Abstract
The nonphototropic hypocotyl 4 (nph4)/auxin response factor 7 (arf7) mutant of Arabidopsis (Arabidopsis thaliana) is insensitive to auxin and has defects in hypocotyl tropism, hook formation, differential leaf growth, and lateral root formation. To understand an auxin-signaling pathway through NPH4, we carried out screening of suppressor mutants of nph4-103 and obtained a dwarf suppressor mutant, suppressor of nph4 (snp2). snp2 had short hypocotyls in the dark condition and dark green and round leaves, short petioles, and more lateral shoots than the wild type in the light condition. The snp2 phenotypes were rescued by adding brassinolide to the growth medium in both light and dark conditions. Genetic mapping, sequence analysis, and a complementation test indicated that snp2 was a weak allele of DWARF4 (DWF4), which functions in brassinosteroid (BR) biosynthesis. snp2, which was renamed dwf4-101, exhibited photo- and gravitropisms of hypocotyls similar to those of the wild type with a slightly faster response in gravitropism. dwf4-101 almost completely suppressed defects in both tropisms of nph4-103 hypocotyls and completely suppressed hyponastic growth of nph4-103 leaves. Treatment with brassinazole, an inhibitor of BR biosynthesis, also partially rescued the tropic defects in nph4-103. Hypocotyls of nph4-103 were auxin insensitive, whereas hypocotyls of dwf4-101 were more sensitive than those of the wild type. dwf4-101 nph4-103 hypocotyls were as sensitive as those of dwf4-101. Auxin inducibility of massugu 2 (MSG2)/IAA19 gene expression was reduced in nph4-103. mRNA level of MSG2 was reduced in dwf4-101 and dwf4-101 nph4-103, but both mutants exhibited greater auxin inducibility of MSG2 than the wild type. Taken together, dwf4-101 was epistatic to nph4-103. These results strongly suggest that BR deficiency suppresses nph4-103 defects in tropic responses of hypocotyls and differential growth of leaves and that BR negatively regulates tropic responses.
The plant hormone auxin acts in diverse processes during the course of plant development. For example, it functions as a mediator of tropic responses (Esmon et al., 2005) and as a positional signal in organ development (Berleth and Sachs, 2001). The molecular basis of the role of auxin in the regulation of a wide range of developmental processes has been intensively studied using molecular genetic approaches for nearly 20 years. Because we have been interested in tropic responses, we have screened Arabidopsis (Arabidopsis thaliana) mutants that do not exhibit hypocotyl bending when auxin-containing lanolin paste is applied to hypocotyls unilaterally (Watahiki and Yamamoto, 1997). This screening led us to the isolation of a recessive mutant, nonphototropic hypocotyl 4/massugu 1 (nph4/msg1; Liscum and Briggs, 1995; Watahiki and Yamamoto, 1997), and a dominant mutant, msg2 (Tatematsu et al., 2004). Both mutants were insensitive to auxin and had defects in gravi- and phototropism in hypocotyls. NPH4 encodes AUXIN RESPONSE FACTOR 7 (ARF7; Harper et al., 2000), and MSG2 encodes a transcriptional repressor, AUX/IAA19 (Tatematsu et al., 2004). Because ARFs bind to AUX/IAAs through their conserved C-terminal domains (Hagen and Guilfoyle, 2002), and because AUX/IAA19 is an auxin early-responsive gene (Abel and Theologis, 1996), NPH4/ARF7 and MSG2/IAA19 have been proposed to constitute a negative feedback loop to regulate differential growth responses of hypocotyls (Tatematsu et al., 2004).
Although we have discovered that NPH4 and MSG2 have a central role in auxin-mediated hypocotyl bending, we know almost nothing about the signaling pathways downstream of them. To better understand these pathways, we screened for suppressor mutants of nph4 in this study. nph4-103/msg1-3 is defective in differential growth of cotyledons and rosette leaves as well as tropic responses of hypocotyls (Watahiki and Yamamoto, 1997). They show hyponastic growth of leaves. Therefore, we selected suppressor mutant lines that restored both the weakly epinastic growth of wild-type leaves and the faster gravitropic response of hypocotyls. We isolated mutant lines of three complementation groups, which were named suppressor of nph4 (snp). Here we describe physiological and molecular characteristics of one of them, snp2.
Genetic mapping and a complementation test in this study revealed that snp2 is a new allele of DWARF4 (DWF4) that encodes C-22-α-hydroxylase in the brassinosteroid (BR) biosynthetic pathway (Choe et al., 1998). The results clearly indicate that BR is involved in the auxin signal transduction pathway of tropic responses. In fact, it has long been known that BR is closely associated with auxin signaling (Mandava, 1988). This work presents molecular genetic evidence that concerted regulation of auxin and BR is necessary for normal tropic responses in etiolated Arabidopsis hypocotyls.
RESULTS
Isolation of Suppressor Mutants of nph4
To isolate suppressor mutants of nph4, we mutagenized about 5,000 seeds of nph4-103 by ethyl methanesulfonate. In the M2 generation, we selected 171 mutant lines that did not show hyponastic growth of cotyledons and rosette leaves observed in nph4-103. In the next generation, we isolated suppressor candidates that restored hypocotyl gravitropism. After examination of the presence of the nph4-103 mutation by a derived cleaved-amplified polymorphic sequence marker, we obtained five suppressor mutant lines in three loci, snp1 to 3.
