Abstract
Ethylene receptors are multispanning membrane proteins that negatively regulate ethylene responses via the formation of a signaling complex with downstream elements. To better understand their biochemical functions, we investigated the membrane topology and subcellular localization of CmERS1, a melon (Cucumis melo) ethylene receptor that has three putative transmembrane domains at the N terminus. Analyses using membrane fractionation and green fluorescent protein imaging approaches indicate that CmERS1 is predominantly associated with the endoplasmic reticulum (ER) membrane. Detergent treatments of melon microsomes showed that the receptor protein is integrally bound to the ER membrane. A protease protection assay and N-glycosylation analysis were used to determine membrane topology. The results indicate that CmERS1 spans the membrane three times, with its N terminus facing the luminal space and the large C-terminal portion lying on the cytosolic side of the ER membrane. This orientation provides a platform for interaction with the cytosolic signaling elements. The three N-terminal transmembrane segments were found to function as topogenic sequences to determine the final topology. High conservation of these topogenic sequences in all ethylene receptor homologs identified thus far suggests that these proteins may share the same membrane topology.
The gaseous hormone ethylene regulates a number of growth and developmental processes in plants, including seed germination, flower senescence, leaf abscission, fruit ripening, sex determination, transition from the vegetative to the reproductive phase, and responses to some environmental signals, such as pathogen attacks, flooding, and drought (Abeles et al., 1992; Ogawara et al., 2003). Ethylene perception and signal transduction involve a multistep pathway (Guo and Ecker, 2004). Signaling is initiated by the binding of ethylene to a family of receptors that are similar to bacterial two-component His kinase (HisKA). Upon ethylene binding, ethylene receptors deactivate downstream CTR1, a Raf-like Ser-Thr kinase that is a negative regulator of the pathway. EIN2 acts downstream from CTR1 to subsequently activate a cascade of transcription factors, including EIN3/EIL, ethylene-responsive element-binding protein families, and ERF1, which then modulate the expression of ethylene-induced genes. In this pathway, ethylene receptors act at the first step and play a crucial role by negatively regulating ethylene responses (Chang and Stadler, 2001). In this respect, ethylene receptors have received considerable attention because knowledge about this system would provide important contributions to our understanding of the mechanisms of ethylene signaling and also the means to develop effective approaches for engineering ethylene sensitivity in plants.
Ethylene receptors are encoded by a small gene family. In Arabidopsis (Arabidopsis thaliana) for example, five members (ETR1, ETR2, ERS1, ERS2, and EIN4) have been identified (Bleecker et al., 1998). All receptor proteins have a similar structural organization: an N-terminal hydrophobic domain that contains three transmembrane (TM) segments in subfamily I (ETR1 and ERS1) and four in subfamily II (ETR2, ERS2, and EIN4), a GAF domain in the middle portion, followed by a HisKA domain. In addition to these domains, ETR-type receptors contain a receiver domain at the C terminus (Bleecker et al., 1998). Ethylene binds to its receptors at a site located within the N-terminal TM domain (Schaller and Bleecker, 1995; Malley et al., 2005). High-affinity binding requires a copper ion as a cofactor (Rodriguez et al., 1999), which is probably delivered by the copper transporter RAN1 (Hirayama et al., 1999; Woeste and Kieber, 2000). Ethylene receptors operate as membrane-associated disulfide-linked homodimers (Schaller et al., 1995; Takahashi et al., 2002). Unbound receptors are thought to be in an active state, which represses ethylene responses through direct activation of the kinase activity of CTR1 (Clark et al., 1998; Hua and Meyerowitz, 1998). Binding of ethylene results in deactivation of the receptors and thereby causes a conformational change in CTR1 that reduces its activity (Gao et al., 2003; Huang et al., 2003).
Despite our current knowledge of the mode of action of ethylene receptors, the biochemical mechanisms related to these events remain largely unknown. For example, how ethylene binding regulates the activity of receptors and what type of signal output passes to the downstream elements remain unclear (Guo and Ecker, 2004). Further characterization of ethylene receptors is required for better understanding of their biochemical functions. Thus, we examined the subcellular localization and membrane topology of ethylene receptors, which are important determinants of the biochemical functions of multispanning membrane proteins (van Geest and Lolkema, 2000).
Ethylene receptors have long been believed to be located at the plasma membrane (PM). However, a recent study indicated that AtETR1 is localized at the endoplasmic reticulum (ER; Chen et al., 2002). Additionally, CTR1 was also found localized at the ER through interaction with ethylene receptors (Gao et al., 2003). These findings imply a central role of the ER in ethylene perception and early signal transduction (Gao et al., 2003). However, whether the other receptor isoforms are localized at this organelle remains unknown. Using computer-based prediction, AtETR1 is thought to be located in the membrane with its N terminus facing the extracytosolic side and its C-terminal domain facing the cytosolic side (Chang et al., 1993; Schaller et al., 1995). Although the finding that the dimerization of AtETR1 is mediated by disulfide linkages at Cys-4 and Cys-6 residues is indicative of an extracytosolic orientation of the N terminus, as indicated by the above model (Schaller et al., 1995), the overall membrane topology has not been clearly demonstrated so far.
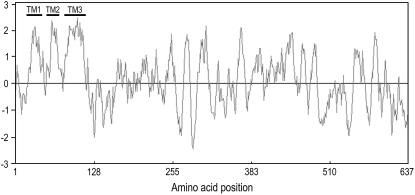
Previously, we identified an ethylene receptor CmERS1 from melon (Cucumis melo; Sato-Nara et al., 1999; Takahashi et al., 2002). This receptor accumulates in melon fruit during enlargement at both message and protein levels. Hydropathy analysis identified three TM domains close to the N terminus (Fig. 1), which are indicative of a membrane-localized protein. In agreement with this observation, CmERS1 is also associated with melon microsomes and forms disulfide-linked homodimers (Takahashi et al., 2002). Here, our data show that CmERS1 is an integral ER membrane protein that spans the lipid bilayer three times, with its N terminus facing the luminal space, and the large C-terminal portion, including the GAF and HisKA domains, on the cytosolic side. High conservation of the topogenic sequences in all ethylene receptor homologs identified thus far suggests that these proteins may share the same membrane topology.
Figure 1.
Hydropathy plot of CmERS1 indicates three N-terminal TM domains. A Kyte-Doolittle plot was generated with a window size of 11 amino acids using the GENETYX-MAC (version 11.0) program. Hydrophobic and hydrophilic regions are indicated above and below the zero line, respectively.
