Abstract
Rhizobia reside as symbiosomes in the infected cells of legume nodules to fix atmospheric nitrogen. The symbiotic relation is strictly controlled, lasts for some time, but eventually leads to nodule senescence. We present a comprehensive transcriptomics study to understand the onset of nodule senescence in the legume Medicago truncatula. Distinct developmental stages with characteristic gene expression were delineated during which the two symbiotic partners were degraded consecutively, marking the switch in nodule tissue status from carbon sink to general nutrient source. Cluster analysis discriminated an early expression group that harbored regulatory genes that might be primary tools to interfere with pod filling-related or stress-induced nodule senescence, ultimately causing prolonged nitrogen fixation. Interestingly, the transcriptomes of nodule and leaf senescence had a high degree of overlap, arguing for the recruitment of similar pathways.
Leguminous plants are capable of growing under nitrogen-limiting conditions because of their ability to establish endosymbiosis with soil bacteria, collectively called rhizobia. During this interaction, new organs, called nodules, are formed wherein the microsymbiont fixes atmospheric nitrogen to supply the plant with ammonium. In return, the microsymbionts obtain photosynthates and a protective niche.
Establishment of symbiosis requires the specific perception of bacterial signal molecules by plant receptor kinase complexes (Oldroyd and Downie, 2004). These recognition events allow the invasion of the host as well as the formation of a nodule primordium; these two processes occur in parallel and eventually merge when infection threads release bacteria into the cytoplasm of the newly formed primordial cells (Schultze and Kondorosi, 1998). The bacteria become enclosed by a plant-derived membrane, the peribacteroid membrane (collectively called symbiosome), and bacteroid differentiation precedes the metabolic phase of symbiosis. At some point, nitrogen fixation ceases, nodule cells senesce, and the symbiotic relationship is lost.
Nodules can be developmentally classified into two main groups. Determinate nodules, as observed in Lotus japonicus and some tropical legumes, are characterized by a nonpersistent meristem that results in round-shaped organs with a homogeneous central fixation zone composed of infected rhizobia-filled cells interspersed with some uninfected cells. In these nodules, senescence develops radially, starting from the inside and slowly spreading toward the outside. In contrast, Medicago truncatula and many other legumes form indeterminate, cylindrical nodules that consist of a gradient of developmental zones with a persistent apical meristem (zone I), an infection zone (zone II), and a fixation zone (zone III). In mature nodules, a senescence zone (zone IV) is established proximal to zone III. Upon aging, this zone gradually moves in a proximal-distal direction until it reaches the apical part and the nodule degenerates.
Nodule senescence can be triggered prematurely by various types of stress (Gogorcena et al., 1997; González et al., 1998; Matamoros et al., 1999) or as a consequence of deficient recognition between partners (Banba et al., 2001). This induced nodule senescence develops much more rapidly than developmental senescence and present features of oxidative stress and plant cell death (Puppo et al., 2005).
Senescence in nodules is visible by a color shift in the nitrogen-fixing zone from pink, associated with the functional leghemoglobin protein, to green, associated with the degradation of its heme group (Roponen, 1970). At the ultrastructural level, changes in the symbiosomes and cellular organelles are observed: The cytoplasm becomes less electron dense (Andreeva et al., 1998) and vesicles and ghost membranes appear as a result of host and symbiosome membrane disintegration (Timmers et al., 2000). A hallmark of nodule senescence is the triggering of a wide range of proteolytic activities that cause large-scale protein degradation (Pladys and Vance, 1993). The final outcome is death of both bacteroids and nodule cells.
For many leguminous crops, nodule senescence coincides with pod filling (Lawn and Brun, 1974; Bethlenfalvay and Phillips, 1977). Exogenous application of nitrogen during pod filling increases both yield and seed protein content (Merbach and Schilling, 1980). Hence, extending the period of active nitrogen fixation by delaying the nodule senescence process might have a beneficial effect on crop yield and seed quality. The existence of soybean (Glycine max) varieties with a delayed nodule senescence and a longer active nitrogen fixation period (Espinosa-Victoria et al., 2000) points to a genetic control of the onsetof senescence, suggesting a molecular basis to modify timing and/or progression of developmental nodule senescence.
Signals that trigger the formation of a senescence zone need to be identified to modulate the active lifespan of a nodule. Developmental nodule senescence has been proposed to result from a decrease in the ascorbate-gluthatione antioxidant pool coupled to decreasing carbon-to-nitrogen ratios inside the nodule (Puppo et al., 2005). These changes would be sensed and transduced via an abscisic acid-mediated signaling pathway mobilizing proteolytic activities. To date, however, signal transduction cascades and regulatory functions controlling nodule senescence are unknown.
We present a comprehensive transcriptomics dataset that demonstrates that nodule senescence is a complex and regulated process. Transcript-profiling analysis of developmental nodule senescence was performed in the legume M. truncatula through a modified cDNA-amplified fragment length polymorphism (AFLP) protocol (Breyne et al., 2003). A very specific sampling method coupled to cluster analysis allowed distinct stages of the nodule senescence process to be identified. A collection was obtained of 508 gene tags that were expressed differentially. Functional clustering of the corresponding genes demonstrated the transition from carbon sink to general source status of the nodule, as well as the activation of a wide variety of defense and stress responses. A high degree of functional overlap was evident with the transcriptome of the leaf senescence process, suggesting that similar mechanisms underpin both processes. Importantly, one cluster contained tags whose expression was enhanced prior to initiation of nodule senescence. The regulatory and signal transduction genes present in this cluster are primary candidates to modulate nodule senescence.
