8.28E-01 | 16.1 | 8.76E-01 | 17.4 | 3.54E-01 | 16.7 | 6.73E-01 | 16.2 | 8.29E-01 | 17.0 | 4.63E-01 | 16.2 | 8.54E-01 | 15.9 | 9.73E-01 | 16.0 |
| Other metabolic processes | 16.6 | 8.69E-01 | 17.7 | 3.75E-01 | 18.0 | 2.87E-01 | 16.6 | 8.31E-01 | 16.9 | 6.72E-01 | 16.9 | 7.13E-01 | 16.4 | 9.83E-01 | 17.0 | 6.91E-01 | 16.4 |
| Other physiological processes | 15.7 | 6.44E-01 | 16.7 | 9.07E-01 | 19.7 | 1.04E-01 | 17.9 | 6.00E-01 | 17.6 | 6.57E-01 | 18.5 | 3.00E-01 | 17.5 | 7.94E-01 | 17.3 | 7.44E-01 | 17.0 |
| Protein metabolism | 3.9 | 2.95E-02 | 3.9 | 4.36E-04 | 5.5 | 7.02E-02 | 5.3 | 7.63E-02 | 5.4 | 2.98E-02 | 6.8 | 4.14E-01 | 6.2 | 4.72E-01 | 5.8 | 2.32E-01 | 7.7 |
| Response to abiotic or biotic stimulus | 7.5 | 1.70E-03 | 6.4 | 4.50E-04 | 4.9 | 6.76E-02 | 4.1 | 3.77E-01 | 4.4 | 1.43E-01 | 3.3 | 9.97E-01 | 5.5 | 6.95E-02 | 5.4 | 3.01E-02 | 3.4 |
| Response to stress | 1.9 | 9.03E-01 | 4.4 | 2.36E-04 | 2.5 | 2.58E-01 | 1.6 | 7.19E-01 | 2.6 | 1.62E-01 | 1.8 | 8.77E-01 | 3.6 | 3.67E-02 | 3.1 | 1.10E-01 | 1.9 |
| Signal transduction | 2.1 | 2.12E-01 | 1.0 | 5.81E-01 | 1.6 | 7.06E-01 | 2.5 | 5.05E-02 | 2.4 | 2.93E-02 | 1.7 | 3.94E-01 | 1.5 | 5.02E-01 | 1.6 | 5.33E-01 | 1.4 |
| Transcription | 4.4 | 2.02E-01 | 2.2 | 2.95E-01 | 2.6 | 5.76E-01 | 4.9 | 4.53E-02 | 3.7 | 3.23E-01 | 3.2 | 6.85E-01 | 2.1 | 4.86E-01 | 3.6 | 5.16E-01 | 3.1 |
| Transport | 2.9 | 6.47E-01 | 5.0 | 4.65E-03 | 3.4 | 3.82E-01 | 2.9 | 6.61E-01 | 5.4 | 7.60E-04 | 3.7 | 1.33E-01 | 2.6 | 9.56E-01 | 4.9 | 1.90E-02 | 2.8 |
| Total | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | ||||||||
Based on 230 preCIM up-regulated genes; 502 preCIM down-regulated genes.
Based on 478 shoot-specific up-regulated genes; 397 shoot-specifc down-regulated genes.
Based on 568 root-specific up-regulated genes; 583 root-specific down-regulated genes.
Based on 241 callus-specific up-regulated genes; 373 callus-specific down-regulated genes.
Based on 22,590 total genes.
A similar analysis was conducted to identify genes down-regulated during preincubation on CIM. Among the most highly down-regulated genes between 0 and 4 d preCIM were seven that encoded class III peroxidases (Table I). These enzymes are involved in a variety of functions including lignification, suberization, auxin catabolism, wound healing, and defense against pathogen infection (Hiraga et al., 2001). Their down-regulation suggests that functions such as vascular lignification are compromised during CIM preincubation. Of the 502 genes down-regulated more than 10-fold, a greater number than anticipated from their frequency in the total genome encoded proteins associated with other membranes, i.e. other than those known to be associated with chloroplast, mitochondrial, plasma, endoplasmic reticulum, and Golgi membranes (Table II).
Gene Expression Programs during Shoot, Callus, and Root Development
Following CIM preincubation, root explants can be transferred to cytokinin-rich SIM to induce shoot formation, to another auxin-rich RIM to form shoots, or further incubated on CIM to promote more callus formation. The morphogenic events that occur during these developmental processes in Arabidopsis tissue culture have been described by Huang and Yeoman (1984).
We examined gene expression programs on the three different developmental pathways, up to 10 d SIM, CIM, or RIM (10 d SIM, for example, means that root explants have been cultured for a total of 10 d, 4 d preincubation on CIM followed by 6 d incubation on SIM; Fig. 1). Ten-day SIM is about the time of shoot commitment, defined as the developmental stage when root explants can be transferred to basal medium and still continue to form shoots (Cary et al., 2002). Thus, for shoot development, the time course involves early developmental events that precede shoot emergence.
We were particularly interested in genes that are specifically up- or down-regulated early in development on one pathway, but not on the others. To identify genes specifically up- or down-regulated during shoot development, we required that the estimated mean level of expression at 10 d SIM be greater for up-regulated genes or lesser in the case of down-regulated genes than the estimated mean at each of 4 d CIM, 7 d CIM, 10 d CIM, 7 d RIM, and 10 d RIM and to have a q value (Storey and Tibshirani, 2003) less than or equal to 0.05 for all five of these comparisons. Similar criteria were used to identify genes specifically up- or down-regulated during root or callus development. For root-specific genes, 10 d RIM mean expression level was compared to 4 d CIM, 7 d CIM, 10 d CIM, 7 d SIM, and 10 d SIM, while for callus-specific genes 10 d CIM mean expression level was compared to 4 d CIM, 7 d RIM, 10 d RIM, 7 d SIM, and 10 d SIM.
By these criteria, 478 genes were specifically up-regulated and 397 were down-regulated during early shoot development, 568 up-regulated while 583 down-regulated during root development, and 241 up-regulated and 373 down-regulated during callus development (Supplemental Table II). The up-regulated genes were categorized with respect to their assigned cellular compartments, molecular function, and biological processes (gene ontology [GO], The Arabidopsis Information Resource). Of the genes up-regulated during shoot development, genes encoding proteins targeted to chloroplasts were found in 2.2-fold excess over their frequency in the total genome, reflecting the fact that greening occurs (green callus formation) during these stages (Table II, GO cellular component). Among shoot development down-regulated genes, those encoding proteins targeted to the nucleus were found 2.1-fold excess over their presence in the total genome. Genes up-regulated during root development encoding proteins with transporter activity occurred in 1.9-fold excess over their expected frequency (Table II, GO molecular function). During callus development, a greater frequency than expected of up-regulated genes were involved in response to stress (1.9-fold excess; Table II, biological process).
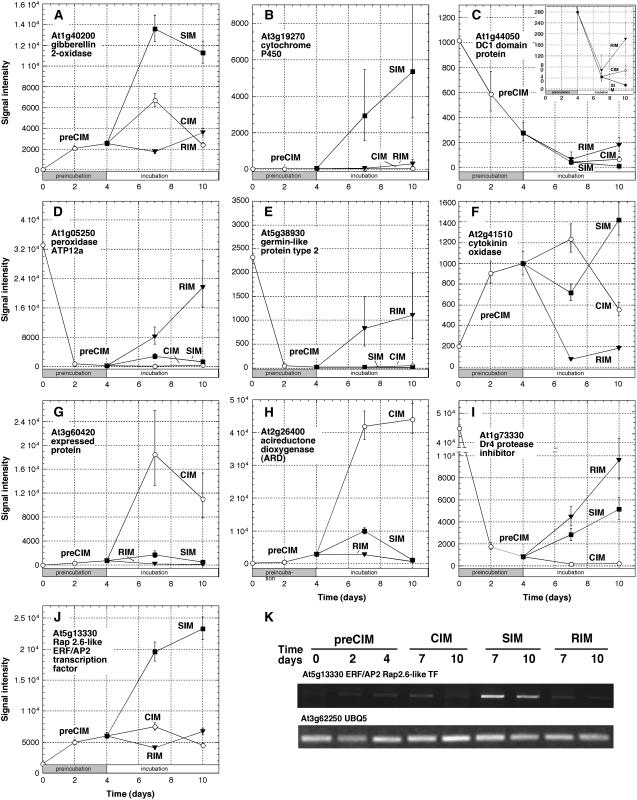
From Supplemental Table II we extracted the top 20 most highly up- or down-regulated genes on the three different developmental pathways (Table III). The genes were rank ordered on the shoot development pathway by fold increase or decrease in comparing estimated mean expression levels (signal intensities) at 7 d SIM with 0 time (Table III). The top three most highly up-regulated genes during shoot development encoded GA 2-oxidase (At1g30040; Fig. 3A), a cytochrome P450 (At3g19270; Fig. 3B), and a GA-regulated protein (At1g74670). Brenner et al. (2005) also found that a number of GA-related genes were up-regulated in seedlings in response to cytokinin. Most of the top 20 genes specifically up-regulated during shoot development (actually 17 out of 18 for which there are data in the AtGenExpress) are genes ultimately expressed most highly in shoots or organs associated with shoots (leaves, floral organs, and so forth). This is important to note because most of the genes most highly up-regulated on SIM are not root genes, but genes likely involved in the formation of the new shoots. The top 20 genes down-regulated during shoot development are dominated by DC1-containing proteins, such as At1g44050 (Fig. 3C, see inset; Table III). These are proteins with Cys/His clusters that coordinate metal ions. Many of the top 20 genes (11 out of 20) specifically down-regulated during shoot development are genes ultimately expressed most highly in the root. Thus, a number of root-specific genes are being turned off during shoot development in root explants.
Table III.
