Summary
The sperm protein fertilinβ, a member of the ADAM family of proteins is implicated in sperm-egg binding in all mammals studied to date. Multivalent inhibitors containing the three-amino acid binding sequence of fertilinβ, ECD, have been shown previously to be more effective inhibitors of fertilization than their monovalent counterparts. In this work, we probed sperm-egg interactions by examining the potency of fertilization inhibition by polymers that contained from 3 to 70 ECD pharmacophores in densities ranging from 10-100%. These polymers were synthesized by ruthenium-catalyzed ring opening metathesis polymerization (ROMP). Evaluation of the polymer potencies, and synthesis of a triblock copolymer from 2 building blocks, revealed that two multivalent contacts are sufficient for maximal inhibition, and that the distance between ECD pharmacophores required is 7-9 monomers spanning 4-5 nm. We conclude that inhibition requires recruitment of two receptors on the egg surface into an inhibitory complex.
Introduction
Fertilization of a mammalian oocyte by a single sperm is an extremely complex event. Many of the protein to protein and protein to carbohydrate interactions in the pathway are still unknown. The organization, localization, and binding properties of receptor-ligand complexes between sperm and egg still need to be characterized. Elucidation of the pathway and the mechanisms involved is critical for the design and synthesis of pharmaceuticals, which target population control and treat infertility. Sperm-egg recognition, binding and fusion events are dictated by a variety of multivalent receptor-ligand contacts. Using ring-opening metathesis polymerization (ROMP), we have developed synthetic ligands which mimic the multivalent display of sperm protein fertilinβ [1]. Here, we demonstrate the value of ROMP chemistry for discerning the optimal fertilinβ ligand presentation.
Fertilinβ is located in the equatorial region of the sperm head and is involved in sperm binding to the egg plasma membrane during fertilization [2-5]. Fertilinβ is a member of the ADAM (A Disintegrin and A Metalloprotease domain) family of proteins [6]. A tripeptide, glu-cys-asp (ECD) is the minimal recognition element necessary for the binding of fertilinβ to its egg receptor [7-10]. This recognition motif is taken from the sperm protein’s disintegrin loop. Peptides containing the ECD motif inhibit sperm-egg adhesion in vitro [1, 7, 8, 11-13]. For example, the IC50 for inhibition of fertilization by the ECD monomer 1 is approximately 500 μM. Peptides containing this sequence have been assayed in a diverse range of species, and the peptides inhibit fertilization in all of them [7, 8, 12, 14-16]. Thus, ECD is a small pharmacophore around which inhibitors of fertilization may be designed.
Regardless of their length or flanking sequence, all monomeric ECD peptides are poor inhibitors. Knowing that multivalent display of ligands often improves their affinity [17-20], we developed multivalent presentations of the ECD ligand [1, 13, 21]. Liposomes presenting 8 copies of the ECD ligand are 100-fold better inhibitors than the corresponding monomer [13, 21]. Polymers containing 10 copies of the ECD motif, e.g., 210, show 50 to 70-fold increased inhibition over the corresponding monomer [1]. These are the highest potencies reported to date for an ECD inhibitor. Incorporation of a fluorophore into the probes established that the target of the polyvalent ECD inhibitors is on the surface of the egg, and not on the sperm surface [21, 22]. Previously, we synthesized an 125I-labeled DECD peptide which both inhibited fertilization, and photoaffinity labeled integrin α6β1 [23]. Several other laboratories have also reported integrin α6β1 as being the receptor [12, 24, 25]. However, Myles and coworkers have established that mice with a conditional β1 integrin knockout in the egg are fertile [26] and that α6 null eggs are still fertilized by wild-type sperm [27]. Little information has been obtained about the differences of the egg surface proteome between the integrin knockout eggs and wild-type eggs because of the extraordinarily small amounts of protein material that may be obtained. Clearly chemical methods that can be used to investigate the proteome are required. In this work, we advance ring-opening metathesis polymerization (ROMP) of peptide bearing monomers to define the optimal presentation of ECD ligands for future development of proteome probes that are of high affinity, and that have useful handles for identification of cell surface receptors, and co-receptors.
We employed ruthenium-catalyzed ROMP [28] for several reasons. Using ROMP, it is possible to prepare polymers of well-defined length. Ruthenium catalysts are very stable, readily available, and functional group tolerant. In addition, ROMP allows for the multivalent presentation of one or more functional groups along a polymer backbone. Monomer building blocks are constructed from strained cyclic alkenes, in our case 5-norbornene-exo-carboxylic acid. The utility of ROMP is that one can easily manipulate the length of a polymer by varying the monomer to catalyst ratio. Moreover, the pharmacophore density of a polymer is readily adjusted by feeding 2 simple monomer building blocks, one a pharmacophore and one a spacer, to the catalyst in varying ratios. The pharmacophore and spacer may be mixed in a random fashion [29-31]. In this work, we take advantage of the living nature of the polymerization catalyst to prepare block copolymer. Thus, using 1 or 2 simple monomer building blocks and ROMP chemistry, a family of polymers may be generated to determine the length and spacing requirements for maximal polymer affinity or avidity.
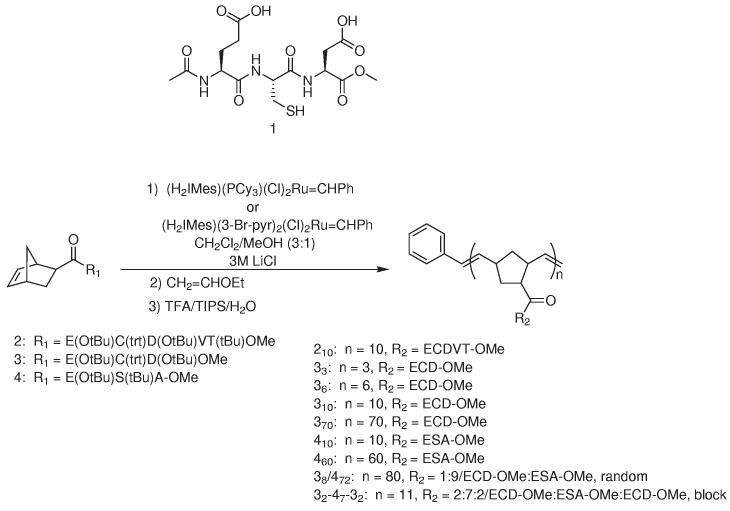
We synthesized a family of structurally diverse fertilinβ peptide polymers derived from 210 using a combination of solution phase peptide synthesis and ROMP to define the best inhibitor structure (Figure 1). Polymers 310, 370, 410, 470, 38/472, 33, 36, and 32-47-32 were tested as inhibitors in a mouse in vitro fertilization assay. These polymers were chosen to present a diversity of size and densities to the egg. With their assay, we explored the role of inhibitor length, and pharmacophore spacing. The chemical approach developed here is general and will be of use for exploring receptor-ligand interactions in many different biological systems.
Figure 1:
Polymers synthesized by ROMP and tested as fertilization inhibitors. The compound number refers to the structure of the ligand. The subscript to the compound number refers to the number of that specific monomer in the polymer. Refer to Figure 2 for a cartoon representation of the polymer structures.
