We have observed that a cis-acting regulatory element, known as site II, is overrepresented in the promoters of nuclear genes encoding components of the oxidative phosphorylation (OxPhos) machinery from both Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa). Site II elements have been described in promoters of rice and Arabidopsis proliferating cell nuclear antigen (PCNA) genes (Kosugi et al., 1995; Trémousaygue et al., 2003) and are also present in the majority of Arabidopsis genes encoding ribosomal proteins (Trémousaygue et al., 2003). Loss-of-function analysis has determined that a pair of site II elements present in the rice PCNA promoter is essential for the expression of a reporter gene in the shoot and root meristems of transgenic tobacco (Nicotiana tabacum; Kosugi et al., 1991, 1995). In fact, a promoter region containing these elements is enough to confer expression in tobacco protoplasts when fused to a minimal promoter (Kosugi et al., 1995). Similar elements in Arabidopsis PCNA-2 produce expression in root primordia and young leaves (Trémousaygue et al., 2003). Accordingly, these elements have been implicated in the expression of genes in meristematic tissues and/or proliferating cells. Site II elements are frequently present in more than one copy and found in combination with another motif, known as telo box or internal telomeric repeat due to its similarity to the repeated unit of plant telomeres (Trémousaygue et al., 2003).
The generation of new mitochondria, which occurs by fission of preexisting ones, is not necessarily linked to cell division. In fact, the number of mitochondria present in a cell is not constant and depends on the tissue and developmental stage and on external conditions (Huang et al., 1994; Griffin et al., 2001). A well-known example in plants is the increase in the number of mitochondria per cell that occurs in anthers during sporogenesis (Lee and Warmke, 1979). It can be postulated, however, that proliferating tissues would require active mitochondrial biogenesis to maintain the mitochondrial population of newly formed cells. In addition, it can be envisaged that active cell division may require additional energy input derived from mitochondrial OxPhos, and that this may require an increase in the amount of proteins involved in this and related processes.
Biogenesis of the OxPhos complexes requires the expression of two separate genomes within one cell. This is because some of the proteins that compose these complexes are encoded within the organelle genome, while the rest are encoded in the nucleus (Mackenzie and McIntosh, 1999). It is generally assumed that the expression of genes encoded in these genomes must be somehow coordinated to ensure correct complex assembly. A recent study suggests that coordination of the expression of genes encoded in the nuclear and the mitochondrial compartments occurs at the posttranslational level (Giegé et al., 2005). Regarding the coordination of nuclear genes themselves, it is more likely that regulation takes place at the transcriptional level, perhaps through the interaction of common sets of transcription factors with cognate binding sites present in the respective promoters.
Studies of promoter elements required for the expression of one of the two genes encoding mitochondrial cytochrome c (Cytc-1) in Arabidopsis revealed the existence of a couple of site II motifs linked to a downstream-located telo box that were required for expression in root and shoot meristems, nascent leaves, and anthers (Welchen and Gonzalez, 2005). Since cytochrome c participates as a shuttle of electrons between complexes III and IV, we analyzed the existence of similar elements in the putative promoters of nuclear genes encoding components of these complexes and found that a high proportion of them contained site II elements (Welchen and Gonzalez, 2005). Here, we present data on the occurrence, arrangement, and positioning of site II elements in the upstream regions of 103 nuclear genes encoding components of the Arabidopsis mitochondrial OxPhos machinery or proteins involved in the biogenesis of the respective complexes (Supplemental Table I). Eighty-three of these genes contain one or more site II elements in the 1,000-bp region located upstream of the predicted transcription start site. The total amount of site II elements in these promoters is 215, indicating that most genes contain two or more of these elements in upstream regions. Enrichment in site II elements was evident for genes encoding different components of the OxPhos machinery, with the only exception being those encoding complex II subunits (six out of 12 genes contain a total number of 13 site II elements). Although they were included in subsequent analyses, it is likely that complex II components show a different behavior since they are also required for the functioning of the TCA cycle. In a similar way, genes encoding nonphosphorylating components of the electron transport chain, such as alternative NAD(P)H dehydrogenases or alternative oxidase isoforms, do not possess a high number a site II elements in their upstream regions (with some exceptions) and were not included in the analysis.