RESULTS
Microscopic Analysis of Nodule Senescence in M. truncatula
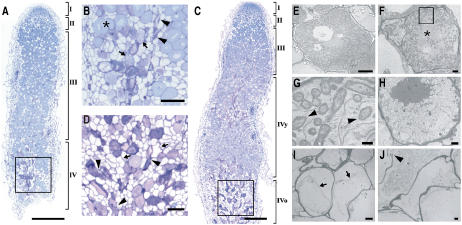
To determine stage-specific traits during developmental nodule senescence, M. truncatula nodules were compared 21, 35, and 61 d postinoculation (dpi) without, with small, and with advanced green senescence zones, respectively. A toluidine blue-stained longitudinal section through a young, fixing nodule had the typical zonation of indeterminate nodules: the meristem, the infection zone, and a large fixation zone (data not shown). In 35-dpi nodules, some infected cells of the proximal part of the fixation zone (type I senescent cells) differed structurally from healthy infected cells (Fig. 1A). Unlike these healthy cells (Fig. 1B, asterisk), type I senescent cells were more purple and presented signs of bacterial degradation, as illustrated by the loss of cell content, but maintained their rigidity (Fig. 1B, arrows). In the same region, a second type of senescent cells was observed that are flattened with symptoms of cell death (type II senescent cells; Fig. 1B, arrowhead). Senescent cells of type I and type II were similarly distributed in the younger senescence zone of 61-dpi nodules (Fig. 1C). The older, proximal part was characterized by a large number of vacuolated cells (type III senescent cells; Fig. 1D, arrows) that corresponded to cells that were completely resorbed. A high number of flattened type II senescent cells (Fig. 1D, arrowhead) were also visible.
Figure 1.
Microscopic analysis of nodule senescence in M. truncatula. A, Toluidine blue-stained longitudinal section through a nodule of 35 dpi, showing a small, young senescence zone. B, Enlargement of the rectangle in A. Asterisks, arrows, and arrowheads indicate fixing, type I, and type II senescent cells, respectively. C, Toluidine blue-stained longitudinal section through a nodule of 61 dpi showing young and old senescence zones. D, Enlargement of the rectangle in C. Arrowheads and arrows indicate type II and type III senescent cells, respectively. E, TEM image of a healthy, nitrogen-fixing cell packed with bacteroids. F, TEM image of a type I senescent cell with a clearly reduced number of bacteroids and prominent presence of endoplasmic reticulum. G, Enlargement of the rectangle in F showing peribacteroid membrane degradation (arrowheads). H, TEM image of a type I senescent cell undergoing complete resorption. I, TEM picture of a type III senescent cell showing signs of plant cell death, such as plasma membrane loosening (arrows). J, TEM image of a type II senescent cell showing loss of cellular integrity and plant cell wall breakage. I, Meristem; II, infection zone; III, fixation zone; IV, senescence zone; y and o, young and old senescence zones, respectively. Bars = 500 μm (A and C), 100 μm (B and D), 10 μm (E), 2 μm (F, H–J), and 500 nm (G).
Using transmission electron microscopy (TEM), bacteroid lysis was seen in type I senescent cells (Fig. 1F) and not in healthy fixing cells (Fig. 1E), as evidenced by degradation of the peribacteroid membrane (Fig. 1G, arrowhead); this lysis concurs with increased vesicular activity and prominent endoplasmic reticulum (Fig. 1F, asterisk). Symbiosomes of these cells were gradually resorbed (Fig. 1H), whereas the plant cell remained intact without signs of death, indicating that at this stage only the microbial partner was targeted for degradation. Finally, these cells became completely devoid of symbiosomes and started to show signs of plant cell death, such as loosening of the plasma membrane from the wall (type III senescent cells; Fig. 1I, arrow). The flattened type II senescent cells also had features of death, such as cell wall breakage (Fig. 1J, arrowhead).
Based on these ultrastructural observations, two consecutive stages could be distinguished during nodule senescence: a first stage, characterized by bacteroid degradation with a few dying plant cells, and a more advanced stage of nodule senescence, during which cells had completely resorbed their symbiosomes and started to decay and collapse.
Transcriptome Analysis
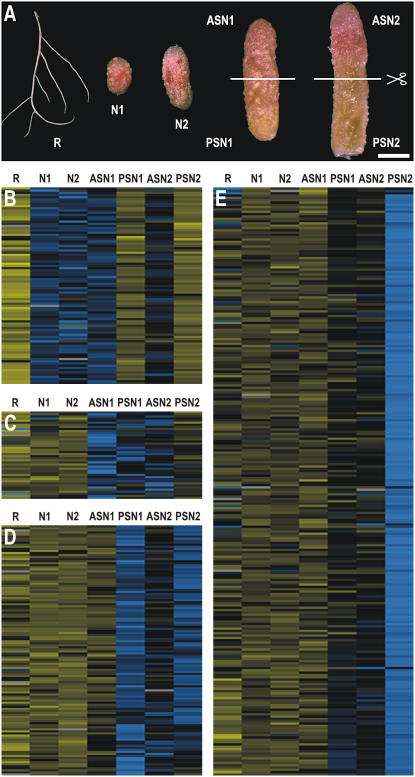
To isolate plant genes that are differentially expressed during the two stages of senescence, the transcriptome of young nodules (without senescence zone; Fig. 2A, samples N1 and N2) was compared with that of middle-aged and old nodules with a small and a large senescence zone, respectively (Fig. 2A). The nodules with a senescence zone were transversally cut in half. Dissection of nodules with a small senescence zone resulted in samples ASN1 (Fig. 2A), containing the meristem, the infection zone, and the apical part of the fixation zone, and PSN1 (Fig. 2A), containing the proximal part of the fixation zone and a young senescence zone. Dissection of nodules with a large senescence zone gave rise to samples ASN2 (Fig. 2A), comprising the meristem, infection zone, a small fixation zone, and the apical part of the senescence zone, and PSN2 (Fig. 2A), comprising senescent tissue only. A nitrogen-starved root sample was included to determine the nodule specificity of transcript profiles (R; Fig. 2A).