Results
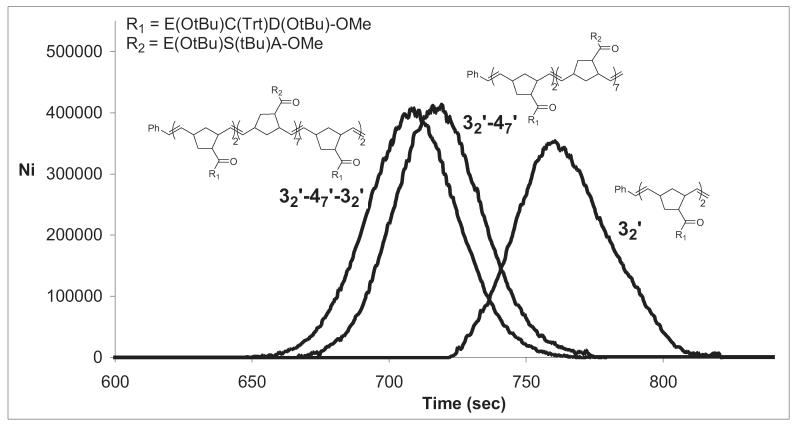
Synthesis of Polymers. The peptide monomers 3 and 4 were synthesized using standard solution phase procedures with t-butyl/trityl based side-chain protection and Fmoc/Cbz α-amino protection. Solution phase methods were used in order to obtain large quantities of pure peptide for ROMP. Polymers were prepared by ROMP using fully protected peptides and (H2IMes)(PCy3) Cl2Ru=CHPh or (H2IMes)(3-BrPyr)2Cl2Ru=CHPh as the catalyst in dichloromethane/methanol (3/1) with 3M LiCl to solubilize the polymer as previously described (Figure 1) [32]. Polymers were deprotected with a trifluoroacetic acid/water/triisopropylsilane cocktail and precipitated with diethyl ether. They were reduced with tris(2-carboxyethyl)phosphine, precipitated with dilute acid, and resuspended in aqueous ammonium hydroxide at pH 7 before use in assays. Isolated yields were 63-95%. Integration of NMR spectra confirmed that the number of monomers incorporated into each polymer was the same as the initial monomer:catalyst ratio. Gel permeation chromatography (GPC) was used to analyze and monitor the molecular weight distribution of the polymer block intermediates in the synthesis of 32-47-32. The synthetic precursors 32’and 32’-47’, and product 32’-47’-32’ were analyzed with side-chain protection intact (Figure 2). For each reaction, the disappearance of monomer was monitored by TLC before addition of the subsequent monomer. Then, a portion of the polymerization reaction was quenched with ethylvinyl ether after each monomer addition for analytical purposes. The three protected polymers were analyzed by 1H NMR spectroscopy and their monomer compositions corresponded to the monomers fed to the polymerization reaction. GPC of 32’, 32’-47’, and 32’-47’-32’ confirmed that none of the precursor block remained after addition of the subsequent monomer and that the number-average molecular weights shifted as expected (Figure 2). The final triblock copolymer 32’-47’-32’ had a narrow polydispersity index of 1.21 consistent with a controlled, living polymerization.
Figure 2:
Comparison of GPC eluted peaks of 32’, 32’-47’, and 32’-47’-32’ using a Phenogel column (5μ, 300 × 4.6 mm, linear mixed bed, 100-106 MW range) and eluting with 0.3 mL/min 10% MeOH in CH2Cl2. Narrowly dispersed polystyrene standards were used to calibrate number-averaged molecular weight. Polymer 32’ eluted at an apparent Mn = 3,372 (PDI: 1.18; expected MW: 3,974); polymer 32’-47’ eluted at apparent Mn = 10,840 (PDI: 1.18; expected MW: 12,780) and the triblock copolymer 32’-47’-32’ has an apparent Mn = 15,013 (PDI: 1.21; expected MW: 18,198).
Assay of Polymers. Polymers were assayed as inhibitors in a mouse in vitro fertilization assay. Sperm fusion was used as an endpoint to measure inhibition of sperm binding. Two measures of fertilization were calculated: fertilization rate (FR) and fertilization index (FI) [7]. The FR is the total number of eggs fertilized, divided by the total number of eggs. The FI is the total number of sperm fused divided by the total number of eggs. Inhibitor concentrations were varied over at least a 400-fold range to determine their IC50’s by both FR and FI. Concentrations were considered both in peptide ligand and in polymer (Table 1). For example, a 10 μM (in polymer) solution of 310 contains 100 μM ECD ligand. Peptide ligand concentrations are useful for comparing the inhibition efficiencies of polymers containing different numbers of ligands.
Table 1:
Polymer IC50’s for inhibition of in vitro fertilization.a
| Polymer | IC50 (μM) in peptide by F.I. | IC50 (μM) in peptide by F.R. | IC50 (μM) in polymer by F.I | IC50 (μM) in polymer by FR |
|---|---|---|---|---|
| 1 (monomer) | >500b | >500b | n.a.c | n.a. |
| 210 | 5.1 ± 1.4 | 5.8 ± 0.3 | 0.51 ± 0.14 | 0.58 ± 0.03 |
| 310 | 3.4 ± 0.3 | 3.2 ± 0.2 | 0.34 ± 0.03 | 0.32 ± 0.02 |
| 370 | 68 ± 11.1 | 99 ± 31 | 0.97 ± 0.16 | 1.4 ± 0.44 |
| 38/472 | 23 ± 5 | 80 ± 24 | 2.9 ± 0.7 | 10 ± 4 |
| 410 | n.i.d | n.i. | n.i. | n.i. |
| 470 | n.i.d | n.i. | n.i. | n.i. |
| 33 | ~500e | ~500e | ~167 | ~167 |
| 36 | ~500e | ~500e | ~83 | ~83 |
| 32-47-32 | 1.1 ± 0.3 | 5.5 ± 2.3 | 0.28 ± 0.08 | 1.4 ± 0.6 |
F.I. (fertilization index) is the average number of fused sperm per egg. The average FI of the control was 1.93. F.R. (fertilization rate) is the percent of eggs fertilized. The average FR of the control was 85.7 %. 200-300 eggs were assayed for each polymer in 8-10 independent experiments. IC50’s were calculated by a 3 parameter fit (GRAFIT software) using the equation: percent fertilization = (100-b) / {1 + ([I]/IC50)}s. Where b is the remaining percent fertilization after saturation with inhibitor, and s is the slope of the fit. Errors are reported as s.e.m.
At 500 μM 1, 29% (FI) and 32% (FR) was observed.
n.a.: not applicable.
n.i.: no inhibition. Negative control polymers 410 and 470 did not inhibit fertilization at 500 μM (peptide).
At 500 μM 33 (peptide), the percent inhibition observed was 66 ± 9 (FI) and 58 ± 2 (FR).
At 500 μM 36 (peptide), the percent inhibition observed was 57 ± 5 (FI) and 52 ± 4 (FR).
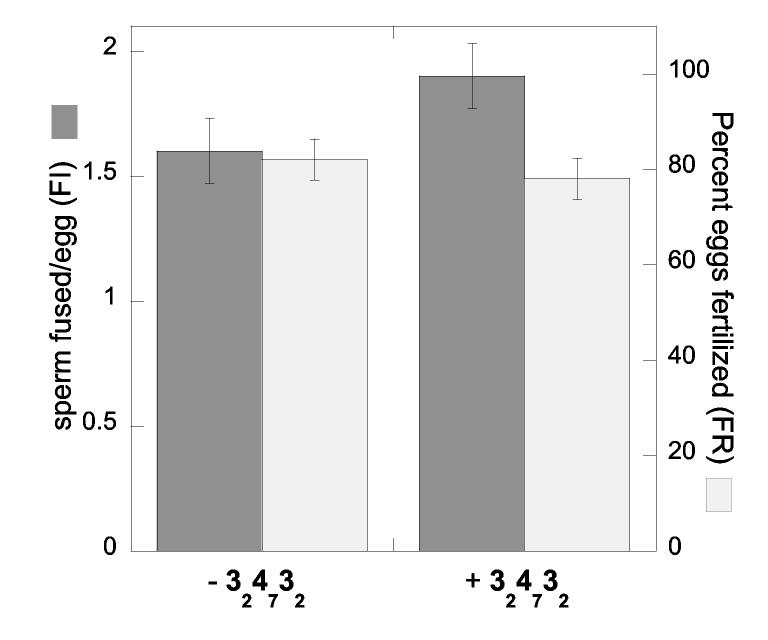
Sperm Susceptibility to Polymers. Sperm were allowed to capacitate and acrosome react then treated with polymer and observed under a light microscope. The percentage of live, motile sperm remaining after 45 min was the same in control incubations, as in 500 μM polymer incubations (70 % ± 7 for 310, 76 % ± 11 for 370, 74 % ± 11 for 32-47-32, and 74 % ± 8 for the control). Sperm were also incubated with 125 μM 32-47-32 for 45 min prior to insemination of eggs. In these experiments the final concentration of 32-47-32 in the sperm-egg incubation buffer was below the IC50. There was no difference in FR or FI with pre-treated sperm compared to fertilization by control sperm that were incubated in buffer alone (Figure 3). Lastly, no inhibition of inhibition was observed in the presence of 500 μM negative control polymers 410 or 470 (Table 1).
Figure 3:
Assay of sperm susceptibility to polymers. Sperm were incubated with 32-47-32 (125 μM in peptide) or M16 buffer only for 45 minutes prior to egg insemination (final 32-47-32 concentration was approximately 2 μM in peptide). The 32-47-32 pretreated sperm and control sperm fertilized eggs equally efficiently as measured by both fertilization rate (FR) and fertilization index (FI) demonstrating that the polymer does not have an adverse affect on the sperm. A total of 60 eggs were used in three independent experiments. Errors are reported as standard error of measurement.