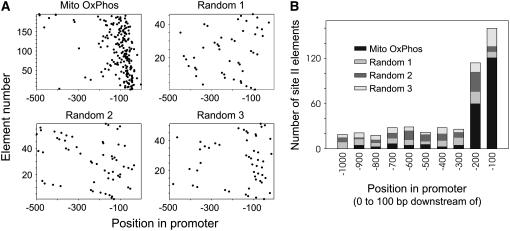
The hypothesis that site II elements are enriched in promoters of nuclear genes encoding OxPhos components was tested by comparison with three different sets of 100 upstream regions from randomly chosen Arabidopsis genes. The sets were generated using the Random ID list generator available at http://bbc.botany.utoronto.ca/, and 1,000-bp regions upstream from the predicted transcription start site were obtained from The Arabidopsis Information Resource (www.arabidopsis.org). Both the proportion of genes with site II elements (42% ± 2%) and the total number of site II elements (83 ± 5) in the random sets are significantly lower when compared with the OxPhos set (Supplemental Table II). In addition, when the distribution of site II elements along the promoters was analyzed, it was observed that the vast majority of these elements are located in proximal regions in the set of genes for OxPhos components, while a more uniform distribution was evident in the random sets (Fig. 1A). A plot of the number of site II elements present in 100-bp regions from −1 to −1,000 in the four different promoter sets (Fig. 1B) showed that the main difference between the OxPhos set and the random sets is due to an enrichment in site II elements in the region from −1 to −200 from the predicted transcription start site (see also Supplemental Table II). Upstream from −200 the number of site II elements is very similar in the four sets analyzed. This selective enrichment in a defined region supports the view that these motifs have a functional role in the expression of genes encoding OxPhos components.
Figure 1.
Number and location of site II elements in upstream regions of Arabidopsis nuclear genes encoding OxPhos components. A, The location of site II elements along the 500-bp upstream region respective to the predicted transcription start site was plotted for 103 genes encoding components of the mitochondrial respiratory chain or for three random sets of 100 genes each. B, The number of site II elements present in 100-bp intervals from −1 to −1,000 was plotted for each of the four sets of genes. The numbers below the bars represent the upstream limit of each interval.
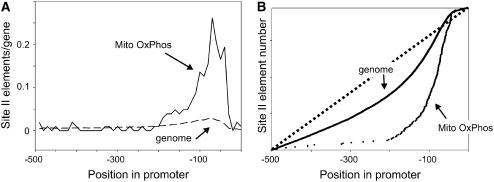
A further analysis of the significance of the presence of site II elements in OxPhos genes was made by comparison with the entire set of 500-bp gene upstream regions from the Arabidopsis genome. The presence of site II elements in the entire Arabidopsis genome set was performed using the program PatMatch from www.arabidopsis.org. The frequency of site II elements (average number of site II elements per gene) present in 10-bp intervals from −1 to −500 was compared for the two sets. Similarly to what has been observed with the random promoter sets, there is no obvious difference in the occurrence of site II elements upstream of −200 from the predicted transcriptional start (Fig. 2A). In proximal promoter regions, however, there is an enrichment of approximately 10-fold in the number of site II elements per gene in OxPhos promoters with respect to the bulk of Arabidopsis promoters (Fig. 2A). The fact that site II elements are not uniformly distributed along the promoters of OxPhos genes is supported by statistical analysis (Supplemental Table III). For the entire genome, enrichment is also observable in proximal regions, similarly to what has been described for the OxPhos set. This deviation from uniform distribution does not reach, however, statistical significance (Supplemental Table III). The data indicate that approximately 5% to 10% of Arabidopsis genes may have a high frequency of site II elements in their promoter proximal regions.
Figure 2.