Figure 2.
Transcriptome analysis of nodule senescence in M. truncatula. A, cDNA-AFLP samples. R, Nitrogen-starved roots; N1, nitrogen-fixing, round, indeterminate nodule without senescence zone; N2, nitrogen-fixing, more elongated, indeterminate nodule without senescence zone; ASN1 and PSN1, apical and proximal part of nodule with small, young green senescence zone, respectively; ASN2 and PSN2, apical and proximal part of nodule with large green senescence zone, respectively. Lines indicate transversal sectioning of senescent nodule samples. Bar = 1 mm. B to E, Results from hierarchical average linkage clustering of gene tags showing a nodule senescence-associated differential expression pattern. Digits at the top of each cluster correspond to the different nodule development and root samples as indicated in A. Yellow and blue correspond to transcriptional repression and activation relative to the average expression level over the different samples, respectively. Gray represents missing data.
cDNA-AFLP analysis across the seven samples revealed a total of 14,860 gene tags. Raw output data were corrected and normalized and gene tags with a coefficient of variance >0.5 were retained as a measure of differential expression. A collection of 508 cDNA-AFLP tags was obtained whose transcript profiles were significantly modulated by nodule senescence (Supplemental Table I).
Hierarchical average linkage clustering analysis (Eisen et al., 1998) of the differentially expressed gene tags revealed three clusters that were up-regulated, designated 1, 2, and 3 (Fig. 2, C–E), and one that was down-regulated, designated cluster 4, with reduced transcript levels in the senescence-associated samples PSN1 and PSN2 (Fig. 2B). In cluster 1, tags had an increased transcript level in samples ASN1, PSN1, and ASN2 (Fig. 2C); in cluster 2, expression was enhanced in samples PSN1, ASN2, and PSN2 (Fig. 2D); and in cluster 3, tags were grouped with an expression primarily up-regulated in sample PSN2 (Fig. 2E).
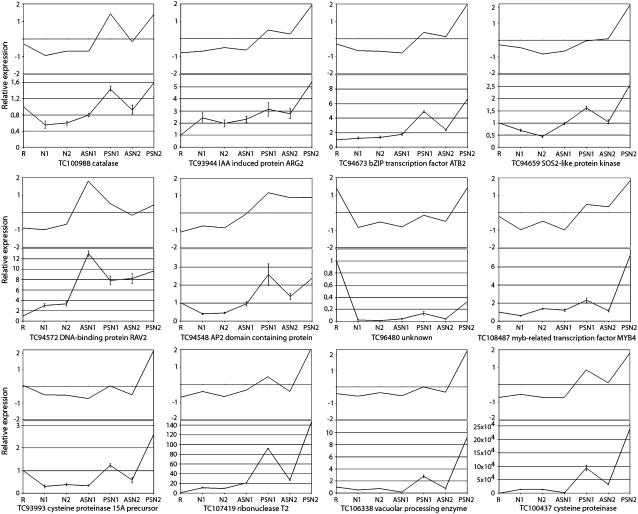
To verify expression profiles, quantitative reverse transcription (qRT)-PCR was performed for a subset of 12 genes. For all of them, qRT-PCR analysis revealed transcript profiles consistent with the results obtained with cDNA-AFLP analysis (Fig. 3). qRT-PCR analysis with cDNA samples from a biological repeat experiment confirmed these results (data not shown).
Figure 3.
qRT-PCR confirmation of cDNA-AFLP transcript profiles. For each of the tags, top and bottom images show cDNA-AFLP and qRT-PCR analysis, respectively, of transcript levels in samples R, N1, N2, ASN1, PSN1, ASN2, and PSN2 (for explanation, see Fig. 2A). For qRT-PCR analysis, values are presented relative to the nitrogen-starved root sample, R. The mean and sd of three repeat experiments are given.
Gene Tag Sequence Analysis
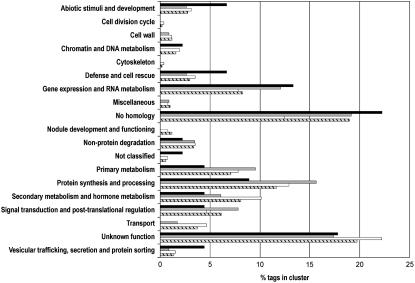
BLASTN and BLASTX algorithms (Altschul et al., 1997) were used to compare sequences of gene tag DNAs and translated proteins with expressed sequence tags (ESTs) of the M. truncatula Gene Index (MTGI; version 7.0) from The Institute for Genome Research (TIGR; www.tigr.org) and a publicly available nonredundant protein database (nr at http://www.ncbi.nlm.nih.gov/BLAST), respectively, with an E-value of 10−3 as the similarity threshold. Of the gene tags, 65% and 16% were significantly similar to genes with a known and an unknown function, respectively, whereas 19% were not homologous to any known sequence in the databases. Most of the latter sequences were found in cluster 3 as well as in cluster 4 (Supplemental Table I). Of the total collection of tags, only 56% were represented in the MTGI by ESTs. Strikingly, for the tags linked to an EST cluster (tentative consensus sequences), only 23% of them had a corresponding EST in the senescent nodule library (Fedorova et al., 2002), indicating that this library is only a limited representation of the nodule senescence transcriptome. The gene tags were grouped into 19 functional classes derived from the Medicago EST Navigation System (Journet et al., 2002; Fig. 4).
