Abstract
We recently reported that SMAP1, a GTPase-activating protein (GAP) for Arf6, directly interacts with clathrin and regulates the clathrin-dependent endocytosis of transferrin receptors from the plasma membrane. Here, we identified a SMAP1 homologue that we named SMAP2. Like SMAP1, SMAP2 exhibits GAP activity and interacts with clathrin heavy chain (CHC). Furthermore, we show that SMAP2 interacts with the clathrin assembly protein CALM. Unlike SMAP1, however, SMAP2 appears to be a regulator of Arf1 in vivo, because cells transfected with a GAP-negative SMAP2 mutant were resistant to brefeldin A. SMAP2 colocalized with the adaptor proteins for clathrin AP-1 and EpsinR on the early endosomes/trans-Golgi-network (TGN). Moreover, overexpression of SMAP2 delayed the accumulation of TGN38/46 molecule on the TGN. This suggests that SMAP2 functions in the retrograde, early endosome-to-TGN pathway in a clathrin- and AP-1–dependent manner. Thus, the SMAP gene family constitutes an important ArfGAP subfamily, with each SMAP member exerting both common and distinct functions in vesicle trafficking.
INTRODUCTION
Clathrin-mediated vesicle formation is an essential step in the intracellular trafficking of membrane compartments. The locations in the cell at which vesicles are formed are regulated by various factors in a highly complex manner. AP-1 and AP-2 are tetrameric adaptors for clathrin and recruit it to sites of vesicle budding (specifically, the trans-Golgi network (TGN)/endosome and plasma membrane, respectively; Hirst and Robinson, 1998; Lewin and Mellman, 1998; Scales et al., 2000). Thereafter, monomeric adaptor proteins such as AP180 and CALM induce clathrin assembly, thereby generating clathrin-coated vesicles (Hao et al., 1999; Tebar et al., 1999; Ford et al., 2001). Subsequently, the budded vesicles fuse to the acceptor membrane where Hsc70 and auxilin disassemble the clathrin coat (Cremona, 2001; Lemmon, 2001; Morgan et al., 2001; Fotin et al., 2004).
Studies on clathrin have mainly focused on secretion from the TGN and endocytosis from the plasma membrane. However, recent studies revealed that clathrin is also an indispensable coat component of vesicles that bud from the endosome (Meyer et al., 2000; Rohn et al., 2000; Hinners and Tooze, 2003). One endosome-to-TGN pathway utilizes AP-1 as a tetrameric adaptor for clathrin. In addition, monomeric adaptor proteins such as EpsinR, TIP47, and PACS-1 are responsible for selecting cargo proteins in endosome-to-TGN transport (Diaz and Pfeffer, 1998; Wan et al., 1998; Saint-Pol et al., 2004). For example, one early endosome-to-TGN pathway that transports TGN38 and Shiga toxin is regulated by EpsinR. In contrast, a late endosome-to-TGN pathway that transports mannose-6-phosphate receptor and furin convertase is regulated by TIP47 and PACS-1.
Vesicle formation not only requires the abovementioned vesicle components (namely, clathrin and its adaptors) but also enzymatic activity. This is supplied by small GTPase ADP-ribosylation factor (Arf; Nie et al., 2003b). Like other small GTPases, Arf cycles between a GTP-bound active state and a GDP-bound inactive state. Its activation and inactivation are regulated by guanine-nucleotide exchange factor (GEF) and GTPase-activating protein (GAP), respectively (Randazzo and Hirsch, 2004; Shin and Nakayama, 2004). The six Arfs are classified into three groups: Arf1, 2, and 3 are class I, Arf4 and 5 are class II, and Arf6 is class III. Among them, Arf1 functions on the Golgi/TGN/endosome. In its active state, Arf1 recruits coat and adaptor proteins such as COPI, AP-1, and AP-3 to the membrane and induces the assembly and nucleation of coated vesicles (Donaldson et al., 1991; Serafini et al., 1991; Traub et al., 1993; Ooi et al., 1998). This localization of the coat/adaptor proteins is sensitive to brefeldin A (BFA) because BFA inactivates Arfs of classes I and II by inhibiting the interaction between Arf and its GEF (Donaldson et al., 1992; Peyroche et al., 1999). Therefore, treatment of cells with BFA causes the disintegration of the Golgi complex and tubulation of the endosomal membrane. In contrast, Arf6 regulates the endocytosis of vesicles from the plasma membrane and the recycling of endosome to the plasma membrane (Donaldson, 2003), and the GEFs for Arf6, ARNO, and EFA are insensitive to BFA (Franco et al., 1999; Peyroche et al., 1999).
The ArfGAP family also contains several members and to date 16 mammalian genes have been reported (Bernards, 2003; Randazzo and Hirsch, 2004). Importantly, it appears that distinct Arf and ArfGAP combinations participate in different intracellular vesicle transport pathways. For example, GAP1 is an Arf1GAP that forms a complex with COPI in an Arf1-dependent manner (Liu et al., 2005) and interacts directly with its cargo protein, a KDEL receptor (Aoe et al., 1999). This interaction is essential for the sorting of cargo proteins into COPI-coated vesicles on the Golgi complex. In contrast, AGAP1, another Arf1GAP, interacts with AP-3 and functions in a late endosome-to-lysosome pathway (Nie et al., 2003a). Moreover, we recently identified SMAP1 as a novel Arf6GAP (Tanabe et al., 2005). SMAP1 interacts with clathrin and regulates the clathrin- and AP-2–dependent endocytosis of transferrin receptors from the plasma membrane.
In this study, we identified a homologue of SMAP1 that we named SMAP2. Like SMAP1, SMAP2 possesses a GAP domain and interacts with clathrin heavy chain (CHC). Furthermore, we show that SMAP2 interacts with the clathrin assembly protein CALM. Unlike SMAP1, however, SMAP2 appears to be a regulator of Arf1 and is involved in clathrin- and AP-1–dependent trafficking in the early endosome-to-TGN pathway. Thus, the SMAP gene family constitutes an important ArfGAP subfamily and each SMAP member exerts common as well as distinct functions in vesicle trafficking.
MATERIALS AND METHODS
Cells, Reagents, and Antibodies
HeLa and Cos-7 cells were cultured in DMEM containing 10% (vol/vol) fetal calf serum. BFA was purchased from Sigma-Aldrich (St. Louis, MO). The following antibodies were purchased: the anti-influenza hemagglutinin (HA) rat monoclonal antibody (mAb) 3F10 was from Roche Diagnostics (Indianapolis, IN), the mouse mAbs against c-Myc (9E10) and AP-1 (γ-adaptin, 100/3) were from Sigma-Aldrich, the mouse mAbs against CHC (X22) and AP-2 (α-adaptin, AP.6), and the rabbit polyclonal antibodies (pAb) against β-COP and furin convertase were from Affinity BioReagents (Golden, CO), the mouse mAb against AP-3 (δ-adaptin) was from Transduction Laboratories (Lexington, KY), the anti-CD25 mouse mAb (7G7/B6) was from Upstate Biotechnology (Lake Placid, NY), the anti-TGN46 sheep pAb was from Serotec (Oxford, United Kingdom), the anti-6His mouse mAb was from Clontech Laboratories (Palo Alto, CA), the Cy3-conjugated goat anti-rat IgG was from Chemicon (Temecula, CA), and the Alexa488-conjugated goat anti-mouse and anti-rabbit IgGs, Alexa488-conjugated donkey anti-sheep IgG and Alexa488-conjugated human-transferrin were from Molecular Probes (Eugene, OR).
