Abstract
Strong evidence exists for polyploidy having occurred during the evolution of the tribe Brassiceae. We show evidence for the dynamic and ongoing diploidization process by comparative analysis of the sequences of four paralogous Brassica rapa BAC clones and the homologous 124-kb segment of Arabidopsis thaliana chromosome 5. We estimated the times since divergence of the paralogous and homologous lineages. The three paralogous subgenomes of B. rapa triplicated 13 to 17 million years ago (MYA), very soon after the Arabidopsis and Brassica divergence occurred at 17 to 18 MYA. In addition, a pair of BACs represents a more recent segmental duplication, which occurred ∼0.8 MYA, and provides an exception to the general expectation of three paralogous segments within the B. rapa genome. The Brassica genome segments show extensive interspersed gene loss relative to the inferred structure of the ancestral genome, whereas the Arabidopsis genome segment appears little changed. Representatives of all 32 genes in the Arabidopsis genome segment are represented in Brassica, but the hexaploid complement of 96 has been reduced to 54 in the three subgenomes, with compression of the genomic region lengths they occupy to between 52 and 110 kb. The gene content of the recently duplicated B. rapa genome segments is identical, but intergenic sequences differ.
INTRODUCTION
Polyploidy is thought to be an important and recurring feature of genome evolution (Soltis and Soltis, 1995; Wendel, 2000; Blanc and Wolfe, 2004a; Maere et al., 2005). The long-term evolution of polyploid genomes is generally a diploidization process occurring through an extensive genome reorganization at both the gene and chromosome levels (Cronn et al., 1999; Wendel, 2000; Feuillet et al., 2001; Lysak et al., 2005). In the genome of Arabidopsis thaliana, which contains ∼29,000 genes, increases in the sizes of gene families have occurred partly via three rounds of apparent whole genome duplication (1R, 2R, and 3R at ∼350 to 300, ∼170 to 156, and ∼26.7 to 25.0 million years ago [MYA], respectively) and via gradual accumulation of small-scale gene/segmental duplications during the last ∼350 million years (Bowers et al., 2003; Maere et al., 2005).
The genus Brassica provides an opportunity to study the genome changes associated with polyploidy by comparative genomics with the model plant Arabidopsis (Paterson et al., 2001). Brassica and Arabidopsis, which diverged 14.5 to 20.4 MYA from a common ancestor (Blanc et al., 2003; Bowers et al., 2003), belong to the same family, Brassicaceae. There is compelling evidence that the tribe Brassiceae, which comprises ∼240 species, descended from a common hexaploid ancestor with a basic genome similar to that of Arabidopsis (Lysak et al., 2005). Chromosome rearrangements, including fusions and/or fissions, resulted in the present-day chromosome number variation for the three diploid Brassica species, B. nigra (B genome; n = 8), B. oleracea (C genome; n = 9), and B. rapa (A genome; n = 10) (Gale and Devos, 1998; Lysak et al., 2005). The genomes of three allotetraploids, B. juncea (AB; n = 18), B. napus (AC; n = 19), and B. carinata (BC; n = 17), were derived by spontaneous hybridization among the three diploid species, followed by chromosome doubling (U, 1935). The genome size of B. rapa is the smallest at ∼529 Mb per haploid, compared with ∼696 Mb in B. oleracea and ∼632 Mb in B. nigra (Johnston et al., 2005).
Before the recognition of the fundamentally triplicated nature of the genomes of all members of the Brassiceae (Lysak et al., 2005), a number of hypotheses had been put forward to explain the results of some of the comparative analyses conducted in Brassica species (Lukens et al., 2003). Some comparative analyses between B. oleracea linkage maps and the A. thaliana genome identified numerous one-to-one segmental relationships and apparent genome duplications, in addition to genome triplications (Lan and Paterson, 2000; Babula et al., 2003; Lukens et al., 2003). However, extensive comparative analysis within the genome of B. napus, conducted at the level of linkage maps, revealed that triplication was extensive within the genomes of the progenitor diploid species (Parkin et al., 2003), and investigations of regions of the B. oleracea genome also revealed various numbers of related genome segments (Quiros et al., 2001; Suzuki et al., 2003). Thus, although analyses conducted using a single gene-specific probe or amplicon will often fail to identify all related genome segments, analyses of comprehensive sets of related genome segments in Brassica species and A. thaliana showed that triplicated segments could generally be identified in the genome of diploid Brassica species (O'Neill and Bancroft, 2000; Rana et al., 2004; Park et al., 2005). The difficulty in drawing conclusions based on the results of experiments with single gene-specific probes or amplicons was caused by the frequent occurrence of interspersed gene loss from duplicated genome segments.