Figure 4.
Functional classification of nodule senescence-associated gene tags. Gene tags belonging to the individual up-regulated clusters (cluster 1, black; cluster 2, gray; cluster 3, white) and all (dashed) were manually assigned to 19 functional classes derived from Journet et al. (2002).
DISCUSSION
A comprehensive transcriptome analysis of the nodule senescence process was performed in the legume M. truncatula. The cDNA-AFLP technology used is a very sensitive method to visualize quantitative expression profiles of genes of low abundance and allows discrimination between highly homologous genes. The analysis resulted in a collection of 508 gene tags that were significantly differentially expressed during nodule senescence. Most gene tags were transcriptionally up-regulated during one or more senescence-associated stages (clusters 1, 2, and 3) and are, therefore, functionally linked to these different stages of nodule senescence. On the other hand, cluster 4 harbors gene tags characterized by reduced transcript levels during these stages. For these tags, it is more difficult to unambiguously assign an active function during nodule senescence because this down-regulation in transcription could also be the result of a halt in the nitrogen fixation metabolism of the nodules. A functional classification demonstrated that an important portion corresponded to potentially regulatory and signal transduction genes. For approximately 19% of the gene tags, no significant homology could be found. These gene tags could be part of divergent untranslated regions, making it difficult to find homologous sequences in other species. A full interpretation of this gene tag collection will have to await completion of the M. truncatula genome annotation, which is currently estimated at 40% (International Medicago Genome Annotation Group).
Nodule Senescence Consists of Consecutive Developmental Stages
Microscopic analysis revealed a young and an old senescence zone in M. truncatula nodules. In the young senescence zone, bacteroids are degraded and only a few plant cells undergo death. In the old zone, after symbiosome resorption, plasma membrane loosening is visible, indicative of plant cell death. This observation demonstrates that nodule senescence is mainly characterized by a slow cell death process, presumably providing time for nutrient mobilization.
The sampling setup, followed by cDNA-AFLP and cluster analyses, allowed us to dissect the nodule senescence process in three consecutive developmental stages. Cluster 1 reflects an early stage of senescence, grouping gene tags that, besides being induced in the senescence zone-containing samples, also show an up-regulation in sample ASN1, which comprises the meristem, infection, and fixation zones of nodules with a young senescence zone. Thus, these healthy tissues encode functions that are not active in the same zones in young nodules and, consequently, are connected to the initiation of nodule senescence. Regulatory genes in this cluster could be part of signal transduction pathways triggered during the very initial stages and therefore could be important targets for future functional research. In this early cluster, two genes code for transcription factors belonging to the APETALA/ETHYLENE RESPONSE FACTOR (AP2/ERF) family known to be involved in developmental as well as disease resistance and stress responses (Kirch et al., 2003; Gutterson and Reuber, 2004; Yi et al., 2004). Interestingly, in this early cluster, a tag is present that is homologous to genes encoding abscisic acid-insensitive/viviparous-related transcription factors and is characterized by an AP2/ERF and B3 DNA-binding domain, which are thus far unique for plants (Kagaya et al., 1999) and without obvious function. Other potential regulatory candidates code for protein kinases and a mitogen-activated protein kinase kinase kinase that is part of a conserved signaling cascade involved in linking several environmental and stress-associated stimuli to downstream cellular responses (Jonak et al., 2002; Teige et al., 2004). In addition, a DEAD-box RNA helicase gene was found whose corresponding product is linked to export of mRNA molecules during stress and plant developmental responses (Gong et al., 2005).
In cluster 2, genes are grouped with induced/enhanced transcript levels in samples PSN1, ASN2, and PSN2, whereas in cluster 3, tags are found that are transcriptionally up-regulated, primarily in sample PSN2. Taking the microscopical data into account, clusters 2 and 3 correspond, respectively, to stage I of senescence, when bacteroids are degraded, and to the later stage II, when host cells are targeted for degradation. Both clusters contain potential regulatory and signal transduction functions. In addition to tight transcriptional control, certain nodule senescence responses may also be translationally controlled because of the up-regulation of gene tags coding for ribosomal proteins, elongation factors, and other proteins involved in regulation of translation (Supplemental Table I).
Macromolecule Degradation and Nutrient Recycling: The Sink-to-Source Transition
Functional classification of tags from clusters 2 and 3 (Fig. 4) reflects the different metabolic events that take place during these consecutive stages. Predominantly present in clusters 2 and 3 is a group of genes encoding representatives of five protease families (Rawlings et al., 2004). Interestingly, a gene coding for a vacuolar processing enzyme (VPE) precursor was also induced. VPEs or legumains are Cys proteinases that process vacuolar-based proteins during seed germination and leaf senescence (Müntz and Shutov, 2002). Recently, certain types of VPEs, known as caspase-like proteases, have been implied in programmed cell death in plants (Hatsugai et al., 2004; Rojo et al., 2004). Similarly, VPEs could play a regulatory role during nodule senescence by activating other proteinases through posttranslational maturation. Also well represented in clusters 2 and 3 are genes coding for components of the Skp1-cullin-F-box-specific ubiquitin/26S proteasome pathway that exerts an important regulatory function in development through specific degradation of regulatory proteins (Smalle and Vierstra, 2004). Five gene tags are homologous to F-box proteins, key players in determining the specificity of the Skp1-cullin-F-box-mediated ubiquitinylation reaction (Gagne et al., 2002), which might hint at active participation of this pathway during nodule senescence.