Arabidopsis genes for mitochondrial OxPhos components show a high frequency of site II elements downstream of −200. A, The number of site II elements per gene present in 10-bp intervals along the 500-bp region located upstream from the predicted transcriptional start was plotted for the set of 103 genes encoding OxPhos components and for the entire set of Arabidopsis nuclear genes. B, The location of site II elements (ordered according to their position in the respective promoters from −500 to −1) was plotted for the two sets of genes. For a random distribution of elements, a straight line (represented by dots) would be obtained.
The observation that site II elements are not evenly distributed in gene upstream regions is clearly observed when they are ordered according to their position respective to the predicted transcription start site (Fig. 2B). Regularly spaced elements would be arranged along a straight line with slope equal to the total number of site II elements in each set divided by distance in base pairs. The graph shown in Figure 2B indicates that in the entire genome set, site II elements are underrepresented upstream of −200 and downstream of −50 (i.e. smaller slope) and overrepresented between −200 and −50. An estimation of the respective slopes in these regions indicates that there is a 4-fold relative enrichment in site II elements between −50 and −200 compared with upstream portions of Arabidopsis promoters. A similar analysis for genes encoding OxPhos components indicates a more pronounced relative enrichment of site II elements (more than 50-fold) in proximal regions. Consequently, both the number and location of these elements in Arabidopsis genes for OxPhos components are considerably different from those observed for the entire set of Arabidopsis genes.
The statistical significance of our observations was also tested using the program Promomer (Toufighi et al., 2005; http://bbc.botany.utoronto.ca/) to analyze sequences that are overrepresented in 1,000 bootstraps of the OxPhos gene set respective to 1,000 randomly generated Arabidopsis promoter sets. The occurrence of the sequence TGGGC (GCCCA), common to both forms of site II elements, was significantly different (P < 0.001) for the two populations, with average numbers of 129.7 and 54.6 for the OxPhos sets and the random sets, respectively. In a similar way, the program oligo-analysis (van Helden et al., 1998) from the Regulatory Sequence Analysis Tools site (http://rsat.ulb.ac.be/rsat/) also returned the sequence GCCCA as the most overrepresented in the OxPhos promoter set, with an E value of 1.5 × 10−36 (indicating that in only one in every 6.7 × 1035 trials would this result be obtained with random promoters).
For genes with more than one site II, we have calculated the distances between adjacent elements. Seventy-eight percent of these distances are of 30 bp or less, suggesting that site II elements usually lie in close proximity to each other. As a consequence, we can define a basic unit composed of two or more site II elements separated by less than 30 bp and located downstream of −200 from the predicted transcriptional start as the most frequently observed in promoters of OxPhos genes. This basic unit is the one present in Arabidopsis promoters for which experimental evidence on the role of site II elements in transcription has been obtained (Trémousaygue et al., 2003; Welchen and Gonzalez, 2005). Site II elements that lie in close proximity to each other occur in tandem and in head-to-head orientation with similar frequencies. This may indicate that the proteins that bind to adjacent elements do not form a complex with a defined spatial arrangement. However, it is noteworthy that the element located downstream in each pair is in the reverse orientation (TGGGCC/T in the noncoding strand) in 81% of pairs, while only 57% of total site II elements are in this orientation. It can be speculated that binding to the downstream element in this orientation favors the interaction of the bound protein with the rest of the transcriptional machinery. This may be related with the fact that mutation of the downstream site II element of the Cytc-1 gene (which is in the reverse orientation) has a greater effect on transcription than mutation of the element located upstream (Welchen and Gonzalez, 2005).
