Abstract
Most extant cyanobacteria contain C16 fatty acids in the sn-2 positions of glycerolipids, which are regulated by lysophosphatidic acid acyltransferase (LPAAT; EC 2.3.1.51). Synechocystis sp. PCC6803 contains sll1848, sll1752, and slr2060 as putative acyltransferase genes. sll1848 was recently reported to encode an indispensable palmitoyl-specific LPAAT; however, here we show that each of the three genes is dispensable. Δ1848 and Δ1848 Δ2060 cells had markedly higher contents of stearate (18:0), oleate (18:1), and linoleate (18:2) in place of palmitate (16:0) in the sn-2 positions, suggesting that Δ1848 Δ2060 cells incorporate 18:0 and 18:1 in the sn-2 positions. The levels of sll1752 transcripts increased in Δ1848 Δ2060 cells. This was accompanied by increased LPAAT activity toward 18:0 coenzyme A and its derivative in the membrane fraction. From these findings, together with the activity of a recombinant sll1752 protein and complementation of the Escherichia coli LPAAT mutant plsC, we conclude that sll1752 encodes a second LPAAT that prefers stearoyl and oleoyl substrates. Δ1848 Δ2060 cells grew slowly at 30°C at lower cell density, and exhibited more severe damage at 20°C than wild-type cells. Furthermore, Δ1848 Δ2060 cells exhibited photoinhibition more severely than wild-type cells. A phycobilisome core-membrane linker protein (slr0335) was also found to be susceptible to protein extraction under our conditions; its content decreased in the membrane fractions of Δ1848 Δ2060 cells. We conclude that C16 fatty acids in sn-2 positions are preferred in the photosynthetic growth of this cyanobacterium, despite sll1752 orthologs being conserved in most cyanobacteria. However, no sll1752 ortholog is conserved among photosynthetic eukaryotes including Cyanidioschyzon merolae.
Phosphatidic acid (PA) is a key intermediate in the biosynthesis of membrane glycerolipids and the storage lipid triacylglycerol. PA is produced predominantly through two subsequent acylation reactions catalyzed by glycerol-3-P acyltransferase (GPAT; EC 2.3.1.15) and lysophosphatidic acid acyltransferase (LPAAT; EC 2.3.1.51). Although the first acylation step by GPAT is partly bypassed by dihydroxyacetone phosphate acyltransferase in some tissues and organisms (Athenstaedt and Daum, 1999), LPAATs are essential to PA synthesis in all cellular organisms expect for archea.
GPATs or LPAATs from distantly related organisms share little sequence homology, except for the shared canonical acyltransferase signatures such as HXXXXD and XXXXXXG signatures separated from each other by 60 to 83 amino acid residues (X stands for arbitrary amino acid residues), despite sharing some homology between acyltransferases from closely related organisms (Frentzen and Wolter, 1998; Lewin et al., 1999). According to this criterion, GPAT and LPAAT genes have been identified in Arabidopsis (Arabidopsis thaliana; Zheng et al., 2003; Kim and Huwang, 2004; Yu et al., 2004) and Synechocystis sp. PCC6803 (Weier et al., 2005).
Ancestral cyanobacteria are generally accepted as the symbiotic origin of plastids (McFadden, 1999). The membranes of cyanobacteria contain the glycolipids monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), and sulfoquinovosyldiacylglycerol (SQDG), and the phospholipid phosphatidylglycerol (PG), which are major lipids in plastid membranes. Most extant cyanobacteria contain C16 fatty acids in the sn-2 positions of glycerolipids (Murata et al., 1992), which are regulated by LPAAT. However, it is not understood how and why the C16 fatty acids have been preferred evolutionarily to C18 fatty acids in the sn-2 positions by a wide range of cyanobacteria.
The cyanobacterium Synechocystis sp. PCC6803 contains three candidate genes for acyltransferases: sll1848, sll1752, and slr2060. Weier et al. (2005) reported recently that sll1848 encodes the major, indispensable LPAAT specific to palmitate (16:0). However, here we show that sll1848 is dispensable and that its disruptants, Δ1848 or Δ1848 Δ2060 cells, exhibit dramatically higher contents of stearate (18:0), oleate (18:1), and linoleate (18:2) in place of 16:0 in the sn-2 positions of glycerolipids. Furthermore, the higher levels of unsaturation of fatty acids in the sn-2 positions appear to be compensated by a decrease in 6,9,12-linolenate content and by an increase in 18:1 content in the sn-1 positions. LPAAT activity toward 16:0 CoA decreased markedly in Δ1848 and Δ1848 Δ2060 cells, whereas LPAAT activity toward 18:0 CoA increased in these cells. We also show that sll1752 functions as the second LPAAT specific to stearoyl and oleoyl substrates. Δ1848 Δ2060 cells showed several growth defects and exhibited photoinhibition more severely than did wild-type cells. We also demonstrated that the recovery of a phycobilisome core-membrane linker protein (Shen et al., 1993) decreased in isolated membrane fractions of Δ1848 Δ2060 cells. We conclude that C16 fatty acids in the sn-2 positions of glycerolipids have some advantages for the photosynthetic growth of this cyanobacterium. The third acyltransferase slr2060, which was recently found to be a lysophospholipid acyltransferase (Weier et al., 2005), exhibited neither GPAT nor LPAAT activities. We discuss the evolutionary significance of the different types of acyltransferases in cyanobacteria and higher photosynthetic eukaryotes.
RESULTS
Disruption of the sll1848 Gene Dramatically Decreases the Content of C16 Fatty Acids in Synechocystis sp. PCC6803 Cells
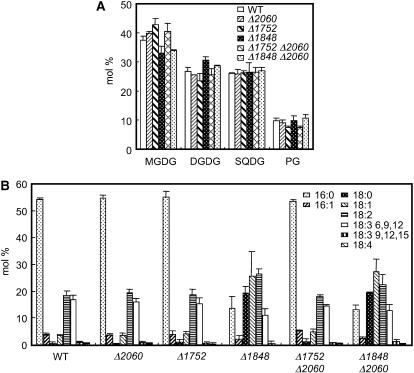
To evaluate the function of acyltransferases in Synechocystis sp. PCC6803, the HXXXXD catalytic signatures of the sll1848, sll1752, and slr2060 genes were replaced with one of a set of gene cassettes conferring drug resistance, as described in “Materials and Methods.” The resultant null mutants for sll1848, sll1752, and slr2060 genes were designated Δ1848, Δ1752, and Δ2060, respectively. We successfully produced the double gene disruptants Δ1752 Δ2060 and Δ1848 Δ2060 but could not succeed in the complete double disruption of sll1848 and sll1752 genes despite repeated experiments. Complete replacement was verified by PCR analyses (Supplemental Fig. 1B). The lipid and fatty acid compositions of Δ1752, Δ2060, and Δ1752 Δ2060 cells did not differ between the wild type and these cells (Fig. 1, A and B). In contrast, although the glycerolipid compositions of Δ1848 and Δ1848 Δ2060 cells were indistinguishable from those of the wild type (Fig. 1A), their fatty acid compositions showed markedly higher content of 18:0 and 18:1 and a reciprocally lower amount of 16:0 (Fig. 1B).
Figure 1.
Compositions of glycerolipids and fatty acids in the gene disruptants of Synechocystis sp. PCC6803. A, Composition of glycerolipids in wild-type and disruptant cells. B, Composition of fatty acids in the total lipid from wild-type and disruptant cells. Each value represents an average of three independent experiments. Bars indicate sd.
Positional analysis of the fatty acids showed that the sn-2 positions of glycerolipids contained exclusively C16 fatty acids in the wild type, whereas 84% to 90% of fatty acids in the sn-2 positions of glycerolipids were occupied by C18 fatty acids in Δ1848 Δ2060 cells (Table I). In particular, Δ1848 Δ2060 cells contained significant levels of 18:0, 18:1, and 18:2 in the sn-2 positions of all glycerolipids, but the wild type contained few C18-unsaturated fatty acids in the sn-2 positions of these glycerolipids. In contrast, the C16/C18 ratios of fatty acids in the sn-1 positions of MGDG and PG were similar in the wild type and the double mutant, whereas that in the sn-1 position of SQDG was significantly lower in the mutant cells (Table I). We also observed significantly lower levels of 6,9,12-linolenate in the sn-1 positions of MGDG and DGDG and of 18:2 in the sn-1 positions of SQDG and PG. In contrast, the content of 18:1 was greater in the sn-1 positions of all glycerolipids. As a result, the double-bond indices of glycerolipids were slightly larger in Δ1848 Δ2060 cells than in the wild-type cells (Table I).