Besides protein degradation machinery, pathways for degradation of nucleic acids, membrane-derived lipids, and sugars are transcriptionally induced (Supplemental Table I). Enhanced expression of a plethora of catabolic genes points to the dismantling of symbiosomes during the young senescent stage and host cell components in the later stage. On the other hand, the induction of genes coding for proteins involved in the transport of a wide variety of molecules, such as ATP-binding cassette proteins and specific transporters of phosphate, amino acids, and metal ions, indicates that degradation of macromolecules and mobilization functions are strongly linked and that nodule senescence involves nutrient mining and recycling. These catabolic events coupled to transportation processes mark the transition of the nodule from strong carbon sink to general nutrient source.
Hormonal Regulation of Nodule Senescence
Functional classification of the gene tags suggests that two hormones, ethylene and jasmonic acid, play a positive role, and GA3 plays a negative role during nodule senescence. The positive role for ethylene is illustrated by the up-regulation of ERF transcription factors and ethylene biosynthetic genes, such as S-adenosyl-Met (SAM) synthetase and 1-aminocyclopropane-1-carboxylate oxidase. Induction of a SAM synthetase gene may also correspond to activation of polyamine (putrescine, spermidine, and spermine) biosynthetic pathways for which SAM is a precursor (Imai et al., 2004). This hypothesis is supported by the induction of a gene encoding a spermidine synthetase enzyme.
Involvement of jasmonic acid, an important signaling factor in plant responses toward several environmental stresses (Creelman and Mullet, 1997) and known to promote leaf senescence (He et al., 2002), is suggested by the induction of lipoxygenase genes during different stages of nodule senescence. Oxygenation of polyunsaturated fatty acids by lipoxygenase enzymes is an essential first step in the biosynthesis of oxylipins, such as jasmonates (Feussner and Wasternack, 2002).
GA might suppress nodule senescence as illustrated by the strong induction of a gene coding for the GA 2-oxidase that converts active GAs to inactive forms (Thomas et al., 1999). No direct abscisic acid-responsive genes were present in our dataset, although abscisic acid has been put forth as an important signal in several types of senescence (Hunter et al., 2004; Buchanan-Wollaston et al., 2005; Puppo et al., 2005).
Nodule Versus Leaf Senescence
Leaf senescence is a highly organized and regulated catabolic process during which leaf mesophyll cells undergo structural redifferentiation followed by recruitment of cell constituents to other plant organs (Buchanan-Wollaston et al., 2003). At first sight, obvious mechanistic parallels exist between nodule and leaf senescence: The respective organs change from sink to general source status with massive degradation of cell material coupled to mobilization to other plant parts.
Interestingly, the primary targets for degradation and sources of nutrients to recycle, which are the abundantly present chloroplast organelles in leaves and symbiosomes in nodules, actually have much in common. From an evolutionary point of view, chloroplasts are derived from the uptake of a prokaryotic ancestor that was able to fix carbon (McFadden and van Dooren, 2004) and that gradually evolved into a plant organelle. Symbiosomes can be considered as horizontally acquired nitrogen-fixing organelles and, therefore, might represent an intermediate phase in an analogous bacteria-to-plant organelle evolution.
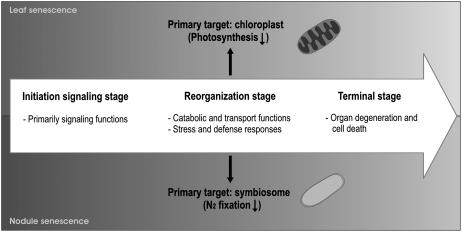
These parallels are also evidenced at the molecular level. A comparison of represented gene families between the different transcriptome datasets of leaf senescence in Arabidopsis (Arabidopsis thaliana; Gepstein et al., 2003; Guo et al., 2004; Zimmermann et al., 2004; Buchanan-Wollaston et al., 2005) and the nodule senescence process reveals a relatively high degree of overlap (Supplemental Table I). Especially in the up-regulated clusters 1, 2, and 3, which are functionally linked to the nodule senescence process, gene families are found that are involved in catabolism, transport, stress, and defense responses, which operate during leaf senescence as well. In contrast, gene family overlap between the down-regulated cluster 4 and the leaf senescence transcriptomes contains more general gene families involved in metabolism and signaling, implying fewer functional similarities. Moreover, the hormones ethylene and jasmonic acid seem to play a role in both senescence programs because genes involved in these two processes are present in both transcriptomes. These observations reflect a similar sequence of events during both senescence programs whereby an initiation signaling stage precedes the reorganization stage during which catabolic nutrients are recycled, finally leading to organ degeneration (Fig. 5).
Figure 5.
Schematic representation of common stages between nodule and leaf senescence.
The most abundantly expressed genes in the nodule senescence dataset are four Cys protease genes that are highly homologous to one of the prominent markers of leaf senescence, sag12 (Lohman et al., 1994), and that belong to a family of Cys proteinases that are synthesized in legume and actinorhizal nodules, especially during senescence (Goetting-Minesky and Mullin, 1994; Naito et al., 2000; Vincent and Brewin, 2000; Asp et al., 2004), as well as during arbuscular mycorrhizal association (Liu et al., 2003). For two of these genes, an open reading frame could be predicted in silico and a putative vacuolar targeting signal (Chrispeels, 1991) was found that might direct these proteins to the bacteroid-containing symbiosome compartment (Vincent and Brewin, 2000), where they could assist in bacteroid protein degradation. Indeed, in situ hybridization analysis indicated that members of this family are specifically transcribed in cells where bacteroids are degraded (W. Van de Velde and S. Goormachtig, unpublished data). As an analogy, SAG12-green fluorescent protein fusions have recently been observed in senescence-associated vacuoles that are only present in chloroplast-containing leaf cells that undergo senescence (Otegui et al., 2005). Whether these SAG12 Cys protease-containing vesicles are involved in degradation of chloroplast proteins is currently unknown.
