Abstract
Plants may shed organs when they have been injured or served their purpose. The differential pattern of organ abscission in different species is most likely the result of evolutionary adaptation to a variety of life styles and environments. The final step of abscission-related cell separation in floral organs of wild-type Arabidopsis thaliana, which only abscises sepals, petals, and stamens, is controlled by INFLORESCENCE DEFICIENT IN ABSCISSION (IDA). Here, we demonstrate that Arabidopsis 35S:IDA lines constitutively overexpressing IDA exhibit earlier abscission of floral organs, showing that the abscission zones are responsive to IDA soon after the opening of the flowers. In addition, ectopic abscission was observed at the bases of the pedicel, branches of the inflorescence, and cauline leaves. The silique valves also dehisced prematurely. Scanning electron microscopy indicated a spread of middle lamella degradation from preformed abscission zone cells to neighboring cells. A transcript encoding an arabinogalactan protein (AGP) was upregulated in the 35S:IDA lines, and large amounts of AGP were secreted at the sites of abscission. AGP was shown to be a constituent of wild-type floral abscission zones during and soon after cell separation had been completed. We suggest that the restricted expression pattern of IDA precludes abscission of nonfloral organs in Arabidopsis.
INTRODUCTION
The shedding, or abscission, of plant organs involves cell separation and plays a key role in the plant life cycle. Perennial species inhabiting latitudes exposed to cold winters most often abscise their leaves in the autumn and store compounds that are retracted from these leaves during their more quiescent life in the winter season. Fruits involved in seed dispersal may abscise at maturity and fall to the ground to be eaten by animals. In other species, pods spring open in a cell separation process to release seeds. Flowers may be shed if they are not fertilized, and floral organs that assist in pollination may abscise after pollination has taken place (Patterson, 2001; Roberts et al., 2002).
Abscission is a developmentally controlled program of cell separation (Bleecker and Patterson, 1997; Patterson, 2001; Roberts et al., 2002), and anatomical studies have revealed that the primary site of wall breakdown is the middle lamella between narrow, specialized bands of cells that form an abscission zone (AZ) (Petersen et al., 1996). The middle lamella is a part of the cell wall that is shared by neighboring cells and functions to cement them together. The dicotyledonous cell wall consists of rigid, inextensible cellulose microfibrils held together by interpenetrating coextensive networks of matrix glycans, pectins, and structural glycoproteins (Yong et al., 2005). One of the key interactions in the primary wall of dicotyledons is formed between cellulose and the hemicellulose xyloglucan, which together typically compose approximately two-thirds of the dry wall mass (Cosgrove, 2005). The cell wall also contains a variety of arabinogalactan proteins (AGPs), which are highly glycosylated proteoglycans that have been implicated in signaling and response systems (Schultz et al., 2000; Gaspar et al., 2001). Elaborate mechanisms exist to disrupt intermolecular bonds in the primary wall and middle lamella, resulting in a loss of cell-to-cell adhesion, including the actions of wall-modifying enzymes such as endo-1,4-β-glucanases, pectinases, expansins, xyloglucan endotransglucosylase/hydrolases (XTHs), and polygalacturonases (Roberts et al., 2002; Rose et al., 2003). Both AGPs and oligosaccharides resulting from the consequent breakdown of complex cell wall polysaccharides have been implicated in the triggering of signaling pathways (Pilling and Hofte, 2003).
Different plants species display distinct patterns of organ loss, most likely adapted during evolution to their diverse life styles (Sexton and Roberts, 1982). Arabidopsis thaliana displays abscission only of floral organs and seeds, in addition to dehiscence of the valves of the siliques; we have found this species to be an excellent model system for the study of floral organ abscission (Butenko et al., 2003; Aalen et al., 2006). In Arabidopsis, this process can be divided into four major developmental steps (Patterson, 2001). After the AZ has formed as a band of small densely cytoplasmic cells at the junction between the flower receptacle and floral organs, this zone acquires the competence to respond to abscission signals. Thereafter, cell separation commences, accompanied by cell elongation at the proximal side of the AZ. The middle lamella between the cells in the AZ is degraded and, finally, a protective layer differentiates on the proximal side of the AZ (Patterson, 2001).
A number of mutants have been identified with changes in floral organ abscission (Aalen et al., 2006; Lewis et al., 2006). Mutants with a defective response to ethylene show a delay in abscission, consistent with the fact that ethylene promotes abscission (Bleecker and Patterson, 1997; Patterson, 2001); however, several mutants show normal sensitivity to ethylene and may represent an ethylene-independent developmentally controlled pathway leading to abscission. The delayed abscission mutants dab1 to dab3 are primarily affected in floral abscission and are defective in earlier steps of the abscission process (Patterson and Bleecker, 2004). RNA interference plants deficient in ACTIN-RELATED PROTEIN7 (ARP7) and ARP4 expression show a delay in abscission in addition to retarded root growth, altered flower development, and reduced fertility (Kandasamy et al., 2005a, 2005b). Similarly, mutations in the AUXIN RESPONSE FACTOR2 (ARF2) gene alone delay the onset of floral abscission and are further enhanced by the loss of ARF1 activity, or by the loss of both ARF7 and ARF19 activities (Ellis et al., 2005; Okushima et al., 2005). In addition, double mutants of the blade-on-petiole1 (bop1) and bop2 genes, encoding NONEXPRESSOR OF PR GENES1–like transcription factors, are totally deficient in abscission and also display a number of other pleiotropic effects, most notably leafy petioles and asymmetric changes in growth (Hepworth et al., 2005; Norberg et al., 2005).
The mutant inflorescence deficient in abscission (ida) has a total block of the abscission process and shows no additional aberrant phenotypes (Butenko et al., 2003). The IDA gene appears to encode a novel putative peptide ligand, based on its small size (77 amino acids), high pI, and N-terminal hydrophobic potential signal peptide that is predicted to direct the mature protein to the secretory pathway (Butenko et al., 2003).
Techniques used to monitor abscission, such as the measurement of petal breakstrength, scanning electron microscopy, and the expression patterns of molecular markers, suggest that the ida mutant is deficient in the later stages of cell separation and that IDA acts either as a promoter of cell separation or as an inhibitor of a repair process that otherwise would be triggered by initial loosening of the middle lamella (Butenko et al., 2003).
