Abstract
In this study, we used gas chromatography-mass spectrometry analysis in combination with flux analysis and the Affymetrix ATH1 GeneChip to survey the metabolome and transcriptome of Arabidopsis (Arabidopsis thaliana) leaves in response to manipulation of the thiol-disulfide status. Feeding low concentrations of the sulfhydryl reagent dithiothreitol for 1 h at the end of the dark period led to posttranslational redox activation of ADP-glucose pyrophosphorylase and major alterations in leaf carbon partitioning, including an increased flux into major respiratory pathways, starch, cell wall, and amino acid synthesis, and a reduced flux to sucrose. This was accompanied by a decrease in the levels of hexose phosphates, while metabolites in the second half of the tricarboxylic acid cycle and various amino acids increased, indicating a stimulation of anaplerotic fluxes reliant on α-ketoglutarate. There was also an increase in shikimate as a precursor of secondary plant products and marked changes in the levels of the minor sugars involved in ascorbate synthesis and cell wall metabolism. Transcript profiling revealed a relatively small number of changes in the levels of transcripts coding for components of redox regulation, transport processes, and cell wall, protein, and amino acid metabolism, while there were no major alterations in transcript levels coding for enzymes involved in central metabolic pathways. These results provide a global picture of the effect of redox and reveal the utility of transcript and metabolite profiling as systemic strategies to uncover the occurrence of redox modulation in vivo.
The cellular redox state is determined by several redox-active species. In addition to being constituents of cellular metabolism per se, redox agents actively participate in metabolic regulation (Bowsher et al., 1992, 1993; Noctor et al., 2004; Foyer and Noctor, 2005). Perhaps the best-characterized example to date is the regulation of the expression and posttranslational regulation of chloroplast proteins by signals triggered by the redox state of photosynthetic electron transport components (Baier and Dietz, 2005). This network allows photosynthesis to adjust to environmental inputs (Dietz and Scheibe, 2004). Light acts via thioredoxins to posttranslationally modulate the activity of a set of chloroplast proteins involved in ATP synthesis and photosynthetic carbon metabolism. Short-term relocation of light-harvesting (LHCII) proteins and redistribution of excitation energy (state transitions) is linked to the redox state of the plastoquinon pool via LHCII protein kinase (Fey et al., 2005). This kinase is also involved in long-term photosynthetic adaptations via a signal that is transferred over the chloroplast envelope to the nucleus where it affects the expression of nuclear-encoded photosynthetic proteins (Bonardi et al., 2005; Fey et al., 2005).
The thioredoxin system was discovered over 20 years ago (Buchanan, 1980). Illumination leads to reduction of ferredoxin at the acceptor site of PSI. Ferredoxin-thioredoxin reductase then leads to reduction of thioredoxins f or m, which activate enzymes in the Calvin cycle and related photosynthetic processes by reducing intramolecular disulfide bonds (Buchanan, 1980; Scheibe, 1990; Schürmann and Jacquot, 2000; Buchanan and Balmer, 2005). Within minutes, this activates the Calvin cycle enzymes Fru-1,6-bisphosphatase (FBPase), sedoheptulose-1,7-bisphosphatase, and phosphoribulokinase, the redox-export enzyme NADP-malate dehydrogenase (MDH), and the ATP synthase coupling factor CF1, and inactivates chloroplast Glc-6-P dehydrogenase (Glc6PDH), a key enzyme of the oxidative pentose phosphate pathway. These changes are reversed in the dark when the thioredoxin system is reoxidized. Metabolites have been shown to influence the rate constants of reduction and oxidation by changing the midpoint potentials of the regulatory sulfhydryls. Fru-1,6-bisP acts as a positive effector for FBPase activation, and thiol activation of NADP-MDH is effectively prevented when the NADPH/NADP ratio is low (Scheibe, 1990).
While the role of redox signals in photosynthesis is relatively well characterized, there is little information about their role in the regulation of other metabolic processes. It was shown recently that thioredoxin regulates ADPGlc pyrophosphorylase (AGPase), a key enzyme of starch biosynthesis (Tiessen et al., 2002; Hendriks et al., 2003). AGPase is rapidly activated upon illumination by reduction of an intermolecular disulfide bond between Cys on the two small subunits of the tetrameric enzyme (Hendriks et al., 2003; Kolbe et al., 2005). Activation is also promoted by sugars, which act additively with light and also on their own in darkened leaves and nonphotosynthetic tissues (Tiessen et al., 2002; Hendriks et al., 2003; Kolbe et al., 2005).
The Arabidopsis (Arabidopsis thaliana) genome contains a small family of thioredoxins, including 19 different isoforms grouped into six subfamilies (Lemaire et al., 2004; Buchanan and Luan, 2005). Thioredoxins f, m, x, and y are located exclusively in the chloroplast, thioredoxin o exclusively in the mitochondria, and thioredoxin h representatives are distributed between the cytosol, nucleus, endoplasmic reticulum (ER), and mitochondria (Schürmann and Jacquot, 2000; Buchanan and Luan, 2005). Thioredoxins can be reduced by two different redox systems, videlicet ferredoxin and ferredoxin:thioredoxin reductase in the chloroplast or NADPH and NADP-thioredoxin reductase in other organelles (Buchanan and Balmer, 2005).
A large number of novel potential thioredoxin targets have been identified using two in vitro strategies: (1) studies of enzymes and proteins after adding dithiothreitol (DTT), a nonphysiological substrate for thioredoxins (Schürmann and Jacquot, 2000), and (2) affinity chromatography with mutated thioredoxins in combination with proteomic analyses (Motohashi et al., 2001; Balmer et al., 2003; Balmer et al., 2004; Lemaire et al., 2004; Marchand et al., 2004; Wong et al., 2004). In total, about 180 putative thioredoxin targets have been identified, distributed in various metabolic processes, different cellular organelles, and diverse photosynthetic and nonphotosynthetic tissues (Buchanan and Balmer, 2005). These results suggest that thioredoxins have a general role in metabolic regulation but in most cases the physiological relevance has not been demonstrated.
In the following experiments we use flux analysis and metabolite and transcript profiling to survey the response of Arabidopsis leaves to incubation with the reductant DTT. This strategy allowed us to identify sites in the metabolic network that are subject to dithiol regulation in vivo. The results are discussed in the context of in vitro findings and previous models for thioredoxin mediated regulation of metabolism.
RESULTS
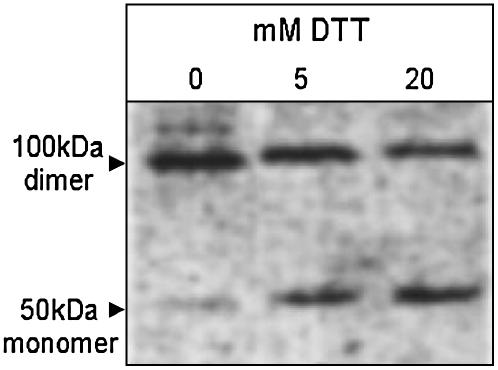
Feeding Dilute DTT Leads to Posttranslational Redox Activation of AGPase and Other Enzymes in Arabidopsis Leaves
DTT is a sulfhydryl reagent that serves as a nonphysiological redox donor and substrate for thioredoxins (Buchanan et al., 1979). We first investigated whether treatment of Arabidopsis leaf discs with 5 mm DTT for 1 h in the dark leads to redox regulation of known target enzymes.