RESULTS
CmERS1 Is Associated with the ER Membrane of Melon Cells
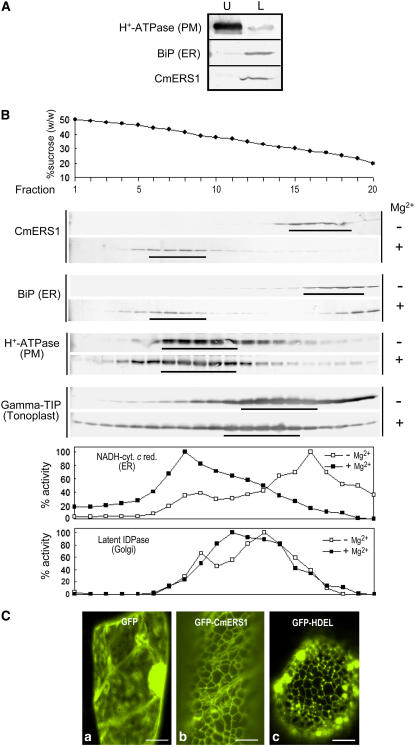
As a first strategy to determine the subcellular localization of CmERS1, we used two-phase partitioning to separate the PM from intracellular membranes according to the surface properties of the membrane vesicles (Larsson et al., 1987). Melon microsomes were subjected to aqueous two-phase partitioning using a 6.4% (w/w) Dextran T500/PEG3350 phase system. Following partitioning, both phases were analyzed by immunoblotting to determine the distributions of CmERS1 and membrane markers (Fig. 2A). As expected, the PM marker H+-ATPase (95 kD) was enriched in the upper phase and the ER marker luminal BiP (78 kD) was located in the lower phase, confirming the efficiency of partitioning. In this condition, CmERS1 (67 kD) was mainly detected in the lower phase, indicating that CmERS1 is not associated with the PM.
Figure 2.
CmERS1 is associated with the ER membrane of melon cells. A, Two-phase separation of melon leaf microsomes. Aqueous two-phase partitioning was performed using a Dextran T500/PEG3350 phase system. Equal amounts of proteins (5–15 μg) from the upper (U) and lower (L) phases were separated using SDS-PAGE and subjected to immunoblot analysis with antibodies specific for H+-ATPase (PM marker; Kinoshita and Shimazaki, 1999), BiP (ER marker), and CmERS1. B, Suc gradient fractionation. Microsomes from melon seedlings were separated on a linear Suc density gradient (20%–50%; w/w) in the absence (−) or presence (+) of Mg2+. Gradient fractions (25 μL) were subjected to SDS-PAGE and analyzed by immunoblotting with antibodies specific for CmERS1, H+-ATPase, BiP, and γ-TIP (tonoplast marker; Suga and Maeshima, 2004). NADH cytochrome c reductase (NADH-cyt c red) and latent IDPase were used as enzymatic markers for the ER and Golgi, respectively. C, GFP imaging of the subcellular localization of CmERS1. GFP fusion constructs were transiently expressed in melon leaf cells by microprojectile bombardment. GFP fluorescence was detected using confocal microscopy. The images were taken from trichomes or leaf vein epidermal cells. Only green channel images are shown here for simplicity. GFP, Negative control expressing only sGFP (S65T; Chiu et al., 1996); GFP-CmERS1, GFP fused with full-length CmERS1; GFP-HDEL, an ER-localized GFP (mGFP5-ER) that contains a C-terminal HDEL motif (Haseloff et al., 1997). Bar = 10 μm.
To identify the intracellular compartment in which CmERS1 is localized, melon microsomes were separated by Suc density gradient centrifugation in the presence or absence of Mg2+. Binding of ribosomes to the ER membrane is Mg2+ dependent; thus, chelation of Mg2+ with EDTA releases the attached ribosomes, lowering ER membrane density, but not that of other organelles (Lord, 1987). In the absence of Mg2+, CmERS1 was abundant in fractions containing 25% to 30% (w/w) Suc, which corresponded to the peak fractions of BiP and NADH-cytochrome c reductase markers for the ER (Fig. 2B). This sedimentation profile was distinguishable from that of the PM marker (H+-ATPase; 37% to 43%), the tonoplast marker (γ-tonoplast intrinsic protein [TIP]; 28% to 35%), and the Golgi marker (latent inosine diphosphatase [IDPase]; 31% to 35%). In the presence of Mg2+, CmERS1 migrated to a higher density, between 39% and 44% Suc, consistent with the Mg2+-induced shift as observed with the ER markers BiP and NADH-cytochrome c reductase. A small amount of BiP detected at the top of the gradient was likely released from the ER lumen during chopping. As controls, H+-ATPase, γ-TIP, and latent IDPase did not exhibit the same extent of shift. These results indicate that CmERS1 is predominantly associated with the ER membrane.
To visualize localization in living cells, we tagged CmERS1 with a green fluorescent protein (GFP) under the control of the cauliflower mosaic virus (CaMV)-35S promoter. The fusion protein GFP-CmERS1 was transiently expressed in melon leaf cells by microprojectile bombardment. GFP fluorescence was detected using confocal microscopy. In the control expressing GFP alone, the GFP signal was seen in the cytosol and nucleus (Fig. 2C, a). In contrast, in cells expressing the GFP-CmERS1 fusion protein, the GFP signal highlighted fluorescence along a reticular network (Fig. 2C, b), which was identical to the typical ER-specific pattern generated by GFP-HDEL (Fig. 2C, c), a well-characterized ER marker protein (Haseloff et al., 1997). A similar localization pattern was observed in all cell types examined, including trichomes, leaf vein epidermal cells, leaf epidermal cells, and stomatal guard cells (data not shown). These observations were consistent with data from membrane fractionation that showed membrane-associated CmERS1 in the ER. To exclude the possibility of mislocalization caused by overexpression that can result in misfolded membrane proteins being retained in the ER, we carried out a time-course analysis using diluted plasmid (Li and Chye, 2003). GFP fluorescence was examined at 2-h intervals and revealed that the fusion protein was consistently associated with the ER from 4 h when fluorescence began to be visualized (data not shown). Therefore, it appears that the images observed were not artifacts caused by overexpression and mislocalization.
CmERS1 Is an Integral Membrane Protein
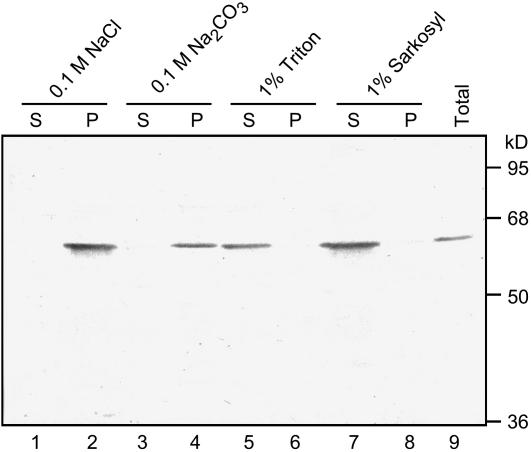
Thus far, we have demonstrated that CmERS1 is associated with the ER membrane. To further determine the nature of the membrane association, melon microsomes were subjected to various extractions to strip nonintegral proteins from the membrane or solubilize integral membrane proteins. Neither high salt nor alkali treatments, which are effective in extracting most peripherally associated proteins, were able to remove CmERS1 from the membranes (Fig. 3, lanes 1–4). In contrast, CmERS1 proteins were solubilized by treatment with the nonionic detergent Triton X-100 or the ionic detergent Sarkosyl, which disrupts the integrity of the lipid bilayer (lanes 5–8). These results indicate that CmERS1 is integrated into the membrane rather than peripherally associated with it.
Figure 3.