An alternative to studies of mutant phenotypes to discern gene function is to investigate phenotypes generated by the overexpression of a given gene product. To elucidate the abscission process in general and test the two hypotheses on IDA function in particular, Arabidopsis plants were transformed with a construct driving IDA expression with the strong constitutive cauliflower mosaic virus 35S promoter. Overexpression of IDA resulted in striking phenotypes, including the ectopic loss of organs that normally do not abscise in Arabidopsis. The shedding of these organs, as well as premature floral organ abscission, was accompanied by the secretion of a white substance that comprised a polysaccharide containing high levels of arabinose and galactose, indicative of an arabinogalactan. The implications of IDA-induced abscission are discussed.
RESULTS
Ectopic Abscission Is Induced by Overexpression of IDA
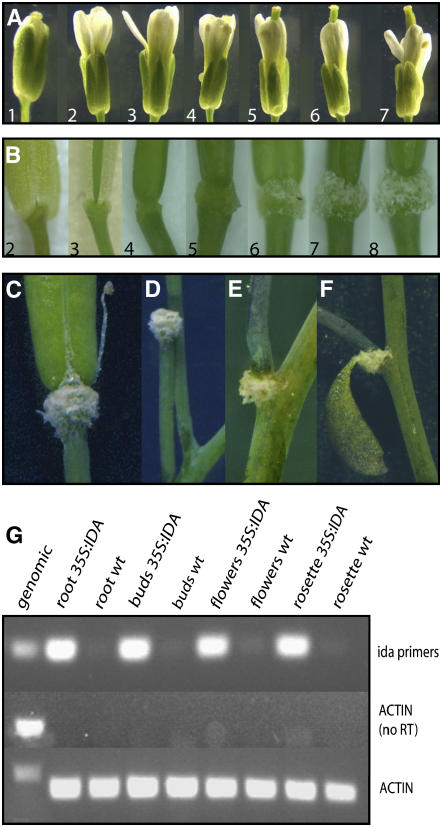
To elucidate the function of IDA, the corresponding cDNA was overexpressed in Arabidopsis driven by the 35S constitutive promoter. After antibiotic selection, 41 primary transformants were retrieved and inspected for aberrant phenotypes. The phenotypes described below were also observed in progeny plants of the second and third generations. Kanamycin selection of seeds was used to identify 35S:IDA lines with single T-DNA loci. The aberrant phenotypes, however, were observed in single- and multiple-locus sublines as well as in homozygous and hemizygous plants. We first investigated whether overexpression of IDA had an effect on floral abscission. The 35S:IDA transgenic plants consistently abscised their floral organs at an earlier stage than wild-type plants: whereas abscission of all floral organs had taken place in wild-type plants by position 10 (counted from the first flower with visible white petals at the top of the inflorescence; Figure 1A) (Butenko et al., 2003), organ separation had occurred by position 4 in all 35S:IDA flowers (Figure 1B). In the subsequent positions, the AZ gradually enlarged, and from position 6 the AZ was covered by a white substance (Figures 1B and 1C).
Figure 1.
Phenotype of 35S:IDA Plants.
(A) Flowers and siliques of a wild-type plant from positions 1 to 7 along the inflorescence, as indicated.
(B) Flowers and siliques of a 35S:IDA plant from positions 2 to 8 along the inflorescence, as indicated. Position 3, abscised sepals; position 4, abscised sepals, petals, and stamens; position 5, enlarged AZ region; positions 6 to 8, secreted white substance.
(C) Full-grown green silique with the white substance covering the AZ, premature opening of the valves, and secretion of the white substance along the dehiscence zone.
(D) Pedicel after shedding of the silique.
(E) Secretion of the white substance and abscission at the base of a branch.
(F) Secretion of the white substance at the base of a cauline leaf about to abscise.
(G) RT-PCR analysis of different tissues from 35S:IDA and wild-type plants, as indicated, using primers amplifying a 237-bp IDA fragment. Primers for ACTIN2-7 were used to amplify a 255-bp fragment from all tissues as a positive control or a 340-bp fragment from genomic DNA. Actin without RT was used as a negative control.
The plants were also inspected for other changes in morphology and development (Figures 1C to 1F) (Aalen et al., 2006). Premature dehiscence of the silique valves and the secreted substance were seen in immature and mature green siliques (Figure 1C). Intriguingly, in many cases (239 of 871 siliques, or 27%), floral abscission continued to the extent that the whole silique was shed and the remaining fracture plane was covered with the white substance (Figure 1D). The percentage of siliques shed increased as the age of the inflorescence increased. In inflorescences with 10 to 15 flowers, 31.5% of the siliques were shed, whereas 42.1 and 51.1% were shed when the inflorescence had developed 16 to 20 and 21 to 25 flowers, respectively. In addition, ectopic abscission occurred in side branches of the inflorescence (Figure 1E), in cauline leaves (Figure 1F), and at the base of the pedicel (Figures 2I to 2M). In wild-type plants, 94 of 97 (97%) branching points had a cauline leaf attached, whereas in 35S:IDA plants, cauline leafs were observed in only 35% of the branching points inspected (n = 60). Surprisingly, seed abscission was unaffected in all plants examined, and the same was the case for rosette leaves, although they showed premature senescence (data not shown).
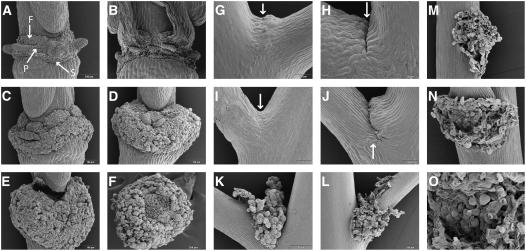
Figure 2.
Scanning Electron Micrographs of Floral AZs and the Base of the Pedicel.
(A) Wild-type AZ of sepals (S), petals (P), and filaments (F) after abscission, as indicated.
(B) Mutant ida AZ with broken cells after forcible removal of floral organs.
(C) Enlarged 35S:IDA AZs.
(D) 35S:IDA AZ with a dramatically greater number of rounded cells.
(E) Gynophore detached from the 35S:IDA AZ.
(F) 35S:IDA AZ showing broken cells where the gynophore was attached.
(G) Wild-type pedicel vestigial AZ with a band of small cells (arrow).
(H) Wild-type pedicel vestigial AZ. A small cleft (arrow) developed in older pedicels.
(I) and (J) Pedicel AZs in 35S:IDA plants at stages comparable to the wild type shown in (G) and (H).
(K) and (L) Pedicel AZs in 35S:IDA plants, later stages.
(M) Base of the pedicel in a 35S:IDA plant showing rounded cells after abscission of the pedicel.