Gross genome organization is highly collinear between the diploid species B. rapa and B. oleracea, which diverged only ∼4 MYA, and B. napus (Rana et al., 2004). In the Brassicaceae, genomic collinearity is conserved, although insertions/deletions and minor rearrangements are common (Kowalski et al., 1994; Lagercrantz, 1998; Paterson et al., 2001; Schmidt et al., 2001; Lukens et al., 2003). By contrast with B. oleracea, in which expansion of genome segments generally seems to have occurred relative to their Arabidopsis homologs (O'Neill and Bancroft, 2000), homologous genome segments generally seem to be more compact in B. rapa than in Arabidopsis (Yang et al., 2005a).
In plants, regulatory genes are believed to be important for the diversification of plant phenotypes, and alleles of several key regulatory genes that control developmental processes are known to interact in an additive manner (Schranz et al., 2002). Replicated FLOWERING LOCUS C (FLC) genes found in B. rapa, FLC1, FLC2, and FLC3, which are found in the syntenic regions of Arabidopsis, and FLC5, which is found in a nonsyntenic region, appear to have a similar function and interact in an additive manner to modulate flowering time (Schranz and Osborn, 2000; Schranz et al., 2002). We describe the consequences of the diploidization process of the triplicated FLC regions from a hexaploid ancestor by comparative analysis of the genome of the paleopolyploid as well as by reference to a proxy for the ancestral genome, that of A. thaliana. We interpret these in terms of the mechanisms involved in genome evolution and the timing of major events.
RESULTS
Genome Triplication and Segmental Duplication in B. rapa
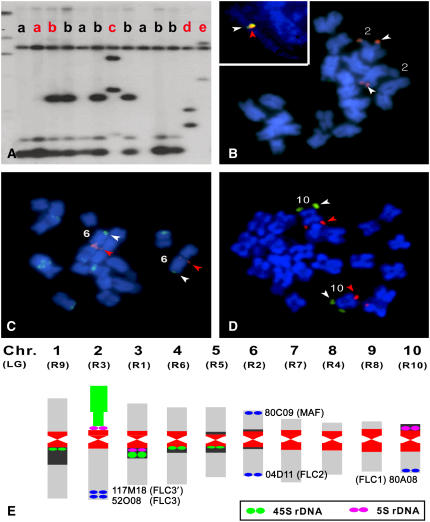
Using the FLC gene, 38 B. rapa BAC clones were identified and classified as five independent groups based on fingerprinting and DNA gel hybridization. The five BAC clone groups were localized on different chromosomes by fluorescence in situ hybridization (FISH) analyses (Figure 1). Two BAC clones, KBrH052O08 (52O08) and KBrH117M18 (117M18), were located in close proximity to each other, near the long-arm terminus of cytogenetic chromosome 2, and three BAC clones, KBrH004D11 (4D11), KBrH080C09 (80C09), and KBrH080A08 (80A08), were identified at the terminus of the long arm of cytogenetic chromosome 6, the terminus of the short arm of cytogenetic chromosome 6, and the long-arm terminus of cytogenetic chromosome 10, respectively. These five BAC clones were sequenced and genetically mapped based on the unique sequence of each BAC. The genetic map of each BAC coincided with the chromosomal localization based on the FISH results (Figure 2).
Figure 1.
Selection and Chromosomal Allocation of the BAC Clones Containing FLC Paralogs.
(A) DNA gel blot hybridization with the FLC gene on the HindIII fingerprint of the FLC-positive BAC clones. Red letters a, b, c, d, and e indicate B. rapa BAC clones, 52O08, 117M18, 4D11, 80A08, and 80C09, respectively.
(B) Metaphase FISH using 52O08. Pachytene FISH using 52O08 (red) and 117M18 (green) is highlighted in the inset.
(C) Metaphase FISH using 4D11 (red) and 80C09 (green).
(D) Metaphase FISH using 80A08 (green) and the chromosome 10–specific BAC clone KBrH053G06 (red).
(E) The relative positions of the five BAC clones are denoted as blue ovals on the cytogenetically defined chromosomes (Lim et al., 2005). Chr. and LG indicate cytogenetic chromosome numbers and their linkage group numbers. Chromosomes of B. rapa are represented as a cartoon based on centromere position (constriction) and major repeat sequences: centromeric repeats (red); 45S rDNA (green); 5S rDNA (magenta); and other pericentromeric heterochromatin (black). FLC homologs are denoted in parentheses near the BAC clones based on the genetic map and similarity to genes reported previously (Schranz et al., 2002).
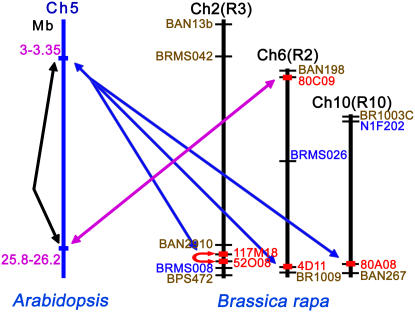
Figure 2.
Comparative Map of the Five B. rapa BAC Clones and Their Counterparts in Arabidopsis.
