Abstract
Several distinct pathways of RNA silencing operate in plants with roles including the suppression of virus accumulation, control of endogenous gene expression, and direction of DNA and chromatin modifications. Proteins of the Dicer-Like and Argonaute (AGO) families have key roles within these silencing pathways and have distinct biochemical properties. We are interested in the relationships between different silencing pathways and have used Nicotiana benthamiana as a model system. While not being an amenable plant for traditional genetics, N. benthamiana is extensively used for RNA-silencing studies. Using virus-induced gene silencing technology we demonstrate that both NbAGO1- and NbAGO4-like genes are required for full systemic silencing but not for silencing directed by an inverted repeat transgene. Phenotypic differences between the virus-induced gene silencing plants indicate that NbAGO1 and NbAGO4 like act at different stages of the silencing pathways. Suppression of NbAGO1 expression recapitulated the hypomorphic mutant phenotype of certain Arabidopsis (Arabidopsis thaliana) ago1 alleles, however, suppression of NbAgo4 like resulted in phenotypes differing in some respects from those reported for Arabidopsis ago4. We suggest that the small interfering RNA amplification step required for full systemic silencing is dependent upon a nuclear event requiring the activity of NbAGO4 like.
RNA silencing is a conserved mechanism among eukaryotes that involves the processing of double-stranded RNA (dsRNA) into small RNA (sRNA) species of 21 to 26 nucleotides (Hannon, 2002; Bartel, 2004). The source of dsRNA can be transcribed perfect or near perfect inverted repeats (IRs), viral RNA, or it can be produced by the action of host-encoded RNA-dependent RNA polymerases (RDRs) on certain RNA templates (Cogoni and Macino, 1999; Dalmay et al., 2000; Mourrain et al., 2000). dsRNA is cleaved by the action of the Dicer class of enzymes of which there are four in plants (Schauer et al., 2002). sRNAs are subclassed according to their origin, with small interfering RNAs (siRNAs) derived from perfect dsRNAs and microRNAs (miRNAs) derived from imperfect endogenously encoded hairpins. sRNAs act posttranscriptionally to direct the cleavage or translational repression of target RNAs, but can also act in the nucleus to drive DNA methylation and/or the formation of heterochromatin (Wasseneger, 2005). Their role as effectors of gene regulation requires the sRNAs to be complexed with effector proteins that may include RNAses that cleave target RNAs or DNA methyltransferases and histone modifiers that modify target DNA templates. Members of the Argonaute (AGO) class of proteins have been demonstrated to act as effectors; with AGO1 of Arabidopsis (Arabidopsis thaliana) and AGO2 of humans both acting as RNA slicers that selectively recruit sRNA species to direct the cleavage of target mRNA in RNA-induced silencing complexes (RISC; Liu, et al., 2004; Baumberger and Baulcombe, 2005; Rivas, et al., 2005; Qi et al., 2005). There are 10 predicted AGO proteins in Arabidopsis, of which only four have been characterized. Arabidopsis AGO1 (AtAGO1) is the founding member of the family and is required for normal plant development (Bohmert et al., 1998). ago1 mutants pleiotropically affect plant architecture and hypomorphic alleles are impaired in RDR-dependent but not IR-mediated RNA silencing (Morel et al., 2002). Null alleles show elevation of miRNA target mRNAs in agreement with AGO1 being the slicer component of RISC (Vaucheret et al., 2004). Since specific AGO and DICER proteins are proposed to have distinct roles within the various RNA-silencing pathways, AtAGO1 may be the slicer component of RISC for RDR-mediated RNA silencing and miRNA targeting but another AGO protein may be active in IR-mediated silencing. Mutants in the closely related PINHEAD/ZWILLE/AGO10 gene show overlapping characteristics with ago1 mutants although defects in RNA silencing or related pathways have not been uncovered (Lynn et al., 1999). AGO4 is required for production of endogenous siRNAs from certain loci and the implication is that it acts in the nucleus to effect epigenetic changes such as DNA methylation or histone modification (Zilberman et al., 2003, 2004). AtAGO7/ZIPPY is a heterochronic gene involved in vegetative phase change although a role for it in RNA silencing has not been established (Hunter et al., 2003).
In plants, RNA silencing can spread systemically, though for silencing to spread throughout the leaf laminar a requirement for the target to be either a transgene or a misregulated endogenous gene appears to be necessary (Palauqui et al., 1997; Voinnet and Baulcombe, 1997; Voinnet et al., 1998; Fagard and Vaucheret, 2000). Systemic silencing of transgenes is associated with transitive spreading of siRNAs and de novo DNA methylation and requires the action of RDR6 for long-range signaling (Vaistij et al., 2002; Himber et al., 2003). In this work we report the use of virus-induced gene silencing (VIGS) to investigate the role of AGO1- and AGO4-like genes in systemic and IR-mediated silencing in Nicotiana benthamiana. N. benthamiana is widely used both for studying the mechanism of RNA silencing and as an experimental organism in VIGS work (Jones et al., 2001; Ratcliff et al., 2001). Expression of NbAGO1 and NbAGO4 genes was suppressed using tobacco rattle virus (TRV) VIGS vectors carrying fragments of these genes. We show that NbAGO1 and NbAGO4 genes are required for full systemic RNA silencing and discuss these findings in the light of what is known of AGO genes in Arabidopsis and other organisms.