To conclude, molecular insight is provided into developmental senescence of nodules whose onset marks the switch from a sink-to-source status of nodule tissue during which cell content is mobilized. Different expression clusters reflect the developmental stages of nodule senescence. An early cluster contains genes that are up-regulated not only in the senescence zone, but also in other zones without visible signs of senescence. The regulatory and signal transduction genes found in this cluster correspond to particularly interesting candidates to modulate nodule senescence. Isolation of the promoters that are specifically activated during one or more stages of the nodule senescence program will provide valuable tools to engineer this agronomically important process in legumes.
MATERIALS AND METHODS
Plant Material, Bacterial Strains, and Growth Conditions
Growth of Medicago truncatula line Jemalong J5, Sinorhizobium meliloti 1021, and plant inoculation were as described (Mergaert et al., 2003). For each nodulation stage, nodules from 30 plants were harvested. Senescent nodules at 35 and 61 dpi were cut transversally under a stereomicroscope (MZFLIII; Leica). Root material was obtained from noninoculated, nitrogen-starved plants grown under the same conditions as nodulated plants.
cDNA-AFLP Transcript-Profiling Analysis
Total RNA was isolated with RNeasy (Qiagen). cDNA synthesis from 1 μg of total RNA and cDNA-AFLP transcript profiling were performed as described (Breyne et al., 2003). In summary, 500 ng of double-stranded cDNA were used for AFLP analysis. For preamplification, an MseI primer without selective nucleotides was combined with a BstYI primer containing either a T or a C at the 3′ end. This preamplification step reduced the complexity of the template mixture by 2-fold. Preamplification PCR conditions were as follows: 25 cycles at 94°C for 30 s, at 56°C for 60 s, and at 72°C for 60 s. The final selective amplification step was done with the MseI and BstYI primers, each with two selective nucleotides (+2/+2) under the following PCR conditions: one cycle at 94°C for 10 min, 13 cycles at 94°C for 30 s, at 65°C for 60 s, and at 72°C for 60 s with a decrease of 0.7°C per cycle, 23 cycles at 94°C for 30 s, at 56°C for 60 s, and at 72°C for 60 s. The use of four selective nucleotides (+2/+2) produced fewer gene tags, avoiding contamination during cutting of fragments and enabled us to visualize more low-abundance transcripts than with a (+2/+1) configuration. The size of the generated AFLP gene tags was roughly between 50 and 650 bp long. Of all the 512 performed primer combinations, the profiles of 478 primer combinations were used for the subsequent analysis.
Quantitative Analysis of cDNA-AFLP Expression Profiles and Cluster Analysis
cDNA-AFLP expression profiles on scanned gel images were quantitatively analyzed with AFLP-Quantar-Pro image analysis software (Keygene N.V.) that scored all visible AFLP fragments. Individual band intensities were measured in each lane. The raw data were corrected for differences in total lane intensities. To that end, the intensity values were summed per lane for each primer combination and each of the sums was divided by the maximal value to yield the correction factors. Subsequently, all raw data were divided by these correction factors. Next, each individual gene expression profile was variance normalized (Tavazoie et al., 1999). For each transcript, the mean expression value across the time course was subtracted from each individual data point, after which the value obtained was divided by the sd. A coefficient of variation was calculated by dividing the sd by the mean. A coefficient of variance threshold of 0.5 was chosen for significantly differentially expressed genes. Hierarchical clustering was performed with the multiple experiment viewer program (TIGR). All the selected tags were isolated, reamplified, and sequenced for further analysis.
Microscopic Analysis
Light and electron microscopy were performed as described previously (D'Haeze et al., 1998). For light microscopy, nodules were fixed in 2.5% gluteraldehyde in 0.05 m cacodylate buffer, washed, dehydrated, embedded in Technovit 7100 (Kulzer Histo-Technik), according to the manufacturer's instructions, and stained with toluidine blue (Van de Velde et al., 2003). Sections of 2 μm were cut on a microtome (Reichert-Jung) and mounted on Vectabond-coated slides (Sigma-Aldrich). For electron microscopy analysis, nodules were cut longitudinally and fixed in 2.5% formaldehyde/3% gluteraldehyde in 0.1 m cacodylate buffer, washed, dehydrated, and embedded in LR White hard grade (London Resin). Serial sections were made with an ultracut microtome (Reichert-Jung) equipped with a diamond knife.
For light microscopy, photographs were taken with a Diaplan microscope fitted with bright-field optics (Leitz). For electron microscopy, sections collected on collodion-coated copper grids were stained with 2% uranyl acetate for 12 min and examined by TEM (Elmiskop 101; Siemens).
qRT-PCR Analysis
qRT-PCR was performed as described (Vlieghe et al., 2005). Total RNA was extracted in the same manner as for the cDNA-AFLP analysis; first-strand cDNA was synthesized from 1 μg of total RNA with the SuperScript II reverse transcriptase kit (Invitrogen) and oligo(dT)18, according to the manufacturer's instructions. Quantitative PCR was performed with the SYBR Green kit (Eurogentec) with 100 nm of primers and 0.125 μL of RT reaction product in a final volume of 25 μL. Reactions were run and analyzed on the iCycler iQ (Bio-Rad), according to the manufacturer's instructions. Relative expression was calculated as the ratio of normalized gene expression against the constitutively expressed 40S ribosomal protein S8 (tentative consensus EST cluster TC100533 of the MTGI from TIGR; Fedorova et al., 2002) by using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Reactions were performed in triplicate and averaged. Primers used (Supplemental Table II) had a calculated melting temperature of 59°C ± 2°C and were unique in the MTGI (TIGR) and the Medicago EST Navigation System database (Journet et al., 2002).