Sequence-based genetic mapping was conducted to localize five BAC clones on the reference map of B. rapa using simple sequence repeat markers and adjacent EST markers (Suwabe et al., 2002; Lowe et al., 2004; J.S. Kim, T.Y. Chung, G.J. King, M. Jin, T.J. Yang, Y.M. Jin, H.I. Kim, and B.S. Park, unpublished data). Double-headed arrows indicate the α duplication block in Arabidopsis (black), counterpart orthologous regions between Arabidopsis and B. rapa (blue and magenta), and a recent segmental duplication in B. rapa (red). The red boxes indicate B. rapa BACs.
A dot-plot sequence comparison of the five Brassica BACs and the two Arabidopsis sequences revealed that all of the sequences were collinear, with a variety of insertions, deletions, or inversions. Four of the BACs were homologous to the 3.0- to 3.35-Mb region of A. thaliana chromosome 5 (At5_3Mb), whereas the fifth was homologous to the 25.8- to 26.2-Mb region (At5_25Mb). The At5_3Mb and At5_25Mb of A. thaliana chromosome 5 are related by the third round (3R) of genome duplication (α duplication) that occurred before the diversification of the Arabidopsis–Brassica clade (Arabidopsis Genome Initiative, 2000; Blanc et al., 2003; Bowers et al., 2003; Blanc and Wolfe, 2004a).
The α duplication blocks in the Arabidopsis–Brassica clade could be distinguished by a large-scale inversion (∼120 kb; Figure 3, green box), a tandem array of four FLC paralogs in At5_25Mb, and an array of three FLC paralogs in Brassica BAC clone 80C09 (green circle in Figure 3), even though there is one FLC homolog in At5_3Mb and four orthologous Brassica BACs. The four BAC clones homologous with At5_3Mb are represented as Brassica paralog A (BrA; 80A08), Brassica paralog B (BrB; 4D11), Brassica paralog C (BrC; 52O08), and Brassica paralog C′ (BrC′; 117M18).
Figure 3.
Dot-Plot Analyses of Seven Homologous Sequences.
Dot plot of At5_3Mb (A), BrA (80A08) (B), and 80C09 (C) versus At5_25Mb (1), 80C09 (2), BrC′ (117M18) (3), BrC (52O08) (4), BrB (4D11) (5), and BrA (80A08) (6). The beginning and end points of collinear alignments are shown as black numerals. Common sequence in four BACs are indicated by red dotted lines. The relative collinearity indexes (collinear nucleotide of Brassica/Arabidopsis) are shown as red and green numerals. A large-scale inversion and an array of four small tandem duplications that differentiate At5_25Mb and 80C09 from the others are indicated by a green dotted box and a green circle, respectively. The green bars indicate the largest deletion (∼23 kb), which corresponds to 3,234,901 to 3,258,580 bp of Arabidopsis chromosome 5.
A comparison of sequence conservation between At5_3Mb and its four orthologous Brassica BACs (the red dotted area in the dot plot in Figure 3) showed that the similarity of BAC clones BrC and BrC′ was much greater than that between any other pair. These two BACs were located in the same chromosomal region, and metaphase and pachytene phase FISH could not differentiate their locations (Figure 1B). The summarized mapping, FISH, and sequence data of four homologous BACs, in agreement with previous reports (Rana et al., 2004; Lysak et al., 2005), indicated that the counterpart of At5_3Mb has surely triplicated into the BrA, BrB, and BrC–BrC′ clade in B. rapa. Additionally, the BrC and BrC′ segments have duplicated very recently.
Deletion of Sequences
All four segments of the B. rapa genome contain the characteristic pattern of conserved subsets of genes in collinear order with A. thaliana (Figure 4), as observed in B. oleracea and another subspecies of B. rapa (O'Neill and Bancroft, 2000; Rana et al., 2004). Deletion processes resulted in a mosaic pattern of the remaining sequence in each triplicate block of B. rapa (Figure 4A). Thirty-five genes were identified in the 124-kb segment of At5_3Mb (At5g10020 to At5g10360, excluding tRNA, At5g10235), and a total of 80 were identified in the collinear blocks of the four Brassica BACs (31, 20, 15, and 14 genes in BrA, BrB, BrC, and BrC′, respectively). Three Arabidopsis genes (At5g10330 to At5g10350) have no Brassica homologs, suggesting that these may be the result of insertions. All of the other 32 genes in the A. thaliana genome segment are represented at least once in B. rapa, although only 4 genes (including FLC, At5g10140) are represented in all four B. rapa paralogous regions. The BrA contains orthologs of the largest subset of these, 24 genes (75%) in 110 kb. The BrB contains orthologs of 17 genes (53%) in 74 kb, and the BrC and BrC′ each contains orthologs of the same 13 genes (41%) in 57 and 52 kb, respectively. Compared with the 32 genes of At5_3Mb, a total of 54 B. rapa orthologs remain as different gene copy numbers in the triplication blocks (Figure 4B), with 14 singlets (orange; 44%), 14 doublets (blue; 44%), and 4 triplets (green; 12%). Overall, the gene complement at triplication has been reduced from a presumptive 96 genes at hexaploidy to 54 (56%) across the three ancient paralogs, with recent duplication of 13 genes in paralog C′. There were no indications of genes having been deleted from the Arabidopsis lineage since divergence from Brassica, which would have been evident by the occurrence of genes with conserved synteny in two or more of the three ancient B. rapa paralogous segments but not in A. thaliana.