RESULTS
Identification of N. benthamiana AGO1- and AGO4-Like Genes
To better understand the process of systemic posttranscriptional gene silencing and the role of AGO genes in N. benthamiana, we searched for potential homologs of defined Arabidopsis AGO genes to suppress their expression using VIGS. Expressed sequence tags (ESTs) showing high levels of sequence identity to AtAGO1 and Arabidopsis AGO4 (AtAGO4) were identified from the The Institute for Genomic Research tomato (Lycopersicon esculentum) EST collection and PCR primers based on these ESTs were used to amplify sequences from N. benthamiana cDNA. Two AGO1-like fragments of 239 and 848 bp and two AGO4-like fragments of 238 and 391 bp were amplified. For both classes, the two sequences were nonoverlapping so it was not clear if they were derived from the same or different genes. RACE-PCR was used to extend the sequence in 5′ and 3′ directions and it became apparent that two AGO1-like and two AGO4-like N. benthamiana sequences had been identified. The N. benthamiana sequences were termed NbAGO1-1, NbAGO1-2, NbAGO4-1, and NbAGO4-2.
NbAGO1-1 and NbAGO1-2 share 85% and 88% identity at the nucleotide and predicted amino acid level, respectively. They also share approximately 80% amino acid identity with AtAGO1 and are more similar to AtAGO1 than to any other AtAGO sequence (Fig. 1). NbAGO4-1 and NbAGO4-2 share 82% and 83% identity with each other at the nucleotide and amino acid levels, respectively. Phylogenetic analysis places both sequences in the same clade as AtAGO4, AtAGO8, and AtAGO9 (S. Baldauf and L. Jones, unpublished data). Although these sequences are most similar to AtAGO4, they may actually represent alleles of a single AGO4-/AGO8-/AGO9-like gene in Nicotiana species as no further ESTs corresponding to genes of this clade have been identified to date. Thus, we conclude that N. benthamiana carries at least two AGO1- and two AGO4-like genes. This can be explained since N. benthamiana is amphidiploid in origin with parents related to Nicotiana sylvestris and Nicotiana obtusifolia and is likely therefore to carry alleles derived from each parental lineage (Chase et al., 2003).
Figure 1.
Alignment of the translated NbAGO1-1 and NbAGO1-2 cDNA sequences with Arabidopsis AGO1 (A) and of the translated NbAGO4-1 and NbAGO4-2 with Arabidopsis AtAGO4, AtAGO8, and AtAGO9 (B).
NbAGO1- and NbAGO4-Like Are Required for Systemic Silencing But Not IR Silencing in N. benthamiana
The amplified (239, 848, 238, and 391 bp) regions of NbAGO1-1, NbAGO1-2, NbAGO4-1, and NbAGO4-2 were each inserted into the TRV-silencing vector and used to initiate VIGS in either N. benthamiana line 16c undergoing systemic RNA silencing of a 35S:green fluorescent protein (GFP) transgene, or in a derivative of 16c termed 16c/GFi, which also carries an IR of the 5′ portion of GFP (GFi; Schwach et al., 2005). The aim was to determine if the NbAgo1- or NbAGO4-like genes were required for systemic silencing or IR-mediated silencing of the 35S-GFP transgene.
Systemic silencing of GFP was initiated in N. benthamiana line 16c by localized Agrobacterium-mediated introduction of a 35S:GFi transgene. Once full silencing was established (after approximately 2 weeks), VIGS was initiated using TRV:NbAGO1-1, TRV:NbAGO1-2, TRV:NbAGO4-1, or TRV:NbAGO4-2 constructs. Control VIGS experiments utilized a TRV empty vector (TRV:00). For VIGS of IR silencing, 16c/GFi N. benthamiana plants with approximately the same leaf number as the fully systemically silenced plants were used.