Functional Comparison of Nodule Senescence cDNA-AFLP Dataset with Arabidopsis Leaf Transcriptomes
The datasets of nodule senescence cDNA-AFLP and different Arabidopsis leaf senescence transcriptomes, consisting either of ESTs (Guo et al., 2004) or of transcriptionally up-regulated genes (Gepstein et al., 2003; Zimmermann et al., 2004; Buchanan-Wollaston et al., 2005), were compared at the level of similar gene families that were represented during both processes and not at the sequence level (therefore, no orthologs or paralogs were defined). To this end, only nodule senescence cDNA-AFLP gene tags with known function were considered for this comparison (65%). When a similar gene family annotation was found for the cDNA-AFLP gene tag in one or more leaf senescence transcriptomic datasets, it was considered an overlap.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ455074 to DQ455556 and DQ458785.
Supplementary Material
Acknowledgments
We thank Wilson Ardiles Diaz and Jan Gielen for sequencing, Ivo Zaenen for bioinformatic analysis, and Martine De Cock and Karel Spruyt for help in preparing the manuscript and figures, respectively.
This work was supported by the Ministerie van de Vlaamse Gemeenschap (grant no. CLO/IWT/020714), by the European Molecular Biology Organization (short-term fellowship to W.V.d.V.), and by the “Bijzonder Onderzoeksfonds” (grant no. 01W00805) of the Ghent University (predoctoral fellowship to J.C.P.G.).
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) are: Sofie Goormachtig (sofie.goormachtig@psb.ugent.be) and Marcelle Holsters (marcelle.holsters@psb.ugent.be).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.078691.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva IN, Kozharinova GM, Izmailov SF (1998) Senescence of legume nodules. Russ J Plant Physiol 45: 101–112 [Google Scholar]
- Asp T, Bowra S, Borg S, Holm PB (2004) Cloning and characterisation of three groups of cysteine protease genes expressed in the senescing zone of white clover (Trifolium repens) nodules. Plant Sci 167: 825–837 [Google Scholar]
- Banba M, Siddique A-BM, Kouchi H, Izui K, Hata S (2001) Lotus japonicus forms early senescent root nodules with Rhizobium etli. Mol Plant-Microbe Interact 14: 173–180 [DOI] [PubMed] [Google Scholar]
- Bethlenfalvay GJ, Phillips DA (1977) Ontogenetic interactions between photosynthesis and symbiotic nitrogen fixation in legumes. Plant Physiol 60: 419–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyne P, Dreesen R, Cannoot B, Rombaut D, Vandepoele K, Rombauts S, Vanderhaeghen R, Inzé D, Zabeau M (2003) Quantitative cDNA-AFLP analysis for genome-wide expression studies. Mol Genet Genomics 269: 173–179 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D (2003) The molecular analysis of leaf senescence—a genomics approach. Plant Biotechnol J 1: 3–22 [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin J-F, Wu S-H, Swidzinski J, Ishizaki K, et al (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42: 567–585 [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ (1991) Sorting of proteins in the secretory system. Annu Rev Plant Physiol Plant Mol Biol 42: 21–53 [Google Scholar]
- Creelman RA, Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48: 355–381 [DOI] [PubMed] [Google Scholar]
- D'Haeze W, Gao M, De Rycke R, Van Montagu M, Engler G, Holsters M (1998) Roles for azorhizobial Nod factors and surface polysaccharides in intercellular invasion and nodule penetration, respectively. Mol Plant-Microbe Interact 11: 999–1008 [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa-Victoria D, Vance CP, Graham PH (2000) Host variation in traits associated with crown nodule senescence in soybean. Crop Sci 40: 103–109 [Google Scholar]
- Fedorova M, van de Mortel J, Matsumoto PA, Cho J, Town CD, VandenBosch KA, Gantt JS, Vance CP (2002) Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol 130: 519–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I, Wasternack C (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53: 275–297 [DOI] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu S-H, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99: 11519–11524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MF, Yariv I, Dor C, Bassani M (2003) Large-scale identification of leaf senescence-associated genes. Plant J 36: 629–642 [DOI] [PubMed] [Google Scholar]
- Goetting-Minesky MP, Mullin BC (1994) Differential gene expression in an actinorhizal symbiosis: evidence for a nodule-specific cysteine proteinase. Proc Natl Acad Sci USA 91: 9891–9895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogorcena Y, Gordon AJ, Escuredo PR, Minchin FR, Witty JF, Moran JF, Becana M (1997) N2 fixation, carbon metabolism, and oxidative damage in nodules of dark-stressed common bean plants. Plant Physiol 113: 1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Dong C-H, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu J-K (2005) A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González EM, Aparicio-Tejo PM, Gordon AJ, Minchin FR, Royuela M, Arrese-Igor C (1998) Water-deficit effects on carbon and nitrogen metabolism of pea nodules. J Exp Bot 49: 1705–1714 [Google Scholar]
- Guo Y, Cai Z, Gan S (2004) Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ 27: 521–549 [Google Scholar]
- Gutterson N, Reuber TL (2004) Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol 7: 465–471 [DOI] [PubMed] [Google Scholar]
- Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I (2004) A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305: 855–858 [DOI] [PubMed] [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DA, Ferrante A, Vernieri P, Reid MS (2004) Role of abscisic acid in perianth senescence of daffodil (Narcissus pseudonarcissus ‘Dutch Master’). Physiol Plant 121: 313–321 [DOI] [PubMed] [Google Scholar]