Figure 4.
Schematic Representation of Sequence Conservation between At5_3Mb and Its Four Orthologous Blocks in B. rapa.
(A) Summarized percentage identity plot of 124 kb from At5_3Mb (chromosome 5: 3,134,987 to 3,258,842) and the collinear sequences in four orthologous B. rapa BACs: 110 kb (1 to 110,219) from BrA; 74 kb (15,001 to 89,318) from BrB; 57 kb (58,254 to 115,292) from BrC; and 52 kb (18,227 to 70,502) from BrC′ (the red dotted area in Figure 3). The 36 genes of At5_3Mb (At5g10020 to At5g10360) are labeled A to Z and a to j, with colored arrows below indicating frequency in the triplicate blocks of B. rapa: green arrows (three paralogs), blue arrows (two paralogs), orange arrows (one paralog), and red arrows (none; the putative insertion). Gene Ontology (GO Slim) categories are shown in parentheses: UN (molecular function unknown); OB (other binding activity); PB (protein binding); HA (hydrolase activity); KA (kinase activity); EA (other enzyme activity); OM (other membrane activity); TR (transporter activity); TA (transferase activity); NB (nucleotide binding); TF (transcription factor activity); SM (structural molecule activity). The summarized percentage identity plot indicates the occurrence of homologous sequence in the frame of 124 kb of At5_3Mb, with collinear sequences from all four BACs (4BACs) and with a collinear sequence from each BAC. Red, green, and white blocks denote conserved, homologous, and nonhomologous sequences, respectively. Repeat denotes regions of low complexity (gray bars) and transposons (magenta bars) based on RepeatMasker results.
(B) Comparative genome alignments based on discontinuous Megablast results between At5_3Mb and the collinear sequence of each BAC. The locations of predicted genes are indicated with colored arrows and labels as in (A). Genes with major changes are denoted as diamonds for a chimera of two adjacent genes, asterisks for deletion of >50% of exons, and triangles for insertion. Nonhomologous genes (putative insertions) are denoted as red arrows. Supporting data and full annotation of genes are available in Supplemental Figure 1 and Supplemental Table 1 online.
Insertion of Sequences
The Arabidopsis sequences with no complement in the four Brassica orthologs might be insertions. Three genes were identified in these insertion regions (Figure 4A, red arrows), and two of them are transposons. The Brassica sequences contain 11 insertions of predicted genes, compared with the complement in A. thaliana: 6 in BrA, 2 each in BrB and BrC, and 1 in BrC′, as shown by red arrows in Figure 4B. None of these are conserved across paralogs. One gene, 4D11_12, is an intact polyprotein gene of a retrotransposon. Two others, 80A08_13 and 80A08_30, showed strong similarity (E < e−32) to expressed genes of Medicago truncatula (GenBank accession number BG586262.1) and B. napus (GenBank accession number CD827307.1), respectively. There is no evidence for the remainder representing functional genes.
Dating of the Evolutionary Events
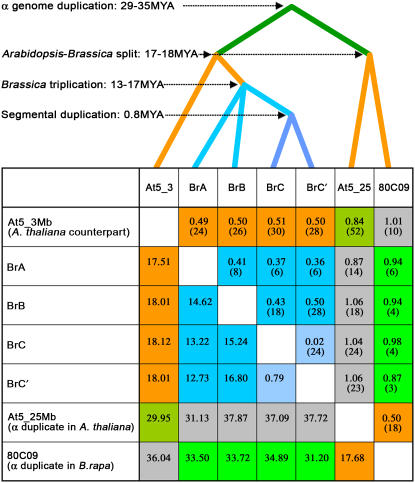
Estimates of the times since divergence of the B. rapa paralogs from each other and from the A. thaliana genome were made by calculation of synonymous base substitution rates (Ks values) and assumption of a mutation rate of 1.4 × 10−8 substitutions per synonymous site per year (Koch et al., 2000). The median Ks values between orthologous genes in the A. thaliana chromosome At5_3Mb region and its four B. rapa counterparts (0.49 to 0.51) are representative of the Brassica–Arabidopsis divergence, which we estimate at 17 to 18 MYA. The Ks values between the four B. rapa paralogs (0.36 to 0.50) are representative of the triplication in Brassica, which we estimate at 13 to 17 MYA. The Ks value between BrC and BrC′ was extremely low, 0.02, supporting the notion that they were duplicated recently, ∼0.8 MYA (Figure 5; see Supplemental Tables 2 and 3 online).