Four chloroplast enzymes that are known to be thioredoxin targets were investigated: Glc6PDH, FBPase, NADP-MDH (Scheibe, 1990), and AGPase (Geigenberger et al., 2005; Kolbe et al., 2005). The redox activation of AGPase was increased by DTT treatment (Fig. 1). This is revealed by a strong increase in the degree of monomerization of the small subunit of the tetrameric enzyme, which runs as a monomer in the reduced and a dimer in the oxidized form in nonreducing SDS gels. Feeding 20 mm DTT led to only a slight further activation of AGPase. Supplying 1 to 10 mm DTT for 2 h to potato (Solanum tuberosum) tuber discs also redox activated AGPase (Tiessen et al., 2002), while feeding 10 mm glutathione had no effect (A. Tiessen and P. Geigenberger, unpublished data). Leaf discs were rapidly extracted and activity measured in nonreductive conditions to investigate the response of Glc6PDH, FBPase, and NADP-MDH. The activities (nmol/g fresh weight [FW] min) in leaf discs treated without or with 5 mm DTT were 174 ± 18 and 115 ± 7 for Glc6PDH and 13.2 ± 6.6 and 27.0 ± 1.1 for FBPase; this matches the response expected based on the known effect of thiol-disulfide reduction on these two enzymes. The values for NADP-MDH were 19.4 ± 3.3 and 21.1 ± 1.1, respectively (mean ± sd, n = 2 independent incubations). Redox activation of NADP-MDH is strongly dependent on a high NADPH/NADP ratio in the chloroplast (Scheibe, 1990). These data indicate that DTT treatment did not strongly increase the NADP redox state of the leaf discs.
Figure 1.
DTT feeding leads to posttranslational redox activation of AGPase in leaves. Leaves of Arabidopsis plants were harvested at the end of the night to prepare discs that were immediately incubated on 0, 5, or 20 mm DTT for 1 h in the dark. Discs were then quickly rinsed in buffer to remove external DTT before freezing in liquid nitrogen to analyze monomerization of the small subunit of AGPase (AGPB) using nonreductive SDS gels. Under these conditions oxidized AGPB runs as a 100 kD dimer while reduced AGPB runs as a 50 kD monomer.
To investigate whether DTT treatment affected the in vivo thiol status of mitochondrial proteins, we analyzed the dimerization state of the mitochondrial alternative oxidase (AOX). This protein is activated by reduction of an intermolecular disulfide bond formed between the Cys-126 residues of the AOX homodimer (Umbach and Siedow, 1993; Vanlerberghe et al., 1998; Vanlerberghe et al., 1999). Reduction of the intermolecular disulfide bridge leads to an increased mobility of the AOX protein during gel electrophoresis because the protein runs as a dimer in the oxidized and a monomer in the reduced form. Leaf discs were rapidly extracted in trichloroacetic acid, proteins separated using nonreducing SDS-PAGE, and AOX protein detected via immunoblot. The monomer/dimer ratio increased from 0.83 in the control to 1.90 in discs treated with 5 mm DTT (Supplemental Fig. 1). Overall, these results highlight the utility of DTT as a tool to manipulate disulfide-bond formation of various enzymes in leaf tissues.
Changes in Carbon Fluxes in Arabidopsis Leaves in Response to Short-Term DTT Treatment
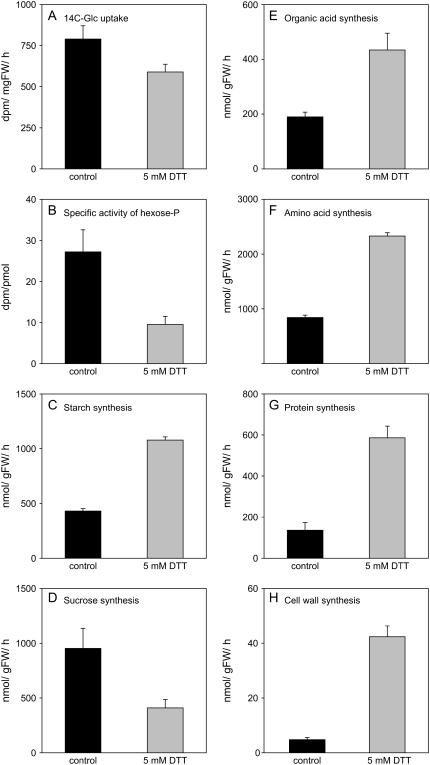
To investigate the effect on carbon fluxes, leaf discs were incubated for 1 h with carrier-free high specific activity 14C-Glc in the presence or absence of 5 mm DTT. DTT treatment resulted in a small inhibition of 14C-Glc uptake (Fig. 2A), which resembles the results obtained with potato tuber tissue (Tiessen et al., 2002). DTT led to major changes in the partitioning of label. An increased percent of the label was incorporated into starch, amino acids, protein, and cell walls, while labeling of organic acids remained unchanged and labeling of Suc decreased (data not shown). Further separation of label in cell walls revealed that DTT treatment led to a preferential increase in labeling of cellulose, with the labeling ratio of cellulose/matrix polysaccharides increasing from 0.75 ± 0.13 in the control to 1.31 ± 0.01 in the 5 mm DTT treatment, respectively (data not shown). To calculate absolute fluxes, the specific activity of the internal hexose-phosphate pool was determined (see Geigenberger et al. [1997] for a full explanation of the assumptions inherent in the estimation of fluxes). The specific activity decreased 2-fold after feeding DTT (Fig. 2B). This was partly due to the decreased uptake of label (Fig. 2A) but also indicates there is dilution of label by increased mobilization of unlabeled endogenous storage carbon such as starch or Suc. DTT treatment led to a 2- to 3-fold increase in the rate of starch (Fig. 2C), organic acid (Fig. 2E), and amino acid synthesis (Fig. 2F), a 4-fold increase in protein synthesis (Fig. 2G), and a 10-fold increase in cell wall synthesis (Fig. 2H), while flux to Suc (Fig. 2D) decreased. The results extend previous studies showing that redox activation of AGPase leads to increased rates of starch synthesis in potato tubers and leaves of various species (Tiessen et al., 2002; Hendriks et al., 2003). In addition, they provide evidence that the thiol-disulfide status is involved in regulating further pathways of carbon metabolism in leaves, including amino acid, protein, and cell wall synthesis. Interestingly, many of these pathways are at least partially located outside the plastid.
Figure 2.
Changes in metabolic fluxes after incubation of Arabidopsis leaf discs in 5 mm DTT. Incubations were performed exactly as described in Figure 1 with the exception that high specific activity 14C-Glc was included in the incubation medium. After 1 h, discs were sampled and frozen in liquid nitrogen to analyze uptake and metabolism of the labeled Glc and the specific activity of the internal hexose-phosphate pool. The specific activity of the hexose-phosphate pool was used to calculate absolute fluxes in nmol gFW−1 h−1. A, Total uptake of label. B, Specific activity of the hexose-phosphate pool. C, Starch synthesis. D, Suc synthesis. E, Organic acid synthesis. F, Amino acid synthesis. G, Protein synthesis. H, Cell wall synthesis. Results are the mean ± se (n = 5).