Detergent treatments of melon microsomal membranes demonstrate that CmERS1 is an integral membrane protein. Microsomes were prepared from melon leaves. The membrane pellet was resuspended in 0.1 m NaCl, 0.1 m Na2CO3 (pH 11), 1% (v/v) Triton X-100, or 1% (v/v) Sarkosyl. After incubation at 4°C for 1 h, the samples were separated into supernatant (S) and pellet (P) fractions by centrifugation at 125,000g for 1 h. Equal amounts of the proteins (15 μg) were subjected to western-blot analyses with anti-CmERS1 antibody. Numbers on the right indicate the molecular mass.
The Large C-Terminal Portion of CmERS1 Is Exposed in the Cytosol
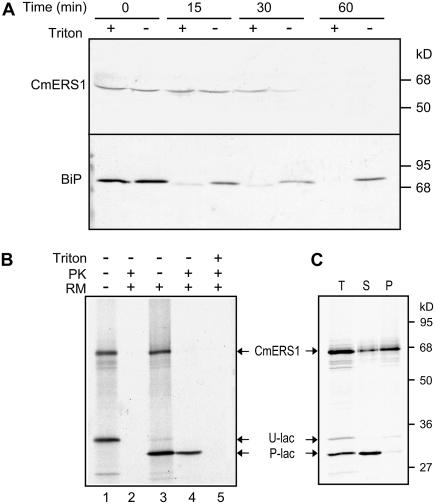
Although all programs that we used for predicting topology identified three N-terminal TM segments ranging from 18 to 28 amino acid residues, which could potentially span the membrane, the predicted membrane orientations were contradictory. For example, PHD 2.1 (Rost et al., 1996) and TMPRED (Hofmann and Stoffel, 1993) predicted an Nlum-Ccyt orientation, whereas HMMTOP 2.0 (Tusnady and Simon, 1998) and TOPPRED 2.0 (Claros and von Heijne, 1994) predicted an opposite orientation. To determine the actual membrane topology of CmERS1, melon microsomes were treated with proteinase K (PK) in the presence or absence of Triton X-100 and then analyzed by immunoblotting (Fig. 4A). In the control assay using BiP, the protein was resistant to protease digestion unless the microsomes were made permeable using detergent, suggesting that the microsomal membranes were fully intact. Using the same conditions, CmERS1 was found to be sensitive to protease digestion even in the absence of detergent. Because the anti-CmERS1 antibody was generated against the GAF domain (Takahashi et al., 2002), the protease sensitivity of CmERS1 detected using this antibody indicates that its large C-terminal portion, including the GAF and HisKA domains, is located in the cytosol.
Figure 4.
The C-terminal region of CmERS1 is located in the cytosol. A, Protease digestion of melon microsomes. Microsomes from melon fruits were treated with PK (100 μg mL−1) in the presence (+) or absence (−) of 0.1% (v/v) Triton X-100. The reactions were performed at 30°C for 0 to 60 min and terminated by adding phenylmethylsulfonyl fluoride to a final concentration of 1 mm. Treated proteins (20 μg) were separated by SDS-PAGE and immunoblotted with anti-CmERS1 or anti-BiP antibody. B, Protease digestion of in vitro translated CmERS1. CmERS1 cDNA was transcribed and translated in vitro in a TNT T7 quick-coupled transcription/translation system supplemented with (+) or without (−) RMs. The membrane pellet isolated from the translation mixture was treated with PK (200 μg mL−1) with (+) or without (−) 1% Triton X-100 (Triton). Samples were incubated on ice for 30 min and then separated by SDS-PAGE and analyzed using autoradiography. C, Membrane association of in vitro translated CmERS1. The membrane pellet isolated from the translation mixture (T) was extracted with 0.1 m Na2CO3 (pH 11), and then reseparated into supernatant (S) and membrane pellet (P) by ultracentrifugation. E. coli β-lactamase mRNA was cotranslated as a control. Bands corresponding to CmERS1, the unprocessed β-lactamase (U-lac), and the processed β-lactamase (P-lac) are indicated.
CmERS1 contains two potential N-glycosylation sites (N258LS and N544KT) in its deduced amino acid sequence. This allowed us to use a glycosylation assay as an alternative approach to assess its membrane topology. The addition of N-linked glycans occurs in the ER lumen; thus, glycosylation of the consensus sites would indicate a luminal orientation (van Geest and Lolkema, 2000). CmERS1 was transcribed and translated in vitro in a rabbit reticulocyte lysate system and processed using canine pancreatic microsomes (RM). When CmERS1 was synthesized in the absence of RM, a major band with the expected molecular mass was observed (Fig. 4B, lane 1). When synthesized in the presence of RM, an identical band was detected (lane 3). Because no signal peptide processing occurred upon membrane insertion, as indicated by the identical size of the TM1-2-3-GFP fusion protein synthesized in the absence or presence of microsomes (data not shown), the results suggest that the native glycosylation sites were not used in our cell-free system. Similar results were obtained using constructs that contained an exogenous site at three different positions (Q201S, I435N, S629N; data not shown). The lack of glycosylation of both native and engineered sites may be because the location of this region is cytosolic or because the protein structure prevents glycosylation. To determine whether the region is cytosolic, posttranslational digestion was performed using PK. To monitor the integrity of the RM, Escherichia coli β-lactamase mRNA (supplied in the kit) was cotranslated as an internal control. The processed β-lactamase, a soluble protein that is known to be translocated into the lumen, was resistant to protease digestion unless the RM was made permeable by the detergent (lanes 4–5), suggesting that the RMs were intact. Under the same conditions as the β-lactamase, CmERS1 was degraded by protease digestion even in the presence of RM (lanes 2 and 4), indicating the cytosolic location of this region. To ensure that the synthesized protein was indeed inserted into the heterogeneous microsomes, the pellet fraction that was isolated from the translation mixture was extracted using 0.1 m Na2CO3 (pH 11.0) and then reseparated into supernatant and pellet fractions by centrifugation. Most of the CmERS1 proteins cosedimented with the membrane, although a small amount of protein was extracted into the supernatant (Fig. 4C). Cotranslated β-lactamase was recovered in the supernatant, suggesting efficient extraction with alkali. These results indicate that in vitro synthesized CmERS1 was inserted into the heterogeneous microsomes, although the efficiency of membrane integration was not as high as that of native proteins (compare with Fig. 3, lanes 3 and 4). Taken together, data from the in vitro expression assay confirm the Ccyt orientation of CmERS1.