Top 20 genes up- and down-regulated during shoot, root, and callus development
Mean signal intensities were derived from two independent replications at each of nine different time points. Genes designated as specifically up- or down-regulated during shoot, root, or callus development, had estimated mean levels of expression at 10 d on SIM, CIM, or RIM, respectively, greater or lesser than the estimated mean levels of expression for five other comparisons as described in the text and had a q value less than or equal to 0.05 for all five comparisons. Genes were rank ordered in each developmental category by FC in expression as indicated. ca, Carpel; ce, early cotyledon; cg, green cotyledon; co, cotyledon; e, embryo; f, flower; h, hypocotyl; l, leaf; lc, cauline leaf; lm, mature leaf; lr, rosette leaf; ls, senescent leaf; ly, young, expanding leaf; pd, pedicel; pe, petiole; po, pollen; pt, petal; r, root; se, seed; sh, shoot; si, shoot internode; sn, shoot node; sp, sepal; ss, shoot seedling; st, stamen; wp, whole plant; *, data not available.
| Locus Identification | Mean Signal Intensitiesa
|
FCb | P Valuesc | q Valuesc | AtGenExpd | Gene Description | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 Time | 2 d CIM | 4 d CIM | 7 d CIM | 10 d CIM | 7 d RIM | 10 d RIM | 7 d SIM | 10 d SIM | ||||||
| Shoot up | ||||||||||||||
| At1g30040 | 59 | 2,061 | 2,529 | 6,682 | 2,391 | 1722 | 3,588 | 13,580 | 11,279 | 228.4 | 1.42E-10 | 4.93E-07 | pd, s, s, cg | GA 2-oxidase/GA2-oxidase |
| At3g19270 | 15 | 11 | 28 | 19 | 37 | 48 | 281 | 2,913 | 5,354 | 188.5 | 3.65E-04 | 1.66E-03 | sn, f, sp, sh | Cytochrome P450 |
| At1g74670 | 10 | 319 | 476 | 595 | 392 | 156 | 127 | 479 | 2,543 | 50.0 | 1.36E-05 | 2.00E-04 | l, sp, pt, l | GA-regulated protein 4 precursor |
| At3g54820 | 761 | 2,767 | 5,833 | 12,545 | 6,681 | 5,946 | 11,676 | 24,111 | 22,045 | 31.7 | 1.79E-08 | 5.77E-06 | f, f, f, sh | Aquaporin MIP-like protein |
| At1g69880 | 478 | 5,241 | 4,634 | 4,238 | 3,792 | 1,635 | 1,537 | 13,027 | 14,565 | 27.3 | 9.76E-09 | 5.06E-06 | sh, r, e, ss | Thioredoxin H-type 2 (TRX-H-2) |
| At2g40610 | 244 | 724 | 2,136 | 1,598 | 804 | 1,688 | 1,617 | 6,538 | 6,333 | 26.8 | 4.40E-06 | 1.03E-04 | f, f, pt, ly | Putative expansin (EXP8) |
| At3g13130 | 15 | 11 | 11 | 43 | 13 | 16 | 10 | 379 | 904 | 25.9 | 4.48E-04 | 1.91E-03 | *, *, *, * | Hypothetical protein |
| At5g25190 | 219 | 1,388 | 2,934 | 3,905 | 2,635 | 1,382 | 1,339 | 5,278 | 6,908 | 24.1 | 1.10E-08 | 5.06E-06 | ly, l, l, l | ERF/AP2 transcription factor subfamily B-6 |
| At2g16005 | 919 | 440 | 213 | 130 | 207 | 1,654 | 3,943 | 21,064 | 9,714 | 22.9 | 3.37E-07 | 2.32E-05 | r, r, r, r | MD-2-related lipid recognition domain-containing protein |
| At3g62950 | 4 | 4 | 8 | 5 | 4 | 7 | 5 | 70 | 119 | 16.2 | 9.30E-03 | 1.74E-02 | l, co, lm, ly | Glutaredoxin-like protein |
| At1g74890 | 214 | 134 | 203 | 310 | 28 | 56 | 71 | 3,039 | 2,656 | 14.2 | 3.28E-04 | 1.54E-03 | sh, sh, ly, h | Response regulator 15 (ARR15) |
| At5g53820 | 90 | 168 | 307 | 360 | 92 | 761 | 600 | 1,279 | 3,275 | 14.1 | 5.98E-07 | 3.21E-05 | st, po, f, f | ABA-inducible protein |
| At2g40200 | 160 | 147 | 262 | 166 | 164 | 440 | 1,347 | 2,240 | 4,377 | 14.0 | 6.54E-05 | 5.36E-04 | pd, si, si, sh | Basic helix-loop-helix (bHLH) family protein |
| At5g13330 | 1,540 | 5,003 | 6,030 | 7,495 | 4,537 | 4,115 | 6,747 | 19,560 | 23,314 | 12.7 | 3.56E-12 | 3.93E-08 | ce, cg, ce, st | ERF/AP2 transcription factor subfamily B-4 |
| At1g29090 | 111 | 75 | 206 | 115 | 66 | 111 | 153 | 1,335 | 3,550 | 12.1 | 2.88E-03 | 7.29E-03 | *, *, *, * | Peptidase C1A papain family protein |
| At3g59060 | 5 | 7 | 16 | 7 | 9 | 10 | 23 | 64 | 251 | 11.7 | 1.28E-03 | 4.01E-03 | lm, ly, lc, co | Basic helix-loop-helix (bHLH) family |
| At5g24780 | 517 | 239 | 67 | 190 | 131 | 405 | 1,026 | 5,892 | 9,905 | 11.4 | 2.05E-05 | 2.56E-04 | f, f, f, st | Vegetative storage protein 1 (VSP1) |
| At2g40670 | 186 | 286 | 162 | 31 | 11 | 50 | 137 | 2,125 | 3,222 | 11.4 | 1.28E-04 | 8.32E-04 | pt, st, st, sp | Response regulator 16 (ARR16) |
| At3g13980 | 799 | 732 | 1,403 | 1,487 | 1,191 | 3,516 | 5,990 | 8,748 | 10,156 | 10.9 | 4.88E-07 | 2.88E-05 | sn,si,h,pe | Hypothetical protein |
| At5g06870 | 80 | 140 | 381 | 1,464 | 768 | 275 | 351 | 835 | 3,360 | 10.4 | 3.18E-05 | 3.38E-04 | pe, l, pt, lm | Polygalacturonase inhibiting protein 2 (PGIP2) |
| Shoot down | ||||||||||||||
| At1g44050 | 1,016 | 588 | 279 | 45 | 67 | 181 | 202 | 43 | 14 | 0.04 | 3.01E-05 | 3.23E-04 | r, r, r, r | DC1 domain-containing protein |
| At1g80240 | 582 | 137 | 130 | 352 | 754 | 906 | 855 | 30 | 36 | 0.05 | 1.28E-04 | 8.35E-04 | r, r, r, r | Expressed protein |
| At5g50200 | 28,175 | 4,812 | 2,163 | 2,942 | 2,091 | 2,866 | 4,191 | 1,652 | 633 | 0.06 | 2.04E-06 | 6.79E-05 | r, ss, r, ss | Expressed protein |
| At1g22160 | 793 | 433 | 933 | 543 | 576 | 521 | 363 | 47 | 68 | 0.06 | 3.40E-04 | 1.58E-03 | lc, ca, f, f | Senescence-associated protein |
| At4g15400 | 1,814 | 918 | 527 | 75 | 85 | 222 | 273 | 121 | 17 | 0.07 | 6.75E-04 | 2.55E-03 | r, r, ca, r | Deacetylvindoline 4-O-acetyltransferase-like protein |
| At2g38940 | 18,789 | 1,376 | 1,263 | 1,055 | 2,183 | 2,594 | 3,685 | 1,264 | 558 | 0.07 | 1.16E-07 | 1.34E-05 | st, r, st, ss | Phosphate transporter (PT2) |
| At2g02850 | 9,273 | 21,046 | 6,326 | 1,938 | 1,661 | 6,970 | 4,949 | 626 | 742 | 0.07 | 1.30E-06 | 5.37E-05 | cg, cg, ce, ca | Plantacyanin (blue copper protein) |
| At2g21540 | 542 | 59 | 39 | 91 | 153 | 137 | 232 | 56 | 13 | 0.10 | 2.71E-04 | 1.37E-03 | po, st, st, e | Phosphoglyceride transfer protein |
| At1g55430 | 276 | 302 | 318 | 302 | 280 | 163 | 154 | 34 | 16 | 0.12 | 1.23E-02 | 2.13E-02 | r, r, r, r | DC1 domain-containing protein |
| At5g40590 | 4,895 | 5,110 | 3,906 | 1,199 | 902 | 1,977 | 1,160 | 609 | 265 | 0.12 | 9.51E-05 | 6.84E-04 | r, r, r, r | DC1 domain-containing protein |
| At3g11370 | 728 | 277 | 319 | 239 | 150 | 173 | 156 | 91 | 60 | 0.12 | 4.77E-05 | 4.36E-04 | r, r, r, r | DC1 domain-containing protein |
| At5g49780 | 823 | 295 | 216 | 107 | 89 | 94 | 204 | 113 | 27 | 0.14 | 3.94E-05 | 3.89E-04 | r, r, r, r | Leu-rich repeat transmembrane protein kinase |
| At1g48750 | 8,530 | 10,396 | 10,238 | 11,174 | 9,289 | 2,561 | 2,475 | 1,358 | 957 | 0.16 | 8.51E-07 | 4.02E-05 | f, f, ca, f | Protease inhibitor/seed storage/lipid transfer protein (LTP) |
| At1g18100 | 1,181 | 8,383 | 7,545 | 658 | 443 | 417 | 178 | 210 | 73 | 0.18 | 3.22E-05 | 3.40E-04 | se, e, e, ce | Mother of FT and TF1 protein (MFT) |
| At4g31875 | 1,870 | 3,083 | 1,080 | 1,104 | 554 | 858 | 741 | 343 | 209 | 0.18 | 2.75E-04 | 1.38E-03 | r, r, r, r | Expressed protein |
| At5g45480 | 2,827 | 696 | 914 | 1,826 | 1,771 | 1,484 | 1,682 | 533 | 473 | 0.19 | 1.88E-05 | 2.42E-04 | r, r, r, r | Expressed protein |
| At3g48080 | 833 | 442 | 334 | 861 | 474 | 213 | 206 | 163 | 113 | 0.20 | 1.20E-05 | 1.89E-04 | ls, ly, lc, lm | Lipase class 3 family protein |