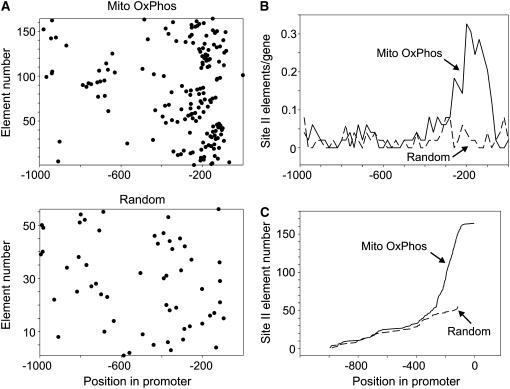
The importance of site II elements for the expression of PCNA genes has been documented in rice as well as in Arabidopsis (Kosugi et al., 1995; Trémousaygue et al., 2003), suggesting the existence of a conserved role of these elements in monocot and dicot plants. We have then analyzed the occurrence of site II elements in 1,000-bp regions located upstream from the translation start sites of 49 rice genes encoding mitochondrial OxPhos components (Supplemental Table IV). Similarly to what we observed in Arabidopsis, many of these genes contain site II elements in their upstream regions (42 of them contain one or more site II element in the 1,000-bp region located upstream of the ATG translation start codon). The total amount of site II elements in these promoters is 164, an average of almost four elements per gene. A plot of the distribution of site II elements in rice OxPhos promoters (Fig. 3A, top) indicates a relative enrichment in proximal regions, around −200. Conversely, a set of 50 random rice promoters contains a lower amount of elements (56) randomly distributed along the 1,000-bp upstream region (Fig. 3A, bottom). Comparison of the respective frequencies in 20-bp intervals demonstrates that the deviation from random behavior occurs exclusively in the region between −100 and −300 respective to the translation start site, resulting in a 10-fold relative increase in the number of site II elements in this region (Fig. 3, B and C).
Figure 3.
Rice genes encoding mitochondrial OxPhos components show an enrichment of site II elements in the region from −100 to −300 from the translation start site. A, The location of site II elements along the 1,000-bp upstream region respective to the translation start site was plotted for 49 rice genes encoding components of the mitochondrial respiratory chain or for a random set of 50 rice genes. B, The number of site II elements per gene present in 20-bp intervals along the 1,000-bp upstream region was plotted for both sets of rice genes. C, The location of site II elements (ordered according to their position in the respective promoters from −1,000 to −1) was plotted for the two sets of rice genes.
The observation that site II elements are significantly enriched in promoters of nuclear genes encoding mitochondrial OxPhos components is a strong indication that these elements may participate in the coordinated expression of this set of genes in response to either internal or external factors. As mentioned above, site II elements are present in promoters of genes preferentially expressed during cell proliferation. More recently, it has been observed that Arabidopsis genes that are up-regulated in response to signals that promote axillary bud outgrowth (like stem decapitation) often contain site II elements in their promoters (Tatematsu et al., 2005). Accordingly, it can be proposed that the biogenesis of the mitochondrial machinery involved in energy production would be linked to cell proliferation and growth through site II elements and the factors that interact with them.
It has been shown that site II elements interact with transcription factors containing the so-called TCP domain (Kosugi and Ohashi, 1997; Trémousaygue et al., 2003; Li et al., 2005; Welchen and Gonzalez, 2005), a plant-specific DNA-binding and dimerization motif related to the bHLH domain (Cubas et al., 1999). The Arabidopsis genome encodes more than 20 different TCP-domain proteins, which can be divided into two classes. Members of class I bind the sequence GTGGGNCC in vitro and are positive regulators of gene expression during cell proliferation (Kosugi and Ohashi, 2002). Members of class II, on the other hand, negatively regulate cell proliferation (Gaudin et al., 2000) and bind the sequence GTGGNCCC (Kosugi and Ohashi, 2002). Mutations in class II members from several species, like teosinte branched 1 in maize (Zea mays) and cycloidea (cyc) in Antirrhinum, produce developmental alterations (Luo et al., 1996; Doebley et al., 1997). The expression patterns produced by Cytc-1 promoter/gus fusions and the observation that mutation of site II elements abolishes Cytc-1 gene expression (Welchen and Gonzalez, 2005) suggests that class I genes are the best candidates in this case. Nevertheless, the sequence of the site II element used here (TGGGCC/T), which is the one defined by Trémousaygue et al. (2003), is present in the preferred target sites of members of both classes, and we have not found a particular enrichment of any of the sequences selected by class I and II TCP proteins in OxPhos promoters. Accordingly, it can be speculated that different TCP-domain proteins may either positively or negatively regulate the expression of OxPhos components, depending on their expression domains and relative amounts in defined cell types. A similar hypothesis has been presented recently for the regulation of the expression of genes encoding cyclin CYCB1;1 and ribosomal protein genes during cell growth and division, through the presence of site II-like motifs (GCCCR) that are recognized by TCP-domain proteins (Li et al., 2005).