Discussion
Understanding protein-protein interactions in fertilization is of fundamental importance for elucidating the molecular display required for biological function. However, this biological system is not amenable to many methods typically used to probe protein interactions, for example phage display or mutagenesis of intact egg proteins, because of the limited quantities of material available to study gametes in mammalian systems and the lack of cell culture models. We have developed ROMP chemistry in order to advance our understanding of fertilization using polymer probes.
Previously in our laboratory, we demonstrated that a norbornyl polymer displaying the peptide sequence ECDVT, 210, is a significantly more potent inhibitor of fertilization than a simple monomeric peptide [1]. The first step to optimize polymer structure was to simplify the ligand appended to the polymer backbone. We prepared and tested 310 and compared its potency to 210. Inhibition by 310 was equipotent to that of 210 (Table 1). These results demonstrate that, as expected, removal of valine and threonine to shorten the ligand sequence to ECD did not impair inhibition. The polymer behaved similarly to 210. The shorter ligand improved solubility of the ROMP monomer precursors, as well as the polymer, and reduced the number of synthetic steps required.
In order to be certain that the inhibition observed was caused by the ECD binding motif, we prepared control polymers 410 and 460 with the mutant sequence ESA. We chose to mutate ECD rather than scramble it because two of the amino acids contain carboxylates. We were concerned that scrambling the peptide might not completely eliminate binding, and multivalent presentation would restore significant inhibition. We chose to mutate cysteine and aspartate because mutagenesis work by White and coworkers [33] and Evans and coworkers [9] showed that these residues were critical for binding. We retained the glutamate residue for solubility reasons. No inhibition of fertilization by polymers 410 and 470 was observed at polymer concentrations of 50 μM and 8 μM, respectively. Thus, the inhibition by 310 observed is due to a specific interaction of the ECD peptide with a cell surface molecule.
Previous work had demonstrated that fluorescently labeled polymer 210 binds to the surface of the egg, and not the sperm surface [22]. We tested whether the inhibition observed could be due to deleterious effects of polymer upon sperm. Polymers that only contained ESA peptide substituants: e.g. 410 and 470 showed no inhibition of fertilization. Incubation of sperm with ECD containing polymers did not affect their motility or viability. Sperm pre-treated with polymer 32-47-32 before insemination in M-16 buffer were able to fertilize eggs normally (Figure 3). Therefore, treatment of sperm with our polymers does not affect sperm viability, motility, or penetrability. Targeting of the ECD polymers for the egg cell surface is responsible for inhibition.
A longer polymer should cluster many more receptors on the oocyte and have a statistical advantage that favors rebinding of ligand over dissociation, and therefore be a more potent sperm inhibitor. For example in studying glycopolymer inhibition of hemagglutination, Kanai and coworkers have shown that inhibition potency (as measured on a per residue basis) saturates at a length of about 50 residues [18]. Inhibition potency does not decrease as the polymer length increases beyond the length required to span binding sites as would be expected if only chelation were important [18]. Hence, we synthesized polymer 370 which contained approximately 70 ECD ligands, and polymer 38/472. Polymer 38/472 was approximately the same length polymer as 370, but only 10% of the peptides presented (randomly) were ECD ligand, the remainder were the ESA mutant sequence. To our surprise, we observed that the longer polymers showed decreased inhibition potency compared to 310 when compared on the basis of peptide concentration (Table 1). When compared on the basis of polymer concentration 310, and 370, are essentially equipotent and 38/472 is less effective.
Previous investigations had demonstrated that if the cysteine residue of the ECD sequence is protected as a disulfide, no inhibition is observed [8, 10]. We investigated the rate of thiol oxidation in the polymers under the conditions of the in vitro fertilization assay. If significant oxidation of the polymer thiols were to occur faster in the longer polymers, their potency would be reduced compared to the shorter polymers. We measured reduced thiol concentration as a function of time using 5,5′-dithiobis(2-nitrobenzoic acid) [34] in M16 buffer at 37 °C. For all of the polymers, less than 20% of the thiols were oxidized over the 45 minute incubation period of the assay, and the oxidation rate of 310 did not differ significantly from the oxidation rates of 370 or 38/472.
We concluded that increasing the number of ligands presented and/or increasing the distance between ligands did not improve inhibitor potency. Our data imply that chelation of the receptor is more important than statistical factors favoring ligand rebinding. A polymer length of 10 represented the maximum length required for efficient inhibition. When modeled in the most extended backbone configuration (trans-syndiotactic), the distance spanned by 310 is at most 5.3 nm. We next sought to determine the minimum length of polymer required for inhibition.
We synthesized fully substituted ECD ligand 3-mers and 6-mers. Polymers 33 and 36 were both poor inhibitors, and showed no more potency than the ECD monomer 1. These results indicated that multivalency was required for effective binding and inhibition, and that the 3-mer and 6-mer did not have sufficient length to span the distance between binding sites of two adjacent receptors. In the most extended configuration, 36 can at most span 3 nm.
The poor inhibition of fertilization by polymer 36 implied that the potency of 310 was due to the ligation of the terminal ECD moieties and that the internal ECD peptides did not engage receptor-binding sites. The living nature of the ROMP polymerization allows the sequential addition of monomer building blocks to the growing polymer chain. We took advantage of this polymer property to construct 32-47-32 which contained on average two ECD ligands at either terminus separated by a 7-mer spacer comprised of biologically inactive ESA peptides. The block nature of the polymer was verified by gel permeation chromatography (GPC) analysis of 32’-47’-32’ as well as the monoblock, and diblock precursors (Figure 2). There were no low molecular weight shoulders on the 32’-47’-32’ peak, confirming that the polymerization was living and that the intact triblock polymer had been prepared.
The inhibition potency of polymer 32-47-32 was equal to polymer 310 in polymer concentration, and slightly better when compared on the basis of peptide concentration. We conclude that the internal ECD ligands in polymer 310 are not required for binding or inhibition (Figure 3). The distance between ECD ligands on an extended 9-mer is approximately 4.7 nm, and on an extended 10-mer is approximately 5.3 nm, indicating that the distance between receptor binding sites can be no greater than 5 nm (Figure 4). Typically membrane protein receptors are 2-3 nm in diameter. Based on the ineffective inhibition observed with polymer 36, fertilinβ receptor binding sites are more than 3 nm apart. Taken together, our polymers appear to engage the egg surface receptor in a bivalent fashion.
Figure 4:
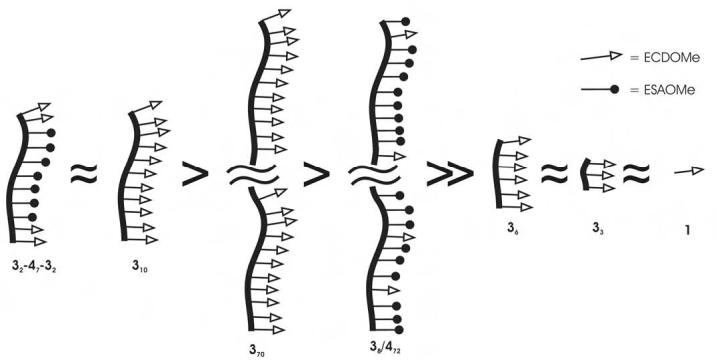
Relative potencies of fertilinβ polymers when compared on the basis of peptide ligand concentration.
Our results are analogous to other systems in which bivalent inhibitors show high selectivity and affinity. For example, Portoghese and coworkers designed bivalent ligands for the opioid receptors [35]. Their results are consistent with a model in which two receptors are recruited into van der Waals contact, approximately 3 nm apart. Potent proteasome inhibitors have been designed using a similar strategy. Moroder and coworkers [36, 37] synthesized inhibitors containing two arg-leu-arg peptides spaced by a 6.5 nm polyethylene glycol linker. The bivalent inhibitor was 124-fold more potent than the monomeric peptide. Potent inhibition required a linker that was flexible and long enough to span the distance between the active β-subunits of the proteosome.
Our polymer assay results are consistent with a bivalent inhibition model like that of the opiod receptor or the proteosome. Using block synthesis of copolymers, we conclude that inhibition of fertilization by fertilinβ mimics requires recruitment of two receptors on the egg surface into an inhibitory complex. The recruitment of additional receptors with longer polymers does not enhance inhibition potency. These experiments establish the parameters for future polymer design and testing. The structural requirements defined here will be incorporated into future polymers that include tagging elements for probing the identity of egg surface fertilinβ receptors.