| At5g59530 | 1,484 | 1,339 | 716 | 606 | 486 | 442 | 749 | 313 | 153 | 0.21 | 6.44E-05 | 5.32E-04 | r, r, r, r | 2-Oxoglutarate-dependent dioxygenase |
| At1g68810 | 1,972 | 574 | 554 | 603 | 616 | 796 | 665 | 429 | 307 | 0.22 | 1.54E-05 | 2.13E-04 | h, r, si, r | Basic helix-loop-helix (bHLH) family protein |
| At1g61360 | 2,187 | 1,100 | 835 | 1,095 | 887 | 181 | 868 | 490 | 391 | 0.22 | 1.06E-04 | 7.34E-04 | ss, r, r, r | S-locus lectin protein kinase family protein |
| Root up | ||||||||||||||
| At1g05250 | 33,194 | 785 | 162 | 134 | 281 | 8,070 | 21,606 | 2,690 | 1,271 | 49.9 | 1.24E-05 | 1.00E-03 | r, r, r, r | Peroxidase ATP12a |
| At5g38930 | 2,326 | 48 | 17 | 17 | 42 | 831 | 1,102 | 16 | 4 | 49.8 | 1.57E-03 | 1.70E-02 | sh, l, sh, r | Germin-like protein type 2 |
| At1g49860 | 12,773 | 179 | 109 | 32 | 96 | 4,751 | 4,616 | 55 | 69 | 43.8 | 7.00E-04 | 1.04E-02 | r, r, r, r | Glutathione S-transferase |
| At1g34510 | 2,479 | 24 | 19 | 12 | 155 | 752 | 3,051 | 357 | 269 | 39.8 | 2.78E-04 | 5.92E-03 | r, r, r, r | Peroxidase ATP13a |
| At3g23800 | 2,330 | 47 | 29 | 17 | 75 | 1,130 | 3,501 | 8 | 41 | 38.8 | 1.18E-03 | 1.44E-02 | r, r, r, r | Selenium-binding protein |
| At2g14900 | 3,661 | 500 | 144 | 736 | 1,701 | 5,362 | 7,729 | 2,621 | 2,487 | 37.3 | 4.94E-08 | 8.25E-05 | st, e, pt, pd | GA-regulated protein |
| At5g23020 | 16,205 | 228 | 121 | 47 | 151 | 4,266 | 9,310 | 190 | 2,059 | 35.2 | 1.62E-05 | 1.09E-03 | r, r, r, r | 2-Isopropylmalate synthase |
| At3g19710 | 21,398 | 133 | 115 | 151 | 223 | 3,566 | 5,598 | 270 | 1,632 | 31.0 | 1.49E-05 | 1.08E-03 | sn, pe, lm, h | Branched-chain amino acid aminotransferase |
| At5g53250 | 12,919 | 88 | 53 | 75 | 104 | 1,564 | 4,074 | 1,099 | 1,541 | 29.6 | 1.55E-06 | 3.46E-04 | r, r, r, r | Arabinogalactan protein |
| At4g02270 | 22,222 | 763 | 250 | 356 | 1,105 | 6,695 | 23,130 | 4,695 | 2,853 | 26.8 | 4.88E-06 | 6.12E-04 | *, *, *, * | Extensin family protein |
| At5g65530 | 1,686 | 238 | 94 | 156 | 70 | 2,219 | 2,790 | 1,215 | 1,298 | 23.6 | 7.39E-08 | 9.94E-05 | po, cg, st, ce | Putative protein kinase |
| At1g01750 | 4,781 | 43 | 69 | 62 | 132 | 1,583 | 4,668 | 956 | 839 | 23.1 | 4.86E-05 | 2.04E-03 | r, r, r, r | Actin depolymerizing factor (ADF) |
| At2g45180 | 3,752 | 161 | 204 | 242 | 375 | 4,600 | 7,334 | 643 | 1,184 | 22.6 | 5.73E-04 | 9.15E-03 | ly, ly, l, lr | Protease inhibitor/lipid transfer protein |
| At4g33730 | 2,351 | 47 | 43 | 23 | 133 | 831 | 3,269 | 776 | 567 | 19.1 | 1.58E-05 | 1.08E-03 | r, r, r, r | Pathogenesis-related protein (PR-1) |
| At5g44020 | 23,250 | 340 | 186 | 140 | 142 | 3,303 | 11,895 | 1,712 | 964 | 17.8 | 1.80E-05 | 1.17E-03 | ss, ss, ly, co | Acid phosphatase class B family protein |
| At2g32300 | 2,436 | 199 | 91 | 110 | 166 | 1,577 | 2,697 | 1,073 | 1,233 | 17.3 | 1.52E-06 | 3.46E-04 | r, r, r, r | Uclacyanin I |
| At5g15830 | 2,512 | 158 | 31 | 26 | 80 | 511 | 1,417 | 41 | 187 | 16.7 | 1.74E-04 | 4.48E-03 | lm, r, r, r | bZIP transcription factor |
| At2g27370 | 2,526 | 167 | 93 | 19 | 84 | 1,410 | 2,019 | 619 | 978 | 15.2 | 6.77E-07 | 2.64E-04 | r, r, r, r | Integral membrane protein |
| At4g14130 | 5,785 | 115 | 342 | 498 | 1,876 | 4,914 | 6,920 | 812 | 713 | 14.4 | 3.23E-06 | 5.20E-04 | r, r, r, r | Xyloglucan endotransglycosylase protein XTR-7 |
| At1g05260 | 12,728 | 430 | 254 | 492 | 477 | 3,588 | 9,862 | 3,628 | 4,257 | 14.2 | 5.25E-06 | 6.16E-04 | r, r, r, r | Peroxidase 3 (PER3) |
| Root down | ||||||||||||||
| At2g41510 | 204 | 908 | 1,001 | 1,237 | 556 | 74 | 184 | 717 | 1,422 | 0.07 | 1.64E-07 | 1.35E-04 | h, r, r, * | Cytokinin oxidase family protein |
| At5g42380 | 1,147 | 231 | 222 | 230 | 80 | 18 | 11 | 306 | 62 | 0.08 | 2.90E-04 | 6.08E-03 | r, r, h, r | Calmodulin-related protein |
| At3g28150 | 159 | 1,492 | 1,973 | 1,176 | 412 | 174 | 178 | 2,179 | 1,575 | 0.09 | 4.70E-05 | 2.00E-03 | po, st, st, se | Expressed protein |
| At1g35910 | 197 | 205 | 426 | 1,967 | 1,495 | 53 | 26 | 1,228 | 2,188 | 0.12 | 1.81E-03 | 1.87E-02 | sp, st, st, f | Rehalose-6-phosphate phosphatase |
| At2g41810 | 6,536 | 1,073 | 2,962 | 2,774 | 1,093 | 387 | 214 | 3,636 | 3,074 | 0.13 | 1.77E-03 | 1.84E-02 | r, r, *, * | Expressed protein |
| At2g14450 | 87 | 29 | 75 | 73 | 84 | 11 | 9 | 100 | 69 | 0.14 | 1.61E-02 | 7.00E-02 | *, *, *, * | Putative replication protein A1 |
| At1g77110 | 271 | 536 | 836 | 1,348 | 642 | 121 | 166 | 851 | 1,071 | 0.14 | 3.10E-03 | 2.60E-02 | f, f, f, f | Auxin transport protein (PIN6) |
| At1g61340 | 2,595 | 1,869 | 2,369 | 1,393 | 1,130 | 347 | 326 | 2,541 | 1,278 | 0.15 | 1.22E-03 | 1.46E-02 | r, r, st, ss | F-box family protein |
| At2g18470 | 19 | 510 | 816 | 1,99 | 288 | 120 | 57 | 753 | 1,058 | 0.15 | 5.80E-03 | 3.76E-02 | po, st, st, f | Protein kinase family protein |
| At1g30100 | 15 | 170 | 475 | 2,066 | 710 | 70 | 64 | 553 | 413 | 0.15 | 9.78E-03 | 5.15E-02 | cg, cg, ce, sp | 9-cis-Epoxycarotenoid dioxygenase |
| At4g25700 | 407 | 7,358 | 4,668 | 1,113 | 949 | 710 | 414 | 1,415 | 1,285 | 0.15 | 1.81E-04 | 4.56E-03 | pt, st, sp, st | β-Carotene hydroxylase |
| At1g69930 | 215 | 1,040 | 1,388 | 4,865 | 2,781 | 220 | 221 | 1,137 | 1,053 | 0.16 | 8.93E-05 | 2.87E-03 | sp, ls, ss, f | Glutathione S-transferase |
| At2g02990 | 893 | 2,454 | 4,352 | 1,272 | 2,720 | 725 | 360 | 1,190 | 791 | 0.17 | 1.30E-05 | 1.01E-03 | ce, cg, e, ce | Ribonuclease 1 (RNS1) |
| At1g71380 | 268 | 5,148 | 8,693 | 11,324 | 4,992 | 1,476 | 1,037 | 6,531 | 7,539 | 0.17 | 2.83E-07 | 1.67E-04 | po, sn, r, r | Glycosyl hydrolase family 9 protein |
| At1g03820 | 2,477 | 14,875 | 18,470 | 22,729 | 10,558 | 3,262 | 2,830 | 14,819 | 18,138 | 0.18 | 1.89E-07 | 1.46E-04 | sh, h, r, r | Expressed protein |
| At2g37770 | 141 | 1,076 | 700 | 849 | 756 | 126 | 132 | 460 | 424 | 0.18 | 5.02E-04 | 8.38E-03 | sp, sp, f, st | Aldo/keto reductase family protein |
| At5g55250 | 553 | 4,123 | 4,618 | 6,680 | 3,305 | 902 | 230 | 1,177 | 1,276 | 0.20 | 1.38E-05 | 1.04E-03 | e, e, e, e | S-Adenosyl-l-methionine:carboxyl methyltransferase protein |
| At2g41800 | 7,123 | 20,620 | 15,298 | 12,361 | 9,313 | 3,008 | 2,349 | 8,003 | 11,524 | 0.20 | 4.16E-07 | 2.17E-04 | r, ca, r, r | Expressed protein |
| At3g27400 | 427 | 10,966 | 16,651 | 21,461 | 8,234 | 3,302 | 2,370 | 17,133 | 23,630 | 0.20 | 1.46E-04 | 3.96E-03 | e, r, sn, h | Pectate lyase family protein |
| At4g27520 | 1,713 | 2,209 | 2,656 | 2,890 | 4,399 | 540 | 1,011 | 3,297 | 3,672 | 0.20 | 3.92E-05 | 1.78E-03 | lr, ly, l, ly | Plastocyanin-like domain-containing protein |
| Callus up | ||||||||||||||
| At3g60420 | 6 | 277 | 672 | 18,501 | 11,030 | 165 | 14 | 1,662 | 485 | 2982.5 | 1.49E-07 | 5.69E-06 | sp,ly,lm,sp | Expressed protein |
| At2g26400 | 245 | 485 | 2,965 | 41,945 | 44,066 | 2,716 | 620 | 9,332 | 1,160 | 171.3 | 4.52E-10 | 7.02E-07 | f,f,f,f | Acireductone dioxygenase (ARD/ARD') family protein |