Changes in Metabolite Profiles in Arabidopsis Leaves in Response to Short-Term DTT Treatment
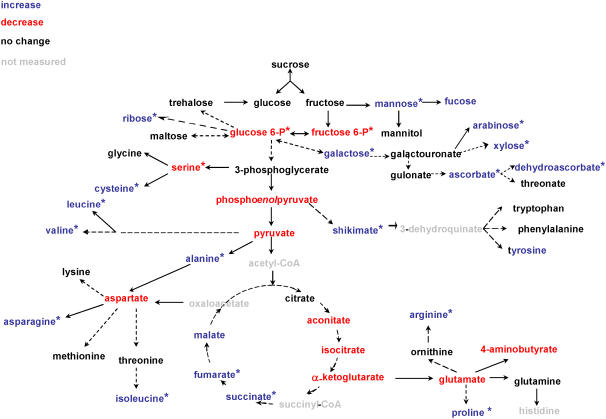
For a more comprehensive analysis, metabolite and transcript profiles were investigated in leaf discs incubated in the presence and absence of DTT in parallel to the incubations described above. After sampling and extraction, metabolite contents were determined and expressed as described in Roessner et al. (2001a, 2001b). Figure 3 presents a schematic overview of the metabolite changes and Supplemental Table I provides a comprehensive list of all metabolites measured and documents the extent and significance of these changes. DTT treatment led to a significant decrease of hexose-phosphates. These represent the entry point of carbon into starch synthesis, glycolysis, and cell wall synthesis. There were significant increases of succinate and fumarate in the second half of the tricarboxylic acid (TCA) cycle and significant increases in the levels of various amino acids such as Arg, Pro, Asn, Ile, Ala, Leu, Val, and Cys, while the level of Ser decreased. There was a significant increase in shikimate, which is an intermediate in the synthesis of aromatic amino acids and secondary metabolites. DTT treatment did not lead to substantial changes in the levels of the major sugar pools such as Suc, Fru, and Glc. There were, however, significant changes in the levels of the minor sugars, such as Man, Gal, Ara, Xyl, and Rib. These sugars are involved in ascorbate, cell wall, and nucleotide metabolism. Moreover, there was a significant increase in ascorbate and a smaller increase in dehydroascorbate levels, showing that total ascorbate pool increased. The changes in metabolite levels were largely consistent with the labeling studies described above in that they suggested an increase in the flux to cell wall and amino acids and proteins (see Fig. 2). The significant decrease in Glc6-P and Fru6-P levels is consistent with a stimulation of the use of hexose-phosphates for starch, cell wall, and amino acid synthesis.
Figure 3.
Changes in metabolite profiles after incubation of Arabidopsis leaf discs in 5 mm DTT. Incubations were performed exactly as described in Figure 1 before material was frozen in liquid nitrogen for metabolite analysis. Changes in metabolite levels were calculated as the ratio between DTT and buffer incubated samples and are listed in Supplemental Table I. To visualize the changes, increases or decreases larger than 10% were indicated with blue or red letters, respectively, within a metabolic scheme. Changes that are significantly different from the control (P < 0.05 using the Student's t test of Microsoft Excel) are indicated with an asterisk. Results are the mean ± se (n = 5).
Changes in Transcript Profile in Arabidopsis Leaves in Response to Short-Term DTT Treatment
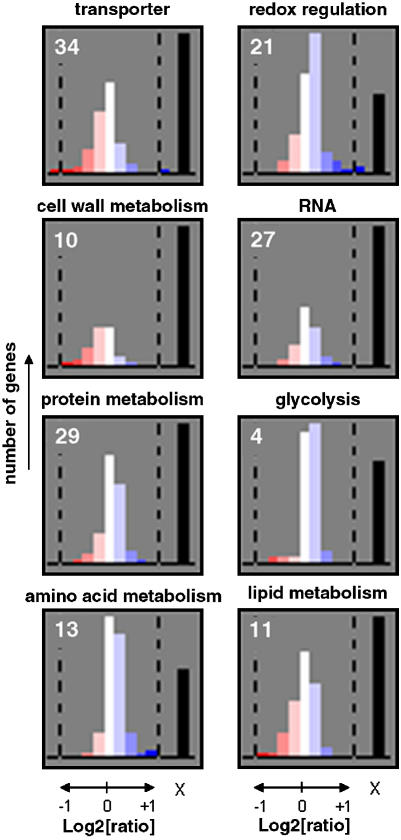
To investigate changes in the transcriptome of Arabidopsis leaves, Affymetrix ATH1 GeneChip analyses were performed on control and DTT-treated samples. Feeding of 5 mm DTT for 1 h led to a relatively small number of changes in transcript levels. Out of the 11,998 genes called present by the software, 15 were up-regulated and 29 were down-regulated >2-fold in both biological replicas (see Supplemental Table II). Using the MapMan software (Thimm et al., 2004; Usadel et al., 2005), genes were categorized in functional bins: It was tested whether the observed expression values of each bin differ from the collection of genes from all other bins using the Wilcoxon Rank Sum test with Benjamini-Hochberg P-value correction. Table I shows the major functional bins in the order of their probability value. The lowest P value (2.28E-29) was attributed to genes encoding transport proteins, revealing that the combined response of transcripts from genes assigned to this functional category is highly significantly different compared to the response of all other expressed genes on the array. As the responses of transcripts for genes assigned to the bins cell wall metabolism (2.57E-27), protein metabolism (4.81E-14), amino acid metabolism (4.47E-07), redox regulation (4.61E-04), RNA metabolism (1,15E-03), glycolysis (0.03), and lipid metabolism (0.04) also had P values < 0.05, it can be assumed that expression of sets of genes assigned to these bins is also being coordinately regulated. Surprisingly, genes assigned to central metabolic pathways such as photosynthesis, Calvin cycle, TCA cycle, starch, and Suc metabolism, in secondary metabolism and in many other categories relating to nutrient assimilation and stress responses showed no significantly different responses. Figure 4 provides an overview of the direction of changes in transcript levels in selected bins showing significantly different responses (P < 0.05). The data show that DTT leads to a preferential decrease in transcripts for genes encoding transporters and enzymes involved in cell wall metabolism, while there was a preferential increase in transcripts for genes assigned to the other categories including glycolysis, amino acid synthesis, protein synthesis, and redox processes.
Table I.
Transcript profiling after incubation of Arabidopsis leaf discs in 5 mm DTT as described in Figure 1
Statistical evaluation of the behavior of transcript levels of genes within a functional category (bin) as compared to all the other remaining bins. Genes that belong to a bin that has a P value below 0.05 are likely to be coregulated. Categorization of genes and subsequent statistic analyses were performed with the MapMan software, using the Wilcoxon Rank Sum test with Benjamini-Hochberg correction.