ssnp2 Shows Dwarf Phenotype That Is Rescued by Addition of Brassinolide to the Medium
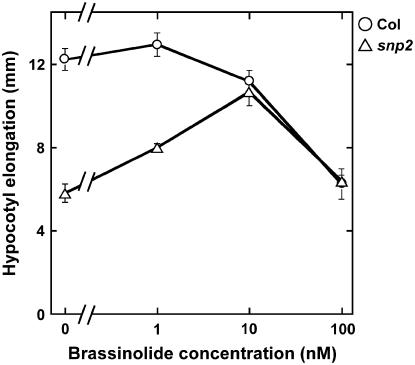
snp2-1 showed a dwarf phenotype; it displayed short stems, dark green and round leaves with short petioles, and more lateral shoots than wild-type Columbia (Col; Fig. 1). It also exhibited short hypocotyls in the dark condition (Fig. 2). These characteristics suggested that snp2 was a BR-related mutant. Because BR-deficient phenotypes can be rescued by exogenous application of BR (Szekeres et al., 1996), we examined whether addition of brassinolide (BL) to snp2-1 restored the wild-type growth of hypocotyls in the dark. In the presence of 10 nm BL, snp2-1 hypocotyls elongated as long as those of Col (Fig. 2). BL treatment also reversed the aberrant phenotype in leaf and petiole length observed in light-grown snp2-1 seedlings (data not shown). These results suggest that snp2-1 is a BR-deficient mutant.
Figure 1.
snp2, a suppressor mutant of nph4-103. Arabidopsis plants of Col (a), nph4-103 (b), snp2-1 nph4-103 (c), and snp2-1 (d) were grown for 2 (A) or 8 (B) weeks in continuous light conditions. Diameter of the pots is 5.5 cm.
Figure 2.
Effects of BL on hypocotyl elongation of Col (○) and snp2-1 (▵) grown in liquid medium for 5 d in the dark. Values shown represent the mean ± se of three independent experiments in which hypocotyl elongation of 20 seedlings was measured.
snp2 Is a Weak Allele of DWF4
The SNP2 locus was mapped by crossing snp2 (Col background) to Landsberg erecta (Ler). By examining 44 chromosomes, snp2-1 was found to be tightly linked to the T20E23 and F18B3 markers in the lower arm of chromosome 3: Only one recombinant was obtained for each marker. The DWF4 gene lies between the two markers, which span 182 kb. DWF4 encodes cytochrome P450 (CYP), which functions as C-22-α-hydroxylase in BR biosynthesis. dwf4 exhibits a dwarf phenotype due to defects in BR biosynthesis (Azpiroz et al., 1998).
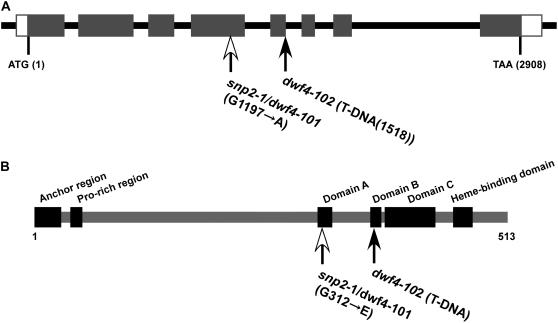
Because the reported dwf4 alleles, dwf4-1 to 4, were from a Wassilewskija (Ws) or Enkheim background (Azpiroz et al., 1998), we searched for other dwf4 alleles in the Col background and found a T-DNA insertion line (SALK_020761), which we named dwf4-102. T-DNA was found in the fifth exon of the DWF4 gene in dwf4-102 (Fig. 3), and its phenotype was as severe as that of dwf4-1 (data not shown). We carried out a complementation test between snp2-1 and dwf4-102. Because dwf4-102 was completely infertile, heterozygous dwf4-102 was crossed with snp2-1. The segregation ratio of F1 and F2 progeny in Table I showed that snp2-1 was allelic to dwf4-102. Thus, we renamed snp2-1 dwf4-101 thereafter. When grown in the dark, dwf4-101/snp2-1 hypocotyls were shorter than those of Col, but longer than those of dwf4-102 (Table II). dwf4-102 appeared to be a null allele and dwf4-101 was a weak allele of dwf4.
Figure 3.
A, Structure of the DWF4/SNP2 gene. White and black arrows indicate mutation position of dwf4-101 and T-DNA insertion position of dwf4-102, respectively. The DWF4 coding sequence (1,542 bp) consists of eight exons (black rectangles). White rectangles are untranslated regions. B, Structure of the DWF4 protein (Choe et al., 1998). White arrow indicates amino acid substitution position in dwf4-101.
Table I.
Complementation test between snp2-1 and dwf4-102
Phenotype was determined when plants were grown in continuous light for 20 d.
| Cross | Wild-Type Phenotype | Dwarf Phenotype |
|---|---|---|
| snp2-1 snp2-1 × dwf4-102 DWF4 F1 | 12 | 8 |
| F2 | 0 | 18 |
Table II.
Effects of the dwf4 and bri1 mutations and Brz treatment on hypocotyl length of seedlings grown for 3 d in the dark
Values shown indicate the mean ± sd of 20 seedlings. ND, Not determined.
| Genotype | Hypocotyl Length
|
|
|---|---|---|
| −Brz | +1 μm Brz | |
| mm | ||
| Col | 9.5 ± 1.4 | 4.9 ± 0.7 |
| dwf4-101/snp2-1 | 4.1 ± 0.6 | ND |
| dwf4-102 | 1.8 ± 0.4 | ND |
| Ws | 7.3 ± 1.3 | 3.5 ± 0.5 |
| bri1-5 | 4.2 ± 0.8 | ND |
The DWF4 gene consists of eight exons and seven introns. A sequence analysis of dwf4-101 showed that a single guanine nucleotide was replaced by an adenine nucleotide in the fourth exon, resulting in substitution of the Gly residue at position 312 by a Glu residue (Fig. 3). The mutation occurred in one of the most highly conserved domains among the CYP superfamily that binds to dioxygen (Chapple, 1998). Of 246 Arabidopsis CYP proteins, 81.7% have Gly at this position, 11.7% have Ala, 3.6% have Ser, 2.0% have Thr, 0.4% have Leu, and the remaining 0.4% have Asp (http://www.p450.kvl.dk/p450.shtml; Paquette et al., 2000).