| At2g04450 | 46 | 220 | 437 | 6,297 | 2,465 | 306 | 101 | 610 | 435 | 138.0 | 2.15E-08 | 2.26E-06 | wp,lm,wp,ly | MutT/nudix family protein |
| At3g57950 | 93 | 1,002 | 1,348 | 4,769 | 3,129 | 705 | 176 | 1,036 | 604 | 51.4 | 3.13E-09 | 1.25E-06 | st,r,f,st | Hypothetical protein |
| At2g45760 | 51 | 37 | 64 | 1,996 | 1,249 | 48 | 27 | 171 | 22 | 39.4 | 4.17E-04 | 9.01E-04 | r,r,r,r | BON1-associated protein 1 BAP1 |
| At4g34970 | 164 | 587 | 976 | 5,652 | 14,020 | 1,236 | 2,926 | 335 | 336 | 34.5 | 5.49E-07 | 1.14E-05 | si, pd, sn, ss | Actin-depolymerizing factor 5 (ADF-5; AtADF5) |
| At5g39110 | 378 | 95 | 51 | 8,487 | 8,400 | 1,946 | 1,307 | 1,206 | 613 | 22.4 | 5.88E-05 | 2.10E-04 | r, r, r, r | Germin-like protein |
| At1g43910 | 958 | 801 | 1,633 | 18,241 | 9,882 | 802 | 612 | 2,736 | 2,442 | 19.0 | 4.55E-09 | 1.25E-06 | pt, st, f, sp | AAA-type ATPase family protein |
| At3g56400 | 638 | 418 | 823 | 10,650 | 7,357 | 393 | 297 | 2,087 | 1,739 | 16.7 | 2.92E-07 | 8.14E-06 | ly, sp, ls, ly | Transcription factor DNA-binding protein 4 (WRKY4) |
| At1g02450 | 178 | 56 | 64 | 2,854 | 1,864 | 31 | 96 | 346 | 149 | 16.0 | 1.07E-03 | 1.89E-03 | pd, f, ca, f | NPR1/NIM1-interacting protein 1 (NIMIN-1) |
| At3g25620 | 96 | 234 | 268 | 1,455 | 991 | 214 | 69 | 219 | 120 | 15.2 | 3.44E-05 | 1.45E-04 | sn, si, st, st | ABC transporter family protein |
| At3g13630 | 225 | 168 | 331 | 3,196 | 1,528 | 171 | 210 | 191 | 224 | 14.2 | 3.73E-05 | 1.53E-04 | *, *, *, * | Hypothetical protein |
| At1g57650 | 17 | 11 | 36 | 237 | 137 | 17 | 13 | 19 | 14 | 13.6 | 1.05E-04 | 3.20E-04 | *, *, *, * | Disease resistance protein RPP1-WsA |
| At1g74710 | 495 | 256 | 490 | 6,302 | 2,845 | 371 | 274 | 1,079 | 605 | 12.7 | 1.63E-09 | 1.02E-06 | ls, sp, lm, f | Isochorismate synthase (icsI) |
| At1g19850 | 709 | 3,688 | 6501 | 8,133 | 10,930 | 6,705 | 5,747 | 7,784 | 7,500 | 11.5 | 2.07E-08 | 2.26E-06 | f, f, f, f | Transcription factor MONOPTEROS (MP) |
| At5g59060 | 35 | 22 | 139 | 392 | 619 | 146 | 174 | 108 | 10 | 11.1 | 3.72E-04 | 8.30E-04 | *, *, *, * | Expressed protein |
| At5g22570 | 291 | 104 | 159 | 3,144 | 1,712 | 201 | 157 | 297 | 237 | 10.8 | 3.28E-07 | 8.84E-06 | r, h, ly, ls | WRKY transcription factor 38 (WRKY 38) |
| At4g00750 | 70 | 148 | 211 | 655 | 1,688 | 267 | 174 | 151 | 34 | 9.3 | 2.16E-03 | 3.30E-03 | *, *, *, * | Dehydration stress ERD3 protein |
| At1g73800 | 532 | 245 | 374 | 4,738 | 2,734 | 308 | 207 | 561 | 470 | 8.9 | 7.90E-05 | 2.58E-04 | ls, wp, lm, co | Calmodulin-binding protein |
| At4g14390 | 534 | 960 | 1,495 | 4,597 | 3,700 | 1,026 | 519 | 723 | 395 | 8.6 | 1.60E-05 | 8.72E-05 | r, cg, po, lm | Ankyrin repeat family protein |
| Callus down | ||||||||||||||
| At1g73330 | 44,803 | 1,754 | 829 | 148 | 209 | 4,414 | 9,558 | 2,804 | 5,125 | 0.003 | 2.90E-08 | 2.36E-06 | r, r, r, r | Dr4 protease inhibitor |
| At1g48690 | 1,382 | 185 | 46 | 5 | 6 | 239 | 703 | 30 | 172 | 0.004 | 7.29E-08 | 3.76E-06 | r, r, r, r | Auxin-responsive GH3 family protein |
| At1g43160 | 3,186 | 328 | 31 | 14 | 5 | 49 | 838 | 188 | 276 | 0.004 | 1.18E-06 | 1.71E-05 | ce, ce, cg, r | ERF/AP2 transcription factor (subfamily B4, RAP2.6) |
| At3g44990 | 37,430 | 2,732 | 1,273 | 237 | 316 | 1,568 | 5,372 | 2,584 | 7,435 | 0.006 | 3.40E-07 | 9.03E-06 | h, r, r, lr | Xyloglucan:xyloglucosyl transferase |
| At1g60680 | 5,511 | 247 | 127 | 36 | 17 | 383 | 1,362 | 254 | 677 | 0.007 | 1.36E-05 | 7.78E-05 | cg, r, cg, r | Aldo/keto reductase family protein |
| At1g51860 | 1,529 | 762 | 271 | 15 | 18 | 233 | 787 | 170 | 81 | 0.010 | 1.97E-05 | 1.00E-04 | r, r, r, r | Leu-rich repeat protein kinase |
| At3g62040 | 6,989 | 797 | 527 | 85 | 188 | 982 | 3,809 | 518 | 841 | 0.012 | 5.27E-08 | 3.30E-06 | r, r, r, r | Haloacid dehalogenase-like hydrolase protein |
| At5g59090 | 16,582 | 1,174 | 436 | 213 | 88 | 2,648 | 6,910 | 4,400 | 11,315 | 0.013 | 8.65E-07 | 1.46E-05 | r, r, r, r | Subtilisin-like Ser protease (subtilase) |
| At5g53980 | 1,142 | 713 | 285 | 18 | 15 | 371 | 1,488 | 301 | 681 | 0.016 | 3.09E-08 | 2.46E-06 | r, r, *, * | Homeodomain Leu zipper class I (HD-Zip I) |
| At1g27030 | 8,147 | 1,526 | 1,040 | 134 | 378 | 4,686 | 8,489 | 6,785 | 8,251 | 0.016 | 1.32E-07 | 5.32E-06 | r, r, r, r | Expressed protein |
| At4g01440 | 2,836 | 716 | 478 | 47 | 20 | 754 | 917 | 237 | 205 | 0.017 | 4.21E-04 | 9.08E-04 | r, r, r, r | Nodulin MtN21 family protein |
| At5g17820 | 25,559 | 10,539 | 6,623 | 445 | 891 | 7,113 | 16,637 | 4,904 | 2,677 | 0.017 | 6.45E-06 | 4.76E-05 | r, r, r, r | Peroxidase 57 (PER57) |
| At4g12480 | 17,572 | 26,080 | 20,771 | 314 | 342 | 1,897 | 5,607 | 3,023 | 917 | 0.018 | 9.36E-09 | 1.68E-06 | lc, sh, ss, r | pEARLI 1; protease inhibitor/lipid transfer protein (LTP) |
| At2g21045 | 19,070 | 7,631 | 1,667 | 369 | 399 | 2,741 | 13,756 | 1,921 | 1,856 | 0.019 | 4.81E-08 | 3.18E-06 | r, r, r, r | Senescence-associated protein |
| At5g66390 | 6,496 | 1,773 | 313 | 129 | 159 | 2,323 | 4,822 | 1,604 | 2,383 | 0.020 | 2.53E-11 | 1.18E-07 | r, r, r, r | Peroxidase 72 (PER72) |
| At1g67330 | 9,150 | 4,544 | 1,832 | 232 | 653 | 2,724 | 5,865 | 3,471 | 3,101 | 0.025 | 3.99E-08 | 2.86E-06 | r, r, r, r | Expressed protein |
| At3g19030 | 11,251 | 2,015 | 772 | 288 | 395 | 4,398 | 9,330 | 4,229 | 4,280 | 0.026 | 1.87E-08 | 2.11E-06 | ss, ss, lm, lm | Expressed protein |
| At4g12470 | 18,137 | 25,547 | 14,123 | 562 | 429 | 1,211 | 9,030 | 1,752 | 4,743 | 0.031 | 2.77E-07 | 7.91E-06 | ss, ss, ss, h | pEARLI 1-like; protease inhibitor/lipid transfer protein (LTP) |
| At1g08325 | 4,919 | 1,167 | 783 | 164 | 104 | 960 | 2,089 | 885 | 821 | 0.033 | 3.27E-06 | 3.12E-05 | *, *, *, * | Leu zipper protein |
| At5g44530 | 1,074 | 203 | 146 | 36 | 29 | 338 | 544 | 279 | 417 | 0.034 | 5.36E-05 | 1.97E-04 | lr, e, sn, ly | subtilisin-like Ser protease (subtilase) |
Means from three independent replications.
FC for SIM 7 d/SIM 0 time, for CIM 7 d/CIM 0 time, and for RIM 7 d/RIM 4 d CIM.
P and q values for the comparison used in computing FC.
Four tissues with the highest expression levels according to the Expression Atlas of Arabidopsis Development in AtGenExpress.
Figure 3.
Examples of genes specifically up- or down-regulated on one developmental pathway. Most highly up-regulated (A and B) and down-regulated (C) shoot development-specific genes, most highly up-regulated (D and E) and down-regulated (F) root development-specific genes, and most highly up-regulated (G and H) and down-regulated (I) callus development-specific genes. Expression pattern of Rap2.6L ERF/AP2 transcription factor, At5g13330, a shoot development-specific gene (J). Data in A to J drawn from Supplemental Table I and Table II. Error bars represent se. Note the insert with the expanded signal intensity scale in C and the broken scale in I. K, Semiquantitative RT-PCR of the expression profiles of Rap2.6L. At3g62250 (UBQ5) was used as a control.