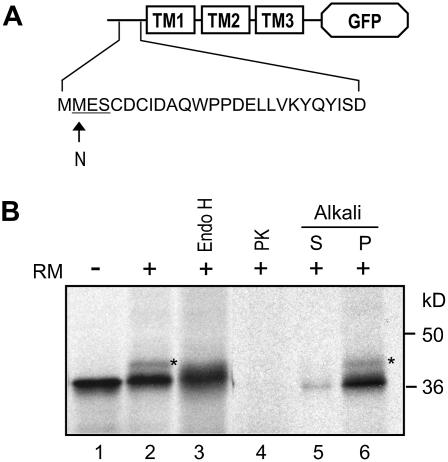
The N-Terminal Region of CmERS1 Is Located in the ER Lumen
To assess the N terminus orientation, the TM region of CmERS1 (amino acids 1–120) was fused to the N terminus of GFP, which was used as a reporter domain. A glycosylation site was then introduced into the extreme N terminus of this fusion construct by substituting Asn for Met-2; the resulting construct was named TM (M2N)-GFP (Fig. 5A). In vitro transcription/translation of the TM (M2N)-GFP construct in the presence of RM resulted in an additional band that was around 2.5 kD larger than that observed in the absence of RM (Fig. 5B, compare lanes 1 and 2). To test whether this increased molecular mass was caused by glycosylation, the protein was treated with endoglycosidase H (Endo H), a glycan-removing enzyme. Endo H treatment converted this protein into the unglycosylated form (Fig. 5B, lane 3), indicating that the M2N site was successfully glycosylated. In addition, the GFP domain was sensitive to protease digestion (Fig. 5B, lane 4), suggesting that the introduction of the glycosylation site did not affect the overall topology of the fusion protein. Alkali extraction showed that the fusion protein was fully integrated into the microsomes (Fig. 5B, lanes 5–6). These results indicate that the N terminus of CmERS1 is located on the luminal side of the membrane. Note that the relatively low glycosylation efficiency in this assay may be caused by the nearby disulfide linkage sites (Cys-5 and Cys-7), which may be involved in dimerization of the receptor (see “Discussion”).
Figure 5.
The N-terminal region of CmERS1 is located in the lumen. A, Schematic of TM (M2N)-GFP construct. An N-glycosylation site was introduced to the N terminus of the TM1-2-3-GFP fusion by substituting Asn for Met-2. B, The plasmid was transcribed and translated in vitro in the absence (−) or presence (+) of RM. Endo H treatment was performed at 37°C for 6 h. PK digestion and alkali extraction were performed as described in Figure 4. The glycosylated form is indicated by an asterisk.
Topogenic Function of the N-Terminal TM Segments
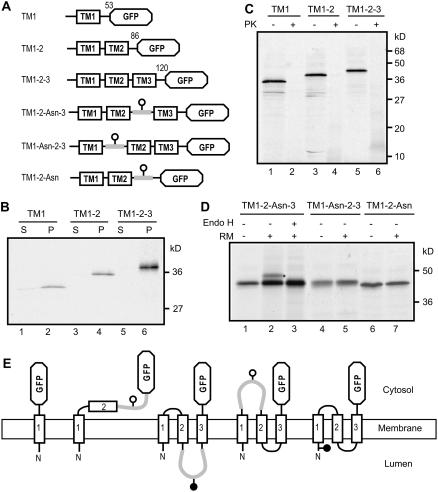
To further elucidate the membrane topogenesis of CmERS1, we assessed the topogenic functions of each TM segment. The integration of a multispanning protein into the membrane is proposed to occur sequentially from the N terminus, with TM segments showing alternative translocation initiation and stop-transfer functions (Blobel, 1980; Friedlander and Blobel, 1985; Wessels and Spiess, 1988). According to this sequential insertion model, to achieve the final Nlum-Ccyt topology of CmERS1, TM1 should function as a signal-anchor (SA)-I sequence responsible for ER targeting and translocation of its preceding N-terminal portion. TM2 would then function as an internal SA-II sequence responsible for reinitiating translocation of the following portion, and TM3 would function as a stop-translocation sequence to interrupt the ongoing transfer initiated by TM2, thereby leaving the C-terminal portion on the cytosolic side of the ER membrane.
To verify these topogenic functions, we constructed a series of GFP fusions containing one, two, or three TM segments, which we called TM1, TM1-2, and TM1-2-3, respectively (Fig. 6A). As a first step, membrane integration of the in vitro synthesized proteins was assayed using alkali extraction. The products were efficiently inserted into the membrane, which was indicated by the fact that the majority of the proteins cosedimented with the membrane (Fig. 6B, lanes 2, 4, and 6). A minor amount of TM1 protein was extracted into the soluble fraction (Fig. 6B, lane 1), possibly because of the saturation of the microsomes by this protein. Next, a protease protection assay was used to monitor the membrane orientation of the products. We expected that TM1 and TM1-2-3 would be sensitive to protease digestion, whereas TM1-2 would be resistant. TM1 and TM1-2-3 were degraded by treatment with protease as expected (Fig. 6C, lanes 2 and 6). However, TM1-2 also exhibited protease sensitivity (Fig. 6C, lane 4). We examined higher dosages and a different lot of RM, and β-lactamase was parallel translated to monitor the integrity of the RM, but we failed to produce the protected fragments. To examine whether the structure of the GFP domain prevented translocation, we used the original context of CmERS1 with deletion of TM3, but obtained a similar result (data not shown). To confirm the results of a protease protection assay, we introduced a glycosylation site between the TM2 and GFP domain and named this construct TM1-2-Asn (Fig. 6A). TM1-2-Asn was not glycosylated (Fig. 6D, lanes 6 and 7). This is consistent with the Ccyt orientation as determined by proteolysis analysis. These observations imply that TM2 did not reinitiate translocation of the following portion of the protein. This unexpected finding raises the possibility that either only TM1 or all three TMs are inserted into the membrane in the context of TM1-2-3. To clarify this, we introduced a glycosylation site in loop 1 (Fig. 6A, TM1-Asn-2-3) or loop 2 (TM1-2-Asn-3). If the three TMs are inserted into the membrane, loop 1 is cytosolic and thus the site is not glycosylated, whereas loop 2 should be translocated into the lumen, and thus the site can be glycosylated. TM1-Asn-2-3 was not glycosylated, as expected (Fig. 6D, lanes 4–5). TM1-2-Asn-3 was glycosylated and the glycosylation was confirmed by treatment with Endo H (lanes 1–3). These results indicate that loop2 was indeed translocated into the lumen. Consequently, it is reasonable to conclude that all three TMs were inserted into the membrane in the context of TM1-2-3 (Fig. 6E).
Figure 6.
Topogenic functions of the N-terminal TM segments of CmERS1. A, Scheme of constructs used in this experiment. The numbers indicate the amino acid positions of CmERS1 at the fusion points. In the TM1-2-Asn-3, TM1-Asn-2-3, and TM1-2-Asn constructs, an N-glycosylation site was introduced into the loops and indicated by a white circle. B, Membrane association of in vitro translated fusion proteins. The constructs were transcribed and translated in vitro in the presence of RM. Alkali treatment was performed as described in Figure 4. S, Supernatant; P, membrane pellet. C, Protease digestion of in vitro translated fusion proteins. Microsomes isolated from the translation mixtures were treated with PK as described in Figure 4. D, Glycosylation analysis. The constructs were transcribed and translated in vitro in the absence (−) or presence (+) of RM. Endo H treatment was performed as described in Figure 5. The glycosylated form is indicated by an asterisk. E, Schematic for the possible orientation of each chimera on the membrane. TM1 was inserted into the membrane in an Nlum-Ccyt orientation. TM2 was not inserted into the membrane and the GFP domain remained in the cytosol. TM3 was inserted into the membrane in the same orientation as that of TM1. In the presence of TM3, TM2 was inserted into the membrane and loop 2 was translocated.