Figure 5.
Chronology of Evolutionary Events and Ks Estimates for Homologous Genes in Duplication Blocks.
Calculated divergence times in millions of years are shown below the self-comparison diagonal (T = Ks/2 × synonymous mutation rate per year [1.4 × 10−8] [Koch et al., 2000]). Median Ks values are shown above the self-comparison diagonal. Colored boxes represent different evolutionary events: the α genome duplication in A. thaliana (dark green); the α genome duplication in B. rapa (light green); Arabidopsis–Brassica divergence (orange); genome triplication in B. rapa (dark blue); segmental duplication in B. rapa (light blue); and comparison between paralogous blocks across species (gray). Numbers in parentheses indicate the number of genes compared between two blocks. All Ks values based on pairwise comparisons of homologous genes are shown in Supplemental Table 2 online. Ks values between tandemly duplicated genes in duplication blocks, such as At5g10220 and At5g10230, are shown in Supplemental Figure 2 and Supplemental Table 3 online.
DISCUSSION
The Diploidization Process
The reduction observed in the overall number of replicated genes can be regarded as part of the diploidization process, whereby the genome tends toward its original gene complement. Analysis of the gene families that have been reduced least by this process shows that they include transcription factor, transporter, and structural molecular activities (see Supplemental Table 1 online). It is notable that six of the seven genes for which function is unknown have returned to a single copy state (Figure 4). These observations are in accordance with previous reports on the genome duplication of Arabidopsis, which showed higher retention for genes with regulatory functions, such as transcription factors, kinases, phosphatases, and calcium binding proteins (Blanc and Wolfe, 2004b; Seoighe and Gehring, 2004).
Duplicated genes are not necessarily deleted, even over long periods of evolution. For example, individual genes can attain a selective advantage by gaining new function by partitioning of the ancestral function between the two duplicates (reviewed in Hurles, 2004). Neofunctionalization and subfunctionalization of duplicate genes may diversify gene products in an organ-, time-, and/or environment-specific manner. Although there is no evidence from sequence data alone of the acquisition of new function, the insertion of small transposons such as terminal repeat retrotransposons in miniature (TRIM) or miniature in transposable element (MITE) elements could confer new function (Bennetzen, 2000). Several genes in the B. rapa triplication blocks are modified by insertion of TRIM or MITE elements (Figure 4B, black triangles) and by insertion of an intact long terminal repeat retrotransposon (Figure 4B, complement of gene K in BrB).
The Process of Genome Expansion in B. rapa
The wide range of nuclear genome sizes, 50 to ∼85,000 Mb of DNA per haploid nucleus, in flowering plants is a consequence of the frequent formation of polyploids and the accumulation of transposons (Plant DNA C-values Database, http://www.rbgkew.org.uk/cval/homepage.html) (Grover et al., 2004; Kellogg and Bennetzen, 2004). In Graminea, genome expansion is attributable to the accumulation of transposons in the collinear sequence (Ilic et al., 2003; Kellogg and Bennetzen, 2004; International Rice Genome Sequencing Project, 2005; Scherrer et al., 2005). The process of genome expansion in Brassica appears to be different. The combined length of the Brassica paralogs is 194% (or 236% if segmental duplicate BrC′ is counted) of the length of their homolog in A. thaliana. However, the gene density in the compared regions is almost the same, with one gene per 3731 and 3440 bp in B. rapa and A. thaliana, respectively. This small difference in gene density is insufficient to account for the difference in genome size, with that of B. rapa (529 Mb) estimated to be 337% that of A. thaliana (157 Mb) (Johnston et al., 2005). We found no significant increase in transposon insertion in the four Brassica BACs except for several small TRIM and MITE elements. Almost 50% of the 92,000 BAC end sequences from three different BAC libraries (HindIII, BamHI, and Sau3AI) showed no homology with any sequence of Arabidopsis (Yang et al., 2005a). Similarly, ∼60% of the whole genome shotgun sequences of B. oleracea (significant match found in 197,344 of 454,274 nuclear DNA reads) did not match any Arabidopsis sequences (Ayele et al., 2005). Thus, the extra genome expansion in B. rapa appears to be the result of amplification of Brassica-specific sequences, many of which are likely to form heterochromatic blocks of transposon or tandem repeats (Zhang and Wessler, 2004; Lim et al., 2005).
The total number of genes is increased 1.7-fold across the genome triplication (54 genes) and 2.1-fold across all four paralogs (67 genes), compared with the corresponding region of Arabidopsis (32 genes). Approximately 9% of the 100,000 Brassica EST sequences were found not to be homologous with any gene in Arabidopsis (http://brassica-rapa.org). These results suggest that some genes occur in Brassica that are not present in A. thaliana. Extrapolation of our data to the whole genome suggests that the estimated range of gene content in B. rapa is from 49,000 (1.7 times the estimated gene number of A. thaliana [i.e., 29,000], assuming that additional segmental duplications are rare and that few of the Brassica-specific ESTs represent truly novel genes) to 63,000 (assuming that additional segmental duplications such as we describe are common and that the 9% of Brassica-specific ESTs do largely represent novel genes).