Table I.
Positional distribution of fatty acids in glycerolipids in the wild-type and Δ1848 Δ2060 cells
Each value represents an average of three independent experiments. The numbers in parentheses denote the sd.
| Lipid | Cell Type | Fatty Acid
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16:0 | 16:1 | 18:0 | 18:1 | 18:2 | 18:3 (6,9,12) | 18:3 (9,12,15) | 18:4 | C16/C18 | Double-Bond Index | ||||
| mol % | |||||||||||||
| MGDG | Wild type | sn-1 | 6.8 (1.4) | 6.1 (0.32) | 0.0 (0.46) | 3.6 (1.2) | 33 (0.32) | 49 (0.13) | 0.25 (0.13) | 1.5 (0.17) | 13/87 | 2.3 | 1.3 |
| sn-2 | 98 (1.4) | 0.0 (0.32) | 0.27 (0.46) | 1.4 (1.2) | 0.37 (0.32) | 0.75 (0.13) | 0.0 (0.0) | 0.0 (0.17) | 98/2 | 0.044 | |||
| Δ1848 Δ2060 | sn-1 | 11 (3.9) | 3.1 (0.19) | 0.0 (0.59) | 17 (3.5) | 33 (7.9) | 39 (0.16) | 1.3 (0.18) | 1.2 (0.31) | 14/86 | 2.1 | 1.5 | |
| sn-2 | 16 (3.9) | 0.11 (0.19) | 24 (0.59) | 46 (3.5) | 13 (7.9) | 0.18 (0.16) | 0.0 (0.18) | 0.0 (0.31) | 16/84 | 0.91 | |||
| DGDG | Wild type | sn-1 | 7.9 (1.4) | 6.4 (2.2) | 0.26 (0.51) | 0.0 (2.1) | 26 (0.24) | 57 (0.24) | 0.75 (0.21) | 3.1 (0.16) | 14/86 | 2.4 | 1.2 |
| sn-2 | 95 (1.4) | 1.2 (2.2) | 1.5 (0.51) | 2.3 (2.1) | 0.27 (0.24) | 0.14 (0.24) | 0.0 (0.21) | 0.0 (0.16) | 96/4 | 0.045 | |||
| Δ1848 Δ2060 | sn-1 | 8.8 (4.1) | 2.5 (0.57) | 3.6 (4.2) | 11 (6.8) | 29 (2.8) | 42 (0.38) | 1.2 (0.16) | 1.9 (0.26) | 11/89 | 2.1 | 1.5 | |
| sn-2 | 14 (4.1) | 0.33 (0.57) | 9.8 (4.2) | 54 (6.8) | 21 (2.8) | 0.55 (0.38) | 0.46 (0.16) | 0.0 (0.26) | 14/86 | 0.99 | |||
| SQDG | Wild type | sn-1 | 29 (3.0) | 14 (0.21) | 0.0 | 13 (2.4) | 42 (1.4) | 1.3 (0.314) | 1.8 (0.12) | 0.0 (0.0) | 43/57 | 1.2 | 0.62 |
| sn-2 | 94 (3.0) | 0.21 (0.21) | 2.7 (1.3) | 2.8 (2.4) | 0.0 (1.4) | 0.0 (0.314) | 0.0 (0.12) | 0.0 (0.0) | 94/6 | 0.030 | |||
| Δ1848 Δ2060 | sn-1 | 23 (1.8) | 10 (0.16) | 11 (3.3) | 22 (2.3) | 30 (0.43) | 0.8 (0.068) | 2.0 (0.13) | 0.0 (0.0) | 33/67 | 1.0 | 0.63 | |
| sn-2 | 9.5 (1.8) | 0.090 (0.16) | 72 (3.3) | 14 (2.3) | 5.1 (0.43) | 0.0 (0.068) | 0.074 (0.13) | 0.0 (0.0) | 10/90 | 0.25 | |||
| PG | Wild type | sn-1 | 10 (2.5) | 1.4 (0.43) | 0.0 (3.4) | 5.7 (1.8) | 68 (0.083) | 0.0 (0.0) | 17 (0.27) | 0.0 (0.0) | 11/89 | 1.9 | 0.96 |
| sn-2 | 95 (2.5) | 0.39 (0.43) | 3.6 (3.4) | 1.0 (1.8) | 0.048 (0.083) | 0.0 (0.0) | 0.0 (0.27) | 0.0 (0.0) | 95/5 | 0.015 | |||
| Δ1848 Δ2060 | sn-1 | 8.1 (3.3) | 1.5 (1.9) | 6.1 (6.6) | 17 (0.76) | 56 (0.37) | 0.0 (1.7) | 12 (2.2) | 0.0 (0.0) | 10/90 | 1.7 | 1.1 | |
| sn-2 | 11 (3.3) | 1.9 (1.9) | 53 (6.6) | 29 (0.76) | 3.6 (0.37) | 1.0 (1.7) | 0.0 (2.2) | 0.0 (0.0) | 13/87 | 0.41 | |||
sll1752 Is Up-Regulated in Δ1848 Δ2060 Cells, and This Up-Regulation Correlates with Enhanced Stearoyl-Dependent LPAAT Activities in the Membrane Fraction
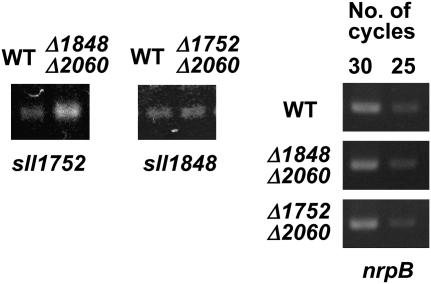
Because Δ1848 and Δ1848 Δ2060 cells have similar fatty acid compositions, we investigated the mechanisms responsible for the changes in the fatty acids in Δ1848 Δ2060 cells. We first used reverse transcription (RT)-PCR to examine whether the level of transcripts for sll1752 are altered in Δ1848 Δ2060 cells. Figure 2 shows that the intensity of an amplified band for sll1752 was stronger when the RNA from Δ1848 Δ2060 cells was used instead of the wild-type RNA. In contrast, the intensity of an amplified band for sll1848 did not differ between RNA samples from wild-type or Δ1752 Δ2060 cells. In all of these RNA samples, a control band for the nrpB transcripts was amplified at a similar intensity (Fig. 2). These results suggest that the level of the sll1752 transcripts is up-regulated in Δ1848 Δ2060 cells relative to the wild-type cells, but that the sll1848 transcripts are not affected. It should be noted that the intensity of bands amplified for sml0005 (PsbK) and sll1635 (Thy1 homolog) gene transcripts, which are located downstream of sll1752, also increased slightly (Supplemental Fig. 1C). However, it has been reported that these genes do not share the same operon as sll1752 (Ikeuchi et al., 1991).
Figure 2.
RT-PCR analysis of the transcripts of sll1752 and sll1848 in wild-type and disruptant cells. Approximately equal amounts of rnpB transcripts were present in each sample, and cycle numbers that ensured the linearity of amplifications are shown.
We then measured LPAAT activities in Δ1848 Δ2060 mutants and other deletion mutants. Although cyanobacterial acyltransferases are likely to use acyl-acyl carrier protein (acyl-ACP) in vivo (Lem and Stumpf, 1984; Weier et al., 2005), acyl-CoA substrates were sufficient to detect LPAAT activities. In most cases, acyltransferase shows similar fatty acid specificity for acyl-CoA substrates as for acyl-ACP (Bertrams and Heinz, 1981; Frentzen et al., 1983). Table II summarizes the specific activity of LPAAT in the membrane fractions of the wild type and mutants of Synechocystis sp. PCC6803. When 16:0 CoA was used as an acyl donor, the specific activities of LPAAT did not differ significantly between the wild-type, Δ1752, Δ2060, and Δ1752 Δ2060 cells. In contrast, LPAAT activities toward 16:0 CoA decreased significantly to 3.9% of wild-type levels in Δ1848 cells and to 0.82% of wild-type levels in Δ1848 Δ2060 cells. The LPAAT activity toward 18:0 CoA of Δ1848 or Δ1848 Δ2060 cells increased several times compared with the wild type and other mutants. These results are similar to our results, described earlier, showing that the sll1752 transcripts are enhanced in Δ1848 Δ2060 cells. Taken together, our data suggest that the expression of sll1752 increases in Δ1848 and Δ1848 Δ2060 cells to compensate for the large decrease in LPAAT activity caused by sll1848 disruption.