Optimal expression of the Cytc-1 gene requires, in addition to site II elements, the presence of a telo box (Welchen and Gonzalez, 2005). Mutation of the Cytc-1 telo box produces a significant decrease in expression, as also observed in other genes that contain both types of motifs (Trémousaygue et al., 2003; Tatematsu et al., 2005). However, we have not observed an overrepresentation of telo boxes in promoters of OxPhos components. This may indicate that site II elements in these promoters are coupled to different responsive elements. As an example, site II elements act in concert with MSA elements (AACGG) to promote high level expression of the CYCB1;1 gene in the G2/M phase of the cell cycle (Li et al., 2005), while PCNA-2, which contains site II elements and a telo box, is expressed during the G1/S transition (Trémousaygue et al., 2003). The rice PCNA gene, in turn, contains two site II and a site I motif, which resembles a G box (Kosugi et al., 1995).
Although it is assumed that the expression of genes encoding mitochondrial components must somehow be coordinated, it is also true that this coordination may only occur in response to specific, either internal or external, signals or factors. Accordingly, different genes or subsets of genes may possess particular expression characteristics in response to other factors. In addition, since many of the mitochondrial components are encoded by more than one gene, it is possible that not all the genes encoding the same component are subject to coordinate control, since gene duplication followed by mutations in regulatory regions may have changed the expression characteristics of different members of gene families. Examples of this are genes encoding cytochrome c (Welchen and Gonzalez, 2005) and the iron-sulfur subunit of complex II (Elorza et al., 2004), which are differentially expressed in Arabidopsis. Based on this, we propose a model in which the promoters of genes encoding OxPhos components are composed of a mix of common and gene-specific regulatory elements. For the Cytc-2 gene, for example, preliminary evidence suggests that expression depends on the presence of site II elements, a G box, and yet unidentified motif(s) (E. Welchen, L. Prendes, and D.H. Gonzalez, unpublished data).
Considering the presence of site II elements in only a subset of gene family members, it can be observed that genes encoding complex II subunits SDH1/2/4/7 and 8 possess proximal site II elements in their promoters. The same applies for genes encoding one of the alternative oxidase isoforms (AOX1c) and the alternative NAD(P)H dehydrogenase NDA2.
We have also analyzed the presence of site II elements in the promoters of genes encoding other mitochondrial components. We have not found enrichment in site II elements in genes encoding TCA-cycle enzymes (15 out of 37 genes contain site II elements in the proximal 500-bp upstream region) or mitochondrial carriers (16 out of 45 genes contain site II elements; Supplemental Tables V and VI). Concerning the mitochondrial import machinery, the presence of site II elements is particularly evident in genes encoding components of the TIM complex (Supplemental Table VII). Out of 18 genes analyzed, 13 of them, representing genes for all components except for TIM10, have a total of 29 site II elements in their upstream regions. Interestingly, 11 genes contained one or more copies of the telo box upstream from the translation start codon, most of them in proximal regions. This represents enrichment in telo boxes compared with the three different random sets analyzed above, since only 12% to 15% of random genes contain telo boxes in their respective upstream regions. For eight of the TIM genes, encoding isoforms of TIM23, TIM17, TIM50, TIM13, TIM14, TIM22, TIM9, and TIM8, the arrangement of site II elements and the telo box is similar to the one observed in the Cytc-1 promoter. This is also valid for genes encoding a plant homolog of metaxin (known to be involved in protein import in animals) and the outer membrane components TOM20 and TOM6. Accordingly, expression of part of the mitochondrial import apparatus may be coordinated with the expression of nuclear-encoded components of the OxPhos machinery, which must be imported into mitochondria to assemble functional complexes.