dwf4-101 Suppresses Gravitropic and Phototropic Defects in nph4
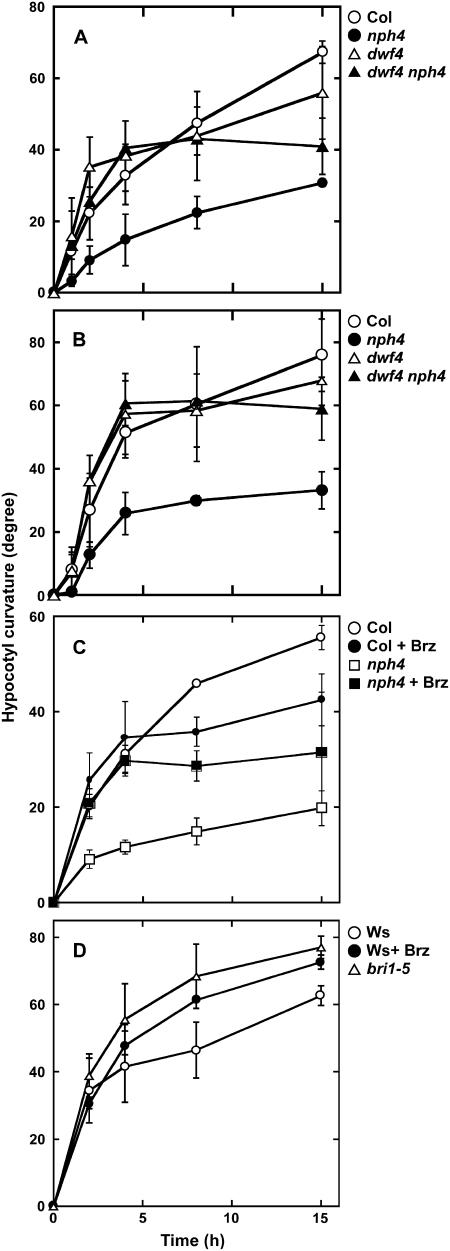
We examined the effects of the dwf4-101 mutation on gravitropism and phototropism of nph4-103. The gravitropic response of dark-grown nph4-103 hypocotyls was slower than that of Col (Fig. 4A). But dwf4-101 nph4-103 hypocotyls bent upward as quickly as those of Col up to 8 h after the start of gravistimulation. At a later stage, their tropic movement was slowed down more readily than that of Col. Consequently, at 15 h, gravitropic bending of dwf4-101 nph4-103 hypocotyls was greater than that of nph4 hypocotyls, but less than that of Col hypocotyls. This result indicates that dwf4-101 partially suppressed the gravitropic defects of nph4-103. We also examined the gravitropic response of a dwf4 single mutant. Gravitropic bending of dwf4-101 hypocotyls was significantly greater than that of Col at 4 h (P = 0.043 in Student's t test) and it became similar to that of Col thereafter.
Figure 4.
Time course of tropic reorientation of etiolated hypocotyls. A and B, Gravi- and phototropic reorientation of hypocotyls of Col (○), nph4-103 (•), dwf4-101 (▵), and dwf4-101 nph4-103 (▴), respectively. C, Gravitropic reorientation of Col (circle) and nph4-103 (square) hypocotyls in the presence (black symbols) or absence (white symbols) of 1 μm Brz. D, Gravitropic reorientation of hypocotyls of wild-type Ws (circle) and bri1-5 (▵) in the presence (•) or absence (○) of 1 μm Brz. Seedlings grown on vertically held plates for 3 d in the dark condition were turned 90° to a horizontal position or subjected to unilateral blue light (0.1 μmol m−2 s−1). The angle of hypocotyl curvature was measured at the indicated times thereafter; 90° represents complete reorientation upward or to direction of the light stimulus. Values shown represent the mean ± se of three independent experiments in which 20 seedlings were measured.
The phototropic response of etiolated nph4-103 hypocotyls was also slower than that of Col hypocotyls (Fig. 4B). The response of dwf4-101 nph4-103 was similar to that of Col. Namely, dwf4-101 completely suppressed phototropic defects of nph4 hypocotyls. The dwf4-101 single mutant also showed essentially the same curvature as Col. Taken together, these results indicate that hypocotyl bending of the dwf4-101 single mutant was similar to that of Col in both gravi- and phototropism, except that it was faster than the hypocotyl bending of Col in the earlier phase of gravitropism. dwf4-101 was almost epistatic to nph4-103 with respect to the tropic responses. dwf4-101/snp2-1 also suppressed the nph4-103 defects in leaf morphology because dwf4-101 nph4-103 exhibited weakly epinastic leaves and cotyledons in contrast to the hyponastic growth observed in nph4-103 (Fig. 1).
Inhibition of BR Biosynthesis Rescues Gravitropic Defects of nph4-103 Hypocotyls
Because dwf4-101 is a BR-deficient mutant, we examined the effects of an inhibitor of BR biosynthesis, brassinazole (Brz; Asami et al., 2000), on gravitropism of hypocotyls (Fig. 4C). Treatment with 1 μm Brz did not affect or sometimes promoted hypocotyl gravitropism of Col in the earlier phase of the response, but partially inhibited it in the later phase. Brz treatment completely rescued the gravitropic defects of hypocotyls of nph4-103 in the earlier phase and reduced the defects by about 50% at 15 h. Thus, the gravitropic time course of Col and nph4-103 hypocotyls in the presence of 1 μm Brz (Fig. 4C) looked similar to that of dwf4-101 and dwf4-101 nph4-103 hypocotyls, respectively (Fig. 4A).