| MapMan Bin Number | MapMan Bin Description | Number of Elements in Bin | P Value |
|---|---|---|---|
| 34 | Transporters | 529 | 2.28E-29 |
| 10 | Cell wall metabolism | 221 | 2.57E-27 |
| 29 | Protein metabolism | 1,856 | 4.81E-14 |
| 13 | Amino acid metabolism | 251 | 4.47E-07 |
| 21 | Redox regulation | 143 | 1.46E-04 |
| 27 | RNA | 1,240 | 1.15E-03 |
| 4 | Glycolysis | 48 | 0.03 |
| 11 | Lipid metabolism | 258 | 0.04 |
| 6 | Gluconeogenese/glyoxylate cycle | 6 | 0.07 |
| 7 | Oxidative pentose phosphate pathway | 24 | 0.07 |
| 23 | Nucleotide metabolism | 108 | 0.08 |
| 8 | TCA/transformation of organic acids | 57 | 0.11 |
| 18 | Cofactor and vitamin metabolism | 32 | 0.15 |
| 9 | Mitochondrial electron transport/ATP synthesis | 84 | 0.17 |
| 25 | C1 metabolism | 21 | 0.19 |
| 30 | Signaling | 613 | 0.33 |
| 19 | Tetrapyrrole synthesis | 28 | 0.35 |
| 31 | Cell | 365 | 0.37 |
| 5 | Fermentation | 11 | 0.41 |
| 2 | Major carbohydrate metabolism | 73 | 0.41 |
| 1 | Photosynthesis | 168 | 0.49 |
| 12 | Nitrogen metabolism | 21 | 0.57 |
| 33 | Development | 231 | 0.59 |
| 28 | DNA | 251 | 0.73 |
| 20 | Stress | 409 | 0.77 |
| 24 | Biodegradation of xenobiotics | 13 | 0.87 |
| 22 | Polyamine metabolism | 8 | 0.90 |
| 15 | Metal handling | 55 | 0.94 |
| 16 | Secondary metabolism | 181 | 0.95 |
| 14 | Sulfur assimilation | 13 | 0.97 |
| 3 | Minor carbohydrate metabolism | 84 | 0.97 |
| 17 | Hormone metabolism | 260 | 1.00 |
Figure 4.
Frequency distribution of DTT induced changes in transcript levels of genes categorized in functional bins that divert significantly from the mean change in expression of the genes from all other bins (see also Table I). Color codes of the bars relate to the level of expression (log2[ratio]) as indicated in the legend (blue, increased expression; red, decreased expression). The numbers refer to the functional bins as listed in Table I and defined by the MapMan software. The bar indicated with X indicates the number of genes within the category that were not expressed. Incubation of leaf discs with 5 mm DTT was performed exactly as described in Figure 1.
In Supplemental Figure 2 changes in transcripts were pasted in a metabolism overview scheme using the MapMan software. For this analysis, a highly sensitive visualization scale was chosen at which a log2[ratio] of plus or minus 1 was assigned the strongest color intensity. This allows visualization of general trends of changes in transcript levels rather than the identification of significantly different changes of individual transcripts. The data again clearly document a preferential decrease in transcripts involved in cell wall degradation and reorganization in response to DTT feeding, while transcripts involved in cell wall synthesis preferentially increased. There were hardly any changes in transcripts of central metabolic pathways such as the TCA cycle or photosynthesis. Further analysis also reveals a general decrease in transcripts coding for different subcategories of transport proteins, with the exception of uridine-sugar transporters that showed a preferential increase (Supplemental Fig. 3). Table II provides an overview of the changes in normalized expression values of genes that encode for proteins involved in redox regulation. There was a preferential increase in transcripts coding for protein disulfide isomerases, thioredoxin family proteins, and glutaredoxin family proteins, while transcripts involved in ROS detoxification (peroxidases, etc.) remained largely unchanged (data not shown). MapMan files for all experiments are available in Supplemental Table III. In Supplemental Table II, individual transcripts are listed showing >2-fold changes (log2[ratio] > 1 or < −1) in both biological replicas. The list includes several transporters and enzymes of amino acid metabolism as well as genes associated with protein folding, disulfide-bond formation, cell wall metabolism, and signaling components.
Table II.
Overview of the normalized expression values of genes that encode for proteins involved in redox regulation with changed expression after feeding 5 mm DTT for 1 h
The average log2[ratio] and its P value are calculated using the AffylmGUI software package from Bioconductor. Raw data can be found in Supplemental Table III. Log2[ratio], log2 transformed value of the ratio between DTT treatment and control; AGI, Arabidopsis Genome Initiative.
| ATH1 Code | log2[ratio]
|
P Value | AGI Code | Gene Description | ||
|---|---|---|---|---|---|---|
| Replica 1 | Replica 2 | Average | ||||
| 259757_at | 2.2607 | 1.0765 | 1.6687 | 0.161 | At1g77510 | Protein disulfide isomerase similar to disulfide isomerase precursor |
| 261167_at | 2.3681 | 0.7527 | 1.5605 | 0.063 | At1g04980 | Thioredoxin family protein, protein disulfide isomerase |
| 262504_at | 1.5834 | 0.7566 | 1.1700 | 0.214 | At1g21750 | Protein disulfide isomerase |
| 251196_at | 0.7872 | 0.8415 | 0.8144 | 0.331 | At3g62950 | Glutaredoxin family protein (thioltransferase) |
| 256922_at | 0.8458 | 0.4128 | 0.6293 | 0.156 | At3g19010 | Hypothetical protein contains similarity to flavonol synthase |
| 265067_at | 0.9204 | 0.2961 | 0.6083 | 0.732 | At1g03850 | Glutaredoxin family protein (thioltransferase) |
| 251840_at | 0.9203 | 0.2491 | 0.5848 | 0.282 | At3g54960 | Thioredoxin family protein similar to protein disulfide isomerase |
| 258665_at | 0.7586 | 0.3845 | 0.5716 | 0.341 | At3g08710 | Thioredoxin family protein similar to thioredoxin H type |
| 245504_at | 0.4907 | 0.5291 | 0.5099 | 0.204 | At4g15660 | Glutaredoxin family protein (thioltransferase) |
The lack of substantial changes in transcript levels of genes encoding for enzymes involved in central metabolic pathways indicates that the changes in fluxes and metabolite levels in these pathways that were obtained following short-term incubation in DTT (see Fig. 2) were most likely due to translational or posttranslational effects of the treatment.
DISCUSSION
DTT Treatment Leads to in Vivo Modification of the Thiol-Disulfide Activation of Known Thioredoxin Targets
Thioredoxin plays a key role in the light regulation of photosynthesis and has recently been implicated in the sugar-dependent regulation of starch synthesis (see introduction). In vitro studies have identified approximately 180 putative thioredoxin targets, indicating that there is a wider role for thioredoxin in the regulation of further pathways and of events in additional metabolic compartments as well as the chloroplast (Buchanan and Balmer, 2005). DTT is a nonphysiological substrate for thioredoxins (Buchanan, 1980). We have used short-term treatment of leaf discs with DTT in the dark as a tool to manipulate the thiol-disulfide status. Metabolite profiling combined with flux analysis and transcript profiling were employed in a systematic approach to identify sites within leaf metabolism that are redox regulated by reversible disulfide-bridge formation in vivo. The sites where the flux through a pathway is stimulated can be identified by a decrease in the substrates and an increase in the products.
The experimental approach was validated by investigating the impact of the DTT treatment on a set of known target enzymes and other metabolic parameters that provide information about the thiol-disulfide status and, more generally, the redox status. Three known chloroplastic targets for DTT and thioredoxin (AGPase, FBPase, and Glc6PDH) showed the expected responses; DTT treatment led to increased monomerization of AGPase in nonreductive SDS-PAGE, increased activity of FBPase, and decreased activity of G6PDH. The DTT treatment also led to an increase in the monomer/dimer ratio of mitochondrial AOX.