Plasmid Construction
A fragment that corresponds to a portion of BC052413 (GenBank/NCBI) was PCR-amplified from cDNA from a murine erythroleukemic cell line. By using this fragment as a hybridization probe, we screened a cDNA library prepared from the same cells and obtained cDNA clones encoding SMAP2. A cDNA encoding CALM was isolated from the same cDNA library by a PCR method. The various mutations described below were introduced into the coding region of SMAP2 by a PCR method. cDNAs encoding rat GAP1, murine SMAP1, Arf1, and Arf6 were isolated as described previously (Tanabe et al., 2005). TGN38 cDNA was isolated from murine liver-derived cDNA by a PCR method. The human CHC, GGA1, and CD25 cDNAs were provided by Drs. T. Nagase, K. Nakayama, and H. Asao, respectively. The human furin convertase (79823) and EpsinR (KIAA0171) cDNAs were obtained from ATCC and the Kazusa DNA Research Institute, respectively. A chimeric gene encoding an extracellular domain of CD25 fused to the transmembrane and cytoplasmic domains of TGN38 was constructed by a PCR method.
To express these cDNAs in mammalian cells, they were inserted into pcDNA3 (Invitrogen, Carlsbad, CA). For immunodetection, each protein was tagged with the epitope at its amino-terminus as follows: HA-SMAP2, Myc-SMAP2, HA-GAP1, Myc-CALM, Myc-EpsinR, Myc-GGA1, and Myc-furin convertase.
Yeast Two-Hybrid Screening
The Matchmaker Two-Hybrid System 3 (Clontech Laboratories) was used according to the instructions in the manufacturer's manual. The bait plasmid was constructed by inserting the murine SMAP2 cDNA next to the GAL4 DNA-binding domain of the vector pGBKT7. A cDNA library prepared from murine brain was fused to the GAL4 DNA-activation domain of the vector pGADT7 and served as the source of prey plasmids. AH109 cells were used as host cells. The selective medium was devoid of Trp, Leu, His, and Ade and added by 2.5 mM 3-amino-1,2,4-triazole. Plasmid DNAs were recovered from positive clones and sequenced by using an ABI PRISM 310 genetic analyzer (Applied Biosystems, Foster City, CA).
Preparation of Recombinant Proteins
To express cDNAs in bacteria, they were inserted into pGEX5X-3 (Amersham Pharmacia Biotech, Piscataway, NJ). The sequences expressed were the amino-terminal 163 residues of SMAP2, the amino-terminal 255 residues of SMAP1, the amino-terminal 246 residues of GAP1, and the entire coding sequences of Arf1 and Arf6. All were fused at their amino-termini to glutathione S-transferase (GST) and tagged with hexahistidines at their carboxy-termini.
Recombinant proteins were prepared as described previously (Tanabe et al., 2005). Briefly, the Escherichia coli strain BL21 was transformed with one of the expression plasmids and cultured in LB medium containing ampicillin. Isopropyl-β-d-thiogalactoside was then added at 0.4 mM to induce protein expression. The cells were harvested, and the GST fusion proteins were purified by using a B-PER GST-spin purification kit (Pierce Chemical, Rockford, IL). The fusion protein was digested with Factor Xa (Novagen, Madison, WI) and the GST-free protein was purified by using a 6*His spin purification kit (Pierce Chemical). The protein concentrations were assayed by a BCA protein assay kit (Pierce Chemical). The purity of the recovered proteins was ∼95% as judged by SDS-PAGE.
GAP Assay
GAP assays were performed as described in our previous study (Tanabe et al., 2005). Briefly, recombinant nonmyristoylated Arf1 or Arf6 proteins were loaded with [α-32P] GTP, and GTP hydrolysis was initiated by the addition of GAP for the indicated time. The reaction was then terminated and unbound nucleotides were removed. GTP and GDP were eluted from the Arf protein and separated by TLC. The plate was exposed to a phosphorimager screen and radioactivity was detected by a Bio Imaging Analyzer (Fuji Photo Film, Tokyo, Japan).
Immunoprecipitation Analysis
Cos-7 cells were transfected with the SMAP2 expression plasmid using Effectene (QIAGEN, Chatsworth, CA) and incubated for 24 h at 37°C. The cells were then lysed with a buffer consisting of 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% (vol/vol) Nonidet P-40, and a mixture of protease inhibitors (Complete; Roche Diagnostics). The cell lysate was precleared by protein G-Sepharose beads (Amersham Pharmacia) and then incubated with the primary antibody, and the immunoprecipitates were adsorbed to the beads. The beads were washed five times with lysis buffer in which the concentration of NaCl was raised to 200 mM. Proteins were eluted by boiling the beads in SDS sample buffer. Gel electrophoresis of the proteins, transfer of the proteins from the gel to a filter, and immunoreaction with the secondary antibody were performed as described previously (Chiba et al., 1997). Products were visualized by use of the ECL Plus reagent (Amersham Pharmacia).
Immunofluorescence Microscopy
Cos-7 or HeLa cells on coverslips were transfected with plasmid DNA using Effectene and incubated for 24 h at 37°C. They were then fixed with 4% (wt/vol) paraformaldehyde in phosphate-buffered saline (PBS) for 15min at room temperature (RT), washed three times with PBS, and blocked with 1% (wt/vol) bovine serum albumin and 0.1% Triton X-100 in PBS for 15 min at RT. In some cases, the transfected cells were treated with 5 μg/ml BFA for 2 min before fixation. The fixed cells were incubated with appropriately diluted primary and then secondary antibodies for 45 min each at RT. After washing with PBS, the coverslips were mounted on slide glasses using Vectashield (Vector Laboratories, Burlingame, CA) and observed with a confocal microscope (LSM-401, Zeiss, Thornwood, NY). To detect the internalization and transport of transferrin or CD25-TGN38, cells were rinsed with PBS and preincubated in 0.1% (wt/vol) bovine serum albumin in DMEM for 30 min at 37°C. The cells were then incubated at 37°C with 25 μg/ml Alexa488-conjugated human transferrin or 10 μg/ml anti-CD25 mAb for the indicated time, rinsed with PBS, fixed, and processed for immunofluorescence as described above.
