GAs form a large family of phytohormones that have long been recognized as plant growth regulators of numerous aspects of growth and development. They are additionally involved in the discernment of environmental stimuli, thus are important not only for a plant's growth and development but also in perception of its environment (Hedden and Kamiya, 1997). The elucidation of its biosynthetic pathway enzymes in a cereal crop plant such as rice (Oryza sativa) would be of interest to the research community. In this month's High Impact, the identification of seven enzymes involved in GA synthesis in rice is discussed along with mutants associated with some of the genes. The article by Sakamoto et al. titled “An overview of gibberellin metabolism enzyme genes and their related mutants in rice” appeared in our April 2004 issue and, as of April 2006, had 40 citations (Thompson ISI Web of Science, http://www.isinet.com).
BACKGROUND
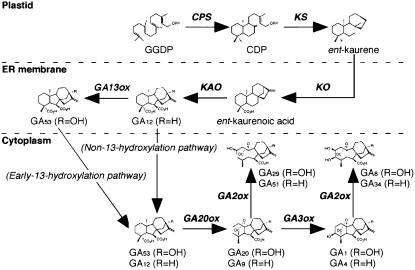
GAs are diterpenoids derived from four isoprenoid units forming a system of four rings containing either 19 or 20 carbons. They are products of the terpenoid pathway that yields not only GA, but also terpenes, terpene-derived compounds, and steroids. Seven enzymes have been identified to be involved in GA biosynthesis: ent-copalyl diphosphate synthase (CPS), ent-kaurene synthase (KS), ent-kaurene oxidase (KO), ent-kaurenoic acid oxidase (KAO), GA 20-oxidase, GA 3-oxidase, and GA 2-oxidase, as well as cytochrome P450 monooxygenase and α-ketoglutarate-dependant dioxygenase (Fig. 1). Biosynthesis occurs in multiple locations within the cell, with the first two steps in the chloroplast (those catalyzed by CPS and KS), the next three involving KO, KAO, and GA13ox on the ER membrane, and the remaining are in the cytoplasm.
Figure 1.
GA biosynthetic pathway in rice. (Figure courtesy of Sakamoto et al. [2004], with permission of American Society of Plant Biologists.)
This important phytohormone was identified during studies of “bakanae” (foolish seedling) disease in rice, responsible for greatly reduced yields in Japan, Taiwan, and the Asian subcontinent in the 1800s. Bakanae is characterized by pale-yellow, elongated seedlings with slender leaves, stunted roots, and little or no seed production. The causative agent of this disease was demonstrated to be a fungus in an 1898 paper published by Shotaro Hori. In 1926, Eiichi Kurosawa determined that a chemical released by the bakanae fungus (Fusarium moniliforme = Gibberella fujikuroi) was responsible for the stimulated shoot elongation, inhibited chlorophyll formation, and suppressed root growth characteristic of bakanae. Kurosawa also observed that the compound had the ability to induce other plants to elongate. This compound was successfully crystallized by Teijiro Yabuta and Yusuke Sumiki in the 1930s and named “gibberellin” after the fungus it was isolated from (http://www.plant-hormones.info/gibberellinhistory.htm). World War II delayed further studies, and it was not until the 1950s that a GA was isolated from plants, demonstrating it is an endogenous compound in plants. Today more than 125 different GAs have been identified and have been found in all plant species examined along with some fungi and bacteria. Although chemically they differ only slightly, their biological activities vary dramatically.
As demonstrated in bakanae, excess GA causes seedlings to become spindly and lodge (fall over), resulting in diminished harvest. However, mutations in GA synthesis or signal perception have the opposite effect, yielding dwarf plants instead (not all dwarf varieties of plants are due to alterations in GA). Dwarfism is a desirable agronomic trait in crop plants as shorter-stature plants are less likely to lodge, thus increasing harvest index. The identification of GA and the use of the resulting dwarf mutations were factors in the success of the “Green Revolution,” which greatly increased agricultural output in developing nations.
WHAT WAS SHOWN
To identify the genes encoding the seven GA metabolic enzymes in rice, Sakamoto et al. (2004) screened (in silico) all available rice DNA databases with the predicted amino acid sequences of known GA metabolic enzymes from a variety of plant species. Using this strategy, 29 candidate genes were identified. Mapping analysis revealed that the genes corresponding to the enzymes involved in the early steps of GA biosynthesis, i.e. CPS-like, KS-like, and KO-like genes, are arranged in small gene clusters across nine of the 12 rice chromosomes, whereas those involved in the later steps are not clustered but rather found separately in the genome. This is in contrast to what has been found in Arabidopsis (Arabidopsis thaliana), in which the CPS, KS, and KO enzymes involved in GA biosynthesis are encoded by single genes.