- Imai A, Matsuyama T, Hanzawa Y, Akiyama T, Tamaoki M, Saji H, Shirano Y, Kato T, Hayashi H, Shibata D, et al (2004) Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiol 135: 1565–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Ökrész L, Bögre L, Hirt HK (2002) Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol 5: 415–424 [DOI] [PubMed] [Google Scholar]
- Journet E-P, van Tuinen D, Gouzy J, Crespeau H, Carreau V, Farmer M-J, Niebel A, Schiex T, Jaillon O, Chatagnier O, et al (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30: 5579–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Ohmiya K, Hattori T (1999) RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res 27: 470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch T, Simon R, Grünewald M, Werr W (2003) The DORNRÖSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem cell fate and lateral organ development. Plant Cell 15: 694–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawn RJ, Brun WA (1974) Symbiotic nitrogen fixation in soybeans. I. Effect of photosynthetic source-sink manipulations. Crop Sci 14: 11–16 [Google Scholar]
- Liu J, Blaylock LA, Endre G, Cho J, Town CD, VandenBosch KA, Harrison MJ (2003) Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell 15: 2106–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lohman KN, Gan S, John MC, Amasino RM (1994) Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol Plant 92: 322–328 [Google Scholar]
- Matamoros MA, Baird LM, Escuredo PR, Dalton DA, Minchin FR, Iturbe-Ormaetxe I, Rubio MC, Moran JF, Gordon AJ, Becana M (1999) Stress-induced legume root nodule senescence: physiological, biochemical, and structural alterations. Plant Physiol 121: 97–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI, van Dooren GG (2004) Evolution: red algal genome affirms a common origin of all plastids. Curr Biol 14: R514–R516 [DOI] [PubMed] [Google Scholar]
- Merbach W, Schilling G (1980) Aufnahme, Transport und Verwertung von 15N-markiertem Mineralstickstoff durch Weißlupinen im Gefäßversuch. Arch Acker-Pflanzenbau Bodenkd 24: 39–46 [Google Scholar]
- Mergaert P, Nikovics K, Kelemen Z, Maunoury N, Vaubert D, Kondorosi A, Kondorosi E (2003) A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol 132: 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müntz K, Shutov AD (2002) Legumains, and their functions in plants. Trends Plant Sci 7: 340–344 [DOI] [PubMed] [Google Scholar]
- Naito Y, Fujie M, Usami S, Murooka Y, Yamada T (2000) The involvement of a cysteine proteinase in the nodule development of Chinese milk vetch infected with Mesorhizobium huakuii subsp. rengei. Plant Physiol 124: 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd GED, Downie JA (2004) Calcium, kinases and nodulation signalling in legumes. Nat Rev Mol Cell Biol 5: 566–576 [DOI] [PubMed] [Google Scholar]
- Otegui MS, Noh Y-S, Martínez DE, Vila Petroff MG, Staehelin LA, Amasino RM, Guiamet JJ (2005) Senescence-associated vacuoles with intense proteolytic activity develop in leaves of Arabidopsis and soybean. Plant J 41: 831–844 [DOI] [PubMed] [Google Scholar]
- Pladys D, Vance PC (1993) Proteolysis during development and senescence of effective and plant gene-controlled ineffective alfalfa nodules. Plant Physiol 103: 379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puppo A, Groten K, Bastian F, Carzaniga R, Soussi M, Lucas MM, de Felipe MR, Harrison J, Vanacker H, Foyer CH (2005) Legume nodule senescence: roles for redox and hormone signalling in the orchestration of the natural aging process. New Phytol 165: 683–701 [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Tolle DP, Barrett AJ (2004) MEROPS: the peptidase database. Nucleic Acids Res 32: D160–D164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Martín R, Carter C, Zouhar J, Pan S, Plotnikova J, Jin H, Paneque M, Sánchez-Serrrano JJ, Baker B, et al (2004) VPEγ exhibits a caspase-like activity that contributes to defense against pathogens. Curr Biol 14: 1897–1906 [DOI] [PubMed] [Google Scholar]
- Roponen I (1970) The effect of darkness on the leghemoglobin content and amino acid levels in the root nodules of pea plants. Physiol Plant 23: 452–460 [Google Scholar]
- Schultze M, Kondorosi A (1998) Regulation of symbiotic root nodule development. Annu Rev Genet 32: 33–57 [DOI] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM (1999) Systematic determination of genetic network architecture. Nat Genet 22: 281–285 [DOI] [PubMed] [Google Scholar]
- Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15: 141–152 [DOI] [PubMed] [Google Scholar]
- Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96: 4698–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers ACJ, Soupène E, Auriac M-C, de Billy F, Vasse J, Boistard P, Truchet G (2000) Saprophytic intracellular rhizobia in alfalfa nodules. Mol Plant-Microbe Interact 13: 1204–1213 [DOI] [PubMed] [Google Scholar]
- Van de Velde W, Mergeay J, Holsters M, Goormachtig S (2003) Agrobacterium rhizogenes-mediated transformation of Sesbania rostrata. Plant Sci 165: 1281–1288 [Google Scholar]
- Vincent JL, Brewin NJ (2000) Immunolocalization of a cysteine protease in vacuoles, vesicles, and symbiosomes of pea nodule cells. Plant Physiol 123: 521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GTS, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inzé D, et al (2005) The DP-E2F-like DEL1 gene controls the endocycle in Arabidopsis thaliana. Curr Biol 15: 59–63 [DOI] [PubMed] [Google Scholar]
- Yi SY, Kim J-Y, Joung Y-H, Lee S, Kim W-T, Yu SH, Choi D (2004) The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in Arabidopsis. Plant Physiol 136: 2862–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.