In root development, genes were rank ordered by comparing 7 d RIM to 4 d CIM. Those times were chosen because the expression pattern most common to up-regulated root development-specific genes was one that declined during CIM preincubation and rose again on transfer to RIM. Several peroxidases numbered among the highly up-regulated root-specific genes (Table III), including peroxidase ATP12a (At1g05250; Fig. 3D). The peroxidases are most likely involved in cell wall or vascular synthesis in root development. The other most highly up-regulated root-specific genes encoded a germin-like protein (At5g38930; Fig. 3E) and glutathione S-transferase (At1g49860). Again, as might be expected, many of the top 20 genes (13 out of 19 for which there are data in AtGenExpress) specifically up-regulated during root regeneration represent genes that are most highly expressed in roots. The most highly down-regulated genes were a fairly heterogeneous group but included a cytokinin oxidase (At2g41510; Fig. 3F) and two genes encoding proteins involved in carotenoid metabolism (At1g30100 and At4g25700; Table III). In callus development, callus development-specific genes were rank ordered by fold increase in the comparison of estimated mean expression levels at 7 d CIM with 0 time (Table III). The most highly up-regulated genes in callus development encoded an unknown, expressed protein (At3g60420; Fig. 3G) and acireductone dioxygenase (At2g26400; Fig. 3H). The top 20 most highly up-regulated genes during callus development were a mixed group of genes with respect to where they are ultimately most highly expressed in plants. The most highly down-regulated genes included a DR4 protease inhibitor (At1g73330; Fig. 3I), two peroxidase genes (At5g17820 and At5g666390), a couple of pEARLI 1 genes (At4g12480 and At4g12470), and two that encoded subtilases (At5g59090 and At5g44530; Table III). Here, the most highly down-regulated genes were root-specific genes (12 out of 19) possibly reflecting a dedifferentiation process in root explants during callus development.
To gain a better understanding of the regulation of large groups of genes on different developmental pathways, we focused on the expression of transcription factors that are specifically up-regulated on one pathway, but not the others. No single class of transcription factors dominated any one pathway (Supplemental Table II), however, several genes specifically up-regulated on the shoot development pathway-encoded A-type ARRs, such as ARR15 (At1g74890) and ARR16 (At2g40670; Table III). Some A-type ARRs are thought to be non-DNA-binding gene expression regulators (Imamura et al., 1999). Examples of more conventional transcription factors in top 20 list of shoot development-specific up-regulated genes include a basic helix-loop-helix protein (At2g40200) and two ERF/AP2 transcription factors (At5g25190 and At5g13330; Fig. 3J). The expression pattern of Rap2.6L, one of the ERF/AP2 transcription factors, was confirmed by RT-PCR analysis (Fig. 3K).
Function of RAP2.6L in Shoot Development
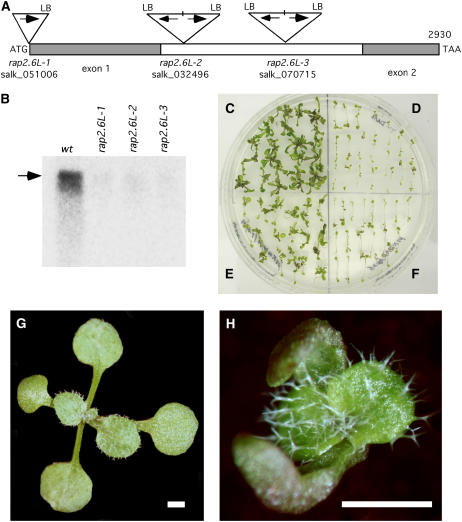
RAP2.6L was selected for further study because preliminary evidence from T-DNA insertion lines indicated that the gene functions during shoot regeneration in culture. Three Salk T-DNA lines (designated here as rap2.6L-1, -2, and -3) available at the time when this study was initiated were made homozygous as determined by PCR analysis (Fig. 4A). The T-DNA in rap2.6L-1 is inserted 1 bp upstream from the start of transcription. T-DNAs in both rap2.6L-2 and -3 appeared to be compound insertions (with two left borders [LBs]) and located in the single, large intron (Fig. 4A). Homozygous lines were recovered and assayed for the presence of transcripts. Full-length transcripts were observed in wild type but only trace amounts, if any, in rap2.6L-1, 2, and 3 when assayed at 10 d SIM during shoot development in root explants (Fig. 4B). Transcript levels were severely reduced in the T-DNA insertion mutants during seedling development as well (data obtained for rap2.6L-2 not shown), however, the mutants had no obvious seedling or mature plant phenotype.
Figure 4.
T-DNA insertion mutations in RAP2.6L. A, Map showing the insertion points and the structure of the T-DNA inserts in RAP2.6L. T-DNAs in rap2.6L-2 and rap2.6L-3 are inverted repeat inserts in the single intron of RAP2.6L. B, Northern-blot of RNA from wild type and rap2.6L-1, 2, and 3 root segments at 10 d SIM hybridized to 32P-labeled RAP2.6L probe. Arrow indicates size of full-length transcript. Lanes were loaded with 10 μg total RNA. C, Shoot regeneration from root explants (13 d SIM) of wild-type seedlings. D, rap2.6L-2 homozygous line. E, Selected T1 population of 35S promoter:RAP2.6L cDNA construct in a rap2.6L-2 line demonstrating the rescue of rap2.6L-2 by RAP2.6L cDNA. F, Selected T1 population of 35S promoter:RAP2.6L-EAR motif construct. G, Fourteen day wild type seedlings. H, Fourteen day 35S promoter:RAP2.6L-EAR motif seedling. Note that cotyledons are misshapen and tend to clasp the apex. Bar = 1 mm.
However, shoot formation in culture in rap2.6L-2 was severely impaired. There were fewer shoots after 17 d SIM (0.63 ± 0.03 shoots/explant; Fig. 4D) compared to wild type (1.64 ± 0.20 shoots/explant; Fig. 4C). In addition, the shoots on rap2.6L-2 explants were smaller and less green giving an overall appearance of much diminished shoot formation in the mutant compared to wild type. The rap2.6L-1 and -3 mutants behaved similarly (data not shown), providing additional evidence that the shoot regeneration phenotype is, indeed, due to the T-DNA mutation. Attempts were made to rescue rap2.6L-2 with a 35S promoter:RAP2.6L-myc cDNA construct. The construct was partially successful in restoring the shoot regeneration phenotype (Fig. 4E).
To further confirm that the T-DNA insertions in the RAP2.6L gene were most likely responsible for the shoot regeneration phenotype, rap2.6L-2 was crossed with wild type and F2s, generated by selfing F1s, were analyzed for the segregation of the T-DNA and the defect in shoot regeneration. The F2 segregants yielded 28 wild type:45 heterozygous T-DNA:23 homozygous T-DNA, which approximated a 1:2:1 pattern (χ2 = 0.895, P = 0.639), consistent with the expected pattern for a single mutant locus. To determine if the shoot regeneration phenotype cosegregated with the T-DNA, root explants from F2 progeny were sorted into categories (wild type, T-DNA homozygotes, and heterozygotes) based on PCR genotyping, and explants groups were scored for percent explants forming shoots and for greenness of shoots. In the wild-type group, 80% (24/30) of explants formed shoots all of which were dark green; in the T-DNA heterozygote group, 96% (28/29) formed shoots, all of which were also dark green; in the T-DNA homozygote group, 45% (15/33) formed shoots, all of which were smaller and light green. We conclude from these observations that the mutant is recessive and the phenotype cosegregates with the T-DNA, indicating that the T-DNA insertion in the RAP2.6L gene is most likely responsible for the shoot regeneration trait.
We also investigated the function of RAP2.6L in shoot development by fusing an ERF-associated amphiphilic repression (EAR) motif to the C terminus of the protein. EAR motifs generally function as transcriptional repressors (Ohta et al., 2001; Hiratsu et al., 2003). Root explants from T1 seedlings bearing the 35S promoter:RAP2.6L-EAR fusion were clearly defective in shoot formation in the standard shoot regeneration system (Fig. 4F). T0 plants bearing the EAR fusion also showed extensive growth defects as seedlings, although transformants differed in the severity of phenotypes. Cotyledons did not fully expand, often forming callus and tending to curl and loosely clasp the apex (compare wild type in Fig. 4G to the seedling bearing the EAR motif fusion in Fig. 4H). The plants with less severe phenotypes were self fertile, and T1 and T2 generations showed the same seedling phenotypes (data not shown).
Localization of Gene Expression
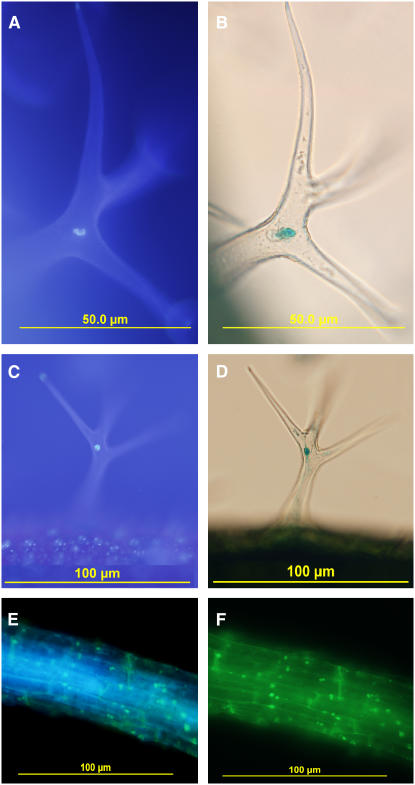
To strengthen the claim that RAP2.6L is a transcription factor, we examined the subcellular localization of RAP2.6L-β-glucuronidase (GUS) translational fusions under the control of the native RAP2.6L promoter in transgenic Arabidopsis plants. RAP2.6L promoter:RAP2.6L-GUS expression was examined in trichomes where it was localized to the large endoreduplicated nucleus of the stalk cell (Fig. 5, A–D). A comparable construct using yellow fluorescent protein (YFP; 35S promoter:RAP2.6L-YFP) showed that the protein was largely localized to nuclei in roots (Fig. 5, E and F). Thus, the subcellular localization of RAP2.6L translational fusions is consistent with its predicted function as a transcription factor.
Figure 5.
Subcellular localization of RAP2.6L in trichomes and root segments from transgenic seedlings bearing the translational fusion construct RAP2.6Lpromoter:RAP2.6L-GUS. A and C, Fluorescence images of trichomes from (7-d-old) seedlings subjected to DAPI staining. B and D, Bright-field image of the same trichomes in transgenic seedlings stained for GUS. E and F, Fluorescent images of root segment from a transgenic seedling expressing a 35S promoter:RAP2.6L-YFP construct stained with and visualized for DAPI (E) and YFP (F).