The presence of multiple CPS-like, KS-like, and KO-like genes raises the question whether all or only a portion of them are GA metabolic enzymes. To determine which ones are involved in GA biosynthesis, the researchers took a genetic approach and screened rice plants exhibiting a GA-deficient phenotype for their ability to respond to exogenous GA application, since plants with a defect in GA synthesis would be “rescued” by the application of the hormone. This approach enabled the identification of 18 lines, half of which were novel recessive mutations. These lines were categorized into three mutant classes based on the severity of the phenotype. The most dramatic mutants, category I, were severe dwarf plants with no flower or seed development. These mutations mapped to one of the four GA metabolic enzymes involved in the early steps of GA biosynthesis, strongly suggesting that only one gene is involved in the biosynthesis of GA. As noted above, in Arabidopsis a single gene also encodes those enzymes involved in the early steps of GA synthesis, and the loss-of-function mutations at these loci result in severe dwarf phenotype. The other two categories of mutants had varying degrees of severity of dwarf phenotype and produced seeds, indicating either separate GA synthesis enzymes involved in vegetative or reproductive development (category II) or gene redundancy (category III).
With only one of the CPS-like and KS-like genes involved in GA synthesis, what is the function of the remaining genes? The biosynthetic pathway of polycyclic diterpenes, including diterpenoid phytoalexins, has the same intermediates as the GA biosynthetic pathway, namely, ent-copalyl diphosphate (CDP) and a kaurene-like intermediate, suggesting a possible role for the other CPS-like and KS-like genes. Phytoalexin synthesis is increased in response to pathogen infection and UV exposure, and the up-regulation of the potential rice genes was tested under these stress conditions. With the exception of OsCPS1 and OsKS1, the CPS-like and half of the other KS-like genes were induced by either UV or fungal elicitor, supporting the hypothesis that the other family members are not involved in GA synthesis; they are more likely involved in the synthesis of diterpenoid phytoalexins. Reminiscent to what is found with the terpenoid biosynthesis genes of Arabidopsis, the rice stress-inducible genes were all contiguously arranged. This clustering of diterpene synthases in rice has since been shown to be functional; CPS-like and KS-like genes, which act sequentially, are located in close proximity to each other (Wilderman et al., 2004).
The findings of this study by Sakamoto et al. on the GA metabolism enzyme genes and their related mutants strongly suggest that only one gene is involved in the early steps of GA biosynthesis in rice and that the biosynthesis of other diterpenoid compounds involves different enzymes. The strong phenotype of the early GA metabolism enzyme mutants demonstrates that mutation cannot be compensated by the similar enzyme. Additionally, the arrangement of small gene families and the presence of transposon-like sequences in these regions raise the possibility of gene duplication.
THE IMPACT
Phytoalexins are low-molecular-mass compounds induced by fungal attack as well as by UV exposure. A large portion of phytoalexins are diterpenoid compounds, which, like GA, are biosynthesized from geranylgeranyl diphosphate. The first branch point between the synthesis of the terpenoid phytoalexins and GA is the cyclization step yielding either ent-CDP or syn-CDP. A question of interest is whether a single cyclase was involved in this step or if there were different enzymes for each pathway. In their study, Sakamoto et al. (2004) isolated four genes encoding labdadienyl copalyl diphosphate (CPP) synthase-like proteins from rice. Of these four, only the mutant from one, OsCPS1, had a severe GA-deficient phenotype, strongly suggesting no overlap in gene function for GA biosynthesis. Of the remaining three genes, one was shown to be a pseudogene, but the expression of the remaining two was induced by UV and elicitor treatment, suggesting a role in the biosynthesis of phytoalexins rather than GA. One of these, OsCPS4, has been identified as a syn-CPP synthase (OsCPSsyn) involved in phytoalexin/allelochemical biosynthesis (Xu et al., 2004), leaving the identity of OsCPS2 open. Two ent-CPP synthases were isolated from rice by Prisic et al. (2004), named OsCPS1ent and OsCPS2ent. Functional studies of these two enzymes demonstrated that they were both ent-CPP synthases capable of converting geranylgeranyl diphosphate to ent-copalyl diphosphate, an early step in the synthesis of diterpenoids (Fig. 1). They further characterized these two enzymes and found OsCPS1ent and OsCPS2ent to correspond to OsCSP1 and OsCPS2 from the Sakamoto et al. (2004) study. While mutant studies showed OsCSP1 to be involved in the synthesis of GA, OsCPS2 was proposed to be involved in the biosynthesis of phytoalexins (Sakamoto et al., 2004). The authors of this study followed up on that hypothesis and found an increase in expression of OsCPS2ent following the application of methyl jasmonate, a signaling molecule shown to increase phytoalexin synthesis, as well as UV exposure. Diterpene cyclase cDNAs were isolated from UV-treated rice leaves in another study by Otomo et al. (2004), one of which, OsCyc2, encoded OsCPS2, lending further support to the involvement of this enzyme in phytoalexin biosynthesis.