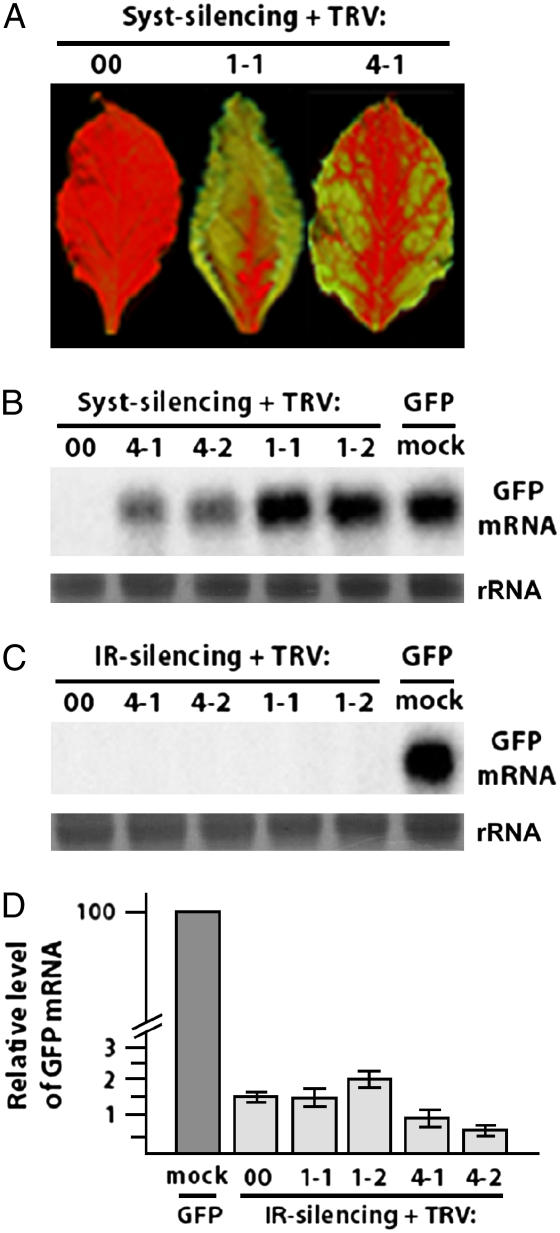
At approximately 21 d post initiation (DPI) of VIGS, reversal of GFP systemic silencing was visible for plants infected with either TRV:NbAGO1-1 or TRV:NbAGO1-2. Green fluorescent patches on leaves and stems were visible under UV light and the extent of reemergence of GFP increased during the remainder of the experimental period. (Fig. 2A). At approximately 28 DPI, reversal of systemic GFP silencing also became visible for plants infected with either TRV:NbAGO4-1 or TRV:NbAGO4-2. Plants undergoing systemic silencing and infected with TRV:00 remained fully silenced for GFP throughout the experiment (Fig. 2A). The appearance of green fluorescence correlated with an increase in GFP mRNA levels as demonstrated by northern-blot analysis of reversed and control tissue (Fig. 2B). Although reversal of systemic silencing was clear, no green fluorescence ever became visible in the 16c/GFi plants infected with TRV:NbAGO or control VIGS constructs. This was confirmed by northern-blot analysis and real-time quantitative PCR that showed no increase in GFP mRNA levels in the 16c/GFi plants undergoing VIGS (Fig. 2, C and D).
Figure 2.
TRV-mediated VIGS of NbAGO genes. A, Representative upper leaves of 16c plants undergoing systemic silencing of GFP and infected with TRV:NbAGO1-1, TRV:NbAGO4-1, or TRV:00. Leaves were photographed at 28 DPI under UV light. Reversal of GFP systemic silencing is visible by the reemergence of green fluorescent patches and was consistently more extensive in TRV:NbAGO1 infections compared with TRV:NbAGO4. B and C, GFP mRNA levels in line 16c plants undergoing systemic silencing (B) or IR silencing (C) and infected with TRV:00, TRV:NbAGO4-1, TRV:NbAGO4-2, TRV:NbAGO1-1, or TRV:NbAGO1-2. RNA from nonsilenced line 16c was included as a positive control (GFP mock). Five micrograms of total RNA extracted at 28 DPI were loaded per lane and analyzed with a GFP-specific probe. D, Quantitative PCR assessing GFP transcript levels in line 16c GF-IR plants undergoing VIGS of NbAGO1-1, NbAGO1-2, NbAGO4-1, or NbAGO4-2 or infected with TRV:00. Values are means of GFP levels based on triplicate values obtained from six individual plants and related to the level of GFP mRNA in 16c that was given a value of 100. Actin mRNA acted as the internal standard for quantification.
Given the high level of nucleotide sequence identity shared between NbAGO1-1 and NbAGO1-2 and also between NbAGO4-1 and NbAGO4-2, we predicted that they are undergoing cosilencing during VIGS. Targeting NbAGO1-1 should cosilence NbAGO1-2 and vice versa. Likewise TRV:NbAGO4-1 and TRV:NbAGO4-2 will each target both NbAGO4-1 and NbAGO4-2. In agreement with these predictions and the observation that the NbAGO1s and NbAGO4s do not share significant stretches of nucleotide identity with each other, northern blotting showed that NbAGO1-1 mRNA levels were suppressed following VIGS with TRV:NbAGO1-1 or TRV:NbAGO1-2 but not by TRV:NbAGO4-1 or TRV:NbAGO4-2 (Fig. 3). Similarly, NbAGO4-1 was suppressed by TRV:NbAGO4-1 or TRV:NbAGO4-2 but not by either of the TRV:NbAGO1s. Attempts were made to target specific NbAGOs by using TRV vectors carrying segments of the 3′ untranslated regions that lack significant sequence identity between the otherwise related sequences. However, this resulted in a lack of consistent reversal of systemic silencing, suggesting that when cosilencing was prevented, functional activity was retained by the partner (data not shown).
Figure 3.
Suppression of NbAGO mRNA levels by VIGS. Northern-blot analysis of NbAGO1-1 (top section) and NbAGO4-1 (middle section) mRNA levels. RNA was prepared from tissue showing reversal of systemic silencing for TRV:NbAGO1-1, TRV:NbAGO1-2, TRV:NbAGO4-1, and TRV:NbAGO4-2 or from systemic silencing plants either infected with TRV:00 or uninfected (mock). The bottom section shows ethidium bromide staining of rRNA.