Table II.
LPAAT activity in the membrane fractions of wild-type and disruptant cells
The activities were measured with [1-14C]palmitoyl-CoA or [1-14C]stearoyl-CoA as described in “Materials and Methods.”
| Cell Type | LPAAT, Specific Activitya
|
|
|---|---|---|
| 16:0 CoA | 18:0 CoA | |
| nmol min−1 mg−1 protein | ||
| Wild-type | 2.1 (0.36) | 0.010 (0.050) |
| Δ2060 | 2.0 (0.30) | 0.033 (0.0070) |
| Δ1752 | 1.1 (0.11) | 0.020 (0.0088) |
| Δ1848 | 0.082 (0.050) | 0.14 (0.053) |
| Δ2060 Δ1752 | 1.5 (0.45) | 0.0087 (0.0071) |
| Δ1848 Δ2060 | 0.017 (0.0028) | 0.15 (0.034) |
Each value represents the average of three independent measurements. The numbers in parentheses denote the sd.
sll1752 Encodes a LPAAT That Prefers Stearoyl and Oleoyl Substrates
To examine the properties of acyltransferases in Synechocystis sp. PCC6803, we produced maltose-binding protein (MBP)-tagged recombinant proteins of sll1848 (designated MBP-1848), sll1752 (designated MBP-1752), and slr2060 (designated MBP-2060) in Escherichia coli TB1 cells. We also produced MBP-tagged E. coli PlsC protein (designated MBP-PlsC) as a control for an authentic LPAAT, and MBP alone as a negative control. The substitution of the D residue of the HXXXXD signature with G significantly decreases the GPAT activity of E. coli PlsB protein (Lewin et al., 1999). Therefore, we used in vitro mutagenesis to produce the D-to-G variants of MBP-tagged proteins, designated MBP-1848D69G, MBP-1752D66G, MBP-2060D76G, and MBP-PlsCD77G, as negative controls, in TB1 cells. MBP-tagged proteins were recovered in the soluble fractions of TB1 cells, partially purified by amylose affinity column chromatography, and subjected to GPAT and LPAAT assays, using 16:0 CoA or 18:0 CoA as an acyl donor.
Unexpectedly, GPAT activity was detected in the amylase-affinity purified fractions of MBP-1848 and MBP-1752 proteins, and this GPAT activity was lower in the corresponding fractions of MBP-1848D69G or MBP-1752D66G proteins. However, repeated experiments by native PAGE revealed that the GPAT activity was derived from E. coli cells, although GPAT from E. coli cells is supposed to be a membrane-bound enzyme (Supplemental Fig. 2).
Table III summarizes the specific activity of LPAAT in the amylose-affinity purified fractions of MBP, MBP-1848, MBP-1752, MBP-2060, and MBP-PlsC proteins, and in the D-to-G substitution variants. When 16:0 CoA was used as a substrate, MBP-1848 and MBP-PlsC proteins exhibited markedly higher levels of LPAAT-specific activity than any other protein samples tested. In contrast, MBP-1848D69G and MBP-PlsCD77G proteins exhibited only background levels of LPAAT activity. Under the same assay conditions, MBP-1752 and MBP-2060 protein fractions exhibited no more than the background levels of LPAAT activities as detected for their D-to-G variant fractions. Nonetheless, when 18:0 CoA was used as a substrate, LPAAT activity was several times higher in the MBP-1752 fraction than in the D-to-G variant fractions. MBP-1752 fractions also exhibited comparable LPAAT activity toward 18:1 CoA as to 18:0 CoA (data not shown). These results suggest that MBP-1752 functions as an LPAAT that prefers 18:0 and 18:1 to 16:0. In support of this, MBP-1752 successfully complemented the temperature-sensitive E. coli plsC mutant strain SM2-1 (Coleman, 1990; Supplemental Fig. 3). In contrast, no colony appeared at 42°C when MBP, MBP-1848, or MBP-2060 was induced in SM2-1 cells. It should be noted that cells harboring the plasmid containing the MBP-1848 gene grew very slowly without isopropyl-β-thiogalactoside, even at the nonrestrictive temperature of 30°C. This result suggests that MBP-1848 might have deleterious effects on the growth of SM2-1 cells.
Table III.
LPAAT activity of the amylose-affinity column purified MBP-tagged proteins
SDS-PAGE analysis showed that MBP, MBP-1848, MBP-1752, MBP-2060, and MBP-PlsC were expressed at high levels as 43-, 67-, 69-, 96-, and 70-kD bands, respectively, in both membrane (or the 100,000g pellet) and soluble (or the 100,000g supernatant) fractions of TB1 cell lysates. Comparable expression profiles were also seen for each of the D-to-G variants of MBP-tagged proteins relative to the corresponding nonmutagenized proteins.
| Protein | LPAAT, Specific Activitya
|
|
|---|---|---|
| 16:0 CoA | 18:0 CoA | |
| nmol min−1 mg−1 protein | ||
| MBP | 0.13 (0.039) | 0.10 (0.044) |
| MBP-2060 | 1.9 (0.98) | 1.9 (0.30) |
| MBP-2060D76G | 3.2 (1.1) | 3.8 (0.74) |
| MBP-1752 | 4.5 (0.55) | 7.0 (0.80) |
| MBP-1752D66G | 4.3 (0.66) | 2.6 (0.22) |
| MBP-1848 | 640 (160) | 5.2 (0.50) |
| MBP-1848D69G | 3.1 (1.2) | 1.9 (0.76) |
| MBP-PlsC | 638 (68) | 348 (120) |
| MBP-PlsCD77G | 2.4 (0.40) | 1.8 (0.38) |
Each value represents the average of three independent measurements. The numbers in parentheses denote the sd.
The Growth, Chlorophyll a Contents, and the Maximum Rates of Photosynthesis Decrease in Δ1848 and Δ1848 Δ2060 Cells Grown at 30°C
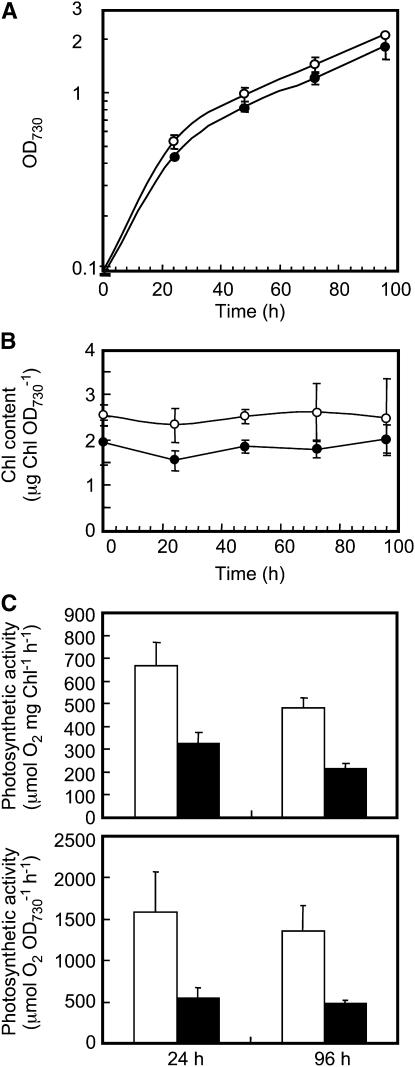
Δ1752, Δ2060, and Δ1752 Δ2060 cells grew indistinguishably from the wild type at 30°C at a photon flux density of 50 to 80 μmol m−2 s−1. In contrast, Δ1848 and Δ1848 Δ2060 cells showed delayed growth at a lower cell density (optical density at 730 mm [OD730] < 0.2) compared with other mutants and the wild type under our photosynthetic growth conditions (see Supplemental Fig. 4). To examine this phenomenon more carefully, portions of the wild-type or Δ1848 Δ2060 cells were collected from late exponential phase cultures, washed with fresh medium, and then inoculated into new culture medium to give an initial OD730 value of 0.1. Cell growth, chlorophyll (Chl) a content per unit OD730, and the maximum rates of photosynthesis were chased for 96 h. Although OD730 values increased at a similar rate for the wild-type and Δ1848 Δ2060 cells after 24 h of new culture (Fig. 3A), the OD730 values at 24 h were slightly lower for the latter cells. However, as shown in Figure 3B, Chl a content per unit OD730 was maintained at a lower level for 96 h in Δ1848 Δ2060 cells than in the wild-type cells, suggesting that Δ1848 Δ2060 cells maintain fewer Chl a-protein complexes than do the wild-type cells. Furthermore, the maximum rates of cellular photosynthesis at the photon flux density of 1,500 μmol m−2 s−1 decreased by more than 50% in the Δ1848 Δ2060 cells relative to the wild type, regardless of the cell basis or Chl a basis. Because the light intensity used for measurements of photosynthetic cells was much stronger than that for growth, the above results suggest that Δ1848 Δ2060 cells could be susceptible to photoinhibition (see below).