DTT treatment did not significantly increase NADP-MDH activity. This is a known target for thioredoxin m, but activation requires a high NADPH/NADP ratio. Activation of NADP-MDH is an indicator for the NADPH/NADP ratio in the chloroplast stroma (Scheibe, 1990). These data indicate that the DTT treatment has not increased the NADPH/NADP ratio, at least in the chloroplast stroma. Independent evidence for this conclusion is provided by our metabolite analyses. The NAD(P)-reduction state of a tissue can be estimated by calculating the in vivo product/substrate ratio of NAD(P)-linked reactions like isocitrate dehydrogenase or MDH, which operate close to their thermodynamic equilibrium (Sies, 1982; Siess et al., 1982; Heineke et al., 1991). When the levels of isocitrate, α-ketoglutarate, malate, Glu, and Asp in leaf discs treated without or with 5 mm DTT were used to calculate these metabolite ratios, only minor changes were found (0.764 and 0.810 for isocitrate/α-ketoglutarate and 27.9 and 22.9 for [malate × Glu]/[Asp × α-ketoglutarate] in the absence and presence of DTT, respectively).
These results provide evidence that DTT treatment led to changes in the thiol status of several known thioredoxin targets in the chloroplast stroma and the mitochondria, while the reduction state of the NAD(P) system was not substantially changed in the discs. However, we cannot rigorously exclude the possibility that some of the changes in the DTT treatment may be due to changes of the ascorbate or glutathione couples.
Thiol-Disulfide Exchange Leads to Global Changes in Leaf Metabolism
A short-term increase in the thiol-disulfide status leads to major alterations in the metabolite profiles and increased flux of carbon into starch, cell wall, the major respiratory pathways, and amino acid synthesis. There are no substantial changes in the levels of major sugars such as Suc, Glc, and Fru (Figs. 2 and 3). This provides evidence that the thiol-disulfide status affects several important metabolic pathways both within and external to the plastid, via a mechanism that is independent of the sugar status.
The stimulation of starch synthesis was accompanied by posttranslational redox activation of AGPase and occurred in the presence of decreased hexose-phosphate and unchanged 3-phosphoglycerate levels. This resembles the response in potato tuber discs (Tiessen et al., 2002). Earlier studies in potato tubers, leaves, and isolated chloroplasts have shown that AGPase is subject to thioredoxin-dependent redox activation (Hendriks et al., 2003; Geigenberger et al., 2005). The redox activation of AGPase is probably due to increased reduction of thioredoxins by DTT, rather than to a general increase in the cellular redox state or in the NADPH/NADP ratio (see above). The Suc-dependent increase of AGPase reduction and monomerization also occurs independently of changes in the NADPH/NADP ratio (Kolbe et al., 2005).
DTT treatment led to a decrease in the levels of organic acids involved in the first part of the TCA cycle (aconitate, isocitrate, and α-ketoglutarate) while succinate and other intermediates of the second part of the TCA cycle increased. This is consistent with stimulation of α-ketoglutarate dehydrogenase (Fig. 3). α-Ketoglutarate dehydrogenase is a key regulatory enzyme of the TCA cycle and is subject in mammals to redox regulation by mitochondrial thioredoxins, which protects against self inactivation during catalysis at low NAD+ (Bunik, 2003). Our data suggest α-ketoglutarate dehydrogenase is also subject to posttranslational redox regulation in plants. Recent proteomic studies identified many plant mitochondrial proteins that interact with thioredoxins in vitro, including the α-ketoglutarate dehydrogenase complex subunit E2 (Balmer et al., 2004; Lemaire et al., 2004).
DTT treatment led to an increased flux of carbon into amino acids and proteins (Fig. 2) and increased levels of many amino acids (Fig. 3), indicating redox activation of enzymes incorporating carbon skeletons into amino acid in leaves. In plants, α-ketoglutarate is an important precursor for ammonia assimilation. Following synthesis in the mitochondria in the TCA cycle, α-ketoglutarate is transported to the chloroplast and used as carbon skeleton for amino acid synthesis via the Gln synthetase (GS)/Gln:oxoglutarate amino transferase (GOGAT) cycle. The decrease of α-ketoglutarate and stimulation of amino acid synthesis after DTT treatment may be indicative for regulation of the GS/GOGAT cycle via posttranslational redox regulation. Fd-GOGAT and GS are subject to thioredoxin-dependent redox regulation in vitro (Choi et al., 1999; Motohashi et al., 2001; Balmer et al., 2003).
Our results identify further redox-regulated steps in the pathways of amino acid synthesis. DTT treatment led to a decrease in Asp, while amino acids deriving from Asp such as Ile and Asn were increased, indicating that one or more enzymes involved in the conversion of Asp to Ile or Asn may be stimulated by increased redox state. A similar situation holds for enzymes involved in the metabolism of pyruvate to Ala, Leu, and Val in the metabolism of Glu to Arg and Pro, conversion of Ser to Cys, and the conversion of phosphoenolpyruvate (PEP) to shikimate and Tyr. The decrease in PEP and increase of shikimate and Tyr is consistent with previous in vitro studies showing that 3-deoxy-d-arabino-heptulosonate 7-P synthase, the first enzyme of the shikimate pathway, is activated by thioredoxin f (Entus et al., 2002). Potential thioredoxin targets captured in proteomic studies using resin-bound thioredoxins (Balmer et al., 2003; Lemaire et al., 2004) include enzymes involved in the synthesis of Ala (Ala aminotransferase), Asn (Asp aminotransferase), Arg (argininosuccinate synthase), valin and isoleucin (dihydroxyacid dehydratase, ketolactid reductoisomerase, and branched chain ketoacid decarboxylase), Leu (3-isopropylmalate dehyrogenase), and Cys (Cys synthase).
Thus, our results combined with published in vitro studies indicate that thioredoxin-dependent regulation may play a major role in the orchestration of amino acid synthesis. Final evidence will require the demonstration that the dithiol-sulfide status of these enzymes changes in vivo in response to physiological treatments like illumination or sugar accumulation, and that mutants lacking thioredoxins or thioredoxin reductase are defective in this response and the accompanying changes of fluxes and metabolites. Probably, the dithiol and further redox couples interact to coordinate N-assimilation, anaplerotic production of carbon skeletons, and amino acid synthesis. While there is no evidence that nitrate reduction is regulated via reversible disulfide-bond formation, it does depend on the NAD(P)H/NAD(P) ratio; nitrate reductase uses NADH as reductant and nitrite reductase requires reduced ferreredoxin, which in the dark is formed from NADPH. In tobacco (Nicotiana tabacum) mutants lacking functional mitochondrial electron transport complex I, an increased reduction state of NAD(P)H leads to a stimulation of nitrate assimilation and an increase of amino acids at the expense of α-ketoglutarate (Dutilleul et al., 2005). Noctor et al. (1998) showed in transgenic poplar (Populus tremula) plants that up-regulation of the synthesis of reduced glutathione in the chloroplast stimulates synthesis of a specific set of amino acids in the chloroplast.
DTT treatment stimulated the flux of carbon into proteins (Fig. 2). This might be due to increased availability of amino acids. Alternatively, DTT might lead to redox activation of translation, protein assembly, and folding. The synthesis, assembly, and transport of proteins are stimulated by DTT and inhibited by SH oxidants (von Stedingk et al., 1997). Several thioredoxin targets involved in translation, protein assembly, and folding were recently identified using proteomics (Balmer et al., 2003). In this context, it is noteworthy that (see below) DTT treatment transcriptionally activated many genes assigned to protein synthesis.