Chronology of Genome Evolution
Our results indicate that the three subgenomes of B. rapa diverged from each other 13 to 17 MYA, very soon after the divergence of the Brassica and Arabidopsis lineages, which we estimate at 17 to 18 MYA. This may have provided the opportunity for evolution of the tribe Brassiceae, which comprises ∼240 highly diverse species. The underlying process of genome evolution has involved, as part of the diploidization process, interspersed gene loss and sequence divergence of retained genes in addition to reorganization at the chromosomal level (Lysak et al., 2005) and the independent accumulation of repeat sequences in heterochromatin blocks in each of the Brassica species (Lim et al., 2005).
METHODS
DNA Sequencing
Using high-density filter hybridization against a Brassica BAC library (KBrH, HindIII library of Brassica rapa inbred line Chiifu) that contains 11× genome coverage (Park et al., 2005), we identified 38 B. rapa BAC clones containing the FLC gene. Fingerprinting and subsequent DNA gel blot hybridization revealed five independent BAC groups (Figure 1A). Five BAC clones, one from each of the five BAC groups, were sequenced as reported previously (Kim et al., 2004; Yang et al., 2004). Sequence reactions using BigDye terminator chemistry version 3.0 (Applied Biosystems) were analyzed using ABI3730 automatic DNA sequencers (Applied Biosystems). Sequence assembly was performed as described previously (Yang et al., 2005b) using Phred for base calling, CROSS_MATCH for removing vector sequences, and Phrap and Consed for assembly (Ewing and Green, 1998; Ewing et al., 1998; Gordon et al., 1998).
Sequence Analysis
Homologous Arabidopsis thaliana sequences were identified by BLAST using the Arabidopsis chromosome database (http://www.ncbi.nlm.nih.gov/BLAST/). Pairwise sequence comparisons were performed using PipMaker (Schwartz et al., 2000) and BLAST2 (http://www.ncbi.nlm.nih.gov/BLAST/). The alignment results were viewed using Gbrowse, a generic genome browser (http://www.gmod.org/ggb/gbrowse.shtml), and SynBrowse, a synteny browser for comparative sequence analysis (http://www.synbrowse.org) based on discontinuous Megablast results. Gene annotation was achieved using the web-based gene prediction program FGENE-SH Arabidopsis (http://www.softberry.com/berry.phtml). Repeats were identified by RepeatMasker (http://www.repeatmasker.org/) followed by manual inspection.
Estimation of Synonymous Substitution Rates and Dating of Duplications
All of the genes in Arabidopsis and the predicted genes in the five Brassica BAC clones were compared through PipMaker (Schwartz et al., 2000). Coding sequences of all of the homologous genes were categorized into subgroups for further analyses. The fraction of synonymous substitutions (Ks) was used to estimate the timing of duplication events between the two sequences, because they are from neutral mutations without amino acid replacements and are not controlled by natural selection (Blanc and Wolfe, 2004a; Maere et al., 2005). Basic methods for Ks estimation were used as described by Maere et al. (2005). Pairwise alignments of the paralogous nucleotide sequences belonging to a homologous gene family were made by ClustalW (Thompson et al., 1994). The Ks values were obtained with the CODEML program (Goldman and Yang, 1994) of the PAML package (Yang, 1997). To estimate absolute dates for the several duplication events, we used a median Ks value for each gene pair between two blocks. Calculations for the dating of the duplication events were completed using a synonymous mutation rate of 1.4 × 10−8 substitutions per synonymous site per year, which was applied to the CHALCONE SYNTHASE gene in eudicots (Koch et al., 2000). Divergence times (T) were estimated using the equation T = Ks/2 × 1.4 × 10−8.
FISH
The basic FISH protocol was described previously (Lim et al., 2005). FISH signals were captured by a charge-coupled device camera and converted into pseudocolored images. The images were optimized for brightness and contrast using Adobe Photoshop image-processing software.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: KBrH080A08, AC155344; KBrH004D11, AC155341; KBrH117M18, AC146875; KBrH052O08, AC155342; and KBrH080C09, AC166741.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Percent Identity Plot of At5_3Mb versus All of the Collinear Sequences in Four Brassica BACs, 80A08, 4D11, 52O08, and 117M18.
Supplemental Figure 2. Ks Estimates between Tandemly Duplicated Genes.
Supplemental Table 1. Pairwise Alignment of Homologous Genes in Seven Duplication Blocks.
Supplemental Table 2. Pairwise Comparisons of Ks Values between Homologous Genes.
Supplemental Table 3. Pairwise Comparisons of Ks Values from Tandemly Duplicated Homologous Genes.