Figure 3.
Growth profiles, Chl a content, and the rates of photosynthesis in wild-type and Δ1848 Δ2060 cells. A, Growth of the wild-type (white circles) and Δ1848 Δ2060 (black circles) cells at a photon flux density of 50 to 80 μmol m−2 s−1. B, Changes in the content of Chl a after the inoculation of new medium with wild-type (white circles) and Δ1848 Δ2060 (black circles) cells. C, The rate of photosynthesis per Chl a (top section) and per OD730 (bottom section) in wild-type (white columns) and Δ1848 Δ2060 (black columns) cells, as measured with an oxygen electrode at a photon flux density of 1,500 μmol m−2 s−1 at 30°C. Each value represents an average of three independent experiments. Bars indicate sd.
Δ1848 Δ2060 Cells Are Susceptible to a Temperature Shift to 20°C
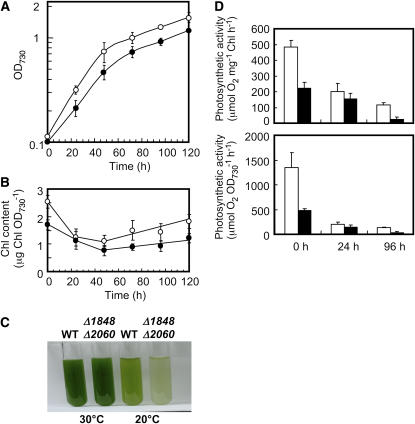
Δ1848 Δ2060 cells grown at 30°C were collected from late exponential phase cultures (96 h), washed with fresh medium at room temperature, and then inoculated into a new culture medium cooled to 20°C to give an initial OD730 value of 0.1. Cell growth, Chl a content per unit OD730, and the maximum rates of photosynthesis were chased for 120 h. Δ1848 Δ2060 cells shifted from 30°C to 20°C grew slower than the wild-type cells (Fig. 4A). After 24 h at 20°C, Chl a content in wild-type cells decreased to 49% of the level observed before the temperature shift, and then began to increase after 48 h (Fig. 4B). Chl a content in Δ1848 Δ2060 cells decreased comparably to 45% of the level observed before the temperature shift and also began to increase after 48 h, but at a much slower rate than in the wild type (Fig. 4B). As a result, the color of Δ1848 Δ2060 cell cultures was much paler than that of the wild-type cell cultures after 120 h at 20°C (Fig. 4C). In contrast, we found little color difference between wild-type and Δ1848 Δ2060 cell cultures after 120 h at 30°C (Fig. 4C).
Figure 4.
Effect of the temperature shift to 20°C on the growth, Chl a content, and the rate of photosynthesis in wild-type and Δ1848 Δ2060 cells at a photon flux density of 50 to 80 μmol m−2 s−1. A, Growth of the wild-type (white circles) and Δ1848 Δ2060 (black circles) cells after the temperature shift to 20°C. B, Changes in the content of Chl a in wild-type (white circles) and Δ1848 Δ2060 (black circles) cells after the temperature shift to 20°C. C, A view of cultures of wild-type and Δ1848 Δ2060 cells at 120 h after the start of new cultures at 30°C or 20°C. D, The rate of photosynthesis per Chl a (top section) and per OD730 (bottom section) in wild-type (white columns) and Δ1848 Δ2060 (black columns) cells grown at 20°C for the indicated time (in hours). The measurement was at the photon flux density 1,500 μmol m−2 s−1. at 30°C. Each value represents an average of three independent experiments. Bars indicate sd.
After 24 h at 20°C, the maximum rates of photosynthesis per Chl a decreased by 59% in wild-type and by 31% in Δ1848 Δ2060 cells compared with the rates before the temperature shift. However, between 24 and 96 h at 20°C, the maximum rates of photosynthesis per Chl a decreased by 42% in wild-type and by 84% in Δ1848 Δ2060 cells. These results suggest that both the construction of the PSI or PSII complexes (Fig. 4B) and the photosynthetic activity (Fig. 3D) are affected more by temperature shifts in Δ1848 Δ2060 cells than in wild-type cells. We observed similar results for Δ1848 cells as for the Δ1848 Δ2060 cells (data not shown). These results suggest that Δ1848 Δ2060 cells and Δ1848 cells could have disadvantages in acclimating to low temperature.
Δ1848 Δ2060 Cells Are More Susceptible to Photoinhibition Than Wild-Type Cells
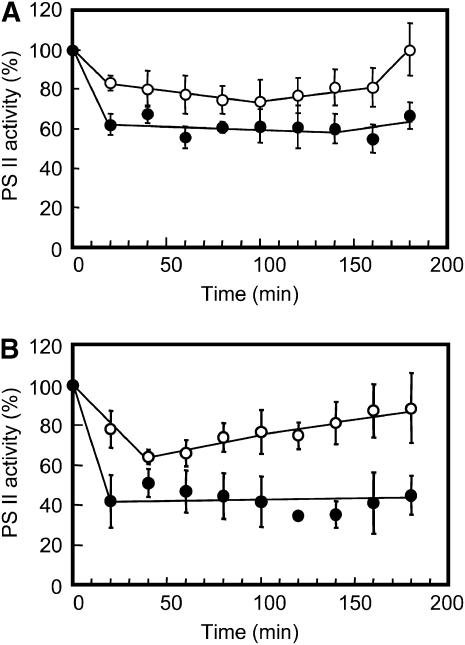
To investigate whether Δ1848 Δ2060 cells are more susceptible to photoinhibition than wild-type cells, cells were grown at a photon flux density of 60 μmol m−2 s−1 and shifted to 220 μmol m−2 s−1 at either 20°C or 30°C. Changes in PSII activities were then monitored with an oxygen electrode using p-benzoquinone as an electron acceptor (Fig. 5). When cells were transferred to 220 μmol m−2 s−1 at 30°C, the PSII activity of wild-type cells decreased by more than 20%. This level was maintained for up to 160 min, and then returned to the level measured before the light shift by 180 min. In contrast, the PSII activity of Δ1848 Δ2060 cells decreased by 38% after 20 min of the light shift at 30°C, and this level was maintained for up to 180 min. When cells were transferred to 220 μmol m−2 s−1 at 20°C, the PSII activity of wild-type cells decreased to 64% after 40 min. This level increased gradually for 180 min to regain 90% of the activity before the light shift. In contrast, the PSII activity of Δ1848 Δ2060 cells decreased to 38% within 20 min of the light shift at 20°C, and this level did not increase for up to 180 min. These results suggest that Δ1848 Δ2060 cells are more susceptible to photoinhibition than wild-type cells and have some defects in recovery from photoinhibition.
Figure 5.
Effect of high light on PSII activity in wild-type and Δ1848 Δ2060 cells. Cells grown at a photon flux density of 60 μmol m−2 s−1 for 48 h at 30°C (A) or 20°C (B) were shifted to 220 μmol m−2 s−1. PSII activities were then measured with an oxygen electrode at a photon flux density of 1,500 μmol m−2 s−1 at 30°C in the presence of 2 mm p-benzoquinone. The PSII activities of wild-type (white circles) and Δ1848 Δ2060 (black circles) cells before the light shift were 838 and 487 μmol mg−1 Chl h−1 for cells grown at 30°C, respectively, and 619 and 349 μmol mg−1 Chl h−1 for cells grown at 20°C, respectively. Each value represents an average of three independent experiments. Bars indicate sd.