RESULTS
Identification of SMAP2 and Its Domain Structure
To identify a homologue of the murine SMAP1 gene, we searched by BLAST the GenBank/NCBI database. We found that the SMAP1-like protein BC052413 was the only one detected in the database as showing a high degree of homology to SMAP1 over the entire length. We then screened a cDNA library prepared from a murine erythroleukemic cell line for the gene encoding BC052413 and obtained several cDNA clones. The longest contained a 2871 base pairs insert and its sequence exactly matched that of BC052413 (unpublished data). We named this gene SMAP2.
The polypeptide encoded by SMAP2 is composed of 428 aa residues and has a predicted molecular mass of 47 kDa. The alignment of the aa sequences of SMAP2 and SMAP1 is shown in Figure 1A. SMAP2 and SMAP1 share 50% overall sequence homology but four distinct regions in SMAP2, namely, aa 16-125, aa 173-212, aa 244-277, and aa 369-428, show a marked degree of conservation with SMAP1 (85, 74, 69, and 48% homology, respectively; Figure 1B). As described in the following sections, we sought to determine the functions of these conserved regions.
Figure 1.
The aa sequence of the SMAP2 protein and depiction of its functional domains. (A) Alignment of the aa sequences of the SMAP2 and SMAP1 proteins. The aa sequence of SMAP2 has been predicted as BC052413, which is registered in GenBank/NCBI. We authenticated the sequence of BC052413 by independently isolating and sequencing cDNA clones. The aa sequence of SMAP1 has been described in our previous study (Tanabe et al., 2005). SMAP2 and SMAP1 encode 428 and 440 aa proteins, respectively. Their identical aa residues are indicated by white letters on a black background. The classical and atypical CHC-binding motifs correspond to LLGLD (aa 187-191) and DLL (aa 212-214), respectively. (B) Functional domains of the SMAP2 protein. These domains include a GAP activity domain (aa 1-163), a clathrin-interacting domain (aa 163-231), and a CALM-interacting domain (aa 339-395). The regions that are highly conserved between SMAP2 and SMAP1, namely, the aa 16-125, aa 173-212, aa 244-277, and aa 369-428 regions, are also indicated (by bars). The aa numbers refer to those in SMAP2.
Detection of GAP Activity Mediated by SMAP2
The amino-terminal region (aa 16-125) of SMAP2 harbors a conserved CX2CX16CX2C zinc finger motif and is strongly homologous to the corresponding region of SMAP1. Therefore, it is possible that SMAP2, like SMAP1, exhibits ArfGAP activity. We tested this possibility by incubating SMAP2 together with Arf1 or Arf6 as a substrate. The SMAP2 protein we used for this purpose was the truncated aa 1-163 segment of SMAP2 that was purified from a bacterial cell lysate (the intact form was insoluble when expressed in bacteria). As shown in Figure 2A, SMAP2 mediated the hydrolysis of GTP that had been loaded onto both Arf1 and Arf6. Similarly, SMAP1, which is an Arf6-specific GAP in vivo, and GAP1, which is an Arf1-specific GAP in vivo, showed GAP activity with both Arf1 and Arf6 in these in vitro assays. The extent of GTP hydrolysis mediated by SMAP2 was found to be dependent on the amount of SMAP2 protein added and the length of the incubation reaction (Figure 2, B and C). The 50th residue of GAP1 (an arginine) is known to be vital for its GAP activity (Szafer et al., 2000) and is also conserved in SMAP2 as the 56th residue. When this R56 residue was replaced with glutamine, it reduced or abolished the GAP activity of SMAP2. Therefore, domain aa 1-163 of SMAP2 was determined to have ArfGAP activity (Figure 1B).
Figure 2.
GAP activity of SMAP2 on Arf1 and Arf6. (A) Comparison of the GAP activity of SMAP2 with that of SMAP1 and GAP1. Six micrograms each of the SMAP2 (aa 1-163), SMAP1 (aa 1-255), or GAP1 (aa 1-246) proteins were incubated in vitro with 1 μg of GTP-loaded Arf1 or Arf6 for 60 min. In “mock,” no GAP was added to the reaction. After separating the GDP from GTP on thin-layer chromatograms, the degree of GTP hydrolysis was calculated. This is shown as a percentage of GDP over GDP+GTP. Each reaction was independently performed three times and the averages as well as SD values are shown. (B and C) Concentration- and incubation time–dependent GAP activity of the SMAP2 protein. Six (○) or 3 μg (•) of SMAP2 protein, or 6 μg of mutated SMAP2R56Q protein (▵) were incubated with 1 μg of GTP-loaded Arf1 (B) or Arf6 (C) for the indicated times. After separating the GDP from GTP on thin-layer chromatograms, the degree of GTP hydrolysis was calculated and is shown as a percentage as in A. Each data point shows the averages and SD values of three independent experiments.
Identification of a Clathrin-interacting Domain in SMAP2
The second conserved region of SMAP2 is contained by aa 173-212. This region harbors both classical and atypical CHC-binding motifs such as LLGLD (aa 187-191) and DLL (aa 212-214), respectively (Figure 1A). We previously reported that the LLGLD motif in SMAP1 functions as a CHC-binding motif (Tanabe et al., 2005; note that a DLL motif is not conserved in SMAP1). We therefore examined whether SMAP2 also binds CHC (Figure 3A). Thus, Cos-7 cells were transfected or not with SMAP2 tagged with a Myc-epitope, and the cell lysate was immunoprecipitated with anti-CHC antibody (top panel). Probing of the filter with anti-Myc antibody revealed that the transduced SMAP2 was coimmunoprecipitated by the anti-CHC antibody. Conversely, when cells were transfected with SMAP2 tagged with HA, CHC was coimmunoprecipitated by the anti-HA but not the irrelevant anti-Myc antibodies (bottom panel). This indicates the SMAP2 and CHC molecules physically associate inside the cells. In addition, replacement of all the residues in both LLGLD and DLL motifs by alanine abolished the coimmunoprecipitation of CHC (see the lane of SMAP2CBm). Thus, the LLGLD and DLL motifs are essential for SMAP2 to interact with CHC. It must be noted that, unlike the case of SMAP1, mutation of the sole LLGLD motif was not sufficient to disrupt the association of SMAP2 with CHC (unpublished data).
Figure 3.
Interaction of SMAP2 protein with the clathrin molecule. (A) Coimmunoprecipitation of SMAP2 and CHC. Cos-7 cells were transfected with Myc-SMAP2, HA-SMAP2, or HA-SMAP2CBm as indicated, and their cell lysates were immunoprecipitated (IP) by anti-Myc, anti-HA, or anti-CHC antibodies as indicated. The precipitates were then subjected to electrophoresis, transferred to a filter, and probed by the indicated antibody (IB). (B) Subcellular localization of the SMAP2 and CHC proteins. Cos-7 cells were transfected with cDNAs encoding the SMAP2 (aa 1-428), with deletion mutants of SMAP2 that corresponded to aa 163-428 or aa 231-428, or with the CBm mutant of SMAP2, respectively. In the CBm mutant, all the aa residues in the LLGLD (aa 187-191) and DLL (aa 212-214) motifs were replaced by alanine. The cells were then fixed and subjected to double immunofluorescence analysis. The HA-tagged SMAP2 and its mutant proteins were detected by anti-HA antibody (red fluorescence), whereas the endogenous clathrin molecules were detected by anti-CHC antibody (green fluorescence). Merged images are shown in the right-hand column. In the insets, portions of the cells are enlarged.