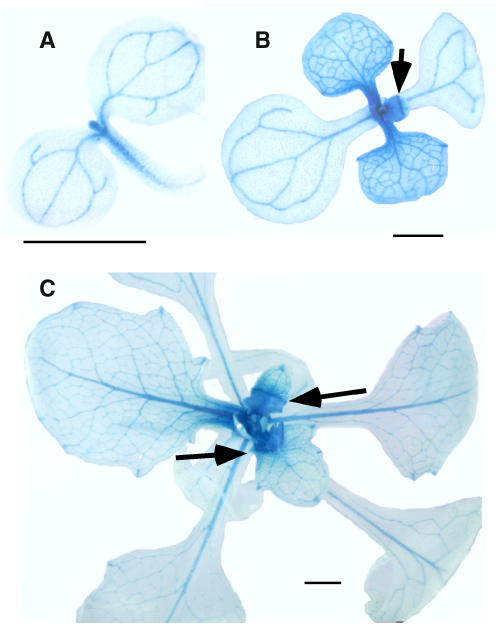
To determine where RAP2.6L is expressed in seedlings, transcriptional fusions (RAP2.6L promoter:GUS constructs) were developed. In untreated seedlings, the construct was largely expressed in the shoot apex and vasculature of roots and leaves (Fig. 6). One very interesting feature was that expression in leaf lamina declined as a frontal wave that traversed down the young leaf as it expanded (see arrows in Fig. 6, B and C). The pattern is very reminiscent of sink-to-source transitions that likewise move as a front down young leaves as they grow (Leisner et al., 1992; Leisner and Turgeon, 1993). It would be interesting to determine if the two fronts correspond.
Figure 6.
Localization of RAP2.6L expression in seedlings. Whole seedling expression patterns are shown for the transcriptional fusion construct RAP2.6Lpromoter:GUS. A, Three day seedlings. B, Seven day seedlings. C, Fourteen day seedlings. Bars = 1 mm.
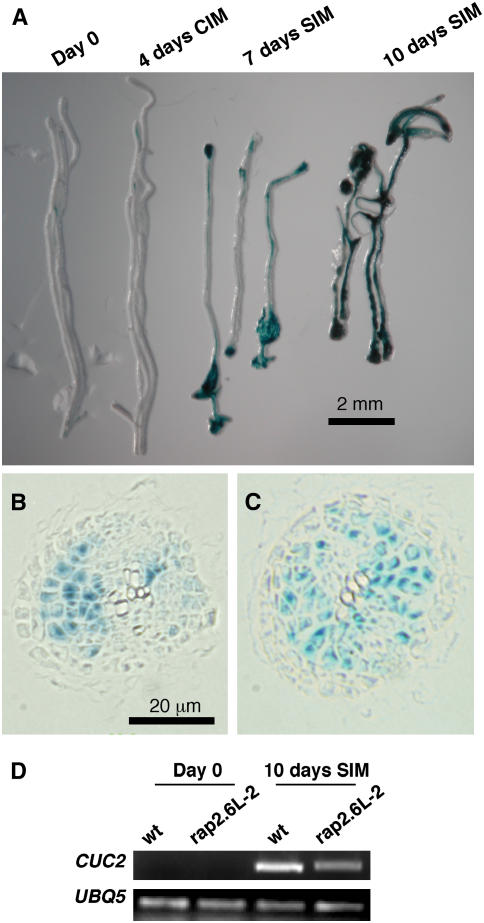
Transcriptional fusions were also used to confirm whether up-regulation in RAP2.6L gene expression in root explants during incubation on SIM is, indeed, a transcriptional phenomenon. GUS was expressed at low levels in root vasculature at day 0 and during preincubation on CIM, however, GUS expression increased dramatically when explants were incubated on SIM (Fig. 7A). Thus, the up-regulation of RAP2.6L has, at least, a strong transcriptional component. RAP2.6L promoter activity (GUS staining) was most intense in regions of the root explant where callus had formed, particularly at the ends of the explanted root segments (Fig. 7A). In cross-sectional view, GUS staining was localized to sites of cell proliferation (Fig. 7, B and C). GUS staining is shown at 7 d SIM, at a time when callus formation is easily recognizable. At this stage the epidermis has deteriorated and the vascular bundle broken apart. Callus tissue, most likely derived from the pericycle and/or vascular parenchyma in the intact root, is heavily GUS stained. It is from this tissue that organs regenerate, however, RAP2.6L expression well precedes any evidence of organ primordia formation (Cary et al., 2002).
Figure 7.
Time course and localization of RAP2.6L expression during shoot regeneration in root explants. A, Root explants from RAP2.6Lpromoter:GUS seedlings were subjected to shoot regeneration conditions and stained for GUS expression at the times indicated. B and C, Cross section of GUS-stained roots at 7 d SIM. At this stage, the epidermal layer and most of the vascular bundle have deteriorated; the cortical and endodermal layers are largely unstained. Callus, most likely derived from the pericycle and vascular parenchyma, show GUS staining. D, Semiquantitative RT-PCR of the induction of CUC2 in wild type and in rap2.6L-2. Expression at 10 d SIM was compared to day 0. At3g62250 (UBQ5) was used as a control.
Downstream Targets of RAP2.6L Expression
Since the T-DNA insertion mutation rap2.6L-2 severely down-regulates the expression of the gene, we attempted to measure the impact of the mutation on the expression of other genes during shoot regeneration as determined by Affymetrix DNA chip analysis. This experiment was performed using three independent wild-type samples and two independent mutant samples. Twenty-four genes showed more than 10-fold down-regulation in rap2.6L-2 compared to wild type when controlling the FDR at the 0.05 level (Table IV; Supplemental Table III). The RAP2.6L gene itself was down-regulated over 30-fold when rap2.6L-2 was compared to wild type. The two most highly down-regulated genes at 10 d SIM are of unknown function; one (At3g05730) is a shoot development-specific gene that is highly up-regulated during shoot development. Others that were significantly down-regulated included cellulose synthetase, subtilisin-like Ser protease, β-glucosidase, and so forth. Further down the list was CUP SHAPED COTYLEDON2 (down 2.4-fold), a gene that acts redundantly with CUC1 to activate SHOOT MERISTEMLESS expression and to specify shoot meristem formation (Aida et al., 1997; Aida et al., 1999; Takada et al., 2001; Daimon et al., 2003; Hibara et al., 2003). CUC2 is highly up-regulated at 10 d SIM (in comparison with day 0) in root explants from wild-type seedlings, but less up-regulated in rap2.6L-2 (Fig. 7D). Of the 478 genes specifically up-regulated during shoot development, 175 (or approximately 35%) were down-regulated more than 1.5-fold in the rap2.6L-2 mutant compared to wild type (Supplemental Table III). Some of these shoot-specific genes might be down-regulated because they are immediate targets of RAP2.6L action. Others may be indirect targets removed several steps from RAP2.6L. Nonetheless, the impact of the rap2.6L-2 mutant demonstrates the pivotal role of RAP2.6L early in the program of gene expression during shoot regeneration.
Table IV.
Down regulation of Arabidopsis genes in rap2.6L-2 mutant
Experiment was performed with three independent wild-type samples and two independent mutant samples. RNA samples were taken at 10 d SIM under standard shoot regeneration conditions as shown in Figure 1. Genes listed down to the break in the table showed more than 10-fold down-regulation in rap2.6L-2 compared to wild type when controlling the FDR at the 0.05 level. The gene below the break, CUC2, showed lower FC, but was included because it is involved in shoot meristem specification.
| Locus Identificationa | FCb | P Valueb | q Valueb | Descriptions |
|---|---|---|---|---|
| At3g05730 | 82.6 | 2.21E-04 | 3.38E-03 | Unknown protein |
| At1g23130 | 40.4 | 7.41E-05 | 2.25E-03 | Bet v I allergen family protein |
| At5g13330 | 33.4 | 3.17E-09 | 3.42E-05 | RAP2.6L |
| At3g55970 | 24.4 | 2.14E-04 | 3.35E-03 | Oxidoreductase, 2OG-Fe(II) oxygenase protein |
| At4g18780 | 22.0 | 1.54E-04 | 2.84E-03 | Cellulose synthase, catalytic subunit (IRX1) |
| At3g16670 | 21.2 | 2.05E-03 | 1.04E-02 | Unknown protein |
| At3g54490 | 21.1 | 6.08E-05 | 2.16E-03 | RNA polymerase II 23-kD polypeptide (rpb5) |
| At1g01900 | 19.9 | 5.94E-06 | 1.09E-03 | Subtilisin-like Ser protease |
| At1g52400 | 16.7 | 2.39E-04 | 3.50E-03 | β-Glucosidase |
| At1g54020 | 16.0 | 9.27E-04 | 6.81E-03 | Myrosinase-associated protein |
| At5g22460 | 15.0 | 2.06E-04 | 3.28E-03 | Esterase/lipase/thioesterase family protein |
| At1g80100 | 14.2 | 2.38E-04 | 3.50E-03 | HPt phosphotransmitter |
| At3g54820 | 13.2 | 4.69E-07 | 8.00E-04 | Aquaporin 2 |
| At5g24420 | 12.5 | 9.23E-04 | 6.81E-03 | Glucosamine/galactosamine-6-P isomerase |
| At4g05110 | 12.0 | 2.78E-04 | 3.81E-03 | Equilibrative nucleoside transporter |
| At1g59500 | 11.8 | 4.93E-03 | 1.72E-02 | Auxin-regulated protein GH3 |
| At1g80130 | 11.0 | 5.19E-04 | 5.13E-03 | Unknown protein |
| At1g73120 | 11.0 | 2.02E-03 | 1.03E-02 | Hypothetical protein |
| At4g37710 | 11.0 | 4.45E-04 | 4.77E-03 | VQ motif-containing protein |
| At3g15720 | 10.9 | 6.68E-07 | 8.00E-04 | Putative polygalacturonase |
| At4g03880 | 10.7 | 3.02E-03 | 1.28E-02 | Putative transposon protein |
| At3g59440 | 10.2 | 1.07E-05 | 1.21E-03 | Calmodulin-like protein calcium-binding protein |
| At4g26150 | 10.2 | 1.16E-03 | 7.70E-03 | GATA-type zinc finger transcription factor |
| At1g23730 | 10.1 | 1.28E-05 | 1.21E-03 | Putative carbonic anhydrase |
| At5g53950 | 2.4 | 1.07E-03 | 7.33E-03 | CUC2 |
Genes showing more than 10-fold down-regulation, q-value threshold for T-DNA comparisons <0.05.
For the comparison of estimated mean expression values between wild type and rap2.6L-2.
DISCUSSION
The developmental system described here is a powerful tool for studying gene expression during organogenesis in plants. By profiling gene expression during CIM preincubation and during early shoot, root, and callus regeneration, we have developed a framework to define molecular signatures for the different developmental processes.