Significance
The mechanisms by which spermatozoa bind and fuse to the oocyte plasma membrane during mammalian fertilization are still unknown. In this work, we utilized the fertilinβ binding site peptide ECD to define the molecular structure required for optimal inhibition of in vitro fertilization, a measure of sperm-egg binding. Using two simple monomeric building blocks, a diverse series of polymers was synthesized by ring-opening polymerization (ROMP) of norbornene. We introduce block copolymer synthesis as a method to optimize polymer potency and to rapidly identify required spacer lengths. In our polymer series, length of spacer between ECD pharmacophores, and numbers of ECD pharmacophores presented to the egg were varied. Polymers tested contained as few as 3 ECD ligands and as many as 70 ECD ligands. The most potent fertilinβ-derived inhibitor of fertilization to date is an 11-mer polymer that contains two ECD ligands at either terminus separated by seven spacer monomers. These results indicate that two ECD ligands bound to the egg surface separated by 4-5 nm are sufficient for maximal inhibition. Definition of the optimal polymer structure for inhibition now allows the design of molecular probes to identify the egg cell surface members of the inhibitory complex.
Experimental Procedures
Materials. Amino acids and coupling agents used were purchased from Advanced Chem Tech. (Louisville, KY) or PerSeptive Biosystems (Framingham, MA). Solvents and chemical reagents were obtained from Fisher Scientific Inc (Springfield, NJ), or Sigma-Aldrich (Milwaukee, WI). Pregnant Mare’s Serum Gonadotropin (PMSG, #367222), hyaluronidase (#H3506), and Hoechst-33342, were purchased from Sigma-Aldrich and human Chorionic Gonadotropin (hCG, #230734) was obtained through NHPP, NIDDK and Dr. A. F. Parlow. (H2IMes)(PCy3)Cl2Ru=CHPh was purchased from Sigma-Aldrich (Milwaukee, WI). (H2IMes)(BrPyr)2Cl2Ru=CHPh was prepared according to the literature [38]. CH2Cl2 was freshly distilled from CaH; CH3OH, and Et2O, were used without further purification. LiCl was oven-dried and stored over P2O5 before use. All reactions were carried out under an Ar atmosphere in oven-dried glassware unless otherwise specified. Moisture and oxygen-sensitive reagents were handled in an N2-filled drybox. 5-Norbornene-exo-carboxylic acid was synthesized according to the literature [19].
General Methods. Analytical thin layer chromatography (TLC) was performed on pre-coated silica gel plates (60F254), and flash chromatography on silica gel-60 (230-400 mesh). TLC spots were detected by UV light and by staining with phosphomolybdic acid (PMA). Peptides were purified by flash column chromatography on silica gel-60. The purities of all peptide monomers were assessed by RPHPLC using a Vydac C18 column. Gradient elution was performed at 1 mL/min with acetonitrile and water (both containing TFA, 0.1%). The purity of the polymers was assessed by aqueous phase gel permeation chromatography (BioSep-SEC-S2000) using 50 mM potassium phosphate, pH 7. Inova400, Inova500, and Inova600 MHz NMR spectrometers were used to perform NMR analysis, and spectra were recorded in CDCl3 unless otherwise noted. 1H NMR spectra are reported as chemical shift in parts per million (multiplicity, coupling constant in Hz, integration). 1H NMR data are assumed to be first order. The usual workup for peptide coupling reactions was three washes of the CH2Cl2 solution with 5% NaHCO3 followed by three washes with 1N HCl and drying of the CH2Cl2 over Na2SO4. After evaporation of solvent, the peptide product was purified by flash silica chromatography.
PMSG and hCG were resuspended in sterile PBS to 10 IU/100 μL. These solutions were stored at -20 °C. Hyaluronidase was re-suspended in sterile water to a final concentration of 30 mg/mL and stored at -20 °C. Hoechst-33342 was stored in the dark at 4 °C. All the steps involving the use of Hoechst dye were performed with minimum exposure to light.
Fmoc-Cys(trt)Asp(OtBu)-OMe. H-Asp(OtBu)-OMe·HCl (16.69 mmol, 4.00 g), Fmoc-Cys(trt)-OMe (18.36 mmol, 10.75 g), EDC·HCl (20.03 mmol, 3.83 g), HOBt·hydrate (20.03 mmol, 3.07 g) were dissolved in 40 mL of dry CH2Cl2 and cooled to 0 °C. DIEA (18.36 mmol, 3.25 mL) was added, and the reaction was stirred for 4 h at rt. The usual workup and chromatography (acetone:CH2Cl2/1:20) yielded Fmoc-Cys(trt)Asp(OtBu)-OMe (12.7 g, 98%) as a white powder. 1H-NMR (400 MHz) δ 7.74 (t, J = 7.6 Hz, 2), 7.57(d, J = 4.8 Hz, 2H), 7.31 (m, 19H), 6.78(d, J = 7.6 Hz, 1H), 5.01(d, J = 6.8 Hz, 1H), 4.71 (m, 1H), 4.34(m, 2H), 4.19 (t, J = 7.0 Hz, 1H), 3.76 (dd, J = 6.8, 6.8 Hz, 1H), 3.66 (s, 3H), 2.89 (dd, J = 17.0, 4.6 Hz, 1H), 2.73 (m, 1H), 2.65 (m, 2H), 1.38(s, 9H). Fmoc-Glu(OtBu)Cys(trt)Asp(OtBu)-OMe. Fmoc-Cys(trt)Asp(OtBu)-OMe, (5.19 mmol, 4.00 g) was dissolved in 10 mL of dry CH2Cl2. 1-Octanethiol (51.9 mmol, 9.04 mL), and DBU (0.52 mmol, 78 μL) were added, and stirred for 15 h at rt under Ar. After evaporation of the solvent, the resulting product was purified by flash chromatography eluting with a step gradient ranging from 2% to 50% EtOAc/CH2Cl2. H-Cys(trt)Asp(OtBu)-OMe (2.09 g, 73%) was obtained as a fine white powder. H-Cys(trt)Asp(OtBu)-OMe, (3.65 mmol, 2.00 g), Fmoc-Glu(OtBu)-OH (4.02 mmol, 1.71 g), EDC·HCl (4.38 mmol, 0.84 g), HOBt·hydrate (4.38 mmol, 0.67 g) were dissolved in 10 mL of dry CH2Cl2 and cooled to 0 °C. DIEA (4.02 mmol, 0.71 mL) was added, and the reaction was stirred for 5 h at rt. The usual workup and chromatography (acetone: CH2Cl2/1:20) yielded Fmoc-Glu(OtBu)Cys(trt)Asp(OtBu)-OMe (3.04 g, 87%) as a white powder. 1H-NMR (400 MHz) δ 7.76 (d, J = 7.6 Hz, 2H) 7.56 (t, J = 7.2 Hz, 2H), 7.40 (m, 8H), 7.28 (m, 8H), 7.18 (m, 3H), 6.90 (d, J = 8.4 Hz, 1H), 6.61(d, J = 7.6 Hz, 1H), 5.84 (d, J = 6.4 Hz, 1H), 4.72 (m, 2H), 4.35 (d, J = 7.2 Hz, 2H), 4.15 (m, 2H), 4.04 (m, 1H), 3.67 (s, 3H), 2.82 (m, 2H), 2.67 (dd, J = 17.0 and 4.8 Hz, 1H), 2.57 (dd, J = 13.2 and 5.2 Hz, 1H), 2.40 (m, 1H), 2.30 (m, 1H), 2.03 (m, 1H), 1.88 (m, 1H), 1.44 (s, 9H), 1.40 (s, 9H).
Ac-Glu(OtBu)Cys(trt)Asp(OtBu)-OMe. Fmoc-Glu(OtBu)Cys(trt)Asp-(OtBu)-OMe, (0.31 mmol, 300 mg) was dissolved in 1 mL of dry CH2Cl2. 1-Octanethiol (3.14 mmol, 546 μL), and DBU (0.031 mmol, 30 μL) were added, and stirred for 16 h at rt. The resulting product was purified by flash chromatography eluting with a step gradient ranging from 2% to 50% EtOAc/CH2Cl2. H-Glu(OtBu)Cys(trt)Asp-(OtBu)-OMe (163 mg, 72%) was obtained as a fine white powder. H-Glu(OtBu)Cys(trt)Asp-(OtBu)-OMe (0.27 mmol, 198 mg) was dissolved in 3 mL of dry CH2Cl2. Acetic anhydride (2.70 mol, 257 μL) was added to the reaction, and followed by DIEA (0.82 mmol, 114 μL) and the reaction stirred for 1 h at rt under Ar. The usual workup and recrystallization of the crude product from CH2Cl2 yielded Ac-Glu(OtBu)Cys(trt)Asp(OtBu)-OMe (163 mg, 78 %) as a white powder. 1H-NMR (400 MHz) δ 7.42 (m, 6H) 7.30 (m, 6H), 7.21 (m, 3H), 6.86 (d, J = 8.4 Hz, 1H), 6.70 (d, J = 7.6 Hz, 1H), 5.53 (d, J = 6.8 Hz, 1H), 4.70 (m, 1H), 4.33 (m, 1H), 4.01 (m, 1H), 3.67 (s, 3H), 2.84 (dd, J = 16.8 and 5.2 Hz, 1H), 2.77 (dd, J = 13.2 and 8.0 Hz, 1H), 2.67 (dd, J = 16.8 and 4.8 Hz, 1H), 2.58 (dd, J = 12.8 and 5.2 Hz, 1H), 2.40 (m, 1H), 2.28 (m, 1H), 2.03 (m, 1H), 1.97 (s, 3H), 1.88 (m, 1H) 1.43 (s, 9H), 1.40 (s, 9H). ESI mass spectrum: Calcd (MNa+) 798.35; Found 798.52.