We next examined the subcellular localization of SMAP2 and CHC by double immunofluorescence analysis (Figure 3B). Thus, Cos-7 cells were transfected with SMAP2 (1-428) tagged with HA and subjected to immunofluorescence staining. The transduced SMAP2 protein was detected by anti-HA antibody, whereas endogenous CHC was detected by anti-CHC antibody. SMAP2 was detected as multiple, aggregate-like structures in the cytoplasm and most of the CHC-derived fluorescence overlapped with that of SMAP2. We then analyzed the two deletion-type mutants of SMAP2 that corresponded to aa 163-428 and aa 231-428, respectively. CHC overlapped with the aggregates formed by the aa 163-428. On the other hand, the aa 231-428 of SMAP2 also formed aggregates but CHC did not overlap with them. In addition, the introduction of CBm mutation abolished the aggregate formation of SMAP2 (1-428) with CHC. Therefore, the aa 163-231 region that harbors the LLGLD and DLL motifs is necessary for SMAP2 to form aggregates and overlap with CHC. The apparent overlap of SMAP2CBm- and CHC-fluorescence at a jutxtanuclear region reflects the TGN as shown below. Probably, SMAP2-CBm and CHC localized to this structure simultaneously and independently. Determination of aa motif, if any, that is responsible for the subcellular localization of SMAP2 needs a separate study.
Collectively, the immunofluorescent results complement those of immunoprecipitation. Thus, based on both, we designate the aa 163-231 region a clathrin-interacting domain (Figure 1B).
Identification of the CALM-interacting Domain in SMAP2
The other two conserved regions, namely, aa 244-277 and aa 369-428, did not show any significant homology to known motifs or domains when we performed BLAST and SMART domain analysis. Therefore, to determine the functions of these regions, we sought to identify molecules that may interact with them by yeast two-hybrid analysis. SMAP2 lacking the amino-terminal aa 1-130 was used as a bait and the prey was a cDNA library prepared from a murine brain. Screening of four million independent clones yielded several positive clones, including a cDNA that encodes a portion of the CALM protein. Consequently, a cDNA clone that harbors the entire CALM protein was reisolated from a library and used in the following studies. This clone was first used to confirm that intact SMAP2 (aa 1-428) and CALM proteins indeed interact in yeast cells (Figure 4A). We then tried to define the CALM-interacting domain in SMAP2 and transduced into yeast cells a portion of SMAP2 that consisted of aa 339-428. This region also interacted with CALM. The observed interaction was specific, because no colony grew on a selective medium if either the bait or prey plasmid harbored no cDNA insert.
Figure 4.
Interaction of SMAP2 with CALM. (A) SMAP2 was tested for its ability to interact with CALM. Yeast cells were cotransformed by the indicated combinations of bait (pGBKT7) and prey (pGADT7) vectors and grown on nonselective (−Trp, −Leu) and selective (−Trp, −Leu, −His, −Ade, 2.5 mM 3-amino-1,2,4-triazole) media, respectively. The cDNAs tested were the intact SMAP2 (aa 1-428), a portion of SMAP2 (aa 339-428), and the intact CALM, respectively. (B) Subcellular localization of SMAP2 and CALM proteins. Cos-7 cells were cotransfected with CALM and SMAP2 (aa 1-428) or deletion mutants of SMAP2 that corresponded to aa 163-395 or aa 339-428, respectively, fixed, and processed for double immunofluorescence analysis. The HA-tagged SMAP2 or deletion proteins were detected by anti-HA antibody (red fluorescence), whereas the Myc-tagged CALM protein was detected by anti-Myc antibody (green fluorescence). Merged images are shown in the right-hand column. In the insets, portions of the cells are enlarged.
We then examined the subcellular localization of the SMAP2 and CALM proteins by double immunofluorescence (Figure 4B). Thus, Cos-7 cells were cotransfected with HA-tagged SMAP2 (aa 1-428) and Myc-tagged CALM and subjected to immunofluroescence analysis. The two proteins colocalized as multiple aggregates in the cytoplasm. Cos-7 cells were transfected as well with deletion-type mutants of SMAP2 that consisted of aa 163-395 and aa 339-428. Both SMAP2 mutants colocalized with CALM. Thus, the aa 339-395 region in SMAP2 is a putative CALM-interacting domain.
In summary, as shown in Figure 1B, the SMAP2 protein harbors three distinct domains that are responsible for GAP activity, clathrin interaction, and CALM interaction, respectively. The functional significance of the remaining conserved region (aa 244-277) is not known at present.
Subcellular Localization of SMAP2 Protein
In the above experiments of cDNA transfection, we used Cos-7 cells in which transduced SMAP2 was excessively produced and appeared to form aggregates. This was convenient to show whether SMAP2 interacts with CHC or CALM by immunohistochemistry. However, aggregate-like structures are not likely to represent the physiological distribution of SMAP2 inside the cell. The anti-SMAP2 antibody we generated was useful to detect the endogenous protein only by immunoblot but not immunofluorescence (Supplementary Figure S1). Therefore, we transfected HeLa cells with an HA-tagged intact SMAP2; in this case, the SMAP2 protein expression level was moderate (Figure 5). This revealed that SMAP2 was distributed on multiple dotlike as well as juxtanuclear structures. Staining for endogenous CHC displayed a similar pattern to that of SMAP2, and at least some of the CHC fluorescence colocalized with that of SMAP2. However, SMAP2 did not overlap with endogenous COPI, which is a coatmer protein that forms vesicles on the Golgi apparatus.
Figure 5.
Subcellular localization of SMAP2 and coat proteins. HeLa cells were transfected with HA-SMAP2 (aa 1-428), fixed, and processed for double immunofluorescence analysis. SMAP2 was detected by anti-HA antibody (red fluorescence), whereas endogenous CHC or COPI were detected by their respective specific antibodies (green fluorescence). Merged images are shown in the right-hand column. In the insets, portions of the cells are enlarged.
SMAP2 and Clathrin Adaptors
Because SMAP2 colocalizes with CHC, we examined which clathrin adaptors are utilized by SMAP2. There are two distinct classes of clathrin adaptors denoted as AP-1 and AP-2. Each adaptor functions in a different pathway of clathrin-mediated vesicle transport. AP-1 functions on the early endosome/TGN, whereas AP-2 does on coated pits on the plasma membrane. AP-3 is a third class of adaptor and it is not known whether AP-3 involves clathrin in vivo. This AP-3 functions on the late endosome. We compared the distribution of transduced intact SMAP2 with that of each endogenous adaptor by double immunofluorescence analysis (Figure 6). The SMAP2 staining significantly overlapped that of AP-1, whereas overlap of SMAP2 with either AP-2 or AP-3 was minimum.