Supplementary Material
Acknowledgments
We thank Maryana Bhak and three anonymous reviewers for valuable comments. This work was supported by the BioGreen 21 Program, the Rural Development Administration, and the National Institute of Agricultural Biotechnology, Korea.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Beom-Seok Park (pbeom@rda.go.kr).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.040535.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Ayele, M., Haas, B.J., Kumar, N., Wu, H., Xiao, Y., Van Aken, S., Utterback, T.R., Wortman, J.R., White, O.R., and Town, C.D. (2005). Whole genome shotgun sequencing of Brassica oleracea and its application to gene discovery and annotation in Arabidopsis. Genome Res. 15 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babula, D., Kaczmarek, M., Barakat, A., Delseny, M., Quiros, C.F., and Sadowski, J. (2003). Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: Complexity of the comparative map. Mol. Genet. Genomics 268 656–665. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J.L. (2000). Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 42 251–269. [PubMed] [Google Scholar]
- Blanc, G., Hokamp, K., and Wolfe, K.H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., and Wolfe, K.H. (2004. a). Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., and Wolfe, K.H. (2004. b). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16 1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers, J.E., Chapman, B.A., Rong, J., and Paterson, A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422 433–438. [DOI] [PubMed] [Google Scholar]
- Cronn, R.C., Small, R.L., and Wendel, J.F. (1999). Duplicated genes evolve independently after polyploid formation in cotton. Proc. Natl. Acad. Sci. USA 96 14406–14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., and Green, P. (1998). Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8 186–194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. (1998). Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8 175–185. [DOI] [PubMed] [Google Scholar]
- Feuillet, C., Penger, A., Gellner, K., Mast, A., and Keller, B. (2001). Molecular evolution of receptor-like kinase genes in hexaploid wheat. Independent evolution of orthologs after polyploidization and mechanisms of local rearrangements at paralogous loci. Plant Physiol. 125 1304–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M.D., and Devos, K.M. (1998). Plant comparative genetics after 10 years. Science 282 656–659. [DOI] [PubMed] [Google Scholar]
- Goldman, N., and Yang, Z. (1994). A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11 725–736. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. (1998). Consed: A graphical tool for sequence finishing. Genome Res. 8 195–202. [DOI] [PubMed] [Google Scholar]
- Grover, C.E., Kim, H., Wing, R.A., Paterson, A.H., and Wendel, J.F. (2004). Incongruent patterns of local and global genome size evolution in cotton. Genome Res. 14 1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles, M. (2004). Gene duplication: The genomic trade in spare parts. PLoS Biol. 2 900–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic, K., SanMiguel, P.J., and Bennetzen, J.L. (2003). A complex history of rearrangement in an orthologous region of the maize, sorghum, and rice genomes. Proc. Natl. Acad. Sci. USA 100 12265–12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436 793–800. [DOI] [PubMed] [Google Scholar]
- Johnston, J.S., Pepper, A.E., Hall, A.E., Chen, Z.J., Hodnett, G., Drabek, J., Lopez, R., and Price, H.J. (2005). Evolution of genome size in Brassicaceae. Ann. Bot. (Lond.) 95 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, E.A., and Bennetzen, J.L. (2004). The evolution of nuclear genome structure in seed plants. Am. J. Bot. 91 1709–1725. [DOI] [PubMed] [Google Scholar]
- Kim, H.R., Yang, T.J., Kudna, D.A., and Wing, R.A. (2004). Construction and application of genomic DNA libraries. In Handbook of Plant Biotechnology, Vol. 1, P. Christou and H. Klee, eds (Hoboken, NJ: John Wiley & Sons), pp. 71–80.