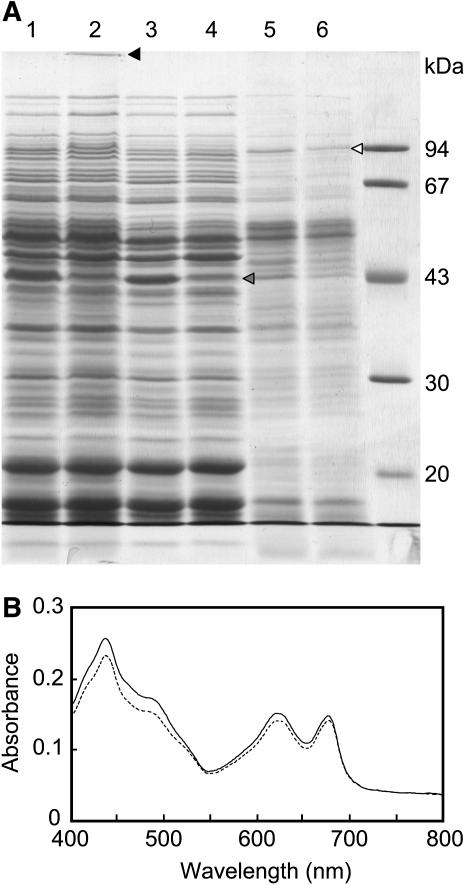
Recovery of a 95-kD Band Corresponding to a Phycobilisome Core-Membrane Linker Protein Decreases in Isolated Membrane Fractions of Δ1848 Δ2060 Cells
Figure 6A shows a photograph of a gel after SDS-PAGE of proteins from the cell-free lysate and soluble and membrane fractions of wild-type and Δ1848 Δ2060 cells grown for 96 h at 30°C. There were several noticeable differences in the polypeptide profiles between wild-type and Δ1848 Δ2060 fractions: In the cell-free lysate fractions, an orange-colored band was stacked at the top of the separating gel for the Δ1848 Δ2060 cell lane (Fig. 6A, lane 2, closed arrowhead); in the soluble fractions, a 45-kD band (gray arrowhead) decreased in the Δ1848 Δ2060 cells; and in the membrane fractions, a 95-kD band (open arrowhead) decreased in the Δ1848 Δ2060 cells (Fig. 6A). Peptide mass fingerprint analysis was successful only for the 95-kD protein, showing that it corresponds to the phycobilisome core-membrane linker protein encoded by slr0335 (apcE; Shen et al., 1993). The 45-kD band gave unidentifiable mixtures of polypeptides. However, the absorption spectrum for whole cells normalized by cell density showed that the peak height at 620 nm, which represented the phycobilisome content, showed only a minor decrease in the Δ1848 Δ2060 cells compared with the wild-type cells (Fig. 6B). Therefore, the phycobilisome complex in Δ1848 Δ2060 cells could be susceptible to protein extraction under our conditions.
Figure 6.
Comparison of the polypeptide composition and the absorption spectrum between wild-type and Δ1848 Δ2060 cells. A, SDS-PAGE of cell-free lysate fractions (lane 1, wild type; lane 2, Δ1848 Δ2060), soluble fractions (lane 3, wild type; lane 4, Δ1848 Δ2060), and membrane fractions (lane 5, wild type; lane 6, Δ1848 Δ2060) on a 10% polyacrylamide gel. Cell-free lysates and membrane fractions equivalent to 2 μg Chl and the corresponding amounts of soluble fractions were applied. The gel was stained with Coomassie Brilliant Blue R-250. B, Absorption spectra for wild-type (line) and Δ1848 Δ2060 (broken line) cells. Wild-type and Δ1848 Δ2060 cells were harvested after 96 h at 30°C at OD730 values of 2.15 and 1.94, respectively. Cells were diluted to an OD730 value of 0.05 before measuring the spectra.
DISCUSSION
Lipid Changes Induced by sll1848 Disruption
As previously reported by Weier et al. (2005), we herein confirmed that sll1848 encodes the major LPAAT of Synechocystis sp. PCC6803 required for maintaining C16 fatty acids in the sn-2 positions of membrane lipids. However, we also showed that sll1848 is not essential for this cyanobacterium. It is interesting that the sn-2 positions of MGDG, DGDG, SQDG, and PG of Δ1848 Δ2060 cells contain 18:0 and a considerable amount of 18:1 and 18:2 fatty acids (Table I). Synechocystis sp. PCC6803 contains four acyl-lipid desaturase genes, desC, desA, desD, and desB, which are responsible for Δ9, Δ12, Δ6, and Δ15 desaturation reactions, respectively. It has been reported that exogenously supplied 18:1(9) is desaturated to 18:1(9,12) in E. coli cells expressing Δ12 desA desaturase (Wada et al., 1993), and 18:1(9) is thought to be incorporated into the sn-2 positions of glycerolipids in E. coli. Therefore, if 18:1(9) is incorporated into the sn-2 position of PA by LPAAT in Synechocystis sp. PCC6803 cells, it could be desaturated to 18:2(9,12) by desA desaturase after conversion to glycerolipids. Alternatively, 18:0 incorporated into the sn-2 position of PA by LPAAT might be desaturated to 18:1 by Δ9 desaturase, and then to 18:2(9,12) by desA desaturase. However, we believe the latter possibility is unlikely because Δ9 desC desaturase is specific to 16:0 and 18:0 esterified fatty acids in sn-1 glycerolipid positions (Sakamoto et al., 1994). There is also no other desC homolog in the genome of Synechocystis sp. PCC6803. Therefore, our results suggest the possibility that 18:1 in sn-1 glycerolipid positions must be reused as a substrate for LPAAT and reincorporated into sn-2 glycerolipid positions. However, this pathway would be negligible in wild-type cells because of the predominant activity and specificity of sll1848 to 16:0, and probably at a lower rate of turnover of 18:1 in the sn-1 position. Therefore, it seems likely that the second LPAAT in Synechocystis sp. PCC6803 transfers 18:0 and 18:1 to the sn-2 position of PA in Δ1848 Δ2060 cells.
The higher unsaturation levels of fatty acids in the sn-2 positions of glycerolipids in Δ1848 Δ2060 cells appear to be compensated by the lower unsaturation levels of C18 fatty acids in the sn-1 positions in Δ1848 Δ2060 cells. This balance of unsaturation levels of fatty acids between sn-1 and sn-2 positions of glycerolipids might be a type of homeoviscous adaptation of membrane lipids. Analyses of lipids from Δ1848 cells led us to draw similar conclusions (data not shown).
We note that SQDG exhibits a relatively lower C16/C18 ratio for the fatty acids in the sn-1 position than other lipids. This suggests that the diacylglycerol moiety of SQDG is selected to decrease the C16/C18 ratios in the sn-1 position. We do not know the exact reason for this change, but cyanobacterial cells may require fewer unsaturated SQDG molecules for photosynthetic growth.
C16 Fatty Acids in the sn-2 Positions of Glycerolipids Are Preferred for Maintaining the Structure and Function of Photosynthetic Protein Complexes
Our peptide mass fingerprint analysis revealed that the recovery of a phycobilisome core-membrane linker protein decreases in isolated membrane fractions of Δ1848 Δ2060 cells. This protein encoded by slr0335 (apcE) attaches on the PSII core complexes in the thylakoid membranes and provides a scaffold for construction of phycobilisome protein complexes (Shen et al., 1993). Because there is little difference in the cellular absorption spectra between wild-type and Δ1848 Δ2060 cells, an altered fatty acid composition could have altered the affinity of the apcE protein to the PSII complex. As such, it was not fully reassembled in vitro into the complex.
Disruption of this gene causes growth retardation under photoautotrophic conditions especially at a lower light intensity (Shen et al., 1993). The growth rate of apcE− mutant cells is significantly lower than that of the wild-type cells at a photon flux density of <50 μmol m−2 s−1, whereas the rates of oxygen evolution by these cells are comparable at a photon flux density of >8,000 μmol m−2 s−1. These results are in contrast to our growth curves for the wild-type and Δ1848 Δ2060 cells in Figure 3A, rates of photosynthetic activities (Fig. 3C), and other data, which show that Δ1848 Δ2060 cells are more susceptible to photoinhibition than wild-type cells (Fig. 5).