Figure 6.
Subcellular localization of SMAP2 and clathrin adaptors. HeLa cells were transfected with HA-SMAP2 (aa 1-428), fixed, and processed for double immunofluorescence analysis. SMAP2 was detected by anti-HA antibody (red fluorescence), whereas AP-1, AP-2, AP-3, EpsinR, and GGA1 were detected by their respective specific antibodies (green fluorescence). AP-1, AP-2, and AP-3 are endogenous molecules while EpsinR and GGA1 are the transfected Myc-EpsinR and Myc-GGA1 molecules, respectively. Merged images are shown in the right-hand column. In the insets, portions of the cells are enlarged. It must be noted that the juxtanuclear structures seen in Figure 5 were out of focus in the SMAP2 photos here.
EpsinR, a monomeric adaptor for clathrin, has been identified as a factor that binds to AP-1 and appears to be utilized in early endosome-to-TGN transport (Saint-Pol et al., 2004). To test whether SMAP2 also overlaps with EpsinR, HeLa cells were cotransfected with HA-SMAP2 and Myc-EpsinR. We found that EpsinR staining colocalized with that of SMAP2. In contrast, HA-SMAP2 did not colocalize with Myc-GGA1, a clathrin-adaptor protein that is utilized in a TGN-to-endosome pathway (Ghosh and Kornfeld, 2004). Thus, SMAP2 is probably involved in AP-1– and EpsinR-mediated vesicle transport. The multiple dot-like fluorescence likely represents the SMAP2 located on the early endosomes, whereas the juxtanuclear fluorescence probably reflects the SMAP2 on endosomes that were located adjacent to the TGN and could not be distinguished from the TGN itself by optical microscopy.
SMAP2 Functions on Arf1 In Vivo
We have shown so far that SMAP2 colocalizes with clathrin, AP-1, and EpsinR on the early endosome/TGN. Judging from its subcellular localization, SMAP2 probably acts as an Arf1GAP in the cell. However, our in vitro GAP assays showed that SMAP2 was equally active with both Arf1 and Arf6 (Figure 2A). Thus, in this section, we examined whether SMAP2 acts on Arf1 in vivo by transfecting HeLa cells with HA-tagged SMAP2R56Q, a GAP-negative mutant. The cells were then processed for double immunofluorescence analysis (Figure 7A). In drug-untreated cells, regardless of the SMAP2R56Q expression, endogenous AP-1 was detected on the endosome/TGN as shown above. The transfected cells were then treated by BFA, an inhibitor of Arf1. In the cells that were not transduced by SMAP2R56Q (indicated by arrowheads), the accumulation of AP-1 on the endosome/TGN was no longer seen, and the AP-1–derived fluorescence was dispersed and abolished. This confirms that the accumulation of AP-1 on the endosome/TGN is an Arf1-dependent process. In contrast, in the SMAP2R56Q-expressing cells (indicated by arrows), the accumulation of AP-1 on membranes persisted and was resistant to BFA treatment. Arf1 in such cells may remain as a GTP-bound form on membranes where SMAP2 is normally the primary ArfGAP. Possible mechanisms how SMAP2R56Q could lead the differently localized AP-1 to become resistant to BFA are discussed later. In any case, the above observations indicate that SMAP2 is likely to function in an Arf1 (and/or the other Arf of classes I and II)-dependent process that involves the endosome.
Figure 7.
Effect of SMAP2R56Q expression and BFA treatment on the subcellular distribution of AP-1 and COPI. HeLa cells were transfected with HA-SMAP2R56Q (aa 1-428; A and B) or HA-GAP1C45A (C), treated with or without BFA, and then fixed and processed for double immunofluorescence analysis. SMAP2R56Q or GAP1C45A were detected by anti-HA antibody, whereas endogenous AP-1 or COPI were detected by their respective specific antibodies. The arrows and arrowheads indicate SMAP2R56Q-expressing and nonexpressing cells, respectively.
Because Arf1 is involved in vesicle transport on the Golgi complex as well, we also examined whether SMAP2 participates in this pathway. COPI is a coat protein that is utilized in Arf1-dependent vesicle transport and is detected on the Golgi complex. Treatment of cells with BFA abolished the accumulation of COPI, but this was not affected by the expression of SMAP2R56Q (Figure 7B). As a control, the cells were transfected with a GAP-negative mutant of GAP1, GAP1C45A. This rendered the accumulation of COP1 on the Golgi complex resistant to BFA (Figure 7C). Thus, SMAP2 is probably not involved in COP1- and Arf1-dependent vesicle transport on the Golgi complex. The accumulation of AP-3 on the late endosome was also abolished by the BFA treatment of cells, irrespective of the SMAP2R56Q expression (unpublished data). This strengthens that SMAP2 is not likely involved in AP-3-dependent transport on the late endosome.
Cargo Protein Candidates in the SMAP2-mediated Pathways
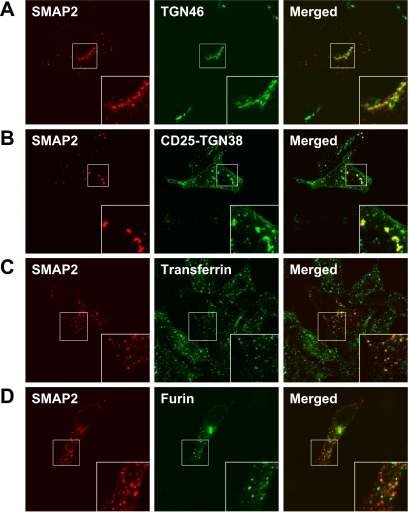
We searched which molecules are likely to be transported by the SMAP2-mediated pathways. Judging from the subcellular localization of SMAP2 on the early endosome/TGN, likely candidates are human TGN46 or murine TGN38 (the two are orthologous proteins). Thus, HeLa cells were doubly stained for endogenous TGN46 and the transfected intact HA-SMAP2 protein (Figure 8A). At least some of the TGN46 proteins colocalized with SMAP2 on the TGN. Because endogenous TGN46 was mainly detected on the TGN and not the endosome, we also analyzed cells cotransfected with SMAP2 and CD25-tagged TGN38 (Figure 8B). The fluorescence of TGN38 on the early endosome overlapped with that of SMAP2. These results suggest that TGN38/46 is a likely cargo protein via a SMAP2-dependent process.
Figure 8.