Ac-GluCysAsp-OMe, 1. Ac-Glu(OtBu)Cys(trt)Asp(OtBu)-OMe, was deprotected in a cocktail of H2O, TIPS, and TFA (2.5:2.5:95) for 5 h. The reaction mixture was concentrated with N2 and precipitated with cold Et2O. The precipitate was dissolved in 10% Acetonitrile in H2O, and reduced with DTT (10 mM) for 1 h with stirring at 37 °C. Pure deprotected product was isolated as a white solid by reversed-phase C18 HPLC (1 cm × 30 cm, 5 μ) using 0.1% TFA/H2O and a linear gradient of CH3CN. ESI mass spectrum: Calcd (MH+) 420.12, Found 420.16.
NB-Glu(OtBu)Cys(trt)Asp(OtBu)-OMe, 3. H-Glu(OtBu)Cys(trt)Asp-(OtBu)-OMe (0.22 mmol, 163 mg), 5-norbornene-exo-carboxylic acid (0.24 mmol, 34 mg), TBTU (0.24 mmol, 78 mg), HOBt·hydrate (0.08 mmol, 13 mg) were dissolved in 2 mL of dry CH2Cl2. DIEA (0.24 mmol, 43 μL) was added, and the reaction was stirred for 1 h at rt. The usual workup and chromatography (acetone:CH2Cl2/1:20) yielded 3 (144 mg, 76%) as a white powder. 1H-NMR (400 MHz) δ 7.41 (m, 6H) 7.29 (m, 6H), 7.21 (m, 3H), 6.94 (d, J = 8.4 Hz, 1H), 6.73 (m, 1H), 6.67 (m, 1H), 6.11 (m, 1H), 6.02 (m, 1H), 4.72 (m, 1H), 4.29 (m, 1H), 4.04 (m, 1H), 3.67 (s, 3H), 2.92 (m, 1H), 2.83 (m, 3H), 2.70 (m, 1H), 2. 48 (m, 2H), 2.29 (m, 1H), 2.02 (m, 2H), 1.88 (m, 4H), 1.64 (d, J = 7.2 Hz, 1H), 1. 44 (d, J = 5.6 Hz, 9H), 1.41 (s, 9H), 1.27 (m, 2H). 13C-NMR (400 MHz) δ 27.15, 27.21, 28.23, 28.28, 30.72, 30.77, 32.19, 32.24, 33.70, 33.77, 37.48, 41.78, 41.81, 44.77, 46.57, 46.63, 47.24, 47.39, 49.10, 52.39, 52.44, 52.69, 53.54, 53.64, 67.40, 81.43, 81.76, 127.11, 128.28, 128.29, 129.79, 136.08, 136.24, 138.34, 138.47, 144.58, 169.67, 169.94, 171.05, 171.36, 171.37, 173.68, 173.69, 176.58.
Cbz-Ser(tBu)Ala-OMe. H-Ala-OMe·HCl (27.20 mmol, 3.80 g), Cbz-Ser(tBu)-OMe (29.91 mmol, 8.84 g), EDC·HCl (32.64 mmol, 6.26 g), HOBt·hydrate (32.64 mmol, 5.00 g) were dissolved in 25 mL of dry CH2Cl2 and cooled to 0 °C. DIEA (29.91 mmol, 5.30 mL) was added, and the reaction was stirred for 12 h at rt. The usual workup and chromatography (acetone:CH2Cl2/1:20) yielded Cbz-Ser(tBu)Ala-OMe (7.81 g, 76%) as a white powder. 1H-NMR (400 MHz) δ 7.34 (m, 5H), 5.73 (m, 1H), 5.12 (m, 2H), 4.57 (m, 1H), 4.23 (m, 1H), 3.81 (m, 1H), 3.74 (s, 3H), 3.38 (t, J = 8.4 Hz, 1H), 1.40 (d, J = 7.2 Hz, 3H), 1.21 (s, 9H).
Cbz-Glu(OtBu)Ser(tBu)Ala-OMe. Cbz-Ser(tBu)Ala-OMe (1.62 mmol, 616 mg) was dissolved in 3 mL of MeOH. 10% Pd/C (0.16 mmol, 17 mg) was added, and stirred for 3 h at rt under H2. After filtration of the catalyst with celite, the resulting amine product was used without further purification. H-Ser(tBu)Ala-OMe (1.62 mmol, 400 mg), Cbz-Glu(OtBu)-OH (1.79 mmol, 604 mg), TBTU (1.34 mmol, 430 mg), HOBt·hydrate (0.45 mmol, 69 mg) were dissolved in 3 mL of dry CH2Cl2. DIEA (2.60 mmol, 460 μL) was added, and the reaction was stirred for 12 h at rt. The usual workup and chromatography (acetone:CH2Cl2/1:20) yielded 8 (627 mg, 68%) as a white powder. 1H-NMR (400 MHz) δ 7.33 (m, 6H) 7.01 (d, J = 6.8, 1H), 5.72 (d, J = 6.4 Hz, 1H), 5.10 (m, 2H), 4.56 (q, 1H), 4.43 (m, 1H), 4.23 (m, 1H), 3.81 (m, 1H), 3.73 (s, 3H), 3.36 (t, J = 7.6 Hz, 1H), 2.38 (m, 2H), 2.12 (m, 1H), 1.95 (m, 1H), 1.43 (s, 9H), 1.39 (d, J = 7.2 Hz, 3H), 1.21 (s, 9H).
NB-Glu(OtBu)Ser(tBu)Ala-OMe, 4. Cbz-Glu(OtBu)Ser(tBu)Ala-OMe (0.53 mmol, 300 mg) was dissolved in 3 mL of MeOH. 10% Pd/C (0.053 mmol, 6 mg) was added, and stirred for 2 h at rt under H2. After filtration of the catalyst with celite, the resulting amine product was used without further purification. H-Glu(OtBu)Ser(tBu)Ala-OMe (0.51 mmol, 220.0 g), 5-norbornene-exo-carboxylic acid (0.56 mmol, 78 mg), TBTU (0.56 mmol, 180 mg), HOBt·hydrate (0.19 mmol, 29 mg) were dissolved in 3 mL of dry CH2Cl2. DIEA (0.61 mmol, 108 μL) was added, and the reaction was stirred for 3 h at rt. The usual workup and chromatography (acetone:CH2Cl2/2:10) yielded 4 (226 mg, 74%) as a white powder. 1H-NMR (400 MHz) δ 7.38 (t, J = 8.0 Hz, 1H) 7.02 (t, J = 7.4 Hz, 1H), 6.82 (d, J = 5.6 Hz, 1H), 6.12 (m, 2H), 4.56 (q, 1H), 4.44 (m, 1H), 4.38 (m, 1H), 3.86 (m, 1H), 3.73 (s, 3H), 3.37 (m, 1H), 2.96 (m, 1H), 2.91 (m, 1H), 2.47 (m, 1H), 2.37 (m, 1H), 2. 13 (m, 1H), 2.07 (m, 1H), 2.00 (m, 1H), 1.90 (m, 1H), 1.65 (m, 1H), 1.45 (d, J = 4.0 Hz, 9H), 1. 41 (dd, J = 7.2 and 0.8 Hz, 3H), 1.35 (m, 2H), 1.19(d, J = 4.8 Hz, 9H) 13C-NMR (400 MHz) δ 18.30, 27.47, 27.52, 28.23, 30.62, 30.79, 32.25, 41.74, 41.76, 44.62, 44.67, 46.51, 46.57, 47.22, 47.37, 48.38, 48.41, 52.45, 52.47, 53.24, 53.87, 53.89, 61.48, 74.13, 74.18, 81.32, 81.33, 136.12, 138.40, 169.86, 169.88, 171.35, 171.38, 173.07, 173.62, 176.54, 176.55.