(N) AZ showing rounded cells after shedding of the cauline leaf in a 35S:IDA plant.
(O) Close-up of the cells in the cauline leaf AZ.
RT-PCR was used to verify that there was high IDA expression in the different organs in 35S:IDA plants (Figure 1G). In IDA:β-glucuronidase (GUS) transgenic plants, GUS is expressed only in the floral AZ (Butenko et al., 2003). Consistent with this, RT-PCR only detected very weak expression in RNA from flowers, whereas the IDA cDNA was easily amplified from all tested organs of 35S:IDA plants with 30 PCR cycles.
35S:IDA Plants Develop a Highly Increased Number of Rounded AZ Cells
Floral AZs and putative vestigial AZs at the base of the pedicel from wild-type and 35S:IDA plants were investigated more closely using scanning electron microscopy. After floral organ separation took place in wild-type plants, the AZ region displayed rounded cells and the actual AZs for each single organ (sepals, petals, and filaments) were clearly visible and distinguishable from the protruding nectaries (Figure 2A). In 35S:IDA, each AZ was substantially larger (Figure 2C), and the AZ region continued to increase in size until it engulfed the base of the silique (Figure 2D). In cases in which the silique was shed (Figures 2E and 2F), it was apparent that the silique was not lost as a result of cell wall loosening and separation of the cell layers at the gynophore. Rather, the broken cells at the fracture plane (Figure 2F), similar to those seen in the petal AZ of the mutant ida (Figure 2B), suggest that the loss of the fruit was an indirect effect of the overdeveloped floral AZ region.
Many plants, albeit not Arabidopsis, can shed their flowers or fruits as a result of active abscission at the base of the pedicel. In wild-type plants, a band of small cells was seen in the upper side of the pedicel base (Figure 2G), whereas in older pedicels, a small cleft was present in this region, suggesting that degradation of the middle lamella may have taken place (Figure 2H). In 35S:IDA plants, the base of the pedicel initially looked no different from that of wild-type plants (Figure 2I); however, a deeper cleft developed and the stem side region on the upper side showed enlarged cells (Figure 2J). At a later stage, rounded cells with a similar appearance to floral AZ cells appeared in this region (Figure 2K). Before pedicel abscission, wall loosening between cells seemed to spread from the upper region to the lower region and outward along the pedicel (Figure 2L). After the pedicel had been shed, rounded AZ cells were seen throughout the fracture plane (Figure 2M). This was also observed in cauline leaf AZs (Figures 2N and 2O). In contrast with the loss of siliques, no broken cells were observed in the fracture plane at the base of the pedicel or in the AZs of cauline leaves (Figures 2M to 2O), indicating that abscission at these positions in 35S:IDA plants resulted from true abscission throughout the pedicel and cauline leaves.
Vestigial AZs Are Found at the Bases of Pedicels, Branches, and Cauline Leaves
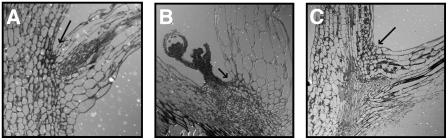
The phenotype of the 35S:IDA plants could possibly be explained by the activation of dormant, vestigial AZs. The AZ cells of the floral organs are characteristically small and densely cytoplasmic, and scanning electron microscopy investigation of the base of the pedicel suggested the presence of differentiated AZ cells with a capacity for cell separation. Light microscopic analysis of thin sections from the regions at the bases of the organs lost in the 35S:IDA lines was used to identify AZ in the wild-type organs. Wild-type plants developed small cells with typical characteristics of AZ cells at the upper region of the organ to be shed (as indicated with arrows in Figure 3). The AZs were seen in branching points (Figure 3A), cauline leaves (Figure 3B), and at the base of the pedicel (Figures 2G, 2H, and 3C).
Figure 3.
Light Microscopic Analysis of Thin Sections of the Regions at the Bases of Organs in Wild-Type AZ Cells (Arrows).
(A) Branching point.
(B) Base of a cauline leaf.
(C) Base of a pedicel.
Arabinogalactan Is Secreted in AZs upon Abscission
The rounded AZ cells seen in 35S:IDA plants (Figure 2) were gradually covered by a white substance as abscission proceeded (Figures 1B and 1C). This substance did not stain with aniline blue, which indicated that the secretion was not attributable to callose deposition (data not shown). Neutral sugar analysis revealed that the substance was highly enriched in arabinose (28%) and galactose (24%) and, to a lesser degree, glucose (Table 1), suggesting that arabinogalactan constitutes a large proportion of the sample. Smaller amounts of xylose, mannose, fucose, and rhamnose were also present.
Table 1.
Sugar Composition of the Secreted Substance
| Sugar | Mol % |
|---|---|
| Rhamnose | 7.83 ± 3.57 |
| Fucose | 2.34 ± 0.62 |
| Arabinose | 28.27 ± 1.17 |
| Xyclose | 11.97 ± 2.18 |
| Mannose | 6.89 ± 0.26 |
| Galactose | 23.61 ± 1.75 |
| Glucose | 19.10 ± 3.35 |
Values are expressed as averages of the molar percentages of three independent experiments ± sd.
AGP Is Present during the Normal Floral Abscission Process and in Excess in 35S:IDA Plants
To confirm the nature of the secreted substance, it was treated with the synthetic chemical reagent β-d-glucosyl Yariv (β-GlcY) (Yariv et al., 1962, 1967), which is considered diagnostic for AGPs and to which it reversibly binds, stains, and precipitates. By contrast, the α-d-glucosyl reagent (α-GlcY) does not bind AGPs and was used as a control. Although the mechanism for the interaction between AGPs and β-GlcY is not fully understood, β-GlcY has been used to perturb the normal functions of AGPs and to detect, quantify, and purify AGPs (Gaspar et al., 2001; Guan and Nothnagel, 2004).
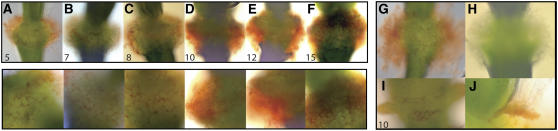
Flowers at different developmental stages from 35S:IDA plants were treated with the β-GlcY, and the presence of AGP was evident from the formation of a red precipitate (Figure 4). Red staining was first observed in position 5 flowers in the periphery of AZ cells (Figure 4A), suggesting that AGP was secreted by the AZ cells. Thereafter, increasing amounts of red-stained precipitate were detected (Figures 4B to 4G), to the point that the AZ cells were fully covered, confirming that at least part of the white secreted substance was AGP. 35S:IDA plants stained with α-GlcY showed no red precipitate (Figure 4H).