Suppression of NbAGO1 But Not NbAGO4 Results in Developmental Abnormalities
Plants undergoing VIGS of NbAGO1-1 and NbAGO1-2 showed developmental abnormalities that were not observed for plants undergoing either VIGS of NbAGO4s or infected with the empty TRV vector. These abnormalities, including the malformation of leaves and flowers, occurred approximately 21 DPI, at the same time that reversal of systemic silencing was becoming apparent. Leaves typically showed narrowing of the lamina and downward curl of leaf margins, outgrowth and detachment of the midrib, and lack of symmetry (Fig. 4). Flowers showed a range of abnormalities including reduced numbers of petals, altered numbers of stamens and styles, stunted and swollen carpels, and ectopic carpel-like tissue. It should be noted that organs on each plant examined had a range of phenotypic severities from minor to gross and this did not always correlate with the extent of reversal of silencing. Some leaves and flowers showed an almost complete reversal of GFP silencing with only minor developmental abnormalities, whereas others showed a more severe developmental phenotype with the same level of GFP reversal.
Figure 4.
Developmental abnormalities associated with VIGS of NbAGO1. Representative leaves (A and B) and flowers (C–E) of plants infected with TRV:NbAGO1-1. Representative leaf (F) and flowers (H and J) of plants infected with TRV:00 and leaf (G) and flower (I) of plants infected with TRV:NbAGO4-1. No phenotypic differences were observed between plants infected with TRV:00 or TRV:NbAGO4 and identical abnormalities were observed in plants infected with TRV:NbAGO1-1 or TRV:NbAGO1-2.
Analysis of Indicators of RNA Silencing in Plants Undergoing VIGS of NbAGO1 and NbAGO4
Both NbAGO1- and NbAGO4-like genes are required for systemic silencing but not for IR-mediated silencing, with suppression of NbAGO1 also being associated with developmental abnormalities. To investigate this further we decided to assess markers of different RNA-silencing events in plants undergoing VIGS. We divided these markers into transgenic and endogenous subclasses: The transgenic markers were GFP siRNAs and associated DNA methylation of the GFP transgene; whereas the endogenous markers were miRNAs mi167 and mi171 and their target mRNAs, and siRNAs from the N. benthamiana TNT1 element.
Transgenic Indicators
The systemic and IR-mediated silencing systems in this study differ in key respects despite both being triggered by an IR and both involving the targeting of GFP mRNA in the background of line 16c. IR-mediated silencing is mediated by the 35S promoter driving expression of the GFi transgene ensuring that dsRNA will be present in most cell types. However, systemic silencing must rely upon a host-encoded, RDR (RDR6) and other factors for further production of dsRNA following the initial trigger by localized introduction of the GFi transgene. In both systems siRNAs will be produced, in systemic silencing these are derived from the GFP transgene itself whereas the siRNAs are expected to derive from both the IR and the GFP transgene in IR silencing. Total RNA was isolated from systemic- and IR-silencing plants undergoing VIGS of NbAGO1-1 or NbAGO4-1 or infected with TRV:00 and analyzed for the presence of GFP siRNAs by northern blotting. It was found that GFP siRNAs were more abundant in plants undergoing IR silencing of GFP compared to those undergoing systemic silencing (Fig. 5A, compare lanes 1 and 2 with lanes 7 and 8). VIGS of NbAGO1-1 or NbAGO4-1 had no effect upon GFP siRNA levels in IR silencing whereas siRNAs associated with systemic silencing were reduced or eliminated by VIGS of NbAGO1-1 or NbAGO4-1 (Fig. 5A). An analysis of the effect of VIGS on transitive siRNAs in IR silencing was attempted by probing specifically for the 3′ portion of GFP that is not present in the GFi transgene. However, the transitive GFP-derived siRNAs in 16c/GFi plants are present in very low levels and although no differences were observed between control and VIGS plants (data not shown), we cannot rule out an involvement of NbAGO1 or NbAGO4 in transitive spreading of siRNAs.
Figure 5.
siRNAs and DNA methylation during VIGS of NbAGOs. A, Assessment of GFP siRNA levels in 16c plants undergoing IR-mediated silencing or systemic silencing of a GFP transgene and infected with either TRV:00 (00), TRV:NbAGO1-1 (1-1), or TRV:NbAGO4-1 (4-1). The two lanes per treatment correspond to RNA samples taken from leaves (left lane) and flowers (right lane). The bottom section shows part of the ethidium bromide-stained gel prior to blotting. B, Structure of the GFP transgene showing the 5′ 453 bp portion (GF) and 3′ 359 (P) portion. The 5′ GF portion is also carried in the GF-IR construct present in the GF-IR plants. Sites for AluI (A) and Sau96 (S) are indicated, as are sizes of expected digestion products in kilobases. C, Southern-blot analysis of DNA samples from 16c nonsilenced plants (lane 1), 16 systemically silenced plants infected with either TRV:00, TRV:NbAGO1-1, or TRV:NbAGO4-1 (lanes 2, 3, and 4, respectively) or 16c GF-IR plants infected with either TRV:00, TRV:NbAGO1-1, or TRV:NbAGO4-1 (lanes 5, 6, and 7, respectively). DNA samples were digested with AluI or Sau96 as indicated and a probe specific for the 3′ 359 bp (P portion) of GFP was used allowing assessment of the methylation status of restriction sites throughout the GFP gene without hybridization to the GF-IR transgene.