DISCUSSION
Because ethylene receptors are multispanning membrane proteins, both the membrane topology and subcellular localization are important determinants of their biochemical function. However, our knowledge of these essential aspects is still limited for these receptors. Here we addressed these cell biological properties for the melon ethylene receptor CmERS1 to provide clues for a better understanding of the biochemical mechanisms of ethylene receptor action in plants, particularly in economically important crop species.
A recent study has shown that AtETR1 is localized to the ER rather than to the PM, as originally assumed, using membrane fractionation and immunoelectron microscopy approaches (Chen et al., 2002). Additionally, the downstream CTR1, a protein that has no obvious TM domains or membrane attachment motifs, was also associated with the ER membrane and this localization was a consequence of direct interaction with the ER-localized receptor (Gao et al., 2003). These findings imply that the ER may play a central role in ethylene perception and early signal transduction (Chen et al., 2002; Gao et al., 2003; Guo and Ecker, 2004). In agreement with this deduction, our results from membrane fractionation and GFP imaging analyses indicate that CmERS1 is predominantly associated with the ER membrane (Fig. 2). This observation provides experimental evidence for ER localization of an ERS-type receptor. Such an endomembrane location is consistent with the hydrophobic nature of ethylene, which enables it to pass freely through the PM into the cell (Kieber, 2002). Mechanisms for the localization of membrane proteins in plants remain largely unknown. CmERS1 has none of the known ER retention signals, such as the cytosolic di-lysine motif KKXX-COOH. Because the determinant for ER localization of AtETR1 was found in its N-terminal TM region (Chen et al., 2002) and because this region is highly conserved in CmERS1, it is likely that localization of CmERS1 to the ER involves a similar TM-mediated mechanism.
Our results provide direct experimental evidence for the membrane topology of the ethylene receptor proposed using computer-based prediction (Chang et al., 1993; Schaller et al., 1995). Ethylene receptors function as negative regulators of ethylene responses via the formation of a signaling complex with downstream CTR1 (Clark et al., 1998; Hua and Meyerowitz, 1998; Gao et al., 2003; Huang et al., 2003). Protein-protein interaction has been detected between the CTR1 N-terminal domain and the HisKA domain of ethylene receptors (Clark et al., 1998). Because CTR1 appears to be a soluble protein that has no obvious ER-targeting signal, these protein interactions should occur on the cytosolic side of the membrane. The membrane topology of CmERS1 established in our study indicates that the large C-terminal portion, including the GAF and HisKA domains, is indeed exposed in the cytosol (Fig. 4). This allows the receptor to interact with cytosolic CTR1. In addition, the HisKA activity of ethylene receptors has been demonstrated in yeast (Saccharomyces cerevisiae; Gamble et al., 1998); however, its function in ethylene signaling in plants is not clear (Wang et al., 2003). The GAF domain, which is involved in cGMP binding or light regulation in other proteins (Aravind and Ponting, 1997), is an unknown function domain in ethylene receptors. The topology of CmERS1 identified here will be useful for future structure-function studies of these domains.
We used an N-glycosylation mutagenesis strategy to determine the precise membrane orientation of the short hydrophilic N-terminal region because this method introduces minimal structural alterations in the wild-type protein and has proven successful in defining the folding pattern of various membrane proteins (Devoto et al., 1999; van Geest and Lolkema, 2000). Our data showed that the N terminus of CmERS1 is located in the ER lumen, as indicated by glycosylation of the engineered site (Fig. 5). A luminal orientation of the N terminus is consistent with the previous finding that CmERS1 formed homodimer links, possibly by disulfide bonds at the Cys-5 and Cys-7 residues (Takahashi et al., 2002) because formation of disulfide bonds is an ER luminal occurrence. Because both the ETR- (Chen et al., 2002) and ERS-type receptors localize to the ER and most likely have similar N-terminal orientations, the possibility of heterodimer formation will be an interesting subject in the study of ethylene receptor activity.
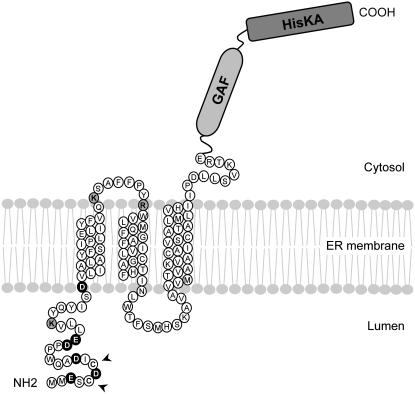
Membrane topology is determined by interactions between topogenic signals and the insertion machinery (van Geest and Lolkema, 2000). In addition to the hydrophobicity of TM segments, the charge distribution in TM-flanking regions appears to be an important determinant of membrane orientation, where positive charges promote cytosolic retention and negative charges promote translocation into the ER lumen; this is commonly referred to as the charge difference rule (Hartmann et al., 1989; Parks and Lamb, 1991; Harley et al., 1998). CmERS1 has a net negative N-terminal charge (Fig. 7). Therefore, it is likely that TM1 functions as a SA-I sequence conferring an Nlum-Ccyt orientation. This prediction is supported by our results, which show that upon insertion into the membrane, the reporter domain of TM1-GFP fusion protein remains on the cytosolic side (Fig. 6). TM2 did not exhibit the predicted translocation initiation function in our cell-free system. We also tried to evaluate it in vivo by introducing the TM1-2-GFP construct into tobacco (Nicotiana tabacum) plants, but failed to get expression of the protein due to unidentified factors. However, despite this, our results indicate that TM2 was inserted into the membrane in the presence of TM3, which functions as a SA-I sequence (Fig. 6). This forced TM orientation has been reported in various multispanning membrane proteins and the reason for this is the weak topogenic function and low hydrophobicity of the corresponding TM segments (Zhang, 1996; Ota et al., 1998a, 1999b; Ukaji et al., 2002). The same appears to be true for CmERS1. Taken together, our data suggest the following membrane topogenesis of CmERS1: TM1 functions as an SA-I sequence that is responsible for ER targeting and translocation of the N terminus, TM2 integrates into the membrane as a consequence of the internal SA-I function of the following TM3, which leaves the C-terminal portion on the cytosolic side.
Figure 7.
A proposed model for CmERS1 topology on the ER membrane. Based on membrane fractionation, GFP imaging, protease protection assay, and glycosylation analysis, we propose that CmERS1 is predominantly localized to the ER and spans the membrane three times with its N terminus facing the luminal space and its C terminus lying on the cytosolic side. The positively and negatively charged residues in TM1-flanking regions are shaded gray and black, respectively. Arrows indicate the residues that are possibly involved in disulfide-linked dimerization of the receptor. The TM helices were identified by PHD 2.1 (Rost et al., 1996).