Figure 4.
Identification of Arabinogalactan by the Yariv Reagent β-GlcY.
(A) to (F) Staining of siliques by β-GlcY, resulting in a red precipitate. Positions are numbered. The bottom row shows greater magnifications of the stained AZs.
(G) Mature silique stained with β-GlcY.
(H) Mature silique stained with the negative control α-GlcY.
(I) Position-10 silique of a wild-type plant stained with β-GlcY.
(J) Separating region of a 35S:IDA filament stained with β-GlcY.
The secretion of AGP in 35S:IDA plants could possibly be an artifact of premature and ectopic abscission, so the wild-type AZs were examined for the presence of AGP. No staining reaction was apparent before organ separation (data not shown). However, at position 10, a low amount of precipitate was detected (Figure 4I). Developmentally, this stage in the abscission process was already reached in the 35S:IDA plants at position 5 (i.e., when a positive β-GlcY reaction was first detected). This suggests that AGP secretion is a normal part of the abscission process, an idea further supported by the presence of an increased accumulation of AGP in the separating region of a 35S:IDA filament (Figure 4J). In the ida mutant, which is blocked in the final stages of the process, β-GlcY precipitate was not detected (data not shown).
AGP24 Is Upregulated in the 35S:IDA Floral AZ
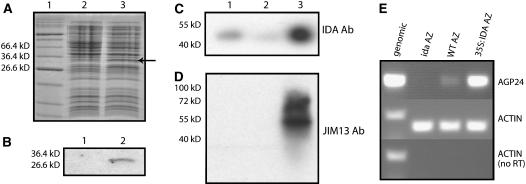
The absence of AGP in the ida mutant could be a direct or an indirect effect, because IDA itself could be an AGP, or the presence of IDA could induce the secretion of an AGP. To investigate these possibilities, an IDA antibody was produced against a short peptide in the middle part of the protein. To confirm that the antibody recognized the IDA protein, a glutathione S-transferase (GST):IDA fusion construct was introduced in Escherichia coli. The antibody detected a protein of the expected size (30 kD) only in E. coli cultures in which expression of the fusion protein had been induced (Figures 5A and 5B). The antibody was used for protein gel blot analysis of protein isolated from AZs of 35S:IDA, wild-type, and ida mutant plants. In the AZ protein samples, the IDA antibody recognized a protein in wild-type plants that was present in significantly higher amounts in 35S:IDA samples but was hardly detectable in samples from the ida mutant (Figure 5C). However, the detected protein was not of the expected size (6.1 kD) but much larger (50 kD), suggesting substantial posttranslational modification.
Figure 5.
IDA and AGP Expression in AZs.
(A) Coomassie blue–stained SDS-PAGE gel of proteins from E. coli cultures harboring a GST:IDA fusion construct, with and without induction with isopropylthio-β-galactoside (lanes 2 and 3, respectively). The induced fusion protein is indicated by an arrow. The sizes of relevant markers of the protein standard (lane 1) are shown at left.
(B) Detection of the GST-IDA fusion protein by protein gel blot analysis of proteins from uninduced (lane 1) or induced (lane 2) E. coli cells using the IDA antibody.
(C) Protein gel blot analysis of 200 μg of protein isolated from AZ tissue of wild-type (lane 1), ida mutant (lane 2), and 35S:IDA (lane 3) plants using the IDA antibody (Ab). Size markers are shown at left.
(D) The same membrane as in (C) probed with JIM13.
(E) RT-PCR analysis with AGP24 primers of cDNA derived from AZs of ida, wild-type (ecotype C24), and 35S:IDA plants, as indicated. Actin primers were used for positive and negative controls with both genomic DNA and cDNA.
Blots first probed with IDA antibody were stripped and reprobed with the JIM13 antibody, which detects AGPs (Knox et al., 1990; Yates et al., 1996). Smeared bands (40 to 100 kD) confirmed the presence of AGPs in the 35S:IDA plants, and no equivalent bands were detected in the wild type or the ida mutant (Figure 5D). Thus, the protein pattern recognized by the JIM13 antibody (Yates et al., 1996) in the ida mutant, wild-type, and 35S:IDA AZ samples does not correlate with the pattern seen with the IDA antibody.
Because there are numerous putative AGP-encoding genes in Arabidopsis, total RNAs from 35S:IDA and wild-type AZ were used in a microarray experiment to search for AGP genes that were upregulated in the IDA-overexpressing line (data not shown). Candidate genes were tested for expression levels in RNA samples from ida mutant, wild-type, and 35S:IDA AZs. Although several AGP genes were expressed in wild-type AZs, only AGP24 showed significant differences in transcript levels in these samples (Figure 5E). Whereas AGP24 RNA could not be detected in the ida mutant, the AGP transcript level was clearly upregulated in 35S:IDA.
DISCUSSION
Floral AZs Are Responsive to IDA When Cell Wall Loosening Is Initiated
We have previously shown that the IDA gene is specifically expressed in floral AZs and is necessary for abscission, as mutant ida plants are totally deficient in abscission (Butenko et al., 2003). In IDA:GUS transgenic plants, expression was observed from position 5, whereas abscission normally takes place at position 10 (Butenko et al., 2003). However, floral AZs must be present and responsive to cell separation signals at earlier positions, as plants exposed to high concentrations of ethylene can shed floral organs at position 1 (Butenko et al., 2003). The 35S:IDA overexpression lines abscise their floral organs earlier than do wild-type plants, at position 4 (Figures 1A and 1B). Petal breakstrength data suggested that cell wall loosening initiates at positions 4 to 6 (Butenko et al., 2003). The premature abscission in 35S:IDA plants shows that the floral AZ is responsive to the expression of IDA at this stage. However, abscission was not seen as early as in plants exposed to ethylene, suggesting that ethylene, in contrast with IDA, also may trigger an earlier onset of the first steps of cell separation.
Our results clearly demonstrate that the differentiation of the AZ and the process and timing of cell separation are under independent control. A differentiated AZ, however, is competent to respond if given the appropriate stimulus.