Subcellular localization of SMAP2 and its possible cargo protein. (A) HeLa cells were transfected with HA-SMAP2 (aa 1-428), fixed, and processed for double immunofluorescence analysis. SMAP2 was detected by anti-HA antibody (red fluorescence), whereas endogenous human TGN46 was detected by anti-TGN46 antibody (green fluorescence). (B) HeLa cells were cotransfected with HA-SMAP2 and the chimeric CD25-murine TGN38 molecule, fixed, and processed for double immunofluorescence analysis. SMAP2 (red fluorescence) and CD25-TGN38 (green fluorescence) were detected by their respective specific antibodies. (C) HeLa cells were transfected with HA-SMAP2, incubated with dye-conjugated transferrin, fixed, and processed for double immunofluorescence analysis. SMAP2 was detected by anti-HA antibody (red fluorescence), whereas transferrin was visible as green fluorescence. (D) HeLa cells were cotransfected with HA-SMAP2 and Myc-furin convertase, fixed, and processed for double immunofluorescence analysis. SMAP2 and furin convertase were detected by anti-HA antibody (red fluorencence) and anti-Myc antibody (green fluorescence), respectively. In A–D, merged images are shown in the right-hand column. In the insets, portions of the cells are enlarged.
Transferrin receptors are constitutively endocytosed from the plasma membrane to the early endosome. Although the residence of transferrin receptors on the early endosome is temporary because of their recycling to the plasma membrane, SMAP2 might partially colocalize with incorporated transferrin receptors. To test this, HeLa cells were transfected with SMAP2 and incubated with dye-conjugated transferrin (Figure 8C). Indeed, we found SMAP2 and transferrin receptors colocalized to some degree on the early endosome.
We also found that furin convertase, which is transported from the late endosome to the TGN (Rohn et al., 2000), is not likely to be a cargo protein carried by a SMAP2-dependent pathway, because cotransfected SMAP2 and furin convertase did not colocalize at all (Figure 8D).
Overexpression of SMAP2 Delays the Transport of CD25-TGN38
Finally, we assessed directly whether overexpression of SMAP2 affects the transport of CD25-TGN38 protein from the early endosome to the TGN. Cos-7 cells were cotransfected with HA-SMAP2 and CD25-TGN38 and incubated with anti-CD25 antibody, which triggers the internalization of the chimeric molecule. After the incubation, the cells were fixed and processed for double immunofluorescence (Figure 9). The photos presented in the panels A to F represent the cells after 15 min incubation. The TGN38 protein appeared to reach and accumulate on the TGN in the untransduced cells, whereas the protein remained on the endosomes and did not reach the TGN in the SMAP2-transduced cells (indicated by arrows in A and B). We counted the number of cells that showed or did not show the TGN accumulation of TGN38, respectively. Among the SMAP2-transduced cells, the inhibition of TGN-accumulation was as high as 35% (see the lane of SMAP2 at 15 min in panel G). In contrast, TGN38 reached the TGN in most of mock-transfected cells (see the inhibition of only 4%). Thus, overexpression of intact SMAP2 significantly impairs the transport of CD25-TGN38 from the early endosome to the TGN.
Figure 9.
Effect of SMAP2 overexpression on the transport of CD25-TGN38 molecule. Cos-7 cells were cotransfected by either one of HA-SMAP2 (A and B), HA-SMAP2R56Q (C and D), or HA-SMAP2CBm (E and F) together with the CD25-TGN38 chimeric cDNA. The cells were then incubated for 15 or 45 min with the anti-CD25 antibody that triggers the internalization of the chimeric molecule. The cells were fixed and processed for double immunofluorescence. In A, C, and E, the SMAP2 protein was detected by the anti-HA antibody, whereas the chimeric protein was detected by the anti-CD25 antibody in B, D, and F. The photos presented in A to F represent the cells after 15 min incubation. The arrows indicate the SMAP2-expressing cells. In G, among the 100 cells that were positive for the SMAP2 staining, the numbers of cells that showed the TGN accumulation or no accumulation of TGN38 were counted, respectively. The percent inhibition corresponds to the cells that were SMAP2-positive but did not accumulate TGN38 at the TGN. Independent transfections were repeated three times, and the averages and SD values of percent inhibition are shown. In a separate experiment, we confirmed by immunoblot analysis that the expression level of HA-SMAP2, HA-SMAP2R56Q, and HA-SMAP2CBm did not vary significantly among the transfected cells.
We then examined the effects of each R56Q or CBm mutation on the above described inhibitory activity of SMAP2. The R56Q mutation reduced the inhibition of TGN-accumulation to 20% (see the lane of SMAP2R56Q at 15 min in panel G) and thus the TGN38 molecule could now reach the TGN (indicated by arrows in the panels C and D). On the other hand, the CBm mutation rather enhanced the inhibitory activity of SMAP2–50% (see the lane of SMAP2CBm at 15 min in panel G) and therefore the TGN38 molecule remained on the endosomes and did not accumulate on the TGN (indicated by arrows in panels E and F). This stronger inhibitory activity of SMAP2CBm on the transport was evident even after the prolonged incubation of cells to 45 min (see the 35% inhibition in the panel G). At this time, most of TGN molecule appeared to reach the TGN in the wild-type as well as R56Q-transduced cells (note that their inhibitions that were <10%).
Possible mechanisms that might explain the differential effects caused by the wild-type as well as mutated SMAP2 proteins are discussed below. At this moment, we conclude that SMAP2 can function in a retrograde transport of CD25-TGN38 from the early endosome to the TGN.
DISCUSSION
In this study, we identified SMAP2 as a novel ArfGAP that shows a high degree of homology with SMAP1. Furthermore, we demonstrated that SMAP2 prefers Arf1 in vivo and interacts with both CHC and CALM. SMAP2 is the first example of an ArfGAP that is located on the early endosome together with AP-1 and EpsinR.
SMAP2, SMAP1, and GAP1 all exhibited a lack of preference for both Arf1 and Arf6 in our in vitro GAP assay. This suggests that the ArfGAP domain itself does not necessarily direct the substrate specificity of an ArfGAP toward a particular isoform of Arf. Therefore, to determine which substrate is preferred by SMAP2, we first assessed the subcellular localization of SMAP2. We found that SMAP2 is localized on the AP-1– and EpsinR-positive subcellular structures, but not on the AP-3- or COPI-positive intracellular organelles. Because the combination of AP-1 and EpsinR indicates a specific adaptor for the clathrin-coat on the early endosome (Saint-Pol et al., 2004), the observed colocalization pattern implies that SMAP2 functions in an early endosome-to-TGN pathway that is known to be regulated by Arf1. Indeed, we then found that SMAP2 colocalizes with the TGN38/46 protein and with transferrin molecules on endosomes. Furthermore, we could demonstrate that overexpression of SMAP2 delays the TGN-accumulation of CD25-TGN38, a cargo protein known to be transported from the early endosome to the TGN. This is probably due to the inactivation of Arf1 by SMAP2 and may reflect the impairment of vesicle budding. The observed effects of R56Q and CBm mutants on the transport need some discussion though. A GAP-negative R56Q mutation should keep Arf1 as an active state, thereby likely promoting the vesicle budding. On the other hand, the lack of GAP activity may not allow vesicles to be uncoated. Thus, the apparent accumulation of TGN38 may represent not the TGN itself but actually the endosomes located near the TGN. In the case of CBm mutation, the clathrin molecules may not be recruited to the membranes, thereby resulting in the significant impairment of vesicle budding. In any case, the present study indicates that SMAP2 can function in a retrograde early endosome-to-TGN pathway. This also substantiates the notion proposed by ourselves as well as others that each ArfGAP member acts specifically in a distinct route of vesicle transport inside the cell.