ROMP. The procedure detailed below for polymer 310 is representative of the procedure followed for synthesis of all of the homopolymers and the random copolymer. Scale, yield, and spectra are presented for each of the individual polymers. Number of ligands (n) is based on the monomer:catalyst ratio used in the synthesis and confirmed by integration in the 1H-NMR spectra.
310. NB-Glu(OtBu)Cys(trt)Asp(OtBu)-OMe 3 (82 μmol, 70 mg) was dissolved in 100 μL of CH2Cl2/MeOH (3/1). To the reaction was added oven-dried LiCl (1.2 mmol, 51 mg) and (H2IMes)(PCy3)Cl2Ru=CHPh (8 μmol, 7 mg) dissolved in 50 μL of CH2Cl2/MeOH (3/1) and an additional 250 μL of CH2Cl2/MeOH (3/1). The reaction was stirred for 3 h. Ethyl vinyl ether (1 mL) was added to quench the reaction and the mixture stirred for an additional 40 min. After removing the solvent, the residue was dissolved in CH2Cl2. The solution was washed three times with H2O, dried with Na2SO4, concentrated by rotary evaporation, and precipitated with cold Et2O. Product was isolated by centrifugation and dried. Crude protected polymer was deprotected with TFA/TIPS/H2O (95/2.5/2.5) for 5 h. The reaction mixture was concentrated with N2 and precipitated with cold Et2O. The precipitate was collected by centrifugation. Polymer was dissolved in H2O (1 mL) at pH 6 and reduced with 10-20 mM tris(2-carboxyethyl)phosphine (TCEP) for 2 h at 37 °C. Reduced polymer was isolated by precipitation with 1N HCl (200 μL). Residual TCEP was removed by repeated washing of the precipitate with H2O (3 × 1 mL). 310, a yellowish white solid, was collected (41 mg, 90%), dried, and stored at -20 °C. 1H-NMR (400 MHz, D2O) δ 7.24 (m) 5.34 (m), 4.65-4.05 (with max at 4.61, 4.42, 4.23), 3.61 (br s), 2.90-2.29 (with max at 2.80, 2.58, 2.45, 2.25), 2.22-1.45 (with max at 2.142, 1.86, 1.78), 1.18 (br s).
33. NB-Glu(OtBu)Cys(trt)Asp(OtBu)-OMe 3 (82 μmol, 70 mg) and (H2IMes)(BrPyr)2Cl2Ru=CHPh (27 μmol, 24 mg) in a total volume of 400 μL yielded 33 as a brownish white solid (30 mg, 82%). 1H-NMR (500 MHz, D2O) δ 7.26 (m) 5.35 (m), 4.65-4.01 (with max at 4.55, 4.26, 4.04), 3.24 (br s), 2.90-2.29 (with max at 2.81, 2.51), 2.22-1.45 (with max at 2.12, 1.93, 1.78), 1.19 (m).
36. NB-Glu(OtBu)Cys(trt)Asp(OtBu)-OMe 3 (82 μmol, 70 mg) and (H2IMes)(BrPyr)2Cl2Ru=CHPh (14 μmol, 12 mg) in a total volume of 400 μL yielded 36 as a brownish white solid (45 mg, 95%). 1H-NMR (500 MHz, D2O) δ 7.26 (m) 5.45-5.10 (with max at 5.36, 5.29, 5.26), 4.65-4.05 (with max at 4.59, 4.45, 4.27), 3.64 (br s), 2.90-2.45 (with max at 2.83, 2.63), 2.22-1.45 (with max at 2.21, 1.94, 1.84, 1.61), 1.21 (m).
370. NB-Glu(OtBu)Cys(trt)Asp(OtBu)-OMe 3 (17 μmol, 15 mg) and (H2IMes)(PCy3)Cl2Ru=CHPh (0.17 μmol, 0.15 mg) in a total volume of 150 μL of CH2Cl2/MeOH (3/1) yielded a yellowish white solid (8 mg, 80%). 1H-NMR (400 MHz, D2O) δ 7.20 (m), 5.23 (m), 4.44-4.09 (with max at 4.39, 4.08), 3.67 (br s), 3.22 (s), 3.15-1.45 (with max at 2.95, 2.80, 2.10, 1.91, 1.81, 1.60), 1.38-1.79 (with max at 1.23, 1.15, 0.90).
410. NB-Glu(OtBu)Ser(OtBu)Ala-OMe 4 (38 μmol, 21 mg) and (H2IMes)(PCy3)Cl2Ru=CHPh (13.8 μmol, 3.3 mg) in a total volume of 200 μL of CH2Cl2/MeOH (3/1) yielded 410 as a brownish white solid (10 mg, 67%). 1H-NMR (600 MHz, D2O) 4.29 (m), 5.23 (br, s), 4.29 (m), 4.00 (m), 3.65 (br, s), 3.20 (s), 3.10-2.30 (with max at 2.80, 2.45), 2.25-1.40 (with max at 2.10, 1.90, 1.79, 1.45), 1.20 (m), 1.00 (m).
460. NB-Glu(OtBu)Ser(OtBu)Ala-OMe 4 (47 μmol, 26 mg) and (H2IMes)(PCy3)Cl2Ru=CHPh (0.47 μmol, 0.40 mg) in a total volume of 200 μL of CH2Cl2/MeOH (3/1) yielded 460 as a brownish white solid (12 mg, 63%). 1H-NMR (600 MHz, D2O) δ 7.20 (m), 5.80-5.20 (m), 3.45 (br, s), 2.2-1.4 (with max at 2.19, 1.99, 1.68), 1.38-0.80 (with max at 1.23, 1.15).
38/472. NB-Glu(OtBu)Cys(trt)Asp(OtBu)-OMe 3 (12 μmol, 10 mg), NB-Glu(OtBu)Ser(OtBu)Ala-OMe 4 (106 μmol, 58 mg) and (H2IMes)(PCy3)Cl2Ru=CHPh (1.17 μmol, 0.99 mg) in a total volume of 400 μL of CH2Cl2/MeOH (3/1) yielded 38/472 as a yellowish white solid (36 mg, 73%). 1H-NMR (600 MHz, D2O) δ 7.22 (m) 5.45-5.10 (with max at 5.34, 5.23, 5.16), 4.65 (br s), 4.39-4.05 (with max at 4.31, 4.04), 3.75 (br s), 3.63 (br s), 2.85 (m), 2.48 (m), 2.22-1.41 (with max at 2.13, 1.90, 1.81, 1.60), 1.40-1.00 (with max at 1.31, 1.23, 1.12), 0.92 (br s).
32’, 32’-47’, 32’-47’-32’, 32-47-32. In three separate flasks, A, B, and C, peptide 3 (23 μmol, 20 mg) was dissolved in 100 μL of CH2Cl2/MeOH (3/1). Then oven-dried LiCl (1.2 mmol, 51 mg) and (H2IMes)(BrPyr)2Cl2Ru=CHPh (12 μmol, 10 mg) dissolved in 50 μL of CH2Cl2/MeOH (3/1) were added to each flask. The reactions were stirred for 1.5 h. To flask A, ethyl vinyl ether (2 mL) was added and the mixture was stirred for 40 min to quench the reaction yielding 32’. To flasks B and C, peptide 4 (82 μmol, 45 mg) dissolved in 250 μL of CH2Cl2/MeOH (3/1) was added, and the reactions were stirred for 1.5 h. Ethyl vinyl ether (2 mL) was added to flask B, and the mixture was stirred for 40 min to yield 32’-47’. Then peptide 3 (23 μmol, 20 mg) dissolved in 250 μL of CH2Cl2/MeOH (3/1) was added to flask C and the reaction was stirred for 2 h. Ethyl vinyl ether (2 mL) was added to quench the reaction and the mixture was stirred for an additional 40 min to yield 32’-47’-32’. For each reaction, TLC was used to confirm complete disappearance of monomer before addition of the next monomer. After removing the solvent from each of the flasks, the residues were dissolved separately in CH2Cl2. The solutions were washed three times with H2O, dried with Na2SO4, and the products 32’, 32’-47’, and 32’-47’-32’ were precipitated with cold Et2O. The precipitates were isolated by centrifugation and dried. Each of the polymers was analyzed by gel permeation chromatography using a Phenogel column (5μ, 300 × 4.60 mm, linear mixed bed, 100-106 MW range). Elution was performed at 0.3 mL/min with 10% MeOH in CH2Cl2. Narrowly dispersed polystyrene standards from Aldrich were used as molecular weight calibrants. The number average and weighted average molecular weights were calculated from the chromatogram to determine the polydispersity index (PDI).