Organs Other Than Petals, Sepals, and Filaments Are Capable of Abscission in Arabidopsis
In many plants, the base of the pedicel contains a functional AZ (Addicott, 1982). It has been argued that in Arabidopsis, this anatomical region is not a physiologically genuine AZ and that this explains why abscission does not spontaneously take place at the pedicel (Cho and Cosgrove, 2000). However, we show that the expression of IDA is sufficient to induce the abscission of pedicels and cauline leaves as well as branches on the inflorescence. This implies that AZ cells are differentiated at the bases of these organs and therefore they are responsive to the presence of IDA. Both scanning electron microscopy and light microscopic inspection of thin sections demonstrated the presence of the expected bands of small cells (Figures 2 and 3).
In floral organs, the initial steps of cell separation start before IDA is expressed, which indicates that such initial steps are needed for IDA action. The small cleft seen in wild-type pedicels (Figure 2H), as well as the expression of abscission-associated genes such as HAESA, which encodes a receptor-like kinase (Jinn et al., 2000), BOP1 (Norberg et al., 2005), and the new abscission marker BASIL (Stangeland et al., 2003), also indicate that elements required for abscission are present. However, other genes that are not expressed in floral AZs may also influence the tendency of the pedicel to abscise at its base (e.g., overexpression of the expansin-encoding gene ATEXPA10 may lead to pedicel separation) (Cho and Cosgrove, 2000).
Overexpression of IDA Induces Cell Wall Breakdown in Cells Neighboring the AZ
Although the floral AZ consists of several layers of cells, not all cells in these cell files behave the same way and cell separation occurs between a subset of cells within the zone. This suggests intercellular communication within the layers, so that only a relatively small proportion of the cells recognize and/or initially respond to the primary abscission signal. For instance, Arabidopsis polygalacturonase PGAZAT expression is restricted to certain cells within the separation layer (Gonzalez et al., 2002; Roberts et al., 2002), and such an expression profile of multiple wall-modifying enzymes may be an important prerequisite for precisely coordinated cell wall degradation.
In the 35S:IDA overexpression lines, cell wall degradation appeared less controlled and restricted. This was noticeable in the floral AZ, where the number of rounded cells, which presumably have experienced cell wall loosening, was increased; in some cases, this even led to loss of the silique. We suspect that the adhesive forces in the gynophore were insufficient to keep the silique in place when cell wall degradation was taking place in additional cell files. In the pedicel, the rounded cells are not restricted to those that initially resemble AZ cells (cf. Figures 2K and 2L with Figure 3C), indicating that the ectopic and prolonged temporal expression of IDA in the overexpressing lines led to an expansion in the region where cell separation occurs. Our hypothesis is that overexpression of IDA not only triggers cell separation in additional AZ cells that are responsive to IDA but that this also leads to prolonged expression of hydrolytic enzymes affecting neighboring cells.
In many plant species, additional cell division takes place during the process of organ shedding (Osborne, 1989); however, it has been suggested that this is not the case for Arabidopsis (Patterson, 2001). The increased number of AZ cells seen in the 35S:IDA overexpression lines (Figure 2) may indicate that cell division could be a component of Arabidopsis floral abscission. A careful reexamination of the wild-type AZ, especially before the formation of the protective layer, would have to be undertaken to clarify this issue.
AGP Is Present in the Cell Wall during the Separation of Floral Organs
In addition to the ectopic abscission, the most dramatic phenotype of the 35S:IDA plants was the secretion of a white substance at the AZs. This material was not callose, which is often synthesized in wounded tissues, but reacted with the Yariv reagent diagnostic of AGPs (Figure 4). Neutral sugar analysis showed that the substance contained polysaccharides that, compared with cell wall extracts from Arabidopsis leaves (Zablackis et al., 1995; Reiter et al., 1997; Burget and Reiter, 1999), were especially enriched in arabinose and galactose (Table 1). These are the main glycosyl components of AGPs and form the type II arabinogalactan backbone, which is attached to multiple sites on the core protein and can then be further substituted with other sugars, such as l-rhamnose, l-fucose, d-xylose, d-mannose, d-glucose, and d-glucosamine, and uronic acids (Showalter, 2001). In some cases, short arabinose oligosaccharides are also attached to the protein backbone (Showalter, 2001). Other neutral monosaccharides, such as xylose, mannose, and fucose, were also found in lesser amounts in the 35S:IDA secretions (Table 1). Two explanations for this could be further substitution of the AGP backbone with such sugars and the presence of other wall polysaccharides in the secreted material.
Interestingly, the cross-reacting band recognized by the IDA antibody in wild-type AZs and at increased levels in 35S:IDA (Figure 5C), which was absent in the ida mutant plants, was much larger than expected, suggesting that the IDA protein is posttranslationally modified. However, IDA is unlikely to be the overexpressed AGP. First, although IDA contains a Pro (P)-rich C-terminal motif (PIP) (Butenko et al., 2003), the Ps are not found in a context typical of AGP core proteins, such as A-P-A-P or (S/T)-P, where the Ps are predicted to be hydroxylated and O-glycosylated (Schultz et al., 2000). Second, the JIM13 antibody recognized a smear of glycosylated proteins only in the AZs of 35S:IDA plants (Figure 5D), but the protein recognized by the IDA antibody was present in wild-type AZ from positions 5 to 8 (Figure 5C). Third, β-GlcY first reacted with AGP in wild-type plants at position 10 (Figure 4I), whereas IDA gene expression, as determined previously using IDA:GUS transgenic plants, occurs in AZ cells from positions 5 to 10 (Butenko et al., 2003).
Therefore, the accumulation of AGP seen in AZ regions in 35S:IDA plants is likely to be caused by the constitutive expression of IDA. In wild-type plants, in which IDA is absent in AZ cells past position 10 (Butenko et al., 2003), AGP overexpression does not occur, indicating that the secreted AGP is under the control of IDA. The presence of AGP as a constituent of the wild-type AZ cell wall during and soon after the completion of cell separation suggests that it is necessary for the separation process.
In Arabidopsis, there are 47 AGPs, which are found either associated with the carbohydrates in the cell wall or embedded in the plasma membrane via a glycosylphosphatidylinositol (GPI) anchor (Schultz et al., 2000). Our results suggest that at least some of the overexpressed AGP is AGP24, because AGP24 transcripts were absent in the ida mutant, present at very low levels in wild-type AZs, and upregulated in 35S:IDA plants (Figure 5E). AGP24 is predicted to be anchored to GPI (Borner et al., 2003) and has been isolated as an arabinogalactan peptide from Arabidopsis seedlings. The mature protein, from which the N-terminal signal peptide and the GPI signal have been removed, is only 17 amino acids long and can exist in vivo in a fully hydroxylated form (HEGHHHHAOAOAOGOAS) (Schultz et al., 2004). AGP24 is most highly expressed in pollen, but it is also present in siliques and roots, according to data compiled by Genevestigator (https://www.genevestigator.ethz.ch/at/).