We also examined gravitropism of a BR-insensitive mutant, brassinosteroid insensitive 1 (bri1), in which a putative BR receptor was disrupted (Li and Chory, 1997). Null mutants of BR biosynthesis and sensitivity, such as dwf4-1 and bri1-1, respectively, did not display significant gravitropism in hypocotyls (data not shown), probably due to the very slow growth of hypocotyls. Therefore, we investigated gravitropism of a leaky allele of bri1, bri1-5 (Ws background; Noguchi et al., 1999; Fig. 4D). Surprisingly, bri1-5 hypocotyls exhibited a stronger gravitropic response than those of Ws. Furthermore, treatment of Ws hypocotyls with 1 μm Brz also enhanced the gravitropic response as much as the bri1-5 mutation did. It was interesting that treatment with 1 μm Brz inhibited hypocotyl growth by about 50% and that similar inhibition of hypocotyl growth was caused in the dwf4-101 or bri1-5 mutation (Table II).
dwf4-101 Restores Auxin Sensitivity Compromised in nph4-103
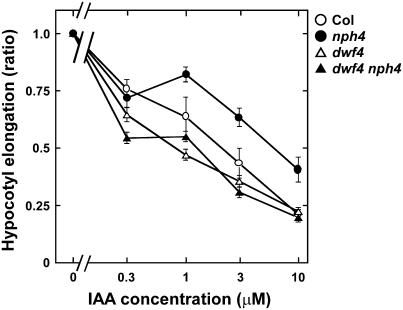
Because nph4 hypocotyls are less sensitive to auxin in terms of auxin-induced growth inhibition (Watahiki and Yamamoto, 1997), we studied the effects of the dwf4-101 mutation on sensitivity of hypocotyl growth to indole-3-acetic acid (IAA) in a liquid medium in the dark (Fig. 5). Growth of hypocotyls was inhibited by IAA in a dose-dependent manner in Col. nph4-103 hypocotyls were more resistant to IAA than those of Col. dwf4-101 was found to be more sensitive to IAA than Col. The sensitivity of dwf4-101 nph4-103 to IAA was similar to that of dwf4-101. Therefore, dwf4-101 restored auxin sensitivity, which was decreased in nph4-103, indicating that dwf4-101 was epistatic to nph4-103 with respect to the auxin sensitivity of hypocotyls.
Figure 5.
Effects of IAA on hypocotyl growth of Col (○), nph4-103 (•), dwf4-101 (▵), and dwf4-101 nph4-103 (▴). Seedlings were grown hydroponically for 5 d at 23°C in darkness and IAA was added after germination. Hypocotyl length is expressed relative to the mean hypocotyl length of the same genotype in the absence of IAA. Each value represents the mean ± se of three independent experiments in which 15 seedlings were measured.
Effects of dwf4 on Gene Expression of MSG2/IAA19
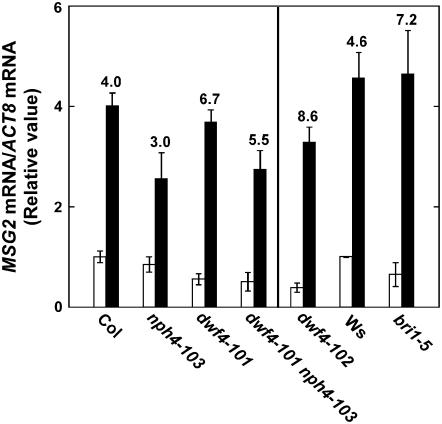
We examined the expression levels of MSG2 in etiolated seedlings of dwf4-101 nph4-103 to know whether dwf4-101 suppressed nph4-103 at the transcriptional level because MSG2 has been shown to play a central role in the tropic responses of hypocotyls (Tatematsu et al., 2004). The reverse transcription (RT)-PCR analyses in Figure 6 show that MSG2 was induced by exogenously applied auxin in Col. The MSG2 expression level of nph4-103 was similar to that of Col, but the auxin-induced level was reduced compared to that of the wild type as reported previously (Tatematsu et al., 2004). The expression levels of dwf4-101 and dwf4-101 nph4-103 were lower than those of the wild type. However, greater auxin induction was observed in these mutants than the wild type. These results show that dwf4-101 restored auxin inducibility, which was reduced in nph4-103, although it might decrease the expression level regardless of auxin treatment. The MSG2 expression of dwf4-102 was similar to that of dwf4-101 with or without auxin treatment. bri1-5, which responded to gravistimulation faster than the wild type, Ws (Fig. 4D), exhibited greater auxin inducibility of the MSG2 gene expression than the wild type.
Figure 6.
RT-PCR analyses of MSG2 gene expression with (black bar) or without (white bar) IAA treatment at 10 μm for 1 h. Induction ratio (fold) by the IAA treatment is written above each set of bars. mRNA level of MSG2 normalized against the ACTIN8 mRNA level is expressed as a ratio to that of wild type (Col and Ws on the left and right, respectively). Each value represents the mean ± sd of three experiments using independently prepared total RNA samples. Seedlings were grown in liquid one-half-strength Murashige and Skoog medium in the dark for 4 d before IAA treatment.