32-47-32. Crude protected polymer 32’-47’-32’ was deprotected with TFA/TIPS/H2O (95/2.5/2.5) for 5 h. The reaction mixture was concentrated with N2 and the polymer precipitated with cold Et2O and isolated by centrifugation. The polymer was dissolved in H2O (1 mL) at pH 6 and reduced with 10-20 mM TCEP for 2 h at 37 °C. Deprotected polymer was isolated by precipitation with 1N HCl (200 μL). Residual TCEP was removed by repeated washing with H2O (3 × 1 mL). 32-47-32, a yellowish white solid (40 mg, 88%), was collected, dried, and stored at -20 °C. 1H-NMR (500 MHz, D2O) δ 7.26 (m) 5.35 (m), 4.40-4.01 (with max at 4.35, 4.27, 4.22, 4.06, 4.04), 3.77 (br s), 3.65 (br s), 3.25 (br s), 2.90-2.29 (with max at 2.85, 2.51), 2.22-1.45 (with max at 2.16, 1.93, 1.84, 1.62), 1.34 (br s), 1.26-1.02 (with max at 1.25, 1.15), 0.94 (br s).
Assay for thiol oxidation. 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) was prepared as a 5 mM stock solution in 100 mM potassium phosphate buffer solution, pH 7.2 containing 0.1 mM EDTA and stored in the dark at 4°C. All polymers were fully reduced with TCEP, washed three times with deionized water, and dissolved in aqueous ammonium hydroxide solution pH 7.2. Aliquots of polymer (< 10 μL) were added to M16 (without BSA or phenol red) to a final volume of 100 μL and incubated at 37 °C. The final concentrations of polymer were 100, 75, and 50 μM and nine aliquots were prepared at each concentration. At five min intervals for 45 min, 100 μL of stock DTNB was added to a polymer aliquot, the solution mixed, and the absorbance at 412 nm measured. The concentration of free sulfhydryl remaining was determined using ε412 = 14,150 M-1cm-1 [34]. A standard curve for sulfhydryl concentration was prepared using cysteine under the same conditions.
Isolation of spermatozoa and oocytes for IVF assay. All experiments performed using mice were in accordance with National Institute of Health and USDA guidelines and the specific procedures performed were approved by the Stony Brook University IACUC (protocol #0616). Sperm for the in vitro adhesion and fusion assay were isolated from the cauda epididymis and vas deferens of 8 month old ICR retired male breeders ICR (Taconic, NJ). Sperm were released from dissected cauda and vas deferens into 3% BSA M16 modified Krebs-Ringer medium. Released sperm were incubated at 37 °C, 5% CO2 for 3 h in the same medium to allow them to capacitate and acrosome react. Eggs were collected from the oviducts of 8-10 week-old super-ovulated female ICR mice (Taconic, NJ). Mice were super-ovulated by injecting 5 IU of PMSG followed 48-52 hours later by an injection of 10 IU of hCG. 14-16 h after hCG injection, oviducts were removed from euthanized mice and incubated in pre-warmed M16 medium with 0.5% BSA. Cumulus-egg complexes were collected and transferred to 500 μL drops of medium containing 30 μg/mL hyaluronidase surrounded by mineral oil. After 5 min incubation at 37 °C, 5% CO2, cumulus free metaphase II eggs (eggs with one polar body) were collected, transferred first to a 80 μL drop of medium and then washed through six, 40 μL drops of medium. Eggs were recovered for 1 h before treating with acid Tyrode’s. Zona pellucida of metaphase II eggs were removed by treating eggs with 100 μL Tyrode’s acid drop for 1 min at RT. Zona free eggs were washed six times with 0.5% BSA medium and recovered for 2 h, then preloaded with HOECHST dye at 10 μg/mL for 30 min. at 37 °C, 5% CO2.
Inhibitor assay. Before use, the polymers were fully reduced with 10 mM TCEP for 1 to 2 h, precipitated with 1N HCl and washed with water, and then redissolved in water adjusted to pH 7 with NH4OH.
Zona free eggs that had been loaded with Hoechst dye were washed and placed in 100 μL drops of 3% BSA M16 medium. Polymer solution was added (no more than 5 μL of stock solution) and incubated with eggs at for 45 min prior to sperm addition. Capacitated and acrosome reacted sperm were added to eggs at a final concentration of 1-5 × 105 sperm/mL. After 45 min at 37 °C, 5% CO2, eggs were gently washed through six 40 μL drops of the M16 medium with 3% BSA. Eggs were mounted onto glass microscope slides and sperm binding and fusion were scored by epi-fluorescence microscopy and DIC microscopy (NIKON Eclipse 400, 40x, 0.75 NA objective). Fusion was scored as the fluorescent labeling of sperm nuclei with HOECHST dye present in the preloaded eggs. Two measures of fusion were used: fertilization index (FI, mean number of fused sperm per egg) and fertilization rate (FR, percentage of eggs fused with at least one sperm). IC50’s were calculated by a 3 parameter fit (GRAFIT software) by the equation:
| (1) |
Where y is the percent FR or FI, b is the remaining percent fertilization after saturation with inhibitor, and s is the slope of the fit. Errors were reported as s.e.m.
Sperm Susceptibility to Polymer Assay. Sperm in 3% BSA M16 modified Krebs-Ringer medium were allowed to capacitate and acrosome react in a 37 °C humidified incubator with 5% CO2 for 2 h 30 min, then treated with 500 μM 310, 370, or 32-47-32 for 45 min. Control sperm were incubated in the same buffer. Samples were transferred to glass slides and sperm motility and viability were checked by light microscope (NIKON TS-100, 10x, 0.25 NA objective). Only sperm that were still swimming actively were counted as viable and motile. In a second experiment, sperm were incubated for 45 min with 125 μM 32-47-32 in 3 % BSA M-16 buffer, or in medium alone. 2 μL of the sperm incubation (1-5 × 105 sperm/mL) was transferred to a 100 μL M-16 drop containing zona free eggs. After 45 min the eggs were analyzed for number of sperm bound and fused as previously described in the assay.
Supplementary Material
Figure 5:
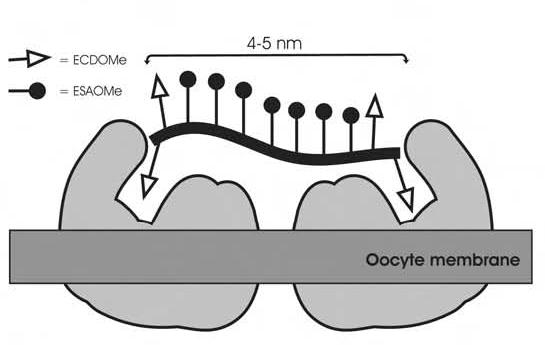
Proposed model for receptor-polymer binding.
Acknowledgments
This work was supported by funding from NIH (R01HD38519, N.S.), NSF (REU CHE0139256, N. F.; CRIF CHE0131146, NMR), and an ACS Cope Scholar Award (N.S.).
Footnotes
Supplementary data including NMR spectra are available on line.