Based on our observations, we propose a model for the membrane topology of CmERS1 (Fig. 7). Because the topogenic sequences identified for CmERS1 (i.e. hydrophobic segments and charge distribution) are highly conserved in all ethylene receptor homologs known to date, it is reasonable to propose that this model may be extended to all receptor members. It should be noted that, although subfamily II members contain four TM segments, because the first one is predicted to be a cleavable signal peptide, the mature forms should also follow this three TM topology model.
MATERIALS AND METHODS
Plasmid Construction
The GFP plasmid, a pUC18-based vector containing CaMV-35S-sGFP (S65T)-NOS3 (Chiu et al., 1996), was kindly provided by Dr. Yasuo Niwa (University of Shizuoka, Japan). To construct the GFP-CmERS1 fusion gene, the CmERS1 coding sequence (accession no. AF037368) was cloned into the GFP plasmid between the XbaI and NotI restriction sites with substitution of the GFP fragment. Because the CaMV-35S promoter was also removed during this cloning procedure because of its flanking XbaI sites, the promoter was reinserted into the vector between the Sse 8387 I and XbaI sites. The GFP coding region was amplified using PCR to remove the stop codon and to introduce XbaI-XbaI sites. The amplified GFP fragments were then digested and cloned into the generated plasmid to fuse in frame with the N terminus of CmERS1 cDNA. To construct the TM1, TM1-2, and TM1-2-3 plasmids for the cell-free expression system, the corresponding TM coding sequences were amplified using PCR to introduce XbaI-BamHI restriction sites, and then cloned into the GFP plasmid to fuse it in frame with the N terminus of GFP. The resulting plasmids were digested with XbaI and NotI restriction enzymes, and the released TM-GFP segments were ligated into the pTNT vector (Promega). Site-directed mutagenesis was used to introduce glycosylation sites (M2N, Q201S, I435N, and S629N) into the CmERS1 protein. To introduce a glycosylation site in loop 1 or loop 2 (TM1-Asn-2-3, TM1-2-Asn-3, and TM1-2-Asn), the strategy described by Campos and Boronat (1995) was used to meet the space requirements for N-glycosylation (van Geest and Lolkema, 2000). PstI-SphI restriction sites were introduced by PCR into the loop 1 coding sequence between nucleotides 148 and 149 or the loop 2 coding sequence between nucleotides 231 and 232. The DNA fragment of Zm-ERabp1 (Hesse et al., 1989), which codes for the glycosylation site and its flanking region (SLKYPGQPQEIPFFQNTTFSIPVNDPHQVWNS; the glycosylation site is underlined) was isolated from maize (Zea mays) using reverse transcription PCR and cloned into the corresponding plasmids between the PstI and SphI restriction sites. All PCR-amplified fragments were confirmed using dye terminator cycle sequencing.
Two-Phase Partitioning
Aqueous two-phase partitioning was performed as described by Larsson et al. (1987), with minor modifications. Melon (Cucumis melo L. cv vedrantais) leaves (60 g) were cut into pieces and homogenized in 150 mL of lysis buffer (50 mm HEPES-KOH, pH 7.5; 0.5 m Suc; 5 mm ascorbic acid; 1 mm dithiothreitol [DTT]; and 0.6% [w/v] polyvinylpyrrolidone) using a mortar and pestle. The homogenate was filtered through two layers of miracloth (Calbiochem) and centrifuged at 10,000g for 10 min to remove chloroplasts and mitochondria. The supernatant was recovered and centrifuged at 100,000g for 30 min at 4°C (type 70.1 rotor; Beckman) to pellet the microsomal membranes. The microsomal pellet was resuspended in 5 mm potassium phosphate buffer (pH 7.8) containing 0.33 m Suc and 3 mm KCl, and then subjected to five cycles of two-phase partitioning using a 6.4% (w/w) Dextran T500/PEG3350 mixture. The resulting upper and lower phases were diluted and centrifuged at 100,000g for 1 h at 4°C. The pellet was solubilized in potassium phosphate buffer.
Sucrose Gradient Fractionation
Suc density gradient centrifugation was performed essentially as described by Ferrol and Bennett (1996). Etiolated seedlings grown at 30°C for 3 d were chopped under 2 volumes of homogenization buffer (50 mm Tris-HCl, pH 8.0; 150 mm NaCl; 5 mm EDTA; 13% [w/v] Suc; 10% (w/v) polyvinylpolypyrrolidone; and 1 mm DTT) using a razor blade and then homogenized using a mortar and pestle. In the presence of Mg2+, the homogenization buffer contained 2 mm EDTA and 7 mm MgCl2. The homogenates were filtered through two layers of miracloth and centrifuged at 12,000g for 15 min at 4°C. The supernatant was layered onto a 58% (w/w) Suc cushion in the homogenization buffer and then centrifuged at 24,000 rpm for 30 min at 4°C (SW28 rotor; Beckman). Microsomes were collected from the interface and diluted with 2 volumes of homogenization buffer without Suc. Diluted microsomes were layered on to a 20-mL linear Suc gradient (20% to 50%, w/w) in a centrifugation buffer (10 mm Tris-HCl, pH 7.5; 2 mm EDTA; and 1 mm DTT). In the presence of Mg2+, 7 mm MgCl2 were added to the centrifugation buffer. The gradient was centrifuged at 24,000 rpm for 12 h at 4°C and 1.0-mL fractions were collected. The Suc concentration of each fraction was measured using a refractometer. Antimycin A-insensitive NADH cytochrome c reductase (Bowles and Kauss, 1976) and Triton-stimulated IDPase (Goubet and Mohnen, 1999) were assayed as markers for the ER and Golgi, respectively. All other membrane markers were detected by immunoblotting.
Microprojectile Bombardment and Confocal Microscopy
Young leaves were cut from melon plants and placed in petri dishes containing solid Murashige and Skoog medium. Leaf samples were then bombarded with gold particles (1 μm) that were coated with 10 μg of plasmid DNA at a distance of 9 cm, using a Bio-Rad PDS-1000/He particle delivery system at 900 psi of He pressure under a vacuum of 700 mm Hg. After bombardment, the melon petioles were placed in the medium and the samples were incubated at 25°C in the dark. After 20 to 24 h, the melon leaves were examined for GFP fluorescence using epifluorescence microscopy and confocal laser-scanning microscopy. At least three independent transient expression experiments were performed for each construct.
For confocal microscopy, leaf pieces showing GFP fluorescence were sliced with a razor blade and mounted in water between a slide and coverslip. The images were taken from trichomes and leaf vein epidermal cells using a confocal microscope (Leica TCS SP2; Leica Microsystems). Excitation wavelengths were set at 488 and 568 nm and images were collected through a fluorescein isothiocyanate filter for GFP fluorescence (green channel) and through a tetramethylrhodamine B isothiocyanate filter for chlorophyll fluorescence (red channel). All images were collected through a water-immersion objective (×63, 1.2 numerical aperture). Optical sections were 0.10-μm thick. Final image assembly was performed using Adobe Photoshop 6.0 software.
Detergent Treatments of Melon Microsomes and Proteolysis
Preparation of microsomes from melon fruits (15 d after pollination) was performed as described by Goubet and Mohnen (1999). Aliquots of membrane pellets were resuspended in 0.1 m NaCl, 0.1 m Na2CO3 (pH 11), 1% (v/v) Triton X-100, or 1% (v/v) Sarkosyl. After incubation at 4°C for 1 h, the samples were centrifuged at 125,000g for 1.5 h at 4°C for separation into pellet and supernatant fractions.