DNA methylation of the GFP transgene was assessed by Southern blotting. DNA was digested with either AluI or Sau96 that cut in the 5′ and 3′ regions of the GFP transgene and are sensitive to nonsymmetrical cytosine methylation typically associated with RNA silencing. To only analyze methylation of the 35S:GFP transgene and not the GF-IR transgene, blots were probed with the 3′ portion of GFP that is not present in the IR (Fig. 5B). For both AluI and Sau96 digests, higher Mr bands corresponding to methylated products were observed in systemic silencing and IR silencing compared to DNA from nonsilenced 16c (Fig. 5C, bands corresponding to methylated products are marked by asterisks). VIGS of NbAGO1-1 resulted in loss of DNA methylation in systemic silencing but not in IR-silencing plants (Fig. 5C, compare lanes 2 and 3 with lanes 5 and 6). VIGS of NbAGO4-1 resulted in loss of DNA methylation associated with systemic silencing and to a lesser extent loss of DNA methylation associated with IR silencing. This was visualized by the enhanced accumulation of unmethylated fragments compared to the higher Mr methylated fragments (Fig. 5C, compare lanes 5 with 7).
Endogenous Indicators
AtAGO1 is reported to be the slicer component of RISC and since VIGS of NbAGO1s showed developmental abnormalities, we postulated that cleavage of the targets of miRNAs is being disrupted. Therefore, total RNA was assessed for accumulation of two miRNAs, miR167 and miR171, and their respective target mRNAs. No differences in accumulation of either miRNA were observed in plants undergoing VIGS of NbAGO1-1, NbAGO1-2, NbAGO4-1, and NbAGO4-2, infected with the empty TRV vector or uninfected controls (Fig. 6A; data not shown). N. benthamiana target sequences of miR167 and miR171 were generated from cDNA by PCR using primers based on known Arabidopsis targets (ARF8, At5g37020 for miR167 and SCL6-IV, At4g00150 for miR171). The sequences were termed NbARF8 and NbSCL6 and showed a high degree of identity to their counterpart in Arabidopsis (data not shown). The sequences had perfectly conserved the miRNA target site giving further confirmation that these were indeed the expected miRNA targets. Using fragments of NbARF8 and NbSCL6 as probes, cleavage products were visible by northern blotting and no significant alteration in this pattern was observed when comparing samples undergoing VIGS of NbAGOs or infected with TRV:00 (Fig. 6B). Thus, despite the observations of developmental abnormalities, no disruption of miRNA-directed mRNA cleavage was observed for the two pairings analyzed in this study. However, these abnormalities may be a result of disruptions to other miRNA-directed cleavage events not analyzed here.
Figure 6.
miRNA and siRNA accumulation during VIGS of NbAGOs. A, Accumulation of miR167 and miR171 in leaves (L) and flowers (F) of N. benthamiana plants infected with TRV:00, TRV:NbAGO1-1, TRV:NbAGO1-2, TRV:NbAGO4-1, or TRV:NbAGO4-2. Plants were systemically silenced for GFP prior to TRV infection and RNA was prepared from tissue showing reversal of GFP silencing apart from for TRV:00 infection where equivalent nonreversed tissue was used. The bottom section shows the ethidium bromide-stained gel prior to blotting. The miR167 probing was performed first followed by stripping and reprobing for miR171. B, Accumulation of full-length mRNAs and stable-cleaved portions of NbARF8 (the target of miR167) and NbSCL6 like (target of miR171) in leaves (L) and flowers (F) of N. benthamiana plants infected with TRV:00, TRV:NbAGO1-1, or TRV:NbAGO4-1 as described above for A. C, Accumulation of siRNAs corresponding to the long-terminal repeat of the TNT1 element. RNA was extracted from plants infected with TRV:00, TRV:NbAGO1-1, or TRV:NbAGO4-1. Three individual infected plants were analyzed per treatment. The bottom section shows the ethidium bromide-stained gel prior to blotting.
Endogenous siRNAs can be derived from repetitive sequences and transposable elements. We therefore assessed levels of endogenous siRNAs derived from the long-terminal repeats of the N. benthamiana TNT1 element (Hamilton et al., 2002). RNA samples were prepared from tissue undergoing VIGS of NbAGO1-1 or NbAGO4-1 and showing reversal of systemic silencing. siRNAs corresponding to the TNT1 element were as abundant in samples undergoing NbAGO VIGS as in control plants infected with TRV:00 (Fig. 6C). Thus, we conclude that VIGS of either NbAGO1s or NbAGO4s had not affected production of siRNAs from this endogenous element.
DISCUSSION
In this work we have used VIGS to demonstrate that both NbAGO1- and NbAGO4-like genes are required for systemic- but not IR-mediated silencing in N. benthamiana. The questions we will focus upon in this “Discussion” include why IR silencing is not affected by reduction in NbAGO1 expression and why NbAGO4 like is required for systemic silencing.