- Koch, M.A., Haubold, B., and Mitchell-Olds, T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17 1483–1498. [DOI] [PubMed] [Google Scholar]
- Kowalski, S.P., Lan, T.H., Feldmann, K.A., and Paterson, A.H. (1994). Comparative mapping of Arabidopsis thaliana and Brassica oleracea chromosomes reveals islands of conserved organization. Genetics 138 499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz, U. (1998). Comparative mapping between Arabidopsis thaliana and Brassica nigra indicates that Brassica genomes have evolved through extensive genome replication accompanied by chromosome fusions and frequent rearrangements. Genetics 150 1217–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan, T.-H., and Paterson, A.H. (2000). Comparative mapping of quantitative trait loci sculpting the curd of Brassica oleracea. Genetics 155 1927–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, K.B., et al. (2005). Characterization of rDNAs and tandem repeats in heterochromatin of Brassica rapa. Mol. Cells 19 436–444. [PubMed] [Google Scholar]
- Lowe, A., Moule, C., Trick, M., and Edwards, K. (2004). Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theor. Appl. Genet. 108 1103–1112. [DOI] [PubMed] [Google Scholar]
- Lukens, L., Zou, F., Lydiate, D., Parkin, I., and Osborn, T. (2003). Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics 164 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak, M.A., Koch, M.A., Pecinka, A., and Schubert, I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere, S., De Bodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., and Van de Peer, Y. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 102 5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, C.M., and Bancroft, I. (2000). Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant J. 23 233–243. [DOI] [PubMed] [Google Scholar]
- Park, J.Y., et al. (2005). Physical mapping and microsynteny of Brassica rapa ssp. pekinensis genome corresponding to a 222 kb gene-rich region of Arabidopsis chromosome 4 and partially duplicated on chromosome 5. Mol. Genet. Genomics 274 579–588. [DOI] [PubMed] [Google Scholar]
- Parkin, I.A., Sharpe, A.G., and Lydiate, D.J. (2003). Patterns of genome duplication within the Brassica napus genome. Genome 46 291–303. [DOI] [PubMed] [Google Scholar]
- Paterson, A.H., Lan, T.H., Amasino, R., Osborn, T.C., and Quiros, C. (2001). Brassica genomics: A complement to, and early beneficiary of, the Arabidopsis sequence. Genome Biol. 2 REVIEWS1011. [DOI] [PMC free article] [PubMed]
- Quiros, C.F., Grellet, F., Sadowski, J., Suzuki, T., Li, G., and Wroblewski, T. (2001). Arabidopsis and Brassica comparative genomics: Sequence, structure and gene content in the ABI-Rps2-Ck1 chromosomal segment and related regions. Genetics 157 1321–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana, D., van den Boogaart, T., O'Neill, C.M., Hynes, L., Bent, E., Macpherson, L., Park, J.Y., Lim, Y.P., and Bancroft, I. (2004). Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 40 725–733. [DOI] [PubMed] [Google Scholar]
- Scherrer, B., Isidore, E., Klein, P., Kim, J.-s., Bellec, A., Chalhoub, B., Keller, B., and Feuillet, C. (2005). Large intraspecific haplotype variability at the Rph7 locus results from rapid and recent divergence in the barley genome. Plant Cell 17 361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, R., Acarkan, A., and Boivin, K. (2001). Comparative structural genomics in the Brassicaceae family. Plant Physiol. Biochem. 39 253–262. [Google Scholar]
- Schranz, M.E., and Osborn, T.C. (2000). Novel flowering time variation in the resynthesized polyploid Brassica napus. J. Hered. 91 242–246. [DOI] [PubMed] [Google Scholar]
- Schranz, M.E., Quijada, P., Sung, S.-B., Lukens, L., Amasino, R., and Osborn, T.C. (2002). Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. (2000). PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 10 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe, C., and Gehring, C. (2004). Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 20 461–464. [DOI] [PubMed] [Google Scholar]
- Soltis, D.E., and Soltis, P.S. (1995). The dynamic nature of polyploid genomes. Proc. Natl. Acad. Sci. USA 92 8089–8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwabe, K., Iketani, H., Nunome, T., Kage, M., and Hirai, M. (2002). Isolation and characterization of microsatellites in Brassica rapa L. Theor. Appl. Genet. 104 1092–1098. [DOI] [PubMed] [Google Scholar]
- Suzuki, G., Kakizaki, T., Takada, Y., Shiba, H., Takayama, S., Isogai, A., and Watanabe, M. (2003). The S haplotypes lacking SLG in the genome of Brassica rapa. Plant Cell Rep. 21 911–915. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U, N. (1935). Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japan. J. Bot. 7 389–452.
- Wendel, J.F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42 225–249. [PubMed] [Google Scholar]
- Yang, T.J., Kim, J.S., Lim, K.B., Kwon, S.J., Kim, J.I., Jin, M., Park, J.Y., Lim, M.H., Kim, H.I., Kim, S.H., Lim, Y.P., and Park, B.S. (2005. b). The Korea Brassica Genome Project: A glimpse of the Brassica genome based on comparative genome analysis with Arabidopsis. Comp. Funct. Genomics 6 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, T.J., Lee, S., Chang, S.B., Yu, Y., de Jong, H., and Wing, R.A. (2005. a). In-depth sequence analysis of the centromeric region of tomato chromosome 12: Identification of a large CAA block and characterization of centromeric retrotransposons. Chromosoma 114 103–117. [DOI] [PubMed] [Google Scholar]
- Yang, T.J., Yu, Y., Frisch, D., Lee, S., Kim, H.R., Kwon, S.J., Park, B.S., and Wing, R.A. (2004). Construction of various copy number plasmid vectors and their utility for genome sequencing. Genomics & Informatics 2 153–158. [Google Scholar]
- Yang, Z. (1997). PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13 555–556. [DOI] [PubMed] [Google Scholar]
- Zhang, X., and Wessler, S.R. (2004). Genome-wide comparative analysis of the transposable elements in the related species Arabidopsis thaliana and Brassica oleracea. Proc. Natl. Acad. Sci. USA 101 5589–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.