DISCUSSION
Here we have shown that gravi- and phototropic defects of nph4-103 are suppressed by a leaky mutation of DWF4/SNP2 that encodes a hydroxylase of the BR biosynthesis pathway. Treatments with Brz, an inhibitor of DWF4 (Asami et al., 2000), also display similar effects on gravitropism of nph4-103, strongly suggesting that a deficiency of BR suppresses the tropic defects of nph4-103. dwf4-101 single mutants exhibit tropic responses that are similar to those of the wild type. However, in the early phase of the responses, dwf4-101 shows slightly faster hypocotyl bending than the wild type. In fact, dwf4-101 hypocotyls bent significantly faster than those of the wild type 2 h after the start of gravistimulation. Furthermore, either bri1-5 mutation or Brz treatment clearly promotes the gravitropic response of hypocotyls in a Ws background. Together, our results strongly suggest that BR is a negative factor of tropic responses and that BR deficiency is epistatic to nph4-103 in terms of tropic responses of hypocotyls. A similar interaction has been reported in the YUCCA gene, which encodes a flavin monooxygenase-like enzyme in auxin biosynthesis. YUCCA is overexpressed by the cauliflower mosaic virus 35S enhancer in the yucca mutant, in which an increase in the auxin level causes long hypocotyls in the light condition (Zhao et al., 2001). Hypocotyls of yucca bri1 double mutants are as short as those of bri1 in the light condition, showing that BR insensitivity is epistatic to auxin action (Nemhauser et al., 2004).
To gain physiological and molecular insight into the epistatic nature of the dwf4 mutation in tropic responses, we examined the auxin sensitivity of dwf4-101 and dwf4-101 nph4-103 with respect to inhibiting activity of auxin on hypocotyl elongation. We find that dwf4-101 is hypersensitive to auxin and that dwf4-101 is epistatic to nph4-103 in this respect. Essentially the same conclusion was drawn when auxin sensitivity was evaluated by gene expression of auxin-inducible MSG2. Greater auxin induction is observed in dwf4-101 than in the wild type and dwf4-101 restores auxin induction of MSG2, which is reduced in nph4-103. Taken together, restoration of tropic responses in dwf4-101 nph4-103 may be brought about by the epistatic effects of dwf4 on auxin insensitivity of nph4.
Increase in auxin sensitivity has already been reported in dwf4 (Azpiroz et al., 1998) and sax1, a putative BR biosynthesis mutant (Ephritikhine et al., 1999a) with respect to root growth. Sensitivity of hypocotyls to auxin is not examined in either case. Although SAX1 has not yet been characterized in molecular terms, feeding experiments with various intermediates of BR biosynthesis suggest that it may encode an enzyme that oxidizes 22-hydroxycampesterol (Ephritikhine et al., 1999b; Fujioka and Yokota, 2003). Interestingly, DWF4 is an enzyme that catalyzes 22-hydroxylation of campesterol derivatives and its products include the oxidized 22-hydroxycampesterol (22-hydroxyergost-4-en-3-one; Choe et al., 1998; Fujioka and Yokota, 2003). Thus, a deficiency in the oxidized 22-hydroxycampesterol may lead to higher sensitivity to auxin. It may be noteworthy that sax1 appears to be leaky when compared to the null mutants of other BR biosynthesis loci.
In contrast to our finding that BR is a negative factor for tropic responses, promotive effects of BR have been reported by a few other authors. BR promotes gravitropism of light-grown tomato (Lycopersicon esculentum) hypocotyl cuttings in the light condition (Park, 1998). A synergistic positive interaction between BR and auxin was reported for gravitropism of partially deetiolated bean (Phaseolus vulgaris) hypocotyl sections (Meudt, 1987) and dark-grown maize (Zea mays) primary roots (Kim et al., 2000). Recently, Li et al. (2005) have also shown promotive effects of BR on root and hypocotyl gravitropism in Arabidopsis. They observed gravitropic curvature of roots and hypocotyls of dark-grown seedlings that were simultaneously irradiated with unilateral white light from above. Judging from the published images of seedlings with large and green cotyledons, photomorphogenetic reactions also appear to occur. Therefore, they actually report promotive effects of BR on a mixture of gravitropism and phototropism caused by much stronger light than is usually used for phototropic measurement and the observed effects may include secondary effects of deetiolation. Because we determined gravi- and phototropisms of etiolated seedlings in a specific manner in the dark, we cannot directly compare our results with theirs. We also found that treatment with 10 nm BL did not significantly affect gravitropism of etiolated hypocotyls under our experimental conditions (data not shown).
Li et al. (2005) also found that BR promotes polar auxin transport through gene expression of PIN-FORMED (PIN) family proteins in agreement with a previous observation by Bao et al. (2004). This seems important because it suggests that BR deficiency caused by dwf4 or Brz treatment may lead to reduction of polar auxin transport. Disruption of MULTIDRUG-RESISTANT1, which encodes an ATP-binding cassette transporter, causes reduction of polar auxin transport in hypocotyls due to the disturbance of PIN1 localization on the basal side of the cells (Noh et al., 2001), resulting in exaggerated bending in gravitropism (Noh et al., 2003). Blakeslee et al. (2004) also observed a disturbance of PIN1 basal asymmetry after unilateral blue-light irradiation during the phototropic response of hypocotyls and suggested that it may contribute to the phototropic lateral redistribution of auxin. These observations suggest that reduced polar auxin transport may enhance tropic bending by strengthening the lateral redistribution of auxin, which could be a cause of the promoted tropic responses of BR-deficient seedlings presented in this study.