It is reported that the PSII is a target of photoinhibition (Aro et al., 1993), and that PG is required for dimerization of the PSII complexes in Synechocystis sp. PCC6803 (Sakurai et al., 2003). Because Δ1848 and Δ1848 Δ2060 cells had unchanged glycerolipid compositions to wild-type cells, it is possible that altered fatty acid compositions of PG could have directly influenced susceptibility to photoinhibition in Δ1848 Δ2060 cells. In addition, the resultant changes in fatty acids in both the sn-1 and sn-2 positions of glycerolipids might influence lipid-protein interactions in the PSII reaction center or other protein complexes. As a result, it is possible that altered higher-order structures of PSII complexes might influence the affinity of apcE proteins to the PSII complex. Therefore, we propose that C16 fatty acids in the sn-2 positions of glycerolipids are preferred to maintain the structure and function of the photosynthetic protein complexes.
slr2060 Is Not Essential for Cyanobacteria and Plants
We showed that MBP-2060 does not exhibit significant levels of GPAT or LPAAT activities in vitro, when acyl-CoAs are used as substrates. Consistent with this finding, Weier et al. (2005) showed that slr2060 encodes acyl-ACP-specific lysophospholipid acyltransferase. The slr2060 protein has 80 amino acid residues between the HXXXXD and the XXXXXXG signatures, and no orthologous acyltransferases are found in Prochlorococcus marinus or Cyanidioschyzon merolae. Therefore, it seems likely that slr2060 is not essential for the evolution of photosynthetic higher eukaryotes. However, we note that the number of amino acids between the acyltransferase signatures of slr2060 is close to that of plastid-targeted GPATs in higher plants encoded by ATS1/ACT1 orthologs (i.e. 82–83). To date, neither the ortholog of plastid-targeted GPAT genes nor a candidate gene for cyanobacterial GPAT has been discovered in the published cyanobacterial databases. These questions should be addressed in future studies, together with a possible link between slr2060 and ATS1/ACT1 genes.
Evolution of Cyanobacterial LPAAT
Most cyanobacterial species have C16 fatty acids in the sn-2 positions of membrane glycerolipids (Murata et al., 1992). The exception is the membrane lipids of Gloeobacter violaceus PCC7421, which conserve many ancestral properties and contain only 32% of 16:0 fatty acids (Selstam and Campbell, 1996), indicating that 36% or more of C18 fatty acids is esterified in the sn-2 position. This suggests that LPAAT from this cyanobacterium should use 16:0 and 18:0 at comparable rates. Because G. violaceus PCC7421 has no orthologs for sll1752, other LPAATs must be responsible for the unique fatty acid composition of this cyanobacterium. Interestingly, G. violaceus PCC7421 exhibits relatively slow growth under photosynthetic conditions and is sensitive to strong light (Rippka et al., 1974). These features are similar to those found in Δ1848 Δ2060 cells, although G. violaceus PCC7421 also lacks SQDG. Because molecular phylogenic analysis shows that G. violaceus diverged at the earliest stage from other cyanobacteria, C16 fatty acids are thought to have become dominant in the sn-2 positions of other cyanobacteria during evolution. This coincides with the ubiquitous presence of sll1848 orthologs within extant cyanobacteria. Therefore, because of its substrate preference, sll1752 may not have been preferred during evolution by most cyanobacterial species. Nevertheless, orthologs of sll1752 are widely conserved in the genomes of cyanobacteria whose databases have been published to date. If their gene products still show a preference for C18 substrates, it remains unclear why sll1752 orthologs are conserved among a wide range of cyanobacterial species. In some cyanobacteria, such as Mastigocladus laminosus, C18 fatty acids account for 10% of fatty acids in the sn-2 positions (Murata et al., 1992), for which sll1752 orthologs could be responsible. However, sll1752 is constitutively transcribed in the wild-type cells of Synechocystis sp. PCC6803, which has few C18 fatty acids in the sn-2 positions (Fig. 5). This raises another possibility that glycerolipids containing C18 fatty acids in the sn-2 positions are covalently bound to some polymers or to proteins, preventing their extraction by conventional lipid-extraction protocols. Future studies should examine the substrate selectivity of proteins encoded by sll1752 orthologs in other cyanobacterial strains and carefully review the importance of these lipids.
Evolution of Higher-Plant LPAAT
Bourgis et al. (1999) suggested that the alignment of the deduced amino acid sequences has the same orthologous relationship in sll1848 and plastidial LPAAT from Brassica napus (BAT2). This idea is confirmed by the observation that both LPAAT1/ATS2 from Arabidopsis and sll1848 from Synechocystis sp PCC6803 show a preference for 16:0 ACP (and 16:0 CoA in vitro). In addition, LPAAT1 protein has 61 amino acid residues between the HXXXXD and the XXXXXXG signatures (Kim and Huwang, 2004; Yu et al., 2004), and this number is close to the corresponding value for sll1848 (i.e. 60 amino acid residues). Therefore, along with the substrate specificity, the number of amino acids between the HXXXXD and the XXXXXXG signatures may also provide information about the gene lineage, although this should be confirmed by future studies.
In this context, LPAATs from several plant species can be classified into several groups as follows: endoplasmic reticulum-located LPAAT, encoded by Limnanthes douglasii LAT2 and Arabidopsis LPAT2, use 18:1 CoA for membrane lipid synthesis, and these LPAAT have 68 or 69 amino acid residues between the conserved motif signatures (Brown et al., 1995; Kim et al., 2005). Another type of endoplasmic reticulum-located LPAAT is involved in triacylglycerol synthesis and uses unusual fatty acids such as 12:0 or 22:1. These LPAAT are seed-specific LPAAT and have 63 amino acid residues between the conserved motif signatures (Brown et al., 1995; Knutzon et al., 1995; Lassner et al., 1995). Because sll1752 has a distance of 64 amino acid residues between the HXXXXD and the XXXXXXG signatures, this number is most similar to that of seed-specific LPAATs. However, these LPAATs show little similarity in their sequences or fatty acid selectivity. Furthermore, no ortholog of sll1752 exists in the genome of C. merolae, the very primitive rhodophyte (Matsuzaki et al., 2004) Chlamydomonas reinhardtii, and Arabidopsis. During the evolution of higher-plant plastids, they came to contain C18/C18 lipid molecular species in MGDG, DGDG, and SQDG. However, the molecular species of plastidial PG always contain C16 fatty acids in the sn-2 position. It would be interesting to test whether introducing C18 fatty acids in the sn-2 positions of PG in plastids has deleterious effects on photosynthesis in higher plants; sll1752 might provide a useful tool for this test.
CONCLUSION
Synechocystis sp. PCC6803 has two LPAATs encoded by sll1848 and sll1752. We showed that sll1848 is dispensable, although it is the major LPAAT specific to 16:0. We also showed that sll1752 is a minor LPAAT that prefers 18:0 and 18:1 as substrates. The third acyltransferase slr2060, which was recently found to be a lysophospholipid acyltransferase (Weier et al., 2005), exhibited neither GPAT nor LPAAT activities, but the number of amino acids between the acyltransferase signatures was close to that for plastid-targeted GPATs in higher plants. The disruption of sll1848 causes enrichment of C18 fatty acids in place of C16 fatty acids in the sn-2 positions of glycerolipids, and growth retardation at lower cell density and at low temperature. The mutant growth phenotypes could be explained by altered lipid-protein interactions in the PSII protein complexes and other photosynthetic protein complexes. Therefore, we conclude that C16 fatty acids in the sn-2 positions of glycerolipids play crucial roles in the photosynthetic growth of Synechocystis sp. PCC6803. To investigate the role of C16 fatty acids of the sn-2 positions of plastidial glycerolipids in higher-plant photosynthesis, it would be interesting to introduce the sll1752 gene into LPAAT1 mutants of Arabidopsis (Kim and Huwang, 2004; Yu et al., 2004).
MATERIALS AND METHODS
Cyanobacterial Strain and Growth
Synechocystis sp. PCC6803 was kindly provided by H. Wada of the University of Tokyo. The cells were grown photoautotrophically at 30°C in BG-11 medium (Stanier et al., 1971) supplemented with 20 mm HEPES-NaOH (pH 7.5) under continuous illumination (50–80 μmol m−2 s−1) in flasks with rotary shaking (120 rpm).