Both SMAP2 and SMAP1 harbor a clathrin box and we showed that both indeed interact with CHC. That both SMAPs interact with clathrin is feasible because the pathways involving SMAP2 (endosome-to-TGN) and SMAP1 (endocytosis from the plasma membrane) are known to utilize clathrin-coated vesicles. The clathrin box in SMAP2 may mediate the indirect interaction between Arf1 and clathrin on the early endosome, similar to the case of SMAP1, as we previously discussed (Tanabe et al., 2005). It must be noted though that, unlike SMAP1, both the classical LLGLD and the atypical DLL motifs appear to contribute to the clathrin-binding in the case of SMAP2. In addition to the SMAPs, Git1 is another example of an ArfGAP that is involved in clathrin-dependent transport. Ligand-dependent endocytosis of G protein–coupled receptors is mediated by Git1 (Claing et al., 2000), and we could detect a canonical clathrin box in Git1 (aa 607-611, unpublished data). A TGN-to-endosome pathway also utilizes clathrin-coated vesicles, but the candidate ArfGAP that may act in this pathway has not been identified so far. Instead, GGA1 is reported to function in this process, and this GGA1 binds not only to clathrin but also to Arf1 (Ghosh and Kornfeld, 2004). Therefore, the presence of clathrin box may be a common feature of the ArfGAPs and GGA1 that are involved in clathrin-dependent vesicle transport.
We could also demonstrate that SMAP2 interacts with CALM and identified a CALM-interacting domain in the carboxy-terminal portion of SMAP2. CALM and AP180 are isoforms and are expressed ubiquitously and specifically in the brain, respectively (Tebar et al., 1999). In cells that are deficient for LAP (an AP180-homologue in Drosophila), the uniformity of clathrin-coated vesicle size is lost (Zhang et al., 1998; Hao et al., 1999). Thus, CALM/AP180 is considered to induce clathrin assembly and to shape the vesicles into an uniform size. Although it is not yet clear whether CALM is involved in vesicle transport from the endosome, it is possible that SMAP2, Arf1, clathrin, and CALM together form a higher order complex at the site of vesicle budding from the endosome.
Treatment of cells with BFA disperses the AP-1 molecules that are accumulated on the early endosomes. We did not see this effect in SMAP2R56Q-transfected cells, although AP-1 and SMAP2R56Q had different localization. One possibility is that the mutant displaces the endogenous SMAP2 from membranes, thereby stabilizing the Arf. An alternative possibility is that the mutant forms a complex with the endogenous SMAP2 and trap it in a nonfunctional state. Observations supporting the latter possibility are described in Supplementary Figure S2. Whichever the mechanisms might be, the observation strongly favors the notion that SMAP2 functions as an Arf1GAP on the early endosomes. In contrast, SMAP1 regulates Arf6-dependent endocytosis from the plasma membrane. Therefore, a reasonable question would be why the homologous SMAP2 and SMAP1 behave so differently in vivo. We found four domains that were conserved between SMAP2 and SMAP1. These correspond to a GAP domain, a clathrin-interacting domain, a domain of unknown function and a CALM-interacting domain. These conserved domains probably do not contain the putative region that is responsible for the different activities of SMAP2 and SMAP1 in vesicle transport. This putative disparate region in the SMAP proteins may be responsible for their different subcellular localizations (SMAP2 on the early endosome and SMAP1 on the plasma membrane) and eventually their specificities for different Arf isoforms (SMAP2 for Arf1 and SMAP1 for Arf6). Thus, these two SMAP proteins, which appear to act in important and distinct pathways of vesicle transport, may help to further dissect the molecular mechanisms involved in these pathways.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to K. Nakayama for providing us with GGA1 cDNA. We also thank M. Kuji for secretarial work. This work was supported in part by Grants in Aid from the Ministry of Education, Science, Sports, Culture, and Technology of Japan. M.S. is a member of the 21st century COE program “Center for Innovative Therapeutic Development Toward the Conquest of Signal Transduction Diseases” at Tohoku University.
Abbreviations used:
- Arf
ADP-ribosylation factor
- BFA
brefeldin A
- CHC
clathrin heavy chain
- GAP
GTPase-activating protein
- GEF
guanine-nucleotide exchange factor
- GST
glutathione S-transferase
- HA
influenza hemagglutinin
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05-10-0909) on March 29, 2006.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Aoe T., Huber I., Vasudevan C., Watkins S. C., Romero G., Cassel D., Hsu V. W. The KDEL receptor regulates a GTPase-activating protein for ADP-ribosylation factor 1 by interacting with its non-catalytic domain. J. Biol. Chem. 1999;274:20545–20549. doi: 10.1074/jbc.274.29.20545. [DOI] [PubMed] [Google Scholar]
- Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- Chiba N., Watanabe T., Nomura S., Tanaka Y., Minowa M., Niki M., Kanamaru R., Satake M. Differentiation dependent expression and distinct subcellular localization of the protooncogene product, PEBP2beta/CBFbeta, in muscle development. Oncogene. 1997;14:2543–2552. doi: 10.1038/sj.onc.1201109. [DOI] [PubMed] [Google Scholar]
- Claing A., Perry S. J., Achiriloaie M., Walker J. K., Albanesi J. P., Lefkowitz R. J., Premont R. T. Multiple endocytic pathways of G protein-coupled receptors delineated by GIT1 sensitivity. Proc. Natl. Acad. Sci. USA. 2000;97:1119–1124. doi: 10.1073/pnas.97.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona O. Live stripping of clathrin-coated vesicles. Dev. Cell. 2001;1:592–594. doi: 10.1016/s1534-5807(01)00079-x. [DOI] [PubMed] [Google Scholar]
- Diaz E., Pfeffer S. R. TIP47, a cargo selection device for mannose 6-phosphate receptor trafficking. Cell. 1998;93:433–443. doi: 10.1016/s0092-8674(00)81171-x. [DOI] [PubMed] [Google Scholar]
- Donaldson J. G. Multiple roles for Arf6: sorting, structuring, and signaling at the plasma membrane. J. Biol. Chem. 2003;278:41573–41576. doi: 10.1074/jbc.R300026200. [DOI] [PubMed] [Google Scholar]
- Donaldson J. G., Finazzi D., Klausner R. D. Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature. 1992;360:350–352. doi: 10.1038/360350a0. [DOI] [PubMed] [Google Scholar]