Although BR may affect hypocotyl bending through modulation of polar auxin transport, it has also been well established that auxin and BR interact at the transcriptional level (Clouse et al., 1992; Goda et al., 2002; Müssig et al., 2002; Yin et al., 2002). Seventeen to 25% of the auxin-induced genes are also activated by BL in microarray analyses (Goda et al., 2004; Nemhauser et al., 2004). Genes responsive to both auxin and BL include those of early auxin-responsive gene families, such as AUX/IAA, SAUR, and GH3, and a putative transcriptional regulator for tropic responses, MSG2/IAA19 (Tatematsu et al., 2004), also responds to both auxin and BL (Nakamura et al., 2003a). The expression of several genes was found to be synergistically promoted by auxin and BL (Nakamura et al., 2003b; Nemhauser et al., 2004). Furthermore, the TGTCTC auxin response element is more enriched in the 5′-flanking region of genes up-regulated by both IAA and BL than those up-regulated specifically by auxin (Goda et al., 2004). In fact, since the discovery of BR, it has long been known that auxin often acts synergistically with BR (Mandava, 1988) and the synergism is observed in hypocotyl elongation of Arabidopsis (Nakamura et al., 2003b; Nemhauser et al., 2004). On the other hand, Wang et al. (2005) show that BL has no obvious effect on auxin-induced gene expression in mesophyll protoplasts and that BL does not induce gene expression through the TGTCTC auxin response element and ARF activators. Hence, cross talk between auxin and BR at the transcriptional level might be observed only when the two hormones are applied to tissues or whole plants. The synergistic interaction might also occur at the level of BR biosynthesis because gene expression of DWF4 is up-regulated by auxin treatment (Choe, 2004).
Our results about tropic responses are inconsistent with the reported synergism between auxin and BR described above. If the synergistic interaction also occurs in our experimental system, nph4 dwf4 double mutants should exhibit smaller curvature of hypocotyls than each single mutant. However, the double mutants actually displayed greater curvature than nph4. Our results show that the decrease in auxin sensitivity that we observed as a result of defects in tropic responses in nph4 was recovered by the presumed decrease in the BR level by the dwf4 mutation. Restoration of auxin sensitivity by a decrease in the BR level suggests that it is not the absolute levels of auxin and BR signals, but the ratio of the auxin-to-BR signal that determines the tropic responses of hypocotyls. This is qualitatively different from the well-documented synergistic interdependency between auxin and BR. This discrepancy appears to arise from the different nature of the observed phenomena. Elongation of hypocotyls in which the auxin-BR interdependency has been observed is a one-dimensional response, whereas tropic responses examined here are two dimensional. In the latter case, the response in each dimension could be independently regulated by auxin and BR.
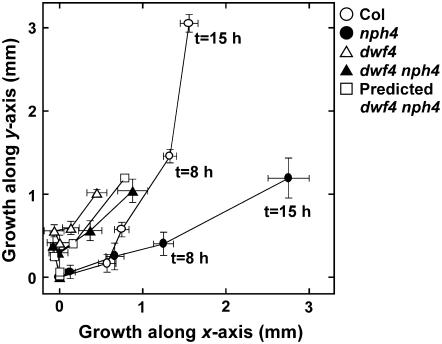
This reasoning prompted us to examine the track of hypocotyl tips in the x-y plane during the gravitropic response (Fig. 7). The x axis is the original growth axis of hypocotyls before gravistimulation and the y axis is vertical, pointing upward. The position of the hypocotyl tip is traced after the start of gravistimulation. This analysis shows that lateral movement of the tip along the y axis is essentially the same in the gravitropic responses of nph4-103 and dwf4-101 nph4-103 and that only growth along the x axis is different between them. These results suggest that growth along the x and y axes is regulated separately by BR and auxin, respectively. In fact, if we transform the trace of the nph4 hypocotyl tip by only reducing growth in the x direction in proportion to the reduction ratio of the x axis growth of dwf4 to that of wild type, the obtained trace (Fig. 7, white squares) is essentially the same as that of dwf4-101 nph4-103 (Fig. 7, black triangles). However, if our rationale is correct, dwf4-101 should exhibit larger curvature than the wild type, which is not the case. This is likely due to the limited growth of dwf4-101 hypocotyls. dwf4 null mutants do not exhibit any curvature upon gravistimulation (data not shown), probably because of the greatly reduced growth of their hypocotyls. The nph4-103 hypocotyl does not lose tropic responses completely and still develops a reduced curvature because of the leaky nature of nph4-103 (Harper et al., 2000) and redundant function of NPH4/ARF7 and ARF19 (Okushima et al., 2004). Therefore, dwf4-101, a weak allele of dwf4, could compensate for the defect in tropic responses by partially reducing growth along the x axis. This interpretation of tropic data seems consistent with the fact that growth of etiolated nph4 hypocotyls is essentially the same as that of wild type and that the tropic response is specifically altered in nph4 hypocotyls (Watahiki and Yamamoto, 1997; data not shown). It would also be worth mentioning that growth of the Arabidopsis leaf is two dimensional, where the longitudinal and lateral growth is independently regulated by ROTUNDIFOLIA3 and ANGUSTIFOLIA, respectively (Tsukaya, 2002). Interestingly, the former is involved in BR biosynthesis, catalyzing a reaction from typhasterol to castasterone, the last step in BR biosynthesis (Fujioka and Yokota, 2003; Kim et al., 2005).
Figure 7.
Track of hypocotyl tips of Col (○), nph4-103 (•), dwf4-101 (▵), and dwf4-101 nph4-103 (▴). White square represents predicted positions of hypocotyl tip of dwf4-101 nph4-103. Each symbol corresponds to position at the measured time, 0, 2, 4, 8, and 15 h after the start of gravistimulation. Position of hypocotyl tips of the same seedlings described in Figure 4A was determined in recorded images. Each position is the mean ± se of three independent experiments in which 20 seedlings were measured. Predicted positions of dwf4-101 nph4-103 hypocotyl tips are calculated as follows: ydwf4 nph4 = ynph4; xdwf4 nph4 = xnph4 × xdwf4/xCol, where xi and yi are growth along the x and y axis of i mutant, respectively. The origin of the x-y plane is the position at the start of gravistimulation for each genotype.