Insertion Mutagenesis
The sll1848, sll1752, and slr2060 genes were disrupted by displacing the HXXXXD signatures with gene cassettes conferring resistance to kanamycin, spectinomycin, and erythromycin, respectively. A 773-bp fragment for sll1848 disruption was amplified by PCR using the primer pairs 5′-AAAGTTAACGGATTCCCGTCACCCTGATTA-3′ and 5′-AAGAATTCACAAAGGATGGCCGCATTCAC-3′, and a 443-bp fragment was amplified using the primer pairs 5′-AAAGAATTCGTTGGCACCAATTCCTGACCA-3′ and 5′-CGGAGAAACGAGAACAGAGGA-3′. A 510-bp fragment for sll1752 disruption was amplified by PCR using the primer pairs 5′-TGCCCCACAAAAAAATTCCCC-3′ and 5′-AAAGAATTCGTAAGCAAAACCCCAACACTG-3′, and a 636-bp fragment was amplified using the primer pairs 5′-AAAGAATTCGTGGGTAGGGGCCAGAATAAT-3′ and 5′-AAACTGCAGGGGCACCATAGACCATTTGAT-3′. A 2,073-bp fragment for slr2060 disruption was amplified by PCR using the primer pairs 5′-AAATCTAGAAAAGCTGCCAGTCCAGACCAG-3′ and 5′-AAACTGCAGCCCTGGGTTACTTTCTCTCC-3′, and a 665-bp fragment was amplified using the primer pairs 5′-AAAGGATCCGCTCCATGCGGCATTAACGGT-3′ and 5′-AAACTGCAGCCCTGGGTTACTTTCTCTCC-3′. Each pair of PCR fragments for gene disruptions was subcloned into the plasmid pBluescript II (Stratagene) in the order of coding sequences. Suitable drug-resistant gene cassettes were then inserted at the EcoRI site for sll1848, the EcoRI for sll1752, and between two BamHI sites for slr2060. The resultant disrupted gene fragments were introduced into the genome of wild-type cells by homologous recombination as described previously (Golden et al., 1987). The resultant gene disruptions were confirmed by PCR, using the following primer pairs: 5′-AAAGAATTCGTGGATTCCGAGATTAATCAT-3′ and 5′-AAAGGATCCTAAGGATGCCATCACTAAT-3′ for sll1848; 5′-AAAGAATTCATGACCAATTCTCCCCTGGCG-3′ and 5′-AAAGGATCCCCCATTAGCCGTTTTGAC-3′ for sll1752; and 5′-AAAGAATTCATGGAATCCCCCATCCAAGCC-3′ and 5′-AAACTGCAGCCCTGGGCTTACTTTCTCTCC-3′ for slr2060.
Lipid Analysis
Lipids were extracted according to the method of Bligh and Dyer (1959). Total lipid extracts (1 mg) were separated into representative lipid classes and the contents of lipids and fatty acids were determined as described previously (Inatsugi et al., 2002). The positional distribution of fatty acids between sn-1 and sn-2 positions of glycerolipids was analyzed as described previously (Sato and Murata, 1988), using a lipase from Rhizopus delemar (Seikagaku Kogyo).
RT-PCR Analyses
RNA was isolated from cells in a midexponential phase using a TRIZOL reagent solution (Invitrogen). The first-strand cDNA was synthesized from 1 μg of total RNA using a Transcriptor First Strand cDNA synthesis kit (Roche Applied Science) according to the manufacturer's protocol. PCR was performed using the gene-specific primer pairs 5′-AGTTAGGGAGGGAGTTGC-3′ and 5′-TAAGCCGGGTTCTGTTCC-3′ for rnpB; 5′-AAAGAATTCGTGGGATTCCGAGATTAAGCAT-3′ and 5′-AAAAATGGGGGGCCAAAATAA-3′ for sll1848; and 5′-AAAGAATTCATGACCAATTCTCCCCTGGCG-3′ and 5′-AAAATGGCGCCCCAACGGGAA-3′ for sll1752. The amounts of template cDNA were optimized to amplify the 417-bp band for rnpB cDNA at a similar intensity between samples. PCR was started with the first denaturation step at 98°C for 2 min, followed by cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 1 min, and the final extension step at 72°C for 2 min. Twenty-five or 30 cycles were used for rnpB cDNA, and 35 cycles were used for sll1848 and sll1752 cDNAs.
Acyltransferase Assay
The LPAAT activity of MBP-tagged proteins was measured in a reaction mixture containing 250 mm HEPES-NaOH (pH 8.2), 1 mg mL−1 bovine serum albumin, 5 mm MgCl2, 1 mm dithiothreitol (DTT), 1 mm oleoyl-lysophosphatidic acid, MBP-tagged protein (2–300 μg), and one of 0.2 mm [1-14C]palmitoyl-CoA, [1-14C]stearoyl-CoA, or [1-14C]oleoyl-CoA. Each reaction was started by adding the radioactive substrates and was terminated after 10 min at 25°C, except that the reactions using [1-14C]palmitoyl-CoA for MBP-1848 and MBP-PlsC were terminated after 2 min. The reaction products were copurified with authentic lysophosphatidic acid (L 7260; Sigma) and phosphatidic acid (A-43; Funakoshi) on a thin-layer silica gel 60 plate (1.05721; Merck) using a solvent mixture of chloroform:methanol:acetic acid:water (65:25:4:2, by volumes) for development. The radioactive spots corresponding to phosphatidic acid were scraped from the plate, and the radioactivity was determined on a scintillation counter (LS6000 [Beckman Coulter] or Tri-Carb 1600TR [Packard]), using a scintisol cocktail (Scintisol EX-H [Wako] or Clear-sol II [Nacalai Tesque]).
Synechocystis sp. PCC6803 cells were fragmented by passing them through a French pressure cell (Ohtake Works) at 1.0 × 108 Pa. Cell lysates were centrifuged at 100,000g for 60 min, and the pellets were resuspended to provide the membrane fraction. LPAAT activity in the membrane fraction was measured using the above reaction mixtures, except that the reactions were performed at 30°C in the presence of 5 mg mL−1 bovine serum albumin.
GPAT activity was measured in the same reaction mixtures as used in LPAAT assays, except that 1 mm [1-14C]glycerol-3-P was used instead of lysophosphatidic acids and 0.2 mm 16:0 CoA instead of labeled acyl-CoA. For assays in the presence of lipids, proteins recovered from native PAGE gel slices were incubated with or without a 2.5 mg mL−1 liposome solution prepared from E. coli membrane lipids.
Expression and Purification of MBP-Tagged Proteins
The coding regions of sll1848, sll1752, and slr2060 were amplified by PCR from DNA extracted from Synechocystis sp. PCC6803 cells. Amplified fragments for sll1848 and sll1752 were digested with EcoRI and BamHI sites, and the fragment for slr2060 was digested with EcoRI and PstI sites. Each of the digested fragments was subcloned in the vector pMAL-c2X (New England Biolabs), and the resultant constructs were expressed in E. coli TB-1 cells. E. coli cells were suspended in a buffer containing 20 mm Tris-HCl (pH 7.5), 1 mm EDTA, 200 mm NaCl, 1 mm DTT, 1 mm NaN3, 1 mm benzamidine HCl, and 5 mm ɛ-aminocaproic acid, and then broken by passing through a French pressure cell (Ohtake Works) at 1.0 × 108 Pa. Cell lysates were centrifuged at 14,000g for 20 min and at 100,000g for 65 min. Supernatants were then recovered to provide soluble fractions. MBP-tagged proteins from the 100,000g supernatants were purified by amylose-affinity column chromatography according to the manufacturer's protocol (New England Biolabs).
In Vitro Mutagenesis
The genes for 1848D69G, 1752D66G, 2060D76G, and PlsCD77G variants were constructed by overlap extension PCR (Ho et al., 1989) and cloned into pMAL-2cX vector for expression in TB1 cells.
Gel Electrophoresis
Native PAGE was performed on a 3.5% polyacrylamide slab gel containing 200 mm Tris-HCl pH 8.3. Samples were made up in 1× loading dye solution (10 mm Tris-HCl, pH 8.0, 6% glycerol, 0.02% bromphenol blue, and 1 mm DTT). Electrophoresis was run at 25 mA for 5.5 h at 4°C. After electrophoresis, a part of the gel was cut off and subjected to denaturing electrophoresis in the second dimension. The remaining part of the gel was sliced into 22 strips, each 5 mm in width, from which proteins were extracted using a buffer containing 20 mm Tris-HCl (pH 7.5), 1 mm EDTA, 200 mm NaCl, and 1 mm DTT for the LPAAT or GPAT assays.