For proteolysis, the microsomes (150 μg of proteins) were solubilized in 50 mm Tris-HCl (pH 6.8) containing 0.4 m Suc and incubated with 100 μg mL−1 of PK in a final volume of 100 μL. The incubation was conducted for 0 to 60 min at 30°C in the absence or presence of 0.1% (v/v) Triton X-100. In the presence of the detergent, a preincubation (15 min, 4°C) was performed to make matrix-luminal protein protease accessible. All digestion was terminated by adding phenylmethylsulfonyl fluoride (Sigma) to a final concentration of 1 mm.
Immunoblot Analysis
SDS-PAGE and immunoblotting were performed as described previously (Takahashi et al., 2002). Briefly, protein samples were mixed with one-third volume of 4× sample buffer and incubated at 70°C for 10 min. Proteins were separated using SDS-PAGE on 10% or 12% gels. The primary antibodies used were as follows: rabbit anti-melon CmERS1 (1:1,000) generated against amino acid residues 117 to 327 (Takahashi et al., 2002), rabbit anti-Vicia faba PM H+-ATPase (anti-VHA antibody; PM marker; 1:1,000; kindly provided by Dr. Ken-ichiro Shimazaki, Kyushu University, Japan), rabbit anti-γ-TIP (tonoplast marker; 1:2,000; kindly provided by Dr. Masayoshi Maeshima, Nagoya University, Japan), and mouse monoclonal antibody against plant-specific BiP (ER marker; 1:2,000; Stressgen Biotechnologies). Incubation with the primary antibodies was performed either overnight at 4°C for anti-BiP antibody or for 1 h at room temperature for the other antibodies. As a secondary antibody, monoclonal anti-rabbit or anti-mouse IgG-horseradish peroxidase (Santa Cruz Biotechnology) was used at 1:10,000 dilution. Signals were detected using a peroxidase stain kit (Nacalai Tesque) according to the manufacturer's instructions.
In Vitro Transcription/Translation
The plasmids were transcribed and translated in a TNT T7 quick-coupled transcription/translation system (Promega) in the presence of [35S]Met (Amersham; AG1094) according to the manufacturer's instructions. Reactions were incubated at 30°C for 90 min in the presence or absence of RMs (Promega). Isolation of microsomal membranes from the translation mixture was performed as described by McCartney et al. (2004). For PK treatments, the pellet fraction was resuspended in 10 mm Tris-HCl (pH 7.5). PK was added to a final concentration of 200 μg mL−1. The samples were incubated on ice for 30 min in the presence of 10 mm CaCl2 to preserve membrane integrity (Devoto et al., 1999). Endo H treatments were performed as described by Vilar et al. (2002). The samples were incubated with 50 milliunits mL−1 of Endo H for 6 h at 37°C. As a control for glycosylation, the α-factor from yeast (Saccharomyces cerevisiae) that is supplied in the kit was parallel translated in the corresponding experiments. Alkali extraction experiments were performed as described by McCartney et al. (2004). Briefly, the microsomal membranes obtained above were dissolved in 50 μL of 0.1 m Na2CO3 (pH 11) and incubated on ice for 30 min; integral membrane proteins were separated from luminal and peripheral proteins by centrifugation.
Treated translation products were separated using SDS-PAGE on 12% or 15% gels that were subsequently fixed in a solution containing 10% acetic acid and 4% glycerol, dried, and analyzed using autoradiography.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF037368.
Acknowledgments
We thank Dr. Yasuo Niwa (University of Shizuoka, Japan) for kindly providing the pUC 18/sGFP (S65T) plasmid, Prof. Jim Haseloff (Cambridge University, UK) for the pBIN/mGFP5-ER plasmid, Dr. Ken-ichiro Shimazaki (Kyushu University, Japan) for the anti-VHA antibody, and Dr. Masayoshi Maeshima (Nagoya University, Japan) for the anti-γ-TIP antibody. We also thank Prof. Shinobu Satoh (University of Tsukuba, Japan), Prof. Mikio Nishimura (National Institute of Basic Biology), and Prof. Harry Klee (University of Florida), as well as Dr. Ken Sato (RIKEN, Japan) and Dr. Willis Owino (University of Tsukuba, Japan) for helpful discussions.
This work was supported by a grant-in-aid from the Research for the Future program of the Japan Society for the Promotion of Science (grant no. JSPS–RFTF00L01601) and by a grant from the 21st Century Centers of Excellence Program of the Ministry of Education, Culture, Sports, Science and Technology, and in part by funds from the National Institute for Basic Biology Cooperative Research Program (grant no. 5–138 to H.E.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Hiroshi Ezura (ezura@gene.tsukuba.ac.jp).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.080523.
References
- Abeles FB, Morgan PW, Saltveit ME (1992) Ethylene in Plant Biology. Academic Press, San Diego
- Aravind L, Ponting CP (1997) The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem Sci 22: 458–459 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Esch JJ, Hall AE, Rodriguez FI, Binder BM (1998) The ethylene-receptor family from Arabidopsis: structure and function. Philos Trans R Soc Lond B Biol Sci 353: 1405–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G (1980) Intracellular protein topogenesis. Proc Natl Acad Sci USA 77: 1496–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles DJ, Kauss H (1976) Characterization, enzymatic and lectin properties of isolated membranes from Phaseolus aureus. Biochim Biophys Acta 443: 360–374 [DOI] [PubMed] [Google Scholar]
- Campos N, Boronat A (1995) Targeting and topology in the membrane of plant 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Cell 7: 2163–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM (1993) Arabidopsis ethylene-response gene ETR1: similarity of product of two-component regulators. Science 262: 539–544 [DOI] [PubMed] [Google Scholar]
- Chang C, Stadler R (2001) Ethylene receptor action in Arabidopsis. Bioessays 23: 619–627 [DOI] [PubMed] [Google Scholar]
- Chen YF, Randlett MD, Findell JL, Schaller GE (2002) Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem 277: 19861–19866 [DOI] [PubMed] [Google Scholar]
- Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330 [DOI] [PubMed] [Google Scholar]
- Clark KL, Larsen PB, Wang X, Chang C (1998) Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc Natl Acad Sci USA 95: 5401–5406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros MG, von Heijne G (1994) TopPred II: an improved software for membrane protein structure predictions. Comput Appl Biosci 10: 685–686 [DOI] [PubMed] [Google Scholar]
- Devoto A, Piffanelli P, Nilsson I, Wallin E, Panstruga R, von Heijne G, Schulze-Lefert P (1999) Topology, subcellular localization, and sequence diversity of the Mlo family in plants. J Biol Chem 274: 34993–35004 [DOI] [PubMed] [Google Scholar]
- Ferrol N, Bennett AB (1996) A single gene may encode differentially localized Ca2+-ATPases in tomato. Plant Cell 8: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander M, Blobel G (1985) Bovine opsin has more than one signal sequence. Nature 318: 338–343 [DOI] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE (1998) Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA 95: 7825–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Chen YF, Randlett MD, Zhao XC, Findell JL, Kieber JJ, Schaller GE (2003) Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complex. J Biol Chem 278: 34725–34732 [DOI] [PubMed] [Google Scholar]