- Donaldson J. G., Kahn R. A., Lippincott-Schwartz J., Klausner R. D. Binding of ARF and beta-COP to Golgi membranes: possible regulation by a trimeric G protein. Science. 1991;254:1197–1199. doi: 10.1126/science.1957170. [DOI] [PubMed] [Google Scholar]
- Ford M. G., Pearse B. M., Higgins M. K., Vallis Y., Owen D. J., Gibson A., Hopkins C. R., Evans P. R., McMahon H. T. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- Fotin A., Cheng Y., Grigorieff N., Walz T., Harrison S. C., Kirchhausen T. Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating. Nature. 2004;432:649–653. doi: 10.1038/nature03078. [DOI] [PubMed] [Google Scholar]
- Franco M., Peters P. J., Boretto J., van Donselaar E., Neri A., D'Souza-Schorey C., Chavrier P. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane cycling and actin cytoskeleton organization. EMBO J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Kornfeld S. The GGA proteins: key players in protein sorting at the trans-Golgi network. Eur. J. Cell Biol. 2004;83:257–262. doi: 10.1078/0171-9335-00374. [DOI] [PubMed] [Google Scholar]
- Hao W., Luo Z., Zheng L., Prasad K., Lafer E. M. AP180 and AP-2 interact directly in a complex that cooperatively assembles clathrin. J. Biol. Chem. 1999;274:22785–22794. doi: 10.1074/jbc.274.32.22785. [DOI] [PubMed] [Google Scholar]
- Hinners I., Tooze S. A. Changing directions: clathrin-mediated transport between the Golgi and endosomes. J. Cell Sci. 2003;116:763–771. doi: 10.1242/jcs.00270. [DOI] [PubMed] [Google Scholar]
- Hirst J., Robinson M. S. Clathrin and adaptors. Biochim. Biophys. Acta. 1998;1404:173–193. doi: 10.1016/s0167-4889(98)00056-1. [DOI] [PubMed] [Google Scholar]
- Lemmon S. K. Clathrin uncoating: auxilin comes to life. Curr. Biol. 2001;11:R49–R52. doi: 10.1016/s0960-9822(01)00010-0. [DOI] [PubMed] [Google Scholar]
- Lewin D. A., Mellman I. Sorting out adaptors. Biochim. Biophys. Acta. 1998;1401:129–145. doi: 10.1016/s0167-4889(97)00130-4. [DOI] [PubMed] [Google Scholar]
- Liu W., Duden R., Phair R. D., Lippincott-Schwartz J. ArfGAP1 dynamics and its role in COPI coat assembly on Golgi membranes of living cells. J. Cell Biol. 2005;168:1053–1063. doi: 10.1083/jcb.200410142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C., Zizioli D., Lausmann S., Eskelinen E. L., Hamann J., Saftig P., von Figura K., Schu P. mu1A-adaptin-deficient mice: lethality, loss of AP-1 binding and rerouting of mannose 6-phosphate receptors. EMBO J. 2000;19:2193–2203. doi: 10.1093/emboj/19.10.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J. R., Prasad K., Jin S., Augustine G. J., Lafer E. M. Uncoating of clathrin-coated vesicles in presynaptic terminals. Roles for hsc70 and auxilin. Neuron. 2001;32:289–300. doi: 10.1016/s0896-6273(01)00467-6. [DOI] [PubMed] [Google Scholar]
- Nie Z., Boehm M., Boja E. S., Vass W. C., Bonifacino J. S., Fales H. M., Randazzo P. A. Specific regulation of the adaptor protein complex AP-3 by the Arf GAP AGAP1. Dev. Cell. 2003a;5:513–521. doi: 10.1016/s1534-5807(03)00234-x. [DOI] [PubMed] [Google Scholar]
- Nie Z., Hirsch D. S., Randazzo P. A. Arf and its many interactors. Curr. Opin. Cell Biol. 2003b;15:396–404. doi: 10.1016/s0955-0674(03)00071-1. [DOI] [PubMed] [Google Scholar]
- Ooi C. E., Dell'Angelica E. C., Bonifacino J. S. ADP-ribosylation factor 1 (ARF1) regulates recruitment of the AP-3 adaptor complex to membranes. J. Cell Biol. 1998;142:391–402. doi: 10.1083/jcb.142.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche A., Antonny B., Robineau S., Acker J., Cherfils J., Jackson C. L. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- Randazzo P. A., Hirsch D. S. Arf GAPs: multifunctional proteins that regulate membrane traffic and actin remodelling. Cell Signal. 2004;16:401–413. doi: 10.1016/j.cellsig.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Rohn W. M., Rouille Y., Waguri S., Hoflack B. Bi-directional trafficking between the trans-Golgi network and the endosomal/lysosomal system. J. Cell Sci. 2000;113:2093–2101. doi: 10.1242/jcs.113.12.2093. [DOI] [PubMed] [Google Scholar]
- Saint-Pol A., et al. Clathrin adaptor epsinR is required for retrograde sorting on early endosomal membranes. Dev. Cell. 2004;6:525–538. doi: 10.1016/s1534-5807(04)00100-5. [DOI] [PubMed] [Google Scholar]
- Scales S. J., Gomez M., Kreis T. E. Coat proteins regulating membrane traffic. Int. Rev. Cytol. 2000;195:67–144. doi: 10.1016/s0074-7696(08)62704-7. [DOI] [PubMed] [Google Scholar]
- Serafini T., Orci L., Amherdt M., Brunner M., Kahn R. A., Rothman J. E. ADP-ribosylation factor is a subunit of the coat of Golgi-derived COP-coated vesicles: a novel role for a GTP-binding protein. Cell. 1991;67:239–253. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- Shin H. W., Nakayama K. Guanine nucleotide-exchange factors for arf GTPases: their diverse functions in membrane traffic. J. Biochem. (Tokyo) 2004;136:761–767. doi: 10.1093/jb/mvh185. [DOI] [PubMed] [Google Scholar]
- Szafer E., Pick E., Rotman M., Zuck S., Huber I., Cassel D. Role of coatmer and phospholipids in GTPase-activating protein-dependent hydrolysis of GTP by ADP-ribosylation factor-1. J. Biol. Chem. 2000;275:23615–23619. doi: 10.1074/jbc.M003171200. [DOI] [PubMed] [Google Scholar]
- Tanabe K., Torii T., Natsume W., Braesch-Andersen S., Watanabe T., Satake M. A novel GTPase-activating protein for ARF6 directly interacts with clathrin and regulates clathrin-dependent endocytosis. Mol. Biol. Cell. 2005;16:1617–1628. doi: 10.1091/mbc.E04-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebar F., Bohlander S. K., Sorkin A. Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol. Biol. Cell. 1999;10:2687–2702. doi: 10.1091/mbc.10.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L. M., Ostrom J. A., Kornfeld S. Biochemical dissection of AP-1 recruitment onto Golgi membranes. J. Cell Biol. 1993;123:561–573. doi: 10.1083/jcb.123.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L., Molloy S. S., Thomas L., Liu G., Xiang Y., Rybak S. L., Thomas G. PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- Zhang B., Koh Y. H., Beckstead R. B., Budnik V., Ganetzky B., Bellen H. J. Synaptic vesicle size and number are regulated by a clathrin adaptor protein required for endocytosis. Neuron. 1998;21:1465–1475. doi: 10.1016/s0896-6273(00)80664-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.