DTT treatment stimulated flux to cell wall components, especially cellulose (Fig. 2). Cellulose synthase has been reported to be subject to posttranslational redox regulation, leading to changes in the activation/oligomerization state of the enzyme (Kurek et al., 2002). It is unknown whether this is mediated by thioredoxin. There was also a general increase of minor sugars that may be involved in cell wall metabolism, like Gal, Ara, Man, Xyl, and Fuc. This increase might also be related to turnover of a cytosolic heteroglycan that is involved in starch degradation (Fettke et al., 2005, 2006). Glucan water dikinase, a key enzyme of starch degradation, is regulated by thioredoxins in vitro (Mikkelsen et al., 2005).
Thiol-Disulfide Exchange Is Accompanied by a Relatively Small Number of Changes in Transcript Levels
DTT treatment did not lead to significant changes in overall transcript levels for genes assigned to central metabolic pathways such as the TCA cycle, starch metabolism, pentose-phosphate pathway, and Calvin cycle. These results indicate that short-term redox regulation of these pathways occurs mainly at the posttranslational rather than the transcriptional level, which is consistent with previous studies showing that these pathways are regulated by the thioredoxin system (Buchanan, 1980; Schürmann and Jacquot, 2000; Bunik, 2003; Balmer et al., 2004; Geigenberger et al., 2005).
Statistical treatment uncovered significant changes of overall transcript levels for some specific segments of metabolism (Table I). This included a preferential increase of transcripts for genes encoding enzymes in amino acid metabolism, a decrease for enzymes involved in cell wall metabolism, an increase for cell wall synthesis, and a decrease for transport proteins (Supplemental Fig. 3) with the exception of ER sugar-nucleotide transporters, which increased. The reason for the negative effect of redox on transcripts involved in transport is unclear but may contribute to the inhibition of sugar uptake (see Fig. 2). The increase in transcripts of ER sugar-nucleotide transporters may provide a strategy to stimulate precursor supply to allow increased cell wall synthesis. There was a significant increase of the transcript levels for genes involved in redox regulation and disulfide-bond formation (Table I), including genes that encode thioredoxin and glutaredoxin family proteins (Table II), but not genes involved in oxidative stress (peroxidase, etc.) or general stress responses. DTT also affected transcripts involved in the synthesis, assembly, and transport of proteins, as seen by von Stedingk et al. (1997). DTT has been reported to increase expression of genes that encode proteins that create the optimal-folding environment, such as protein disulfide isomerase, calreticulin, clanexin, and binding protein (Martinez and Chrispeels, 2003). It is not clear if these changes of transcripts are accompanied by changes in the levels of the encoded proteins. In view of the short duration of the DTT treatment, they might just represent an early step in a response that is triggered in parallel at the transcriptional and posttranslational level. It also remains an open question if there is an additional level of response via regulation of translation.
The response to DTT treatment contrasts to that seen by Ball et al. (2004) after manipulation of glutathione biosynthesis when mainly stress defense genes were induced. Increased levels of reduced glutathione in plants due to feeding or transgenic approaches led to specific changes in the release of intracellular calcium to the cytosol, whereas this effect was absent after feeding DTT (Gomez et al., 2004). These results suggest that DTT and glutathione act via different signaling pathways.
Redox-Based Changes in Carbon Metabolism Are Interlinked with Sugar Signaling
There is a surprising similarity between the stimulating effect of DTT on biosynthetic fluxes documented in this study (Fig. 2) and previously published results on the effect of sugars (Geiger et al., 1998; Morcuende et al., 1998; Hendriks et al., 2003; data not shown). This suggests that sugar and thiol-disulfide signals may share common metabolic targets. In fact, sugars lead to stimulation of starch synthesis via posttranslational redox activation of AGPase (Tiessen et al., 2002; Hendriks et al., 2003). Redox activation of AGPase in potato tubers in response to Glc and Suc is mediated by separate sugar signaling pathways, which differ in their dependency on a SNF1-like protein kinase and in their effect on the reduction state of the NADP system (Tiessen et al., 2003; Kolbe et al., 2005). Glc leads to an increase in the NADPH/NADP ratio that may subsequently lead to an increase in the reduction of thioredoxin targets via NADP-thioredoxin reductase. The signaling pathway that links Suc to redox activation of AGPase does not require an increase of the NADPH/NADP ratio, and involves a SNF1-like protein kinase in the cytosol (Tiessen et al., 2003) and trehalose-6-P as signal molecule that links the sugar status to the plastid (Kolbe et al., 2005; Lunn et al., 2006). More studies are needed to elucidate the mechanisms and signaling components that are involved in this regulatory network and their interaction with dithiol signaling.
In conclusion, this paper complements recent proteomic studies by providing a comprehensive overview of global changes in fluxes, metabolites, and transcripts in response to thiol-disulfide manipulation of leaf tissue. Results show that major biosynthetic processes in plants are redox modulated in vivo and allow various redox-regulated sites to be identified in vivo that have previously been found in proteomic studies to interact with thioredoxins. The large alterations in metabolites and fluxes in central metabolic pathways were accompanied by a relatively small number of changes in transcript levels, suggesting that regulation occurred preferentially at the posttranslational level.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) var. ecotype Columbia-0 (wild type) was grown in a short-day phytotron (with an 8-h light/16-h dark cycle; 180 μE, 20°C/16°C, and 60%/75% humidity). Leaves were harvested in the chamber at the end of the dark period and used immediately for incubation experiments or quenched in liquid nitrogen and stored at −80°C until use. Only source leaves that were not shaded by other leaves were collected.
Incubation Experiments
Leaf discs were floated for on 2 mm MES, pH 6.5, and various concentrations of DTT. The experiment was carried out in a growth cabinet in identical conditions to the growth phytotron, except the light was switched off. After 1 h, the leaf discs were immersed shortly in water to remove external DTT and then frozen in liquid nitrogen.
Enzyme Analysis
Frozen leaf material was homogenized and extracted in trichloroacetic acid as in Hendriks et al. (2003). Redox activation was determined by analyzing the monomerization degree of the protein in nonreductive SDS-PAGE for AGPase as in Hendriks et al. (2003), and for AOX using antibodies raised against thermogenic Sauromatum gutarum AOX obtained from Dr. Thomas Elthon (University of Nebraska, Lincoln). The size of the AOX monomer (approximately 35 kD) was consistent with previous studies (Umbach and Siedow, 1993). Enzyme activities were extracted and analyzed under nonreducing assay conditions for Glc6PDH as in Graeve et al. (1994), FBPase as in Leegood (1990), and NADP-MDH as in Ashton et al. (1990).