IDA Is a Positive Activator of Cell Separation
IDA has been proposed to act either in a positive signaling pathway, in which IDA expression triggers the final separation step of abscission, or in a negative signaling pathway, in which IDA blocks a repair process (Butenko et al., 2003). The ectopic abscission induced in Arabidopsis lines overexpressing IDA, as well as the premature floral abscission in these lines, support the first hypothesis.
IDA-mediated induction of AGP implies a role for AGP in the abscission signaling pathway. Because different AGP family members have been shown to be expressed in tissue-specific patterns, it has been speculated that AGPs have crucial functions in plant growth and development (Kohorn, 2000). Cleaved AGPs, with their highly complex and variable glycan chains, are ideal candidates for diffusible signaling molecules (Pilling and Hofte, 2003). The discovery that chitinases can cleave the N-acetyl-d-glucosaminyl residues present in AGPs (van Hengel et al., 1998) raises the possibility that AGPs act as a signal in AZ cells to coordinate cell wall separation. AGP24 was shown to be expressed in both 35S:IDA plants and wild-type plants before organ shedding but after the initial cell wall loosening had started, suggesting that this specific AGP may be involved in regulating cell separation in floral AZs.
Another consideration is that AGPs may play an indirect role in wall remodeling during or after abscission, because they have been shown to increase the activity of the wall-modifying proteins XTHs (Takeda and Fry, 2004). These enzymes cut and rejoin xyloglucan polymers in the cell wall, which, as part of the load-bearing cellulose–xyloglucan network, are attributed a vital structural role (Cosgrove, 2005). XTH action may thus be important for wall restructuring after enlargement of AZ cells after cell separation. Thus, an alternative hypothesis is that an important final step in the abscission process is IDA induction of AGP24 expression, leading to the breakdown and restructuring of xyloglucans in the walls of AZ cells.
In the 35S:IDA lines, strong IDA expression was found in all organs tested (Figure 1G). Obviously, the presence of IDA is not sufficient to induce middle lamella breakdown between any cells, because leaves, stems, and other organs are intact in these plants. The lack of abscission of rosette leaves in 35S:IDA plants suggests that in Arabidopsis, rosette AZs are not developed or that other indispensable components are not in place for IDA-induced abscission to occur. Leaf abscission may be dispensable in plants such as Arabidopsis, which has a short life cycle, and the ability to abscise rosette leaves may have been lost during evolution. The lack of premature seed dehiscence in 35S:IDA plants suggests the existence of crucial differences between abscission and seed dehiscence mechanisms. The prerequisites for IDA-dependent cell separation in Arabidopsis, however, were present in floral organ AZs, at the bases of pedicels and cauline leaves, and in dehiscence zones of the valves. An intriguing possibility is that plant species that shed organs other than petals, sepals, and anthers have a different expression pattern of putative IDA orthologs.
METHODS
Plant Material and Generation of Transgenic 35S:IDA Plants
Wild-type (C24) and transgenic Arabidopsis thaliana plants were cultivated in growth chambers at 22°C for 8 h of dark and 16 h of light (100 μE·m−2·s−1). The 35S:IDA construct was made using Gateway cloning technology (Invitrogen). The pK7WG2 Agrobacterium tumefaciens binary vector is based on the backbone of the pPZP200 vector (Karimi et al., 2002). The IDA gene was amplified using the primers idapKWG2ATTB1 (5′-CAATGGCTCCGTGTCGT-3′) and idapKWG2ATTB2 (5′-TCAATGAGGAAGAGAGTTAACAAAAGAG-3′), with additional Gateway att sequences at the 5′ ends, and introduced into the pDONR201 Gateway donor vector. Thereafter, the PCR product was recombined through the entry clone into the pK7WG2 destination vector, generating pK7IDA2. The construct was transferred to A. tumefaciens strain C58C1 pGV2260, and Arabidopsis (ecotype C24) plants were transformed using the A. tumefaciens–mediated floral dip method (Clough and Bent, 1998). Transformants were selected on Murashige and Skoog (1962) medium with 50 μg/mL kanamycin.
Reverse Transcriptase–Mediated Expression Analyses
Total RNA was isolated from 100 mg of different Arabidopsis tissues using the Qiagen RNAeasy kit according to the manufacturer's recommendations. An optional on-column DNase digestion step was included. First-strand cDNA synthesis with SuperScript III reverse transcriptase (Invitrogen) was performed in a total volume of 20 μL using 850 ng of total RNA as a template and incubated at 42°C for 60 min. Experiments in which the reverse transcriptase was omitted were used as negative controls. RT-PCR was performed, and the open reading frame sequence of Arabidopsis ACTIN2 (locus number At3g18780) was used as a positive internal control with primers that span inton 2 (5′-CCGCAAGATCAAGACGAAGGATAGC-3′ and 5′-CCCTGAGGAGCACCCAGTTCTACTC-3′). The primers used for PCR amplification were positioned in the coding region of the IDA gene: IDAF (5′-CAATGGCTCCGTGTCGT-3′) and IDAR (5′-TCAATGAGGAAGAGAGTTAACAAAAGAG-3′). The PCR was run for 30 cycles.
Total RNA was also isolated from 100 mg of ida mutant and C24 wild-type AZs (positions 5 to 8) and 35S:IDA AZs (positions 3 to 6). The primers used for RT-PCR amplification of AGP24 were AGP24 antisense (5′-TTATTCAAGAAATCAGATCAACA-3′) and AGP24 sense (5′-TTGAAGATAATGATGATGATGACG-3′).
Scanning Electron Microscopy and Semithin Sectioning
Plant tissues for electron microscopy were fixed, dehydrated, critical point dried, and sputter coated as described previously (Butenko et al., 2003). Samples were viewed at a 10 K accelerating voltage on a JEOL JSM-6400 scanning electron microscope. Tissues for sectioning were fixed in 4% glutaraldehyde (w/v) and 2% paraformaldehyde (w/v) in 100 mM sodium phosphate buffer, pH 7.4, rinsed four times in buffer, dehydrated in an ethanol series, and embedded in Epon (Fluka). Semithin (3 μm thick) sections were cut with glass knives on a LKB microtome (LKB Fabriken) and heat fixed to glass slides. Sections were stained with basic fuchin and toluidine blue as described (Alsop, 1974). Sections were permanently mounted in Eukitt (Canemco and Marivac) and examined with a Zeiss Axioplan2 imaging microscope equipped with differential interference contrast optics and a cooled Axiocam camera imaging system.