Additionally, Prisic et al. (2004) compared the OsCPS1ent, OsCPS2ent, and OsCPSsyn sequences with An1/ZmCPS1ent from maize (Zea mays). The authors found that OsCPS1ent was more similar to An1/ZmCPS1ent (64% identity) than either one of its paralogs (44%), suggesting a possible gene duplication event occurring before the divergence rice and maize.
A transposon insertion mutant in rice with a severe dwarf phenotype was isolated by Margis-Pinheiro et al. (2005) and found to correspond to OsKS1, another enzyme in the early steps of GA biosynthesis (Fig. 1). Despite a severe dwarf phenotype with a lack of flowering, the mutant plants had normal seed germination and root growth. With exogenous GA application, the mutant seedlings grew to normal height and flowered but remained male-sterile. Similar to what was found in the study by Sakamoto et al. (2004), the transcripts of OsKS1 were not detected in germinating seeds or roots but were found only in stems and leaves, an explanation for the mutant phenotype and GA response. This expression pattern also suggests the possibility of a different KS expressed in roots and germinating seeds. A search of the rice expressed sequence tag and genome database identified four additional OsKS1-like sequences, all of which were expressed differently throughout the seedlings. Margis-Pinheiro et al. (2005) also did a phylogenetic analysis of the OsKS1-like sequences with possible orthologous gene products from maize, Arabidopsis, pumpkin (Cucurbita maxima), and other identified plant terpene cyclases. Their results demonstrated that the dicot KSs clustered in one group, whereas the cereal KSs clustered in another.
CONCLUSION
The GA metabolic pathway genes have been identified in fungi, bacteria, and the model plant Arabidopsis, but, before the study by Sakamoto et al. (2004), only individual pathway members from cereal plants had been isolated. The identification of the rice GA metabolism enzyme genes can also aid in the elucidation of those genes in other cereal crops.
The GA biosynthesis pathway always shares common steps with the synthesis of diterpenoid compounds involved in defense. The identification of multiple CPS-like, KS-like, and KO-like genes in rice raised the possibility that different enzymes may be involved in the different pathways. Through the study of GA mutants and response to elicitors, this hypothesis was confirmed, setting the groundwork for further studies into both GA and diterpenoid biosynthesis. Reuben J. Peters, researcher in the field of diterpenoid biosynthesis at Iowa State, feels that this work by Sakamoto et al. (2004) truly is high impact and that “the results of this study also provided a clear framework for investigating diterpenoid biosynthesis in rice more broadly.”
References
- Hedden P, Kamiya Y (1997) Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Physiol Plant Mol Biol 48: 431–460 [DOI] [PubMed] [Google Scholar]
- Margis-Pinheiro M, Zhou XR, Zhu QH, Dennis ES, Upadhyaya NM (2005) Isolation and characterization of a Ds-tagged rice (Oryza sativa L.) GA-responsive dwarf mutant defective in an early step of the gibberellin biosynthesis pathway. Plant Cell Rep 23: 819–833 [DOI] [PubMed] [Google Scholar]
- Otomo K, Kenmoku H, Oikawa H, Konig WA, Toshima H, Mitsuhashi W, Yamane H, Sassa T, Toyomasu T (2004) Biological functions of ent- and syn-copalyl diphosphate synthases in rice: key enzymes for the branch point of gibberellin and phytoalexin biosynthesis. Plant J 39: 886–893 [DOI] [PubMed] [Google Scholar]
- Prisic S, Xu M, Wilderman PR, Peters RJ (2004) Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol 136: 4228–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Miura K, Itoh H, Tatsumi T, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Agrawal GK, Takeda S, Abe K, et al (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol 134: 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ (2004) Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol 135: 2098–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hillwig ML, Prisic S, Coates RM, Peters RJ (2004) Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J 39: 309–318 [DOI] [PubMed] [Google Scholar]