Many of the genes up-regulated during early stages of shoot development were genes that respond to cytokinin induction, most notably the A-type ARRs (Brandstatter and Kieber, 1998; D'Agostino et al., 2000; Sakai et al., 2000; Hwang and Sheen, 2001; Sakai et al., 2001; Hutchison and Kieber, 2002; Hwang et al., 2002; Rashotte et al., 2003; To et al., 2004). A HK (AHK1) associated with osmotic responses (Urao et al., 1999), GA metabolism and response genes, and a variety of transcription factors were also prominently up-regulated during early shoot development. Because this period in shoot regeneration is also characterized by the formation of green callus, many genes involved in the development of the photosynthetic apparatus were up-regulated.
The genes associated with the acquisition of competence and those that are unique to callus formation were more difficult to categorize. During CIM preincubation, cells in the explants are thought to dedifferentiate and acquire competence to respond to subsequent shoot induction signals. As pointed out, a gene involved in chromatin remodeling, a GNAT, and several transcription factors were highly up-regulated at that time. At later stages of CIM incubation (such as 10 d CIM), cells proliferate and form undifferentiated callus tissue. Many genes that were specifically up-regulated are stress-related factors such as genes encoding a AAA-type ATPase family protein, ATP-binding cassette (ABC) transporter, and a WRKY 38 stress-response transcription factor. Similar stress-related genes form the molecular signature for pluripotent animal cells (Ramalho-Santos et al., 2002), which like plant callus tissue retain their stemness or ability to give rise to other more differentiated tissues.
Many of the genes specifically up-regulated during root development on RIM were expressed at high levels on day 0, in the mature root. It might be expected that the molecular signatures for early callus and root regeneration would be quite similar since both represent growth on auxin-rich medium. The most obvious difference was the number of root development-specific genes associated with cell wall and vascular development: peroxidases, extensin, arabinogalactan protein, xyloglucan endotransglycosylase, and so forth.
In this study, we also began to dissect the control of the large-scale gene expression changes that take place during early shoot development. We focused on RAP2.6L (At5g13330), a gene encoding an ERF/AP2 transcription factor, because it was one of the transcription factor genes specifically and highly up-regulated during early shoot development. T-DNA mutations in RAP2.6L reduced the efficiency of shoot regeneration in culture and significantly knocked down the expression of approximately 35% of the 478 genes that are specifically up-regulated on SIM. This would tend to indicate that RAP2.6L acts early and plays a pivotal role during the shoot regeneration process. High on the list of genes impacted by the rap2.6L-2 mutation were genes such as those encoding the catalytic subunit of cellulose synthase, a RNA polymerase II subunit, a subtilisin-like Ser protease, a HPt phosphotransmitter, and a GATA-binding transcription factor. Further down the list, but still significantly down-regulated in the mutant was CUC2, a gene along with CUC1 that is important for shoot regeneration and meristem specification (Aida et al., 1997; Aida et al., 1999; Takada et al., 2001; Daimon et al., 2003; Hibara et al., 2003). Although many genes are down-regulated during shoot regeneration by the rap2.6L-2 mutation, we do not know what genes are the direct targets of RAP2.6L action.
We found that a RAP2.6L promoter:GUS construct was expressed in seedlings, primarily in the shoot apex and the vasculature. Tissue-specific microarray expression data from AtGenExpress (http://www.arabidopsis.org/info/expression/ATGenExpress.jsp) confirms that RAP2.6L transcripts are present at highest levels in germinating seedlings and in the developing cotyledons. In RAP2.6L constructs bearing the transcriptional repressor EAR motif (35S promoter:RAP2.6L-EAR), the most obvious phenotype is a defect in cotyledon development. The RAP2.6L-EAR-expressing seedlings bore curled, not fully expanded cotyledons, often intercalated with nonchlorophyllous callus.
RAP2.6L is up-regulated when root explants are transferred onto cytokinin-rich SIM, however, there are conflicting observations whether cytokinin alone is sufficient to up-regulate the expression of the gene. For example, we have treated seedlings with various concentrations of cytokinin and at various times (usually hours) and not observed up-regulation in RAP2.6L promoter:GUS expression. Also, microarray data at AtGenExpress indicate that RAP2.6L is not significantly up-regulated in seedlings of a similar age treated with transzeatin (1 μm). On the other hand, Brenner et al. (2005) reported that RAP2.6L transcripts increase 2-fold after 15 min BA treatment of 5-d-old seedlings. In our study, we looked at much later time points, and furthermore, root explants were subject to culture conditions (CIM preincubation) that may precondition tissues to respond to cytokinin signals. Thus, the cytokinin signal may also require the appropriate developmental context in which to activate RAP2.6L.
Finally, the observation that seedlings develop normally, but shoots do not efficiently regenerate in the rap2.6L-2 mutant argues that the gene malfunction is less well compensated during shoot regeneration in culture than during shoot formation in seedling development. However, the huge loss in expression of many shoot-specific genes during shoot regeneration in rap2.6L-2 demonstrates the key role for this gene in shoot regeneration.
MATERIALS AND METHODS
Plant Material and Culture Conditions
Arabidopsis (Arabidopsis thaliana) seedlings (ecotype Columbia-0) were grown for 7 d on plant nutrient solution medium (Che et al., 2002). Five millimeter root segments were cut and transferred to CIM: Gamborg's B5 medium (Gamborg et al., 1968) with 0.5 g/L MES, 2.2 μm 2,4-dichlorophenoxyacetic acid, 0.2 μm kinetin, and 0.8% agarose. Explants were preincubated on CIM for 4 d and then transferred to SIM containing 5.0 μm isopentenyladenine and 0.9 μm IAA, fresh CIM, or RIM containing 0.9 μm IAA.
RNA Extraction and Profiling
Total RNA was isolated from plant tissues by TRIzol (Life Technologies, Gibco-BRL) extraction. Precipitated RNA was solubilized in water treated with 0.1% (v/v) diethyl pyrocarbonate and purified with a RNeasy kit (Qiagen). Purified RNA was assessed for integrity using an Agilent 2100 Bioanalyzer. Gene expression patterns were profiled using Affymetrix Arabidopsis 22K GeneChips according to procedures described by Che et al. (2002). Expression data were analyzed with SAS version 9.1 (Inc SI), R version 1.9.0 (R Development Core Team RFfSC; R: A language and environment for statistical computing, http://www.R-project.org [Vienna, Austria]).
Semiquantitative RT-PCR analysis was used to confirm various expression patterns determined by microarray. Two micrograms of total RNA were reverse transcribed using Ready-To-Go You-Prime first-strand beads (Amersham) in a 33 μL reaction. PCR was carried out using 2 μL of the RT reaction as template. Cycle numbers were optimized for each sample to obtain data in the exponential range. Amplified DNA fragments were separated on 2% agarose gel and stained with ethidium bromide. The primers used for amplification were as follows.
Ubiquitin 5 (At3g62250): UBQ5F (5′-CTTGAAGACGGCCGTACCCTC-3′), UBQ5R (5′-CGCTGAACCTTTCAAGATCCATCG-3′); At2g23170: IAAaseF TCCTCACAAGCTCTGGGACA, IAAaseR CGTTAGGGCTCGTGTACACG; At2g23060: AcetF CCTCATGCTGGTGGCTGAGA, AcetR ATTGACGGAAGCGTGATTGT; At2g03850: LEAF ATGATGCCTCACAGAAAGCT, LEAR TGGAGGCATTATAGCTTCTT; At1g75880: EXL1F GATATTGTAGCGGAAGAGCT, EXL1R CTGAGCAAAAGAACGAGCATTRAP; RAP2.6-like transcription factor (RAP2.6L; At5g13330): Rap2.6-likeF (5′-ACCAGACCAAGATCAACCAAGA-3′), Rap2.6-likeR (5′-TTATTCTCTTGGGTAGTTATAA-3′); CUC2 (At5g53950): CUP3 (5′-CAGCCAATATCTTCCACCGGG-3′), CUP11 (5′-GGAGAGGTGGGAGTGAGACGGA-3′).
Microarray Experimental Design and Statistical Analysis
Microarray experiments to identify shoot, root, and callus-specific genes were designed as a randomized complete block design with two independent replications. Within each replication, 10 plates of root explants were randomly assigned to each of the nine time points as indicated. A total of 18 Affymetrix ATH1 GeneChips were used to measure expression in pools of root explants, with one GeneChip per combination of time point and replication.
SAS software was used to conduct a separate ANOVA for each of 22,810 probe sets. Signals were normalized by scaling all GeneChips to a target intensity of 1,500 (see Affymetrix GeneChip GCOS manual: http://www.affymetrix.com/products/software/specific/gcos.affx). The natural logarithm of the scaled signal measure was used as the response variable for each ANOVA. The ANOVA model for each gene included replication and time point effects (with 1 and 8 degrees of freedom, respectively) along with an error term (8 degrees of freedom) essential for testing the statistical significance of observed time point differences. The F test for differences among time point means was used to identify genes in which expression was not constant across all conditions.
To identify genes specifically up- and down-regulated during shoot, root, and callus development, several contrasts of time point means were implemented for each gene as part of our ANOVA. Specific contrasts included a comparison of 10 d SIM with each of 4 d CIM, 7 d CIM, 10 d CIM, 7 d RIM, and 10 d RIM to identify shoot development-specific genes; a comparison of 10 d RIM with 4 d CIM, 7 d CIM, 10 d CIM, 7 d SIM, 10 d SIM to identify root development-specific genes; and a comparison of 10 d CIM to 4 d CIM, 7 d RIM, 10 d RIM, 7 d SIM, and 10 d SIM to identify callus development-specific genes.
A set of 22,810 P values was obtained for the F test for time point differences and each of the other seven contrasts of time point means. Each of these sets of P values was converted to q values using an R implementation (R Development Core Team RFfSC; R: A language and environment for statistical computing, http://www.R-project.org [Vienna, Austria]) of the algorithm of Storey and Tibshirani (2003). These q values can be used to obtain lists of differentially expressed genes while controlling FDR at a specified level. For example, the set of genes whose q values from a particular test are less than or equal to 0.05 form a list of differentially expressed genes for which the FDR is estimated to be 5%.
The microarray analysis to identify the downstream targets of RAP2.6.L (expression in rap2.6L-2 compared to wild type) were identical to the analysis of the developmental time course experiment except that block terms were excluded from the model and gene-specific variance estimates were obtained by pooling across two experiments to obtain sufficient error degrees of freedom (five per gene) for the contrast of interest (rap2.6L-1 mutant versus wild type).