Arabidopsis AGO1 was first identified as a locus controlling leaf development, and mutant phenotypes vary depending upon the allele studied (Bohmert et al., 1998; Morel et al., 2002). Null mutants show severe developmental abnormalities and are sterile and it is believed that these phenotypes can be explained by disruption of miRNA-mediated gene regulation of key developmental genes including AGO1 itself (Vaucheret et al., 2004). AGO1 hypomorphic mutants are less severely affected in development, are completely defective in RDR-dependent RNA silencing, but are wild type for silencing conferred by an IR transgene (Béclin et al., 2002). Our observations showing that VIGS of NbAGO1s is able to reverse systemic- but not IR-mediated silencing are therefore in agreement with data from the Atago1 hypomorphic mutants since extensive systemic silencing of GFP requires the action of RDR6 (Himber et al., 2003; Schwach et al., 2005). VIGS results in a knockdown of gene expression rather than a complete knockout, thus explaining why we have recapitulated the ago1 hypomorphic phenotype.
There is significant genetic and biochemical evidence that AGO1 is the slicer component of RISC and that it uses the sequence of associated sRNAs to guide the cleavage of homologous RNAs. Therefore, the question remains as to why suppression of AGO1 activity by VIGS in N. benthamiana or by mutation in Arabidopsis apparently does not impact upon IR-mediated silencing. Both Qi et al. (2005) and Baumberger and Baulcombe (2005) showed that AtAGO1 interacts selectively with distinct classes of sRNAs. siRNAs derived from IR transgenes coprecipitated with AtAGO1, suggesting that suppression of AGO1 should have had an effect on IR-mediated silencing. However Qi et al. (2005) also found that although miR172 coimmunoprecipitated with AtAGO1, the target of miR172 was not cleaved in in vitro assays. This suggests that other factors may influence whether an AGO1-sRNA RISC is active for cleavage and that data from immunoprecipitations should be considered alongside data from cleavage assays. We can suggest several possible explanations for the apparent lack of effect on IR-mediated silencing in our experiments. AGO1 may distinguish between siRNAs derived from IRs and those generated via RDR activity and only actively target mRNAs for cleavage using the latter. However, care must be taken since in the genetic experiments performed in Arabidopsis, a weak hypomorphic allele of AGO1 (ago1-27) rather than a null mutant was used to demonstrate the involvement of AGO1 in RDR-dependent but not IR-mediated transgene silencing (Béclin et al., 2002). In assays of miRNA-target cleavage, ago1-27 mutants showed only a small increase in miRNA target levels whereas null alleles had significantly elevated levels, suggesting that ago1 null alleles may indeed be affected in IR silencing.
Lack of an effect on IR-mediated silencing may also be due to the nature of the silencing systems being analyzed. Extensive systemic silencing relies upon spread of a signal and the subsequent production of further siRNAs via the activity of an RDR (Himber et al., 2003). If RDRs are only active upon templates that have been cleaved by RISC then this cycle will be intrinsically sensitive to perturbations in AGO1 activity. In IR silencing there will be a transgenically encoded dsRNA present in each cell and since hypomorphic rather than null mutants have been tested for loss of silencing, the remaining RISC activity may be sufficient to maintain silencing.
In our experiments, elevated levels of two miRNA targets were not detected even though the NbAGO1 VIGS plants showed significant developmental abnormalities. It is possible that we did not assess sufficient miRNA targets to report conclusively on the effect of NbAGO1 VIGS. Alternatively, the nature of VIGS may be problematic in that the suppression system is dynamic, often transient, and not present in every cell. VIGS may indeed have suppressed miRNA targeting during early stages of organ formation leading to developmental abnormalities but as this is transient and mosaic in nature, recovery of target levels but not phenotypic rescue may occur in mature organs. Clearly there is differential sensitivity of RNA silencing and development during VIGS of NbAGO1.
The requirement for NbAGO1 in systemic silencing and not IR silencing agrees with previous studies in Arabidopsis showing that RDR6-dependent silencing systems require AGO1 (Béclin et al., 2002). Here we show that effective full systemic silencing of GFP transgenes in N. benthamiana also requires AGO4-like genes. Systemic silencing can be subdivided into transitivity-dependent and transitivity-independent classes with transitivity involving the amplification of the silencing signal (Himber et al., 2003). Short-range transitivity-independent spread of silencing can occur in the absence of either a target mRNA or siRNA amplification as demonstrated in rdr6 mutants. Long-distance spread of systemic silencing requires an amplification mechanism to produce further siRNAs and this has been postulated to involve the copying of RNA templates by RDR6 (Voinnet et al., 1998; Vaistij et al., 2002; Himber et al., 2003; Schwach et al., 2005). Extensive systemic silencing of GFP is associated with both transitivity-dependent siRNA amplification and DNA methylation of the corresponding transcribed region. Whether such DNA methylation is a consequence of the silencing process or is contributing to its amplification is a question of long-standing interest (Jones et al., 1998, 1999; Morel et al., 2000). The amplification