Analysis of Metabolic Fluxes
Leaf discs were incubated in the dark for 1 h at 20°C (humidity of 60%) in medium containing 2 mm MES-KOH (pH 6.5) and 0.66 mm or 0.33 mm [U-14C]Glc (specific activity, 111 kBq/mmol; Amersham-Buchler) without (control) or together with 5 mm DTT in petri dishes (5-mL volume). After incubation, leaf discs were immersed shortly in water to wash them from incubation medium and frozen in liquid nitrogen. Frozen material was extracted in two subsequent steps with 80% (v/v) ethanol (250 μL per 10 mg FW and 150 μL per 10 mg FW), mixed for 20 min at 80°C, centrifuged at 14,000 rpm for 5 min, the pellet reextracted in with 50% (v/v) ethanol (250 μL per 10 mg FW), the supernatants combined and transferred into a small tube, dried under an air stream at 45°C, and taken up in 1 mL water (soluble fraction). The soluble fraction was separated into sugars, anionic, and cationic fractions by ion exchange and thin-layer chromatography (Geigenberger et al., 1997). The insoluble pellet was analyzed to determine label in starch, protein, and cell walls as in Merlo et al. (1993). Label in the hexose phosphate pool was analyzed as in Geigenberger et al. (1997) and total carbon in the hexose phosphate pool was determined in ethanol extracts as in Gibon et al. (2002) using nonradioactive replicates incubated in parallel.
Metabolite Analysis
Metabolites were determined in derivatized methanol extracts by gas chromatography-mass spectrometry analysis as in Roessner et al. (2001a), except that the machine parameters and time reference standards were as in Roessner-Tunali et al. (2003). The values of each metabolite are normalized to those determined in wild type (Roessner et al., 2001b). Recoveries of metabolites through extraction to detection are documented in Roessner et al. (2000). The levels of PEP and pyruvate were determined as in Gibon et al. (2002).
Gene Expression Analysis with 22 K Affymetrix Arrays
Total RNA was isolated from frozen material using Trizol Reagent (Invitrogen) according to the manufacturer's protocol. Synthesis of cDNA, cRNA labeling, and hybridization on the Arabidopsis GeneChip ATH1 was performed at the Deutsches Ressourcenzentrum für Genomforschung (Berlin) using Affymetrix recommended protocols. Two independent biological replicate hybridizations were performed for all samples. Probe signal intensities were processed with the Affymetrix MicroArray Suite software package (MAS 5.0) and the resulting CEL files imported in the Bioconductor software package in R (Gentleman et al., 2004) for data quality control. Subsequent normalization of the raw data and estimation of signal intensities was done using the Robust Multichip Average methodology (Bolstad et al., 2003). Average expression values and their P values are calculated using the affylmGUI package in R (Irizarry et al., 2003). Affymetrix probe set identifiers were assigned to the Arabidopsis TIGR5 gene index database as obtained from The Arabidopsis Information Resource (www.arabidopsis.org). Genes were classified in functional categories and visualized using MapMan, mapping file version 1.7.0 (http://gabi.rzpd.de/projects/MapMan; Thimm et al., 2004; Usadel et al., 2005). Plastidial and mitochondrial encoded transcripts were omitted from the visualization output. Coordinate gene expression was analyzed with the build-in Wilcoxon Rank Sum test with Benjamini-Hochberg P-value correction. Further data analysis and processing was done using Microsoft Excel and Access.
Raw data of the transcript profiles can be found in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo/) with the accession code GSE4021.
Supplementary Material
Acknowledgments
We thank Thomas Elthon for kindly providing the AOX antibody. We are grateful to Janneke Hendriks and Axel Tiessen for valuable discussions, to Oliver Bläsing and Matthew Hannah for their help with microarray analysis software, and to Justyna Michalska for help with AOX analysis.
This work was supported by the Deutsche Forschungsgemeinschaft, with the grants Ge 878/1–4 (to P.G.), SFB 429 TP–B7 (to P.G. and A.K.), and SFB 429 TP–A11 (to P.G., J.T.v.D., and A.R.F.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Peter Geigenberger (geigenberger@mpimp-golm.mpg.de).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.081208.
References
- Ashton AR, Burnell JN, Furbank RT, Jenkins CD, Hatch MD (1990) Enzymes of C4 photosynthesis. In PM Dey, JB Harborne, eds, Methods in Plant Biochemistry, Vol 3. Academic Press, San Diego, pp 39–72
- Baier M, Dietz KJ (2005) Chloroplasts as source and target of cellular redox regulation: a discussion on chloroplast redox signals in the context of plant physiology. J Exp Bot 56: 1449–1462 [DOI] [PubMed] [Google Scholar]
- Ball L, Accotto GP, Bechthold U, Creissen G, Funck D, Jimenez A, Kular B, Leyland N, Mejia-Carranza J, Reynolds H, et al (2004) Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16: 2448–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schürmann P, Buchanan BB (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc Natl Acad Sci USA 101: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Tanaka CK, Hurkman WJ, Gelhaye E, Rouhier N, Jaquot J-P, Manieri W, Schürmann P, Droux M, et al (2004) Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc Natl Acad Sci USA 101: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on bias and variance. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D (2005) Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437: 1179–1182 [DOI] [PubMed] [Google Scholar]
- Bowsher CG, Boulton EL, Rose J, Nayagam S, Emes MJ (1992) Reductant for glutamate synthase is generated by the oxidative pentose-phosphate pathway in nonphotosynthetic root plastids. Plant J 2: 893–898 [Google Scholar]
- Bowsher CG, Hucklesby DP, Emes MJ (1993) Induction of ferredoxin-NADP+ oxidoreductase and ferredoxin synthesis in pea root plastids during nitrate assimilation. Plant J 3: 463–467 [Google Scholar]
- Buchanan BB (1980) Role of light in the regulation of chloroplast enzymes. Annu Rev Plant Physiol 31: 341–374 [Google Scholar]
- Buchanan BB, Balmer Y (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Luan S (2005) Redox regulation in the chloroplast thylakoid lumen: a new frontier in photosynthesis research. J Exp Bot 56: 1439–1447 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Schürmann P, Wolosiuk RA, Jacquot J-P (1979) Thioredoxin and enzyme regulation. Trends Biochem Sci 7: 93–96 [Google Scholar]
- Bunik VI (2003) 2-oxo acid dehydrogenase complexes in redox regulation: role of the lipoate residues and thioredoxin. Eur J Biochem 270: 1036–1046 [DOI] [PubMed] [Google Scholar]
- Choi YA, Kim SG, Kwon YM (1999) The plastidic glutamine synthetase activity is directly modulated by means of redox change at two unique cysteine residues. Plant Sci 149: 175–182 [Google Scholar]
- Dietz KJ, Scheibe R (2004) Redox regulation: an introduction. Physiol Plant 120: 1–3 [DOI] [PubMed] [Google Scholar]
- Dutilleul C, Lelarge C, Prioul JL, De Paepe R, Foyer CH, Noctor G (2005) Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism. Plant Physiol 139: 64–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entus R, Poling M, Herrmann KM (2002) Redox regulation of Arabidopsis 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase. Plant Physiol 129: 1866–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettke J, Eckermann N, Poeste S, Pauly M, Tiessen A, Geigenberger P, Steup M (2006) Analysis of cytosolic heteroglycans from leaves of transgenic potato (Solanum tuberosum L.) plants that under- or overexpress the Pho 2 phosphorylase isozyme. Plant Cell Physiol 46: 1987–2004 [DOI] [PubMed] [Google Scholar]
- Fettke J, Eckermann N, Tiessen A, Geigenberger P, Steup M (2005) Identification, subcellular localisation and biochemical characterisation of water-soluble heteroglycans (SHG) in leaves of Arabidopsis thaliana L.: distinct SHG reside in the cytosol and in the apoplast. Plant J 43: 568–585 [DOI] [PubMed] [Google Scholar]