Genotyping
Segregation analysis was performed by genotyping progeny of the rap2.6L-1, -2, or -3 mutants in various crosses. Progeny (usually 7-d-old seedlings) were genotyped by extracting DNA from seedlings (usually from a single cotyledon) using the DNA Quick-prep procedure (see http://www.biotech.wisc.edu/NewServicesandResearch/Arabidopsis/FindingYourPlantIndex.html) and using the following primers for the rap2.6L-1 insertion site. RAP26T3F: 5′-TTGCGATCCCCACTTGTTGT-3′; Rap26T3R: 5′-TGAAAGATGCATTGAACTTG-3′; for the Rap2.6L-2 and -3 insertion sites, AP226F: 5′-TTCGTCTTGGAACGAGACTG-3′; AP226R: 5′-AAAACTGATTCGACCAACAATAA-3′ and for the LB of the T-DNA insert (LB: 5′-TGGTCCACGTAGTGGGCCATC-3′).
Localization Studies
Translational fusions were constructed between RAP2.6L and GUS to examine the subcellular location of RAP2.6L. RAP2.6L with its own promoter was fused in frame to the GUS reporter gene by insertion into the PstI and BamI sites of the multicloning site in the pCAMBIA3300 GUS vector (see http://www.cambia.org/pCAMBIA_vectors.html#Description). RAP2.6L promoter and coding region were amplified using a genomic DNA template and the following primers: Ap2.6GUSF 5′-AACTGCAGCTGATTTTCCTCTTTAAAACGGAAAACA-3′ and Ap2.6tGUSR 5′-CGGGATCCTCTCTTGGGTAGTTATAATAATTGTAACC-3′. A transcriptional fusion linking the RAP2.6L promoter to GUS was created by inserting the RAP2.6L promoter into the multicloning site of pCAMBIA3300 GUS. The RAP2.6L promoter was amplified using the following primers: AP2.6proF 5′-AACTGCAGTTGTTCTTCCTTGGTTTT-3′ and AP2.6proR CGGGATCCGGCGGTGACATCAGTCTC. The resulting constructs were introduced into Agrobacterium tumefaciens strain C58, which was used to generate transgenic plants by the floral dip method (Clough and Bent, 1998).
Histochemical staining for GUS activity was conducted as described by Jefferson (1987) with minor modifications. Whole seedlings or excised plant organs and tissues were incubated in 5-bromo-4-chloro-3-indole glucuronide (X-gluc) solution (0.5 mg/mL X-gluc in 50 mm Tris/HCl buffer [pH 7.0], 0.5% [v/v] Triton X-100, 0.5 mm K3[Fe(CN)6], 0.5 mm K4[Fe(CN)6], 10 mm Na2EDTA). A total of 100 mm X-gluc stock solution was prepared by dissolving 26.1 mg X-gluc in 0.5 mL dimethyl sulfoxide just before use. Vacuum infiltration was carried out for 10 min. Tissue was then incubated at 37°C in the dark for 16 h or until color developed. Chlorophyll was cleared by extracting with several changes of 70% (v/v) ethanol, and whole mounts were examined under bright-field microscopy using a Nikon SMZ1000 microscope.
For sectioned material, samples stained for GUS were fixed with formaldehyde/paraformaldehyde, dehydrated in a graded ethanol series, cleared with xylene, infiltrated, and embedded using Paraplast X-Tra paraffin (Fisher Scientific). Sections were made using an A/O 820 rotary microtome (Fisher Scientific). After GUS staining was visualized and photographed, the sections were further stained with 1 μg mL−1 4′-6-diamidino-2-phenylindole (DAPI) for 30 min, and sections were visualized by epifluorescence with an Olympus IX71 microscope.
Other Constructs
RAP2.6L constructs bearing the EAR repressor motif (LDLDLELRLGFA) were developed by excising YFP from pSKY by cutting with SpeI and NotI and replacing it with a DNA fragment containing the EAR motif to create pSKEAR. The DNA fragment was assembled from two single-stranded oligomers: EARF 5′-GACTAGTTTAGATCTAGATCTTGAGTTGAGACTGGGTTTCGCCTGAGCGGCCGCTAAACTAT-3′ and EARR 5′-ATAGTTTAGCGGCCGCTCAGGCGAAACCCAGTCTCAACTCAAGATCTAGATCTAAACTAGTC-3′. The RAP2.6L coding region was inserted into the AscI and SpeI sites of pSKEAR by amplifying RAP2.6L using the primers described above, 35S promoter:RAP2.6L-myc and 35S promoter:RAP2.6L-YFP constructs were generated by amplifying the insert from a full-length RAP2.6L cDNA clone obtained from the Arabidopsis Biological Resource Center using the primers Ap2RAP2.6LF 5′-TAGCGGCGCGCCATGGTCTCCGCTCTCAGCCG-3′ and Ap2RAP2.6LR 5′-GACTAGTTTCTCTTGGGTAGTTATAAT-3′ and inserting into the AscI and SpeI sites of pSKM (myc) and pSKY (YFP). The primers were also used to generate 32P-labeled probes used for Northern-blot analysis of RAP2.6L transcripts.
Supplementary Material
This work was supported by the National Science Foundation (IBN-0236060) and by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education and Extension Service (grant no. 2003–35304–13363).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Stephen H. Howell (shh@iastate.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.081240.
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Ishida T, Tasaka M (1999) Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126: 1563–1570 [DOI] [PubMed] [Google Scholar]
- Allison DB, Cui X, Page GP, Sabripour M (2006) Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet 7: 55–65 [DOI] [PubMed] [Google Scholar]
- Banno H, Ikeda Y, Niu QW, Chua NH (2001) Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13: 2609–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry WT, Nobel AB, Wright FA (2005) Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics 21: 1943–1949 [DOI] [PubMed] [Google Scholar]
- Brandstatter I, Kieber JJ (1998) Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10: 1009–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Kollmer I, Burkle L, Schmulling T (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Cary AJ, Che P, Howell SH (2002) Developmental events and shoot meristem gene expression patterns during shoot development in Arabidopsis thaliana. Plant J 32: 867–877 [DOI] [PubMed] [Google Scholar]
- Che P, Gingerich DJ, Lall S, Howell SH (2002) Global and cytokinin-related gene expression changes during shoot development in Arabidopsis. Plant Cell 14: 2771–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- D'Agostino IB, Deruere J, Kieber JJ (2000) Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol 124: 1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daimon Y, Takabe K, Tasaka M (2003) The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol 44: 113–121 [DOI] [PubMed] [Google Scholar]
- Fisher RA (1934) Statistical Methods for Research Workers, Ed 14. Oliver and Boyd, Edinburgh
- Gamborg OJ, Miller RA, Ojima K (1968) Nutrient requirement of suspension cultures of soybean root cells. Exp Cell Res 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Gautheret RJ (1966) Factors affecting differentiation of plant tissue grown in vitro. In W Beerman, PD Nieuwkoop, E Wolff, eds, Cell Differentiation and Morphogenesis. North-Holland Publishing, Amsterdam, pp 55–95
- Hibara K, Takada S, Tasaka M (2003) CUC1 gene activates the expression of SAM-related genes to induce adventitious shoot formation. Plant J 36: 687–696 [DOI] [PubMed] [Google Scholar]
- Hicks GS (1980) Patterns of organ development in plant tissue culture and the problem of organ determination. Bot Rev 46: 1–23 [Google Scholar]
- Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H (2001) A large family of class III plant peroxidases. Plant Cell Physiol 42: 462–468 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Huang BC, Yeoman MM (1984) Callus proliferation and morphogenesis in tissue cultures of Arabidopsis thaliana L. Plant Sci Lett 33: 353–363 [Google Scholar]
- Hutchison CE, Kieber JJ (2002) Cytokinin signaling in Arabidopsis. Plant Cell (Suppl) 14: S47–S59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Chen HC, Sheen J (2002) Two-component signal transduction pathways in Arabidopsis. Plant Physiol 129: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Kiba T, Ueguchi C, Sugiyama T, Mizuno T (1999) Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol 40: 733–742 [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409: 1060–1063 [DOI] [PubMed] [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387–405 [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Leisner S, Turgeon R (1993) Movement of virus and photassimilate in the phloem: a comparative analysis. Bioessays 15: 741–748 [DOI] [PubMed] [Google Scholar]
- Leisner SM, Turgeon R, Howell SH (1992) Long distance movement of cauliflower mosaic virus in infected turnip plants. Mol Plant Microbe Interact 5: 41–47 [Google Scholar]
- Loidl P (2004) A plant dialect of the histone language. Trends Plant Sci 9: 84–90 [DOI] [PubMed] [Google Scholar]
- Mason MG, Li J, Mathews DE, Kieber JJ, Schaller GE (2004) Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol 135: 927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M (2001) Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13: 1959–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A, Sakai H, Iwakoshi S (2002) His-Asp phosphorelay signal transduction in higher plants: receptors and response regulators for cytokinin signaling in Arabidopsis thaliana. Genes Genet Syst 77: 383–391 [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA (2002) “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science 298: 597–600 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, Carson SD, To JP, Kieber JJ (2003) Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 132: 1998–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379: 633–646 [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Bono H, Oka A (1998) Two-component response regulators from Arabidopsis thaliana contain a putative DNA-binding motif. Plant Cell Physiol 39: 1232–1239 [DOI] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24: 703–711 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Schmulling T (2001) CREam of cytokinin signalling: receptor identified. Trends Plant Sci 6: 281–284 [DOI] [PubMed] [Google Scholar]
- Sheen J (2002) Phosphorelay and transcription control in cytokinin signal transduction. Science 296: 1650–1652 [DOI] [PubMed] [Google Scholar]
- Skoog F, Miller CO (1957) Chemical regulation of growth and organ formation in plant tissue cultured in vitro. Symp Soc Exp Biol 11: 118–131 [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ishikawa K, Yamashino T, Mizuno T (2002) An Arabidopsis histidine-containing phosphotransfer (HPt) factor implicated in phosphorelay signal transduction: overexpression of AHP2 in plants results in hypersensitiveness to cytokinin. Plant Cell Physiol 43: 123–129 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T (2001) The Arabidopsis sensor His-kinase, AHk4, can respond to cytokinins. Plant Cell Physiol 42: 107–113 [DOI] [PubMed] [Google Scholar]
- Takada S, Hibara K, Ishida T, Tasaka M (2001) The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128: 1127–1135 [DOI] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K (1999) A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11: 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Lijsebettens MV (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Suzuki T, Terada K, Takei K, Ishikawa K, Miwa K, Mizuno T (2001) The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol 42: 1017–1023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.