Protein Expression, Isolation, and Protein Gel Blotting
The Gateway DNA cassette B (Invitrogen) digested with BamHI was subcloned into the unique BamHI site in the multiple cloning site of the vector pGEX-2TK (Amersham Biosciences), creating a pGEX-AB GAW. The IDA coding sequence minus the predicted signal sequence was amplified by PCR with Advantage2 polymerase mix (Clontech) from genomic plant DNA with the att primers ida-proteinC (5′-TGGTAGCGGCTGCAAGAATTG-3′) and ida-proteinB (5′-TGATCTAATCTACAAAACCAAACCT-3′) and recombined into pDONOR201 (Invitrogen), creating the pDONOR01–IDA construct. This construct was used to recombine the IDA sequence into pGEX-AB GAW to give pGEX-AB-IDA, from which IDA can be expressed in fusion with GST.
The pGEX-AB-IDA vector was used for high-level expression of IDA protein in BL21 SI cells. Cultures were grown ampicillin (100 μg/mL) up to OD600 = 0.8 to 1 and divided in two. In one, isopropylthio-β-galactoside (Sigma-Aldrich) was added to a final concentration of 0.2 mM to induce the fusion protein. The two cultures were incubated for 2.5 h at 28°C with shaking. Bacterial pellets were dissolved in loading buffer (62 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 0.7 M 2-β-mercaptoethanol, and 0.0012% bromphenol blue) and used for protein SDS-PAGE. The SDS gel was stained with Coomassie Brilliant Blue (0.1% Coomassie Brilliant Blue G 250 in 40% methanol and 10% acetic acid) for 30 min.
Isolation of total protein from AZs, ida and C24 AZ positions 5 to 8 and 35S:IDA positions 3 to 6, SDS-PAGE, and protein gel blotting were performed as described by Stacy et al. (1999). Protein samples were separated by SDS-PAGE on 12% gels and subjected to protein gel blot analysis using anti-IDA and JIM13 antibodies (Yates et al., 1996). The IDA antibody was made against amino acids 32 to 46 (TMEMKKNIKRLTFKN). Both primary antibodies were diluted 1:1500. The secondary antibodies, anti-rabbit for IDA and anti-rat for JIM13, were diluted 1:5000. Antibody binding was detected using Pierce SuperSignal West Femto Maximum as specified by the manufacturer.
Glycosyl Composition Analysis
Approximately 0.5 mg of the secreted substance was hydrolyzed in 1 mL of 2 M trifluoroacetic acid at 120°C for 90 min. The supernatant was then evaporated at 42°C in a stream of air. The sugars were reduced with NaBH4, and alditol acetates were prepared according to Gibeaut and Carpita (1991). The alditol acetate derivatives were separated and analyzed by gas chromatography (Agilent 6890 system) with a 30-m × 0.25-mm SP-2330 column (Supelco). The temperature program was as follows: 80 to 175°C at 25°C/min, and then 175 to 240°C at 5°C/min, with a 10-min hold at the higher temperature.
Staining with β-GlcY
β-GlcY (Biosupplies) was used to test for the presence of AGPs, whereas α-GlcY (Biosupplies) was used as a negative control. Plant tissue was incubated in a solution containing 2 mg/mL Yariv reagent in 0.15 M NaCl for 1 h at room temperature and then washed five times with water. The samples were inspected with a Zeiss Axioplan2 imaging microscope.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT1G68765 (IDA) and AT5G40730 (AGP24).
Acknowledgments
We are grateful for the technical assistance of Tove Bakar and Torill Rolfsen at the Electron Microscopy Facility at the Department of Molecular Biosciences, University of Oslo. We thank Sara Patterson for the photographs in Figures 1B to 1E. Thanks also to Roy Falleth and Solveig H. Engebretsen for technical assistance and to Trine Hvolsef-Eide for providing anti-rat secondary antibody. The Norwegian Research Council supported this work (Grants 129525/420 and 158872/I10). This work was facilitated by the Norwegian Arabidopsis Research Center, which is part of the Norwegian Research Council's National Program for Research in Functional Genomics.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Reidunn B. Aalen (reidunn.aalen@imbv.uio.no).
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.106.042036.
References
- Aalen, R.B., Butenko, M.A., Stenvik, G.-E., Tandstad, N.M., and Patterson, S.E. (2006). Genetic control of floral abscission. In Floriculture, Ornamental and Plant Biotechnology: Advances and Topical Issues, Vol. 1, J.T. da Silva, ed (London: Global Science Books), pp. 101–108.
- Addicott, F.T. (1982). Abscission. (London: University of California Press).