In conclusion, we have shown here that dwf4-101/snp2-1 is epistatic to nph4 with respect to tropisms of hypocotyls and the differential growth of leaves. BR, in particular, appears to be a negative factor for the tropisms. This epistatic relationship could be brought about by increased auxin sensitivity of hypocotyls or reduced polar auxin transport due to BR deficiency as suggested by a few previous studies. Alternatively, we propose a new hypothesis that auxin and BR regulate hypocotyl growth along the growth axis and the lateral axis independently in tropic responses, which may be the physical basis of the epistatic relationship observed in the tropic responses of hypocotyls.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of Arabidopsis (Arabidopsis thaliana) were first imbibed in water in the dark at 4°C for 3 d. They were surface sterilized with 1% hypochlorite and sown on nutrient agar plates that contained one-half-strength Murashige and Skoog salts (Murashige and Skoog, 1962), 1% (w/v) Suc, and 1% (w/v) agar. Plants were grown at 23°C under continuous illumination at a fluence rate of 8.2 W m−2 obtained from three 40-W white fluorescent tubes (FL40SW; Mitsubishi-Osram). In some experiments, plants were grown on a 1:1 (v/v) mixture of vermiculite:Metromix 350 (Scotts-Sierra).
For mutagenesis, 100 mg of nph4-103/msg1-3 seeds (Watahiki and Yamamoto, 1997) were treated with 0.3% ethyl methanesulfonate for 16 h. After extensive washing with water, the mutagenized seeds were sown in soil and their seeds collected in four batches. The snp2 mutant was backcrossed twice to Col before phenotypic characterization.
Measurement of Tropic Responses
To determine gravitropism of hypocotyls, seedlings were grown on vertically oriented agar plates for 3 d in the dark and then turned 90° to a horizontal position. For second-positive phototropism, 3-d-old etiolated seedlings grown as above were irradiated with unilateral blue light at a fluence rate of 0.1 μmol·m−2·s−1 obtained by blue light-emitting diodes (λmax = 470 ± 30 nm; Stick-B16; Tokyo Rikakikai). An image of the seedlings was taken with a digital camera (C-4040 Zoom; Olympus) at different times under dim green light and hypocotyl curvature was measured with Image Pro-Plus (Media Cybernetics). To determine effects of BL or Brz on gravitropism, seedlings were grown on vertically oriented agar plates supplemented with BL or Brz for 3 d in the dark and then turned 90° to a horizontal position.
Phytohormone Treatments
To examine the effects of IAA and BL on the growth of hypocotyls, seeds were placed in the above-mentioned nutrient medium without agar under continuous white light at 23°C for 24 h to induce germination after cold treatment and surface sterilization. After the medium was exchanged for a medium supplemented with various concentrations of IAA or BL, seedlings were further grown in darkness for 5 d. Hypocotyl length was determined with Image Pro-Plus after taking photographs.
Genetic Mapping
The genetic location of SNP2 was established by determining the linkage between the mutant allele and simple sequence length polymorphism markers (Bell and Ecker, 1994). Simple sequence length polymorphism markers, T20E3 and F18B3, were generated in the northern and southern ends of bacterial artificial chromosome clones T20E23 and F18B3, respectively. For the T20E3 marker, PCR was performed using a forward primer, 5′-GGAGGATCATGAAAGAAACTCTAT-3′, and a reverse primer, 5′-CCACCACAAGAGAATATATCTCTTC-3′. The PCR product was 244 bp long in Col, whereas it was 230 bp long in Ler. For the F18B3 marker, PCR was performed with a forward primer, 5′-AGCGTTTCAAATATTTGCGG-3′, and a reverse primer, 5′-GGATTTACCTCGAGCGC-3′. The PCR product was 243 and 228 bp long in Col and Ler, respectively.
nph4-103 mutation was distinguished by using a derived cleaved-amplified polymorphic sequence marker. The PCR product amplified with a forward primer, 5′-GTCTCAACAACACAGCAACAACAAT-3′, and a reverse primer, 5′-AGATGCTTGTTGCGACTGATGCAGC-3′, was digested by PvuII into 250- and 24-bp-long fragments in Col; it was not cleaved in nph4-103.
Cloning and Sequencing of DWF4 Gene in snp2 Mutants
The DWF4 gene (Choe et al., 1998) was divided into four parts, each of which was amplified by PCR from snp2 mutants. The oligonucleotide primers used were 5′-AAACCATAACATGAAATTTTGTTGC-3′ and 5′-GTGTTGGTGTAAGTGTACG-3′ for the first part; 5′-CTAATAATAAATCAACGGTCACGA-3′ and 5′-TGCAAGCTTGCCCTAAA-3′ for the second part; 5′-GATGCTCTTCCTTCAAAAGTAT-3′ and 5′-TGGGATTCTTGGGAAATGG-3′ for the third part; and 5′-TGTCTCTCAACATCTTTAAGTAGA-3′ and 5′-CATAACGAGGCAACAAAAGTA-3′ for the fourth part.