mechanism could be purely RNA based, with RDRs only copying certain transcripts and the resulting siRNAs directing DNA methylation as a consequence. However, it is significant that a self-reinforcing silencing mechanism involving distinct plant-specific RNA polymerase IV complexes, Dicer-like 3, and RDR2 has been identified recently. This mechanism is proposed to maintain production of siRNAs associated with repetitive elements and heterochromatin (Xie et al., 2004; Herr et al., 2005; Kanno et al., 2005; Onodera et al., 2005). It is suggested that methylated DNA is transcribed by polymerase IV and that these transcripts act as templates for RDR2 and Dicer-like 3 to produce siRNAs that are used by the RNA-directed DNA methylation machinery to target homologous loci. It is therefore possible that the methylation occurring during the systemic silencing of GFP is indeed required for the generation of further siRNAs by a similar mechanism and that NbAGO4 like is involved in this process. AtAGO4 is known to have a nuclear localization and is required for production of endogenous siRNAs at highly repeated loci and for maintenance of methylation associated with certain endogenous loci or targets of IR transgenes (Zilberman et al., 2003, 2004; Xie et al., 2004). Despite only observing a minor effect on IR-directed GFP methylation during VIGS of NbAGO4 like (Fig. 5C), we propose that NbAGO4 like acts in the nucleus to promote methylation and/or chromatin modification during systemic silencing and that this is required for amplification of the systemic signal. This may occur by a similar mechanism to the amplification loop associated with production of siRNAs from certain endogenous elements and indeed earlier data has indicated a requirement for nuclear events in RNA silencing signal amplification. In transgenic pea (Pisum sativum) lines that differed only in the position of transgene integration, there was a difference in the ability of individual lines to undergo signal amplification of the transgene mRNA. This ability did not correlate with transgene expression levels but it did correlate with the ability to undergo RNA-directed DNA methylation (Jones et al., 1998). These observations could not distinguish cause from consequence but are difficult to explain with a purely RNA-based amplification mechanism. Additionally, Arabidopsis plants defective in the Met-1 methyltransferase also show loss of RDR6-dependent silencing, indicating that methylation may play a key role in the amplification step of RNA silencing (Morel et al., 2000).
MATERIALS AND METHODS
Plant Material and Induction of Systemic Silencing
Transgenic Nicotiana benthamiana line 16c and the derivative 16c GFi (carrying a 35S promoter-driven transgene consisting of an IR of the 5′ portion of GFP) have been described previously (Ruiz et al. 1998; Schwach et al., 2005). For initiation of systemic silencing, whole 16c seedlings were infiltrated with Agrobacterium tumefaciens strain GV3101 carrying the GFi binary vector construct. Seedlings were transferred to a glasshouse at 20°C for 2 weeks until full systemic silencing was established.
Generation of NbAGO Sequences and Recombinant Viruses
PCR primers used to amplify fragments of NbAGO genes were: 5′-CATACCCAGTGGCCTTGTCT-3 and 5′-ATTCGATTGCCAAACTCC-3′ (NbAGO1-1); 5′-GAGTCGTCTCACAGTGCTGT-3′ and 5′-AATATGCAGGGGGAACG-3′ (NbAGO1-2); 5′-GAGCATATTGTAATATTCAG-3′ and 5′-CTAACAAGGAACCTACTG-3′ (NbAGO4-1); and 5′-CAGGAGAGGATGCAAGTCCT-3′ and 5′-ATTCGATTTCAACCCTCAC-3′ (NbAGO4-2). Smart Race cDNA amplification (BD CLONTECH) was used to extend the transcript sequences in 5′ and 3′ directions according to manufacturer's instructions. GenBank accession numbers are DQ321488 (NbAGO1-1), DQ321489 (NbAGO1-2), DQ321490 (NbAGO4-1), and DQ321491 (NbAGO4-2). TRV:NbAGO constructs were generated by inserting the PCR-generated fragments into the SmaI site of pBinTra6 and TRV infections were generated as reported previously (Ratcliff et al., 2001).
RNA Extraction and Analysis
Total RNA was prepared using Tri-reagent (Sigma) according to manufacturer's instructions. For the detection of low Mr RNAs, electrophoretic separation and blotting to HybondNX membranes (Amersham) were performed as described previously (Hamilton et al., 2002). 32P-labeled oligonucleotides (5′-TAGATCATGCTGGCAGCTTCA-3′ for miR167 and 5′-GATATTGGCGCGGCTCAATCA-3′ for miR171) were used to detect miRNAs and 32P-labeled transcripts corresponding to GFP or the long terminal repeat of N. benthamiana TNT1 were used to detect GFP or TNT1 siRNAs, respectively. Detection of higher Mr RNAs was performed as described previously (Jones et al., 2001) with specific 32P-labeled DNA probes used to detect GFP-, NbARF8-, and NbSCL6-like mRNAs. N. benthamiana ARF8- and SCL6-like sequences were obtained by searching The Institute for Genomic Research EST data sets for homology to Arabidopsis (Arabidopsis thaliana) ARF8 and SCL6-II/III and IV. PCR amplification and DNA sequencing confirmed that homologs had been identified and that the miRNA target sequences were conserved.
DNA Extraction and Southern-Blot Analysis
DNA extraction used Qiagen midi plant DNA extraction kits (Qiagen). Ten micrograms of DNA was digested using 10 units of AluI or Sau96. Separation, blotting, and hybridization conditions were as described previously with the modification of using PerfectHyb-Plus (Sigma) as a hybridization buffer (Jones et al., 2001). A 32P-labeled probe corresponding to the 3′ 359 bp of GFP was used to assess the methylation status of restriction enzyme sites located within both the 5′ and 3′ regions of GFP.