SDS-PAGE was performed on a 10% polyacrylamide slab gel. Synechocystis sp. PCC6803 cell lysates were centrifuged at 14,000g for 20 min, and the supernatants were recovered to provide cell-free lysate fractions. The cell-free lysates were then centrifuged at 100,000g for 60 min, and the supernatant was recovered as the soluble fraction. Pellets were resuspended to provide the membrane fraction. Cell-free lysates and membrane fractions corresponding to 2 μg Chl and the corresponding amount of soluble fraction were applied to the gel. The gel was stained with Coomassie Brilliant Blue R-250 (Wako) or Coomassie Brilliant Blue G-250 (Wako) for peptide mass fingerprint analyses.
Determination of Chl Content and Photosynthesis
Chl a was extracted from cells with methanol and determined using a previously published method (Arnon et al., 1974). Photosynthetic O2 evolution was measured in intact cells in BG-11 medium containing 10 mm NaHCO3, using a Clark-type oxygen electrode (Rank Brothers). PSII activity was measured with the oxygen electrode using 2 mm p-benzoquinone as an electron acceptor.
Peptide Mass Fingerprint Analysis
Peptide mass fingerprint analysis was conducted by Shimadzu-Biotech (http://www.shimadzu-biotech.jp/).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NP_440396 (sll1752), NP_441186 (slr2060), and NP_441924 (sll1848).
Acknowledgments
We thank Hajime Wada for the gift of Synechocystis sp. PCC6803, Masahiko Ikeuchi for the erythromycin-resistant gene cassette, and Masayuki Ohmori for providing the oxygen electrode. We also thank Atsuhiko Aoyama for his technical assistance and Yoshibumi Komeda for discussions.
This work was supported by the Itoh Science Foundation and in part by the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN), and a Grant-in-Aid for the Promotion of Priority Areas (17051004) from the Ministry of Education, Culture, Sports, Science and Technology.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Ikuo Nishida (nishida@molbiol.saitama-u.ac.jp).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.075796.
References
- Arnon DI, McSwain BD, Tsujinomo HY, Wada K (1974) Photochemical activity and components of membrane preparations from blue-green algae. Biochim Biophys Acta 357: 231–245 [DOI] [PubMed] [Google Scholar]
- Aro EM, Virgin I, Andersson B (1993) Photoinhibition of photosystem II: inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Athenstaedt K, Daum G (1999) Phosphatidic acid, a key intermediate in lipid metabolism. Eur J Biochem 266: 1–16 [DOI] [PubMed] [Google Scholar]
- Bertrams M, Heinz E (1981) Positional specificity and fatty acid selectivity of purified sn-glycerol 3-phosphate acyltransferases from chloroplasts. Plant Physiol 68: 653–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Bourgis F, Kader J-C, Barret P, Renard M, Robinson D, Robinson C, Delseny M, Roscoe TJ (1999) A plastidial lysophosphatidic acid acyltransferase from oilseed rape. Plant Physiol 120: 913–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AP, Brough CL, Kroon JTM, Slabas AR (1995) Identification of a cDNA that encodes a 1-acyl-sn-glycerol-3-phosphate acyltransferase from Limnanthes douglasii. Plant Mol Biol 29: 267–278 [DOI] [PubMed] [Google Scholar]
- Coleman J (1990) Characterization of Escherichia coli cells deficient in 1-acyl-sn-glycerol-3-phosphate acyltransferase activity. J Biol Chem 265: 17215–17221 [PubMed] [Google Scholar]
- Frentzen M, Heinz E, Mckeon TA, Stumpf PK (1983) Specificities and selectivities of glycerol-3-phosphate acyltransferase and monoacylglycerol-3-phosphate acyltransferase from pea and spinach chloroplasts. Eur J Biochem 129: 629–636 [DOI] [PubMed] [Google Scholar]
- Frentzen M, Wolter FP (1998) Molecular biology of acyltransferases involved in glycerolipid synthesis. In Harwood JL, ed, Plant Lipid Biosynthesis. Cambridge University Press, Cambridge, pp 247–272
- Golden S, Brusslan J, Hslkorn R (1987) Genetic engineering of the cyanobacterial chromosome. Methods Enzymol 153: 215–231 [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Ikeuchi M, Eggers B, Shen G, Webber A, Yu J, Hirano A, Inoue Y, Vermaas W (1991) Cloning of the psbK gene from Synechocystis sp. PCC6803 and characterization of photosystem II in mutants lacking PSII-K. J Biol Chem 266: 11111–11115 [PubMed] [Google Scholar]
- Inatsugi R, Nakamura M, Nishida I (2002) Phosphatidylcholine biosynthesis at low temperature: differential expression of CTP:Phosphorylcholine cytidyltransferase isogenes in Arabidopsis thaliana. Plant Cell Physiol 43: 1342–1350 [DOI] [PubMed] [Google Scholar]
- Kim H, Huwang A (2004) Plastid lysophosphatidyl acyltransferase is essential for embryo development in Arabidopsis. Plant Physiol 134: 1206–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Li Y, Huang HC (2005) Ubiquitous and endoplasmic reticulum-located lyzophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell 17: 1073–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutzon DS, Lardizabal KD, Nelsen JS, Bleibaum JL, Davies HM, Metz JG (1995) Cloning of a coconut endosperm cDNA encoding a 1-acyl-sn-glycerol-3-phosphate acyltransferase that accepts medium-chain-length substrates. Plant Physiol 109: 999–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassner MW, Levering CK, Davies HM, Knutzon DS (1995) Lysophosphatidic acid acyltransferase from meadowfoam mediates insertion of erucic acid at the sn-2 position of triacylglycerol in transgenic rapeseed oil. Plant Physiol 109: 1389–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lem NW, Stumpf PK (1984) In vitro fatty acid and complex lipid metabolism in the cyanobacterium, Anabaena variabilis. Plant Physiol 75: 700–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin TM, Wang P, Coleman RA (1999) Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry 38: 5764–5771 [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, et al (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428: 653–657 [DOI] [PubMed] [Google Scholar]
- McFadden GI (1999) Endosymbiosis and evolution of the plant cell. Curr Opin Plant Biol 2: 513–519 [DOI] [PubMed] [Google Scholar]
- Murata N, Wada H, Gombos Z (1992) Modes of fatty-acid desaturation in cyanobacteria. Plant Cell Physiol 33: 933–941 [Google Scholar]
- Rippka R, Waterbury J, Cohen-Bazire G (1974) A cyanobacterium which lacks thylakoids. Arch Microbiol 100: 419–436 [Google Scholar]
- Sakamoto T, Wada H, Nishida I, Ohmori M, Murata N (1994) Δ9 acyl-lipid desaturases of cyanobacteria. J Biol Chem 269: 25576–25580 [PubMed] [Google Scholar]
- Sakurai I, Hagio M, Gombos Z, Tyystjarvi T, Paakkarinen V, Aro E-M, Wada H (2003) Requirement of phosphatidylglycerol for maintenance of photosynthetic machinery. Plant Physiol 133: 1376–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Murata N (1988) Membrane lipids. Methods Enzymol 167: 245–251 [Google Scholar]
- Selstam E, Campbell D (1996) Membrane lipid composition of the unusual cyanobacterium Gloeobacter violaceus sp. PCC7421, which lacks sulfoquinovolsyl diacylglycerol. Arch Microbiol 166: 132–135 [Google Scholar]
- Shen G, Boussiba S, Vermaas FJ (1993) Synechocystis sp PCC 6803 strains lacking photosystem I and phycobilisome function. Plant Cell 5: 1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier RY, Kunisawa R, Mandel M, Cohen-Bazire G (1971) Purification and properties of unicellular blue-green algae (order Chrococcales). Bacteriol Rev 35: 171–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Avelange-Macherel MH, Murata N (1993) The desA gene of the cyanobacterium Synechocystis sp. strain PCC6803 is the structural gene for Δ12 desaturase. J Bacteriol 175: 6056–6058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weier D, Muller C, Gaspers C, Frentzen M (2005) Characterisation of acyltransferases from Synechocystis sp. PCC6803. Biochem Biophys Res Commun 334: 1127–1134 [DOI] [PubMed] [Google Scholar]
- Yu B, Wakao S, Fan J, Benning C (2004) Loss of plastidic lysophosphatidic acid acyltransferase causes embryo-lethality in Arabidopsis. Plant Cell Physiol 45: 503–510 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Xia Q, Dauk M, Shen W, Selvaraj G, Zou J (2003) Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15: 1872–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]