RNA Extraction and RT-PCR
Total RNA was extracted from whole seedlings of Arabidopsis that were grown in the one-half-strength Murashige and Skoog liquid medium described above in the dark for 4 d using the RNeasy plant mini kit (Qiagen). RT and DNA amplification were carried out for three independently prepared total RNA samples by using Moloney murine leukemia virus reverse transcriptase RNase H− (ReverTra Ace; Toyobo) and Taq polymerase (New England Biolabs). In some experiments, RT-PCR was conducted using the AccessQuick RT-PCR system (Promega). The PCR primers, 5′-CAAGAGAAGTGTAGGAGAAG-3′ and 5′-ATATAGCTGTCTTTCTGAAG-3′, were used to amplify the MSG2 cDNA. ACTIN8 gene was used as an internal control for the RT-PCR with two primers, 5′-TGCTTCTAAACTAAAGAGACATCG-3′ and 5′-GCTACAAACAAACAAACAAATGGA-3′. PCR products were fractionated on 3% agarose gel and stained with ethidium bromide. Images were taken with a digital camera (AE-6905H; Atto) and intensity of fluorescence was quantified with ImageJ (National Institutes of Health).
Acknowledgments
We wish to thank Dr. K. Feldmann (Ceres) for bri1-5 seeds, the Arabidopsis Biological Resource Center for a T-DNA insertion line, and Dr. T. Nakano (RIKEN) for valuable discussions.
This work was supported in part by a Grant-in-Aid for Scientific Research in Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (grant no. 14036201 to K.T.Y.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Kotaro T. Yamamoto (kty@sci.hokudai.ac.jp).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.076273.
References
- Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami T, Min YK, Nagata N, Yamagishi K, Takatsuto S, Fujioka S, Murofushi N, Yamaguchi I, Yoshida S (2000) Characterization of brassinazole, a triazole-type brassinosteroid biosynthesis inhibitor. Plant Physiol 123: 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA (1998) An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10: 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao F, Shen J, Brady SR, Muday GK, Asami T, Yang Z (2004) Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol 134: 1624–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Berleth T, Sachs T (2001) Plant morphogenesis: long-distance coordination and local patterning. Curr Opin Plant Biol 4: 57–62 [DOI] [PubMed] [Google Scholar]
- Blakeslee JJ, Bandyopadhyay A, Peer WA, Makam SN, Murphy AS (2004) Relocalization of the PIN1 auxin efflux facilitator plays a role in phototropic responses. Plant Physiol 134: 28–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapple C (1998) Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Physiol Plant Mol Biol 49: 311–343 [DOI] [PubMed] [Google Scholar]
- Choe S (2004) Brassinosteroid biosynthesis and metabolism. In PJ Davies, ed, Plant Hormones. Biosynthesis, Signal Transduction, Action! Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 156–178
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA (1998) The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22-α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse S, Zurek DM, Mcmoriss TC, Backer ME (1992) Effect of brassinolide on gene expression in elongating soybean epicotyls. Plant Physiol 100: 1377–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephritikhine G, Fellner M, Vannini C, Lapous D, Barbier-Brygoo H (1999. a) The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J 18: 303–314 [DOI] [PubMed] [Google Scholar]
- Ephritikhine G, Pagant S, Fujioka S, Takatsuto S, Lapous D, Caboche M, Kendrick RE, Barbier-Brygoo H (1999. b) The sax1 mutation defines a new locus involved in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J 18: 315–320 [DOI] [PubMed] [Google Scholar]
- Esmon CA, Pedmale UV, Liscum E (2005) Plant tropisms: providing the power of movement to a sessile organism. Int J Dev Biol 49: 665–674 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Yokota T (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S (2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134: 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E (2000) The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12: 757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Fujioka S, Kozuka T, Tax FE, Takatsuto S, Yoshida S, Tsukaya H (2005) CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J 41: 710–721 [DOI] [PubMed] [Google Scholar]
- Kim SK, Chang SC, Lee EJ, Chung WS, Kim YS, Hwang S, Lee JS (2000) Involvement of brassinosteroids in the gravitropic response of primary root of maize. Plant Physiol 123: 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li L, Xu J, Xu ZH, Xue HW (2005) Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell 17: 2738–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Briggs WR (1995) Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7: 473–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandava NB (1988) Plant growth-promoting brassinosteroids. Annu Rev Plant Physiol Plant Mol Biol 39: 23–52 [Google Scholar]
- Meudt WJ (1987) Investigations on the mechanism of the brassinosteroid response. Plant Physiol 83: 195–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 472–497 [Google Scholar]
- Müssig C, Fischer S, Altmann T (2002) Brassinosteroid-regulated gene expression. Plant Physiol 129: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S (2003. a) Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol 133: 1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Shimada Y, Goda H, Fujiwara MT, Asami T, Yoshida S (2003. b) AXR1 is involved in BR-mediated elongation and SAUR-AC1 gene expression in Arabidopsis. FEBS Lett 553: 28–32 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE (1999) Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol 121: 743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh B, Bandyopadhyay A, Peer WA, Spalding EP, Murphy AS (2003) Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature 423: 999–1002 [DOI] [PubMed] [Google Scholar]
- Noh B, Murphy AS, Spalding EP (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13: 2441–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al (2004) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette SM, Bak S, Feyereisen R (2000) Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol 19: 307–317 [DOI] [PubMed] [Google Scholar]
- Park WJ (1998) Effect of epibrassinolide on hypocotyl growth of the tomato mutant diageotropica. Planta 207: 120–124 [DOI] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H (2002) Leaf development. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD, doi: 10.1199/tab.0072, http://www.aspb.org/publications/arabidopsis/
- Wang S, Tiwari SB, Hagen G, Guilfoyle TJ (2005) AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell 17: 1979–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watahiki MK, Yamamoto KT (1997) The massugu1 mutation of Arabidopsis identified with failure of auxin-induced growth curvature of hypocotyl confers auxin insensitivity to hypocotyl and leaf. Plant Physiol 115: 419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]