Taqman Quantitative PCR
Total RNA was extracted using TRI-reagent (Sigma) from six individual plants per treatment. cDNA was synthesized using the Omniscript kit (Qiagen) and oligo dT primers with control reactions performed that lacked reverse transcriptase or template. Real-time quantitative PCR reactions were performed in triplicate using 2× Taqman Universal PCR master mix (PE Applied Biosystems), 5′-FAM fluorescent reporter, and 3′-Tamra quencher probes (Sigma Genosys). Actin was used as an internal standard for quantification. For GFP amplification the primer sequences were 5′-CCTGTCCTTTTACCAGACAACCA-3′ and 5′-CCCAGCAGCTGTTACAAACTCA-3′ and the probe sequence was 5′-ACCTGTCCACACAATCTGCCCTTTCG-3′. For actin amplification the primer sequences were 5′-CCATTGGTGCTGAGAGATTCC-3′ and 5′-GCAGCTTCCATTCCGATCA-3′ and the probe sequence was 5′-CTGCCCAGAAGTTCTGTTCCAACCATCA-3′.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ321488 (NbAGO1-1), DQ321489 (NbAGO1-2), DQ321490 (NbAGO4-1), and DQ321491 (NbAGO4-2).
Acknowledgments
We thank Professor David Baulcombe and Professor Dianna Bowles for their generosity, Colin Abbot and coworkers for excellent plant care, Katherine Fawcett for assistance in generating the NbAGO sequences, Professor Sandie Bauldauf for discussions, and Phil Roberts for photography. Work with recombinant viruses was performed under license from DEFRA (PHL 198/4356).
This work was supported by the Biological and Biotechnology Research Council (grant no. 87/G18736), the Gatsby Charitable Foundation, and the Garfield Western Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Louise Jones (alj2@york.ac.uk).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.076109.
References
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102: 11928–11933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béclin C, Boutet S, Waterhouse P, Vaucheret H (2002) A branched pathway for transgene-induced RNA silencing in plants. Curr Biol 12: 684–688 [DOI] [PubMed] [Google Scholar]
- Bohmert K, Camus I, Bellini C, Bouchez D, Caboche M, Benning C (1998) AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J 17: 170–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase MW, Knapp S, Cox AV, Clarkson JJ, Butsko IY, Joseph J, Savolainen V, Parokonny AS (2003) Molecular systematics, GISH, and the origin of hybrid taxa in Nicotiana (Solanaceae). Ann Bot (Lond) 92: 107–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G (1999) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399: 166–169 [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for post-transcriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553 [DOI] [PubMed] [Google Scholar]
- Fagard M, Vaucheret H (2000) Systemic silencing signal(s). Plant Mol Biol 43: 285–293 [DOI] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe DC (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21: 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ (2002) RNA interference. Nature 418: 244–251 [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120 [DOI] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O (2003) Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J 22: 4523–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Sun H, Poethig RS (2003) The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr Biol 13: 1734–1739 [DOI] [PubMed] [Google Scholar]
- Jones AL, Thomas CL, Maule AJ (1998) De novo methylation and cosuppression induced by a cytoplasmically replicating plant RNA virus. EMBO J 17: 6385–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC (1999) RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11: 2291–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L, Ratcliff F, Baulcombe DC (2001) RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr Biol 11: 747–757 [DOI] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJM (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37: 761–765 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Lynn K, Fernandez A, Aida M, Sedbrook J, Tasaka M, Masson P, Barton MK (1999) The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE gene. Development 126: 469–481 [DOI] [PubMed] [Google Scholar]
- Morel J-B, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel J-B, Mourrain P, Beclin C, Vaucheret H (2000) DNA methylation and chromatin structure affects transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr Biol 10: 1591–1594 [DOI] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, et al (2000) Arabidopsis SGS2 and SGS3 genes are required for post-transcriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pikaard CS (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622 [DOI] [PubMed] [Google Scholar]
- Palauqui JC, Elmayan T, Pollien JM, Vaucheret H (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J 16: 4738–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA-silencing pathways. Mol Cell 19: 421–428 [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernendez AM, Baulcombe DC (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25: 237–245 [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L (2005) Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12: 340–349 [DOI] [PubMed] [Google Scholar]
- Ruiz MT, Voinnet O, Baulcombe DC (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell 10: 937–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer SE, Jacobsen SE, Meinke DW, Ray A (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7: 487–491 [DOI] [PubMed] [Google Scholar]
- Schwach F, Vaistij FE, Jones L, Baulcombe DC (2005) An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic signal. Plant Physiol 138: 1842–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij FE, Jones L, Baulcombe DC (2002) Spreading of RNA targeting and DNA methylation regulation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14: 857–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crete P, Bartel DP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Baulcombe DC (1997) Systemic signaling in gene silencing. Nature 389: 553. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Vain P, Angell S, Baulcombe DC (1998) Systemic spread of sequence-specific transgene RNA degradation is initiated by localized introduction of ectopic promoterless DNA. Cell 95: 177–187 [DOI] [PubMed] [Google Scholar]
- Wasseneger M (2005) The role of the RNAi machinery in heterochromatin formation. Cell 122: 13–16 [DOI] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE (2004) Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol 14: 1214–1220 [DOI] [PubMed] [Google Scholar]