- Alsop, D.W. (1974). Rapid single-solution polychrome staining of semithin epoxy sections using polyethylene glycol 200 (PEG 200) as a stain solvent. Stain Technol. 49 265–272. [DOI] [PubMed] [Google Scholar]
- Bleecker, A., and Patterson, S.E. (1997). Last exit: Senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9 1169–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G.H., Lilley, K.S., Stevens, T.J., and Dupree, P. (2003). Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 132 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burget, E.G., and Reiter, W.D. (1999). The mur4 mutant of Arabidopsis is partially defective in the de novo synthesis of uridine diphospho l-arabinose. Plant Physiol. 121 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko, M.A., Patterson, S.E., Grini, P.E., Stenvik, G.E., Amundsen, S.S., Mandal, A., and Aalen, R.B. (2003). INFLORESCENCE DEFICIENT IN ABSCISSION controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15 2296–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H.T., and Cosgrove, D.J. (2000). Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97 9783–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6 850–861. [DOI] [PubMed] [Google Scholar]
- Ellis, C.M., Nagpal, P., Young, J.C., Hagen, G., Guilfoyle, T.J., and Reed, J.W. (2005). AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132 4563–4574. [DOI] [PubMed] [Google Scholar]
- Gaspar, Y., Johnson, K.L., McKenna, J.A., Bacic, A., and Schultz, C.J. (2001). The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol. Biol. 47 161–176. [PubMed] [Google Scholar]
- Gibeaut, D.M., and Carpita, N.C. (1991). Tracing the biosynthesis of the cell wall in intact cells and plants. Selective turnover and alteration of cytoplasmic and cell wall polysaccharides of proso millet cells in liquid culture and Zea mays seedlings. Plant Physiol. 97 551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, C.Z.H., Whitelaw, C.A., Swarup, R., and Roberts, J.A. (2002). Temporal and spatial expression of a polygalacturonase during leaf and flower abscission in oilseed rape and Arabidopsis. Plant Physiol. 128 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, Y., and Nothnagel, E.A. (2004). Binding of arabinogalactan proteins by Yariv phenylglycoside triggers wound-like responses in Arabidopsis cell cultures. Plant Physiol. 135 1346–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth, S.R., Zhang, Y., McKim, S., Li, X., and Haughn, G.W. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17 1434–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn, T.L., Stone, J.M., and Walker, J.C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14 108–117. [PMC free article] [PubMed] [Google Scholar]
- Kandasamy, M.K., Deal, R.B., McKinney, E.C., and Meagher, R.B. (2005. a). Silencing the nuclear actin-related protein AtARP4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J. 41 845–858. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M.K., McKinney, E.C., Deal, R.B., and Meagher, R.B. (2005. b). Arabidopsis ARP7 is an essential actin-related protein required for normal embryogenesis, plant architecture, and floral organ abscission. Plant Physiol. 138 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Knox, J.P., Linstead, P.J., King, J., Cooper, C., and Roberts, K. (1990). Pectin esterification is spatially regulated both within walls and between developing tissues of root apices. Planta 181 512–521. [DOI] [PubMed] [Google Scholar]
- Kohorn, B.D. (2000). Plasma membrane-cell wall contacts. Plant Physiol. 124 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, M.W., Leslie, M.E., and Liljegren, S.J. (2006). Plant separation: 50 ways to leave your mother. Curr. Opin. Plant Biol. 9 59–65. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Norberg, M., Holmlund, M., and Nilsson, O. (2005). The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132 2203–2213. [DOI] [PubMed] [Google Scholar]
- Okushima, Y., Mitina, I., Quach, H.L., and Theologis, A. (2005). AUXIN RESPONSE FACTOR 2 (ARF2): A pleiotropic developmental regulator. Plant J. 43 29–46. [DOI] [PubMed] [Google Scholar]
- Osborne, D.J. (1989). Abscission. CRC Crit. Rev. Plant Sci. 8 103–129. [Google Scholar]
- Patterson, S.E. (2001). Cutting loose. Abscission and dehiscence in Arabidopsis. Plant Physiol. 126 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, S.E., and Bleecker, A.B. (2004). Ethylene-dependent and -independent processes associated with floral organ abscission in Arabidopsis. Plant Physiol. 134 194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., Sander, L., Child, R., van Onckelen, H., Ulvskov, P., and Borkhardt, B. (1996). Isolation and characterisation of a pod dehiscence zone-specific polygalacturonase from Brassica napus. Plant Mol. Biol. 31 517–527. [DOI] [PubMed] [Google Scholar]
- Pilling, E., and Hofte, H. (2003). Feedback from the wall. Curr. Opin. Plant Biol. 6 611–616. [DOI] [PubMed] [Google Scholar]
- Reiter, W.D., Chapple, C., and Somerville, C.R. (1997). Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. Plant J. 12 335–345. [DOI] [PubMed] [Google Scholar]
- Roberts, J.A., Elliott, K.A., and Gonzalez-Carranza, Z.H. (2002). Abscission, dehiscence, and other cell separation processes. Annu. Rev. Plant Biol. 53 131–158. [DOI] [PubMed] [Google Scholar]
- Rose, J.K.C., Catalá, C., Gonzalez-Carranza, Z.H., and Roberts, J. (2003). Plant cell wall disassembly. In The Plant Cell Wall, J.K.C. Rose, ed (Oxford, UK: Blackwell Publishing), pp. 264–324.
- Schultz, C.J., Ferguson, K.L., Lahnstein, J., and Bacic, A. (2004). Post-translational modifications of arabinogalactan-peptides of Arabidopsis thaliana. Endoplasmic reticulum and glycosylphosphatidylinositol-anchor signal cleavage sites and hydroxylation of proline. J. Biol. Chem. 279 45503–45511. [DOI] [PubMed] [Google Scholar]
- Schultz, C.J., Johnson, K.L., Currie, G., and Bacic, A. (2000). The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12 1751–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton, R., and Roberts, J.A. (1982). Cell biology of abscission. Annu. Rev. Plant Physiol. 33 133–162. [Google Scholar]
- Showalter, A.M. (2001). Arabinogalactan-proteins: structure, expression and function. Cell. Mol. Life Sci. 58 1399–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy, R.A.P., Nordeng, T.W., Culianez, M.F.A., and Aalen, R.B. (1999). The dormancy-related peroxiredoxin anti-oxidant, PER1, is localized to the nucleus of barley embryo and aleurone cells. Plant J. 19 1–8. [DOI] [PubMed] [Google Scholar]
- Stangeland, B., Salehian, Z., Aalen, R., Mandal, A., and Olsen, O.A. (2003). Isolation of GUS marker lines for genes expressed in Arabidopsis endosperm, embryo and maternal tissues. J. Exp. Bot. 54 279–290. [DOI] [PubMed] [Google Scholar]
- Takeda, T., and Fry, S.C. (2004). Control of xyloglucan endotransglucosylase activity by salts and anionic polymers. Planta 219 722–732. [DOI] [PubMed] [Google Scholar]
- van Hengel, A.J., Guzzo, F., van Kammen, A., and de Vries, S.C. (1998). Expression pattern of the carrot EP3 endochitinase genes in suspension cultures and in developing seeds. Plant Physiol. 117 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yariv, J., Lis, H., and Katchalski, E. (1967). Precipitation of arabic acid and some seed polysaccharides by glycosylphenylazo dyes. Biochem. J. 105 1C–2C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yariv, J., Rapport, M.M., and Graf, L. (1962). The interaction of glycosides and saccharides with antibody to the corresponding phenylazo glycosides. Biochem. J. 85 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates, E.A., Valdor, J.F., Haslam, S.M., Morris, H.R., Dell, A., Mackie, W., and Knox, J.P. (1996). Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology. 6 131–139. [DOI] [PubMed] [Google Scholar]
- Yong, W., et al. (2005). Genomics of plant cell wall biogenesis. Planta 221 747–751. [DOI] [PubMed] [Google Scholar]
- Zablackis, E., Huang, J., Muller, B., Darvill, A.G., and Albersheim, P. (1995). Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol. 107 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]