- Goubet F, Mohnen D (1999) Subcellular localization and topology of homogalacturonan methyltransferase in suspension-cultured Nicotiana tabacum cells. Planta 209: 112–117 [DOI] [PubMed] [Google Scholar]
- Guo HW, Ecker JR (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7: 40–49 [DOI] [PubMed] [Google Scholar]
- Harley CA, Holt JA, Turner R, Tipper DJ (1998) Transmembrane protein insertion orientation in yeast depends on the charge difference across transmembrane segments, their total hydrophobicity, and its distribution. J Biol Chem 273: 24963–24971 [DOI] [PubMed] [Google Scholar]
- Hartmann E, Rapoport TA, Lodish HF (1989) Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci USA 86: 5786–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse T, Schell J, Puype M, Palme K, Vandekerckhove J, Feldwisch J, Klambt D, Bauw G, Balshusemann D, Lobler M (1989) Molecular cloning and structural analysis of a gene from Zea mays (L.) coding for a putative receptor for the plant hormone auxin. EMBO J 8: 2453–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Kieber JJ, Hirayama N, Kogan M, Guzman P, Nourizadeh S, Alonso JM, Dailey WP, Dancis A, Ecker JR (1999) RESPONSIVE-to ANTAGONIST1, a Menkers/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 97: 383–393 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W (1993) TMbase—a database of membrane spanning proteins segments. Biol Chem Hoppe Seyler 374: 166 [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Huang YF, Li H, Hutchison CE, Laskey J, Kieber JJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33: 221–233 [DOI] [PubMed] [Google Scholar]
- Kieber J (2002) Ethylene: the gaseous hormone. In L Taiz, E Zeiger, eds, Plant Physiology, Ed 3. Sinauer Associates, Sunderland, MA, pp 519–538
- Kinoshita T, Shimazaki K (1999) Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J 18: 5548–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C, Widell S, Kjellbom P (1987) Preparation of high-purity plasma membranes. Methods Enzymol 148: 558–568 [Google Scholar]
- Li HY, Chye ML (2003) Membrane localization of Arabidopsis acyl-CoA binding protein ACBP2. Plant Mol Biol 51: 483–492 [DOI] [PubMed] [Google Scholar]
- Lord JM (1987) Isolation of endoplasmic reticulum: general principles, enzymatic markers, and endoplasmic reticulum-bound polysomes. Methods Enzymol 148: 577–584 [Google Scholar]
- Malley RCO, Rodriguez FI, Esch JJ, Binder BM, Donnell PO, Klee HJ, Bleecker AB (2005) Ethylene-binding activity, gene expression levels, and receptor system output for ethylene receptor family members from Arabidopsis and tomato. Plant J 41: 651–659 [DOI] [PubMed] [Google Scholar]
- McCartney AW, Dyer JM, Dhanoa PK, Kim PK, Andrews DW, McNew JA, Mullen RT (2004) Membrane-bound fatty acid desaturases are inserted co-translationally into the ER and contain different ER retrieval motifs at their carboxy termini. Plant J 37: 156–173 [DOI] [PubMed] [Google Scholar]
- Ogawara T, Higashi K, Kamada H, Ezura H (2003) Ethylene advances the transition from vegetative growth to flowering in Arabidopsis thaliana. J Plant Physiol 160: 1335–1340 [DOI] [PubMed] [Google Scholar]
- Ota K, Sakaguchi M, Hamasaka N, Mihara K (1998. a) Assessment of topogenic functions of anticipated transmembrane segments human band 3. J Biol Chem 273: 28286–28291 [DOI] [PubMed] [Google Scholar]
- Ota K, Sakaguchi M, von Heijne G, Hamasaka N, Mihara K (1998. b) Forced transmembrane orientation of hydrophilic polypeptide segments in multispanning membrane proteins. Mol Cell 2: 495–503 [DOI] [PubMed] [Google Scholar]
- Parks GD, Lamb RA (1991) Topology of eukaryotic type II membrane proteins: importance of N-terminal positively charged residues flanking the hydrophobic domain. Cell 64: 777–787 [DOI] [PubMed] [Google Scholar]
- Rodriguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB (1999) A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 283: 996–998 [DOI] [PubMed] [Google Scholar]
- Rost B, Fariselli P, Casadio R (1996) Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci 5: 1704–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato-Nara K, Yuhashi K, Higashi K, Hosoya K, Kubota M, Ezura H (1999) Stage- and tissue-specific expression of ethylene receptor homolog genes during fruit development in muskmelon. Plant Physiol 120: 321–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bleecker AB (1995) Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270: 1809–1811 [DOI] [PubMed] [Google Scholar]
- Schaller GE, Ladd AN, Lanahan MB, Spanbauer JM, Bleecker AB (1995) The ethylene receptor mediator ETR1 from Arabidopsis forms a disulfide-linked dimer. J Biol Chem 270: 12526–12530 [DOI] [PubMed] [Google Scholar]
- Suga S, Maeshima M (2004) Water channel activity of radish plasma membrane aquaporins heterologously expressed in yeast and their modification by site-directed mutagenesis. Plant Cell Physiol 45: 823–830 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Kobayashi T, Sato-Nara K, Tomita K, Ezura H (2002) Detection of ethylene receptor protein Cm-ERS1 during fruit development in melon (Cucumis melo L.). J Exp Bot 53: 415–422 [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I (1998) Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol 283: 489–506 [DOI] [PubMed] [Google Scholar]
- Ukaji K, Ariyoshi N, Sagaguchi M, Hamasaki N, Mihara K (2002) Membrane topogenesis of the three amino-terminal transmembrane segments of glucose-6-phosphatase on endoplasmic reticulum. Biochem Biophys Res Commun 292: 153–160 [DOI] [PubMed] [Google Scholar]
- van Geest M, Lolkema JS (2000) Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol Mol Biol Rev 64: 13–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar M, Sauri A, Monne M, Marcos JF, von Heijne G, Perez-Paya E, Mingarro I (2002) Insertion and topology of a plant viral movement protein in the endoplasmic reticulum membrane. J Biol Chem 277: 23447–23452 [DOI] [PubMed] [Google Scholar]
- Wang W, Hall AE, O'Malley R, Bleecker AB (2003) Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transduction. Proc Natl Acad Sci USA 100: 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels HP, Spiess M (1988) Insertion of a multispanning membrane protein occurs sequentially and requires only one signal sequence. Cell 55: 61–70 [DOI] [PubMed] [Google Scholar]
- Woeste K, Kieber JJ (2000) A strong loss-of-function allele of RAN1 results in constitutive activation of ethylene response as well as a rosette-lethal phenotype. Plant Cell 12: 443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JT (1996) Sequence requirements for membrane assembly of polytopic membrane proteins: molecular dissection of the membrane insertion process and topogenesis of the human MDR3 P-glycoprotein. Mol Biol Cell 7: 1709–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]