- Fey V, Wagner R, Bräutigam K, Pfannschmidt T (2005) Photosynthetic redox-control of nuclear gene expression. J Exp Bot 56: 1491–1498 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G (2005) Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Kolbe A, Tiessen A (2005) Redox regulation of carbon storage and partitioning in response to light and sugars. J Exp Bot 56: 1469–1479 [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Reimholz R, Geiger M, Merlo L, Canale V, Stitt M (1997) Regulation of sucrose and starch metabolism in potato tubers in response to short-term water deficit. Planta 201: 502–518 [Google Scholar]
- Geiger M, Stitt M, Geigenberger P (1998) Metabolism in potato tuber slices responds differently after addition of sucrose and glucose. Planta 206: 245–252 [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Vigeolas H, Tiessen A, Geigenberger P, Stitt M (2002) Sensitive and high throughput metabolite assays for inorganic pyrophosphate, ADPGlc, nucleotide phosphates, and glycolytic intermediates based on a novel enzymic cycling system. Plant J 30: 221–235 [DOI] [PubMed] [Google Scholar]
- Gomez LD, Noctor G, Knight MR, Foyer CH (2004) Regulation of calcium signalling and gene expression by glutathione. J Exp Bot 55: 1851–1859 [DOI] [PubMed] [Google Scholar]
- Graeve K, von Schaewen A, Scheibe R (1994) Purification, characterization, and cDNA sequence of glucose-6-phosphate dehydrogenase from potato (Solanum tuberosum L.). Plant J 5: 352–361 [DOI] [PubMed] [Google Scholar]
- Heineke D, Riens B, Grosse H, Hoferichter P, Peter U, Flügge U-I, Heldt HW (1991) Redox transfer across the inner chloroplast membrane. Plant Physiol 95: 1131–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geigenberger P (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Kolbe A, Tiessen A, Schluepmann H, Paul M, Ulrich S, Geigenberger P (2005) Trehalose-6-phosphate regulates starch synthesis via post-translational redox-activation of ADP-glucose pyrophosphorylase. Proc Natl Acad Sci USA 102: 11118–11123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurek I, Kawagoe Y, Deborah JW, Doblin M, Delmer D (2002) Dimerization of cotton fiber cellulose synthase catalytic subunits occurs via oxidation of the zinc-binding domains. Proc Natl Acad Sci USA 99: 11109–11114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood RC (1990) Enzymes of the Calvin cycle. In PM Dey, JB Harborne, eds, Methods in Plant Biochemistry, Vol 3. Academic Press, San Diego, pp 15–37
- Lemaire SD, Guillon B, Le Marechal P, Keryer E, Miginia-Maslow M, Decottignies P (2004) New thioredoxin targets in the unicellular photosynthetic eukaryote Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 101: 7475–7480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Feil R, Hendriks JHM, Gibon Y, Morcuende R, Osuna D, Scheible WR, Carillo O, Hajirezaei MR, Stitt M (2006) Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J (in press) [DOI] [PMC free article] [PubMed]
- Marchand C, Le Marechal P, Meyer Y, Miginiac-Maslow M, Issakidis-Bourguet E, Decottignies P (2004) New targets of Arabidopsis thioredoxins revealed by proteomic analysis. Proteomics 4: 2696–2706 [DOI] [PubMed] [Google Scholar]
- Martinez IM, Chrispeels MJ (2003) Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo L, Geigenberger P, Hajirezaei M, Stitt M (1993) Changes of carbohydrates, metabolites and enzyme activities in potato tubers during development, and within a single tuber along a stolon-apex gradient. J Plant Physiol 142: 392–402 [Google Scholar]
- Mikkelsen R, Mutenda KE, Mant A, Schürmann P, Blennow A (2005) Alpha-glucan, water dikinase (GWD): a plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc Natl Acad Sci USA 102: 1785–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Krapp A, Hurry V, Stitt M (1998) Sucrose feeding leads to increased rates of nitrate assimilation, increased rates of oxoglutarate synthesis, and increased synthesis of a wide spectrum of amino acids in tobacco leaves. Planta 206: 394–409 [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci USA 98: 11224–11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Arisi ACM, Jouanin L, Foyer CH (1998) Manipulation of glutathione and amino acid biosynthesis in the chloroplast. Plant Physiol 118: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Dutilleul C, De Paepe R, Foyer CH (2004) Use of mitochondrial electron transport mutants to evaluate the effects of redox state on photosynthesis, stress tolerance and the integration of carbon/nitrogen metabolism. J Exp Bot 55: 49–57 [DOI] [PubMed] [Google Scholar]
- Roessner U, Luedemann A, Brust D, Fiehn O, Linke T, Willmitzer L, Fernie AR (2001. a) Metabolic profiling allows comprehensive phenotyping of genetically and environmentally modified plant systems. Plant Cell 13: 11–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U, Wagner C, Kopka J, Trethewey RN, Willmitzer L (2000) Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Roessner U, Willmitzer L, Fernie AR (2001. b) High-resolution metabolic phenotyping of genetically and metabolically diverse potato tuber systems: identification of phenocopies. Plant Physiol 127: 749–764 [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali U, Ubanczyk-Wochniak E, Czechowski T, Kolbe A, Willmitzer L, Fernie AR (2003) De novo amino acid biosynthesis is regulated by sucrose in potato tubers. Plant Physiol 133: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibe R (1990) Redox-modulation of chloroplast enzymes: a common principle for individual control. Plant Physiol 96: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann P, Jacquot J-P (2000) Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol 51: 371–400 [DOI] [PubMed] [Google Scholar]
- Sies H (1982) Nicotinamide nucleotide compartmentation. In H Sies, ed, Metabolic Compartmentation. Academic Press, London, pp 205–231
- Siess EA, Brocks DG, Wieland OH (1982) Subcellular distribution of adenine nucleotides and of metabolites of the tricarboxylate cycle and gluconeogenesis in hepatocytes. In H Sies, ed, Metabolic Compartmentation. Academic Press, London, pp 235–257
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tiessen A, Hendriks JHM, Stitt M, Branscheid A, Gibon Y, Farré EM, Geigenberger P (2002) Starch synthesis in potato tubers is regulated by post-translational redox-modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14: 2191–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiessen A, Prescha K, Branscheid A, Palacios N, McKibbin R, Halford NG, Geigenberger P (2003) Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J 35: 490–500 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN (1993) Covalent and non-covalent dimers of the cyanide resistant alternative oxidase protein in higher plant mitochondria and their relationship to enzyme activity. Plant Physiol 103: 845–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L, Yip JYH (1998) Molecular localization of a redox-modulated process regulating plant mitochondrial electron transport. Plant Cell 210: 1551–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Yip JYH, Parsons HL (1999) In organello and in vivo evidence of the importance of the regulatory sulfhydryl/disulfide system and pyruvate for alternative oxidase activity in tobacco. Plant Physiol 121: 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Stedingk EM, Pavlov PF, Grinkevich VA, Glaser E (1997) Mitochondrial protein import: modification of sulfhydryl groups of the inner mitochondrial membrane import machinery in Solanum tuberosum inhibits protein import. Plant Mol Biol 35: 809–820 [DOI] [PubMed] [Google Scholar]
- Wong JH, Cai N, Balmer Y, Tanaka CK, Vensel WH, Hurkman WJ, Buchanan BB (2004) Thioredoxin targets of developing wheat seeds identified by complementary proteomic approaches. Phytochemistry 65: 1629–1640 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.