In conclusion, we provide evidence that TGGGCC/T (site II) motifs are frequently present in several copies in proximal regions located upstream of nuclear genes encoding OxPhos components from both Arabidopsis and rice. We propose that these elements interact with a group of transcription factors that participate in the coordination of the expression of this set of genes and that link the biogenesis of the plant mitochondrial respiratory chain to events related to cell proliferation and growth. Detailed analysis of the promoters of several of these genes will be useful to evaluate the validity of this proposition and to uncover the existence of other, either common or gene-specific, responsive elements.
This work was supported by Consejo Nacional de Investigaciones Científicas y Técnicas, by Agencia Nacional de Promoción Científica y Tecnológica, by Fundación Antorchas, and by Universidad Nacional del Litoral.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Daniel H. Gonzalez (dhgonza@fbcb.unl.edu.ar).
The online version of this article contains Web-only data.
References
- Cubas P, Lauter N, Doebley J, Coen E (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386: 485–488 [DOI] [PubMed] [Google Scholar]
- Elorza A, León G, Gómez I, Mouras A, Holuigue L, Araya A, Jordana X (2004) Nuclear SDH2-1 and SDH2-2 genes, encoding the iron-sulfur subunit of mitochondrial complex II in Arabidopsis, have distinct cell-specific expression patterns and promoter activities. Plant Physiol 136: 4072–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin V, Lunness PA, Fobert PR, Towers M, Riou-Khamlichi C, Murray JA, Coen E, Doonan JH (2000) The expression of D-cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol 122: 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé P, Sweetlove LJ, Cognat V, Leaver CJ (2005) Coordination of nuclear and mitochondrial genome expression during mitochondrial biogenesis in Arabidopsis. Plant Cell 17: 1497–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin KL, Anderson OR, Gastrich MD, Lewis JD, Lin G, Schuster W, Seemann JR, Tissue DT, Turnbull MH, Whitehead D (2001) Plant growth on elevated CO2 alters mitochondrial number and chloroplast fine structure. Proc Natl Acad Sci USA 98: 2473–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Struck F, Matzinger DF, Levings CS III (1994) Flower-enhanced expression of a nuclear-encoded mitochondrial respiratory protein is associated with changes in mitochondrion number. Plant Cell 6: 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (1997) PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Suzuka I, Ohashi Y (1995) Two of three promoter elements identified in a rice gene for proliferating cell nuclear antigen are essential for meristematic tissue-specific expression. Plant J 7: 877–886 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Suzuka I, Ohashi Y, Murakami T, Arai Y (1991) Upstream sequences of rice proliferating cell nuclear antigen (PCNA) gene mediate expression of PCNA-GUS chimeric gene in meristems of transgenic tobacco plants. Nucleic Acids Res 19: 1571–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SLJ, Warmke HE (1979) Organelle size and number in fertile and T-cytoplasmic male-sterile corn. Am J Bot 60: 141–148 [Google Scholar]
- Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P (2005) Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci USA 102: 12978–12983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E (1996) Origin of floral asymmetry in flowers of Antirrhinum. Nature 383: 794–799 [DOI] [PubMed] [Google Scholar]
- Mackenzie S, McIntosh L (1999) Higher plant mitochondria. Plant Cell 11: 571–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E (2005) Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol 138: 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Trémousaygue D, Garnier L, Bardet C, Dabos P, Hervé C, Lescure B (2003) Internal telomeric repeats and “TCP-domain” protein binding sites co-operate to regulate gene expression in Arabidopsis thaliana cycling cells. Plant J 33: 957–966 [DOI] [PubMed] [Google Scholar]
- van Helden J, Andre B, Collado-Vides J (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J Mol Biol 281: 827–842 [DOI] [PubMed] [Google Scholar]
- Welchen E, Gonzalez DH (2005) Differential expression of the Arabidopsis cytochrome c genes Cytc-1 and Cytc-2: evidence for the involvement of TCP-domain protein binding elements in anther- and meristem-specific expression of the Cytc-1 gene. Plant Physiol 139: 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]