References
- 1.Roberts SK, Konkar S, Sampson NS. Comparison of fertilinβ peptide-substituted polymers and liposomes as inhibitors of in vitro fertilization. ChemBioChem. 2003;4:1229–1231. doi: 10.1002/cbic.200300672. [DOI] [PubMed] [Google Scholar]
- 2.Evans JP. Fertilin beta and other ADAMs as integrin ligands: insights into cell adhesion and fertilization. Bioessays. 2001;23:628–639. doi: 10.1002/bies.1088. [DOI] [PubMed] [Google Scholar]
- 3.Evans JP, Schultz RM, Kopf GS. Roles of the disintegrin domains of mouse fertilins α and β in fertilization. Biol. Reprod. 1998;59:145–152. doi: 10.1095/biolreprod59.1.145. [DOI] [PubMed] [Google Scholar]
- 4.Primakoff P, Hyatt H, Tredick-Kline J. Identification and purification of a sperm surface protein with a potential role in sperm-egg membrane fusion. J. Cell Biol. 1987;104:141–149. doi: 10.1083/jcb.104.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Primakoff P, Myles DG. The ADAM gene family: surface proteins with adhesion and protease activity. Trends Genet. 2000;16:83–87. doi: 10.1016/s0168-9525(99)01926-5. [DOI] [PubMed] [Google Scholar]
- 6.Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- 7.Myles DG, Kimmel LH, Blobel CP, White JM, Primakoff P. Identification of a binding site in the disintegrin domain of fertilin required for sperm-egg fusion. Proc. Natl. Acad. Sci. U. S. A. 1994;91:4195–4198. doi: 10.1073/pnas.91.10.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pyluck A, Yuan R, Galligan E, Jr., Primakoff P, Myles DG, Sampson NS. ECD peptides inhibit in vitro fertilization in mice. Bioorg. Med. Chem. Lett. 1997;7:1053–1058. [Google Scholar]
- 9.Zhu X, Bansal NP, Evans JP. Identification of key functional amino acids of the mouse fertilin beta (ADAM2) disintegrin loop for cell-cell adhesion during fertilization. J. Biol. Chem. 2000;275:7677–7683. doi: 10.1074/jbc.275.11.7677. [DOI] [PubMed] [Google Scholar]
- 10.Gupta S, Li H, Sampson NS. Characterization of fertilinβ-disintegrin binding specificity in sperm-egg adhesion. Bioorg. Med. Chem. 2000;8:723–729. doi: 10.1016/s0968-0896(00)00005-5. [DOI] [PubMed] [Google Scholar]
- 11.Evans JP, Schultz RM, Kopf GS. Mouse sperm-egg plasma membrane interactions: analysis of roles of egg integrins and the mouse sperm homologue of PH-30 (fertilin) β. J. Cell Sci. 1995;108:3267–3278. doi: 10.1242/jcs.108.10.3267. [DOI] [PubMed] [Google Scholar]
- 12.Almeida EAC, Huovila APJ, Sutherland AE, Stephens LE, Calarco PG, Shaw LM, Mercurio AM, Sonnenberg A, Primakoff P, Myles DG, White JM. Mouse egg integrin α6β1 functions as a sperm receptor. Cell. 1995;81:1095–1104. doi: 10.1016/s0092-8674(05)80014-5. [DOI] [PubMed] [Google Scholar]
- 13.Gupta S, Sampson NS. Dimyristoylated peptides incorporated into liposomes are polyvalent fertilinβ mimics. Org. Lett. 2001;3:3333–3335. doi: 10.1021/ol016573d. [DOI] [PubMed] [Google Scholar]
- 14.Yuan R, Primakoff P, Myles DG. A role for the disintegrin domain of cyritestin, a sperm surface protein belonging to the ADAM family, in mouse sperm-egg plasma membrane adhesion and fusion. J. Cell Biol. 1997;137:105–112. doi: 10.1083/jcb.137.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bronson RA, Fusi FM, Calzi F, Doldi N, Ferrari A. Evidence that a functional fertilin-like ADAM plays a role in human sperm-oolemmal interactions. Mol. Hum. Reprod. 1999;5:433–430. doi: 10.1093/molehr/5.5.433. [DOI] [PubMed] [Google Scholar]
- 16.Gichuhi PM, Ford WC, Hall L. Evidence that peptides derived from the disintegrin domain of primate fertilin and containing the ECD motif block the binding of human spermatozoa to the zona-free hamster oocyte. Int. J. Androl. 1997;20:165–170. doi: 10.1046/j.1365-2605.1997.00058.x. [DOI] [PubMed] [Google Scholar]
- 17.Choi SK. Synthetic multivalent molecules: concepts and biomedical applications. 1 Edition John Wiley and Sons, Inc; Hoboken, NJ: 2004. [Google Scholar]
- 18.Kanai M, Mortell KH, Kiessling LL. Varying the size of multivalent ligands: the dependence of concanavalin A binding on neoglycopolymer length. J. Am. Chem. Soc. 1997;119:9931–9932. [Google Scholar]
- 19.Strong LE, Kiessling LL. A general synthetic route to defined, biologically active multivalent arrays. J. Am. Chem. Soc. 1999;121:6193–6196. [Google Scholar]
- 20.Mammen M, Choi S-K, Whitesides GM. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 1998;37:2754–2794. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2754::AID-ANIE2754>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 21.Konkar S, Gupta S, Sampson NS. Fertilinβ liposomes inhibit in vitro fertilization by steric blockage. Bioorg. Med. Chem. Lett. 2004;14:1381–1384. doi: 10.1016/j.bmcl.2003.09.097. [DOI] [PubMed] [Google Scholar]
- 22.Roberts KS, Sampson NS. A facile synthetic method to prepare fluorescently labeled ROMP polymers. Org. Lett. 2004;6:3253–3255. doi: 10.1021/ol048935y. [DOI] [PubMed] [Google Scholar]
- 23.Chen H, Sampson NS. Mediation of sperm-egg fusion: evidence that mouse egg α6β1 integrin is the receptor for sperm fertilinβ. Chem. Biol. 1999;6:1–10. doi: 10.1016/S1074-5521(99)80015-5. [DOI] [PubMed] [Google Scholar]
- 24.Evans JP, Kopf GS, Schultz RM. Characterization of the binding of recombinant mouse sperm fertilin β subunit to mouse eggs: evidence for adhesive activity via an egg β1 integrin-mediated interaction. Dev. Biol. 1997;187:79–93. doi: 10.1006/dbio.1997.8611. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi Y, Yamakawa N, Matsumoto K, Toyoda Y, Furukawa K, Sato E. Analysis of the role of egg integrins in sperm-egg binding and fusion. Mol. Reprod. Dev. 2000;56:412–423. doi: 10.1002/1098-2795(200007)56:3<412::AID-MRD12>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 26.He Z-Y, Brakebusch C, Fässler R, Kreidberg JA, Primakoff P, Myles DG. None of the integrins known to be present on the mouse egg or to be ADAM receptors are essential for sperm-egg binding and fusion. Dev. Biol. 2003;254:226–237. doi: 10.1016/s0012-1606(02)00043-x. [DOI] [PubMed] [Google Scholar]
- 27.Miller BJ, Georges-Labouesse E, Primakoff P, Myles DG. Normal fertilization occurs with eggs lacking the integrin α6β1 and is CD9-dependent. J. Cell Biol. 2000;149:1289–1296. doi: 10.1083/jcb.149.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trnka TM, Grubbs RH. The development of L2X2Ru = CHR olefin metathesis catalysts: An organometallic success story. Acc. Chem. Res. 2001;34:18–29. doi: 10.1021/ar000114f. [DOI] [PubMed] [Google Scholar]
- 29.Cairo CW, Gestwicki JE, Kanai M, Kiessling LL. Control of multivalent interactions by binding epitope density. J. Am. Chem. Soc. 2002;124:1615–1619. doi: 10.1021/ja016727k. [DOI] [PubMed] [Google Scholar]
- 30.Maynard HD, Okada SY, Grubbs RH. Inhibition of cell adhesion to fibronectin by oligopeptide-substituted polynorbornenes. J. Am. Chem. Soc. 2001;123:1275–1279. doi: 10.1021/ja003305m. [DOI] [PubMed] [Google Scholar]
- 31.Maynard HD, Okada SY, Grubbs RH. Synthesis of norbornenyl polymers with bioactive oligopeptides by ring-opening metathesis polymerization. Macromolecules. 2000;33:6239–6248. [Google Scholar]
- 32.Roberts KS, Sampson NS. Increased polymer length of oligopeptide-substituted polynorbornenes using LiCl. J. Org. Chem. 2003;68:2020–2023. doi: 10.1021/jo0265737. [DOI] [PubMed] [Google Scholar]
- 33.Bigler D, Takahashi Y, Chen MS, Almeida EA, Osbourne L, White JM. Sequence-specific interaction between the disintegrin domain of mouse ADAM 2 (fertilin beta) and murine eggs. Role of the alpha(6) integrin subunit. J. Biol. Chem. 2000;275:11576–11584. doi: 10.1074/jbc.275.16.11576. [DOI] [PubMed] [Google Scholar]
- 34.Riener CK, Kada G, Gruber HJ. Quick measurement of protein sulfhydryls with Ellman’s reagent and with 4,4′-dithiodipyridine. Anal. Bioanal. Chem. 2002;373:266–276. doi: 10.1007/s00216-002-1347-2. [DOI] [PubMed] [Google Scholar]
- 35.Portoghese PS. From models to molecules: opioid receptor dimers, bivalent ligands, and selective opioid receptor probes. J. Med. Chem. 2001;44:2259–2269. doi: 10.1021/jm010158+. [DOI] [PubMed] [Google Scholar]
- 36.Loidl G, Groll M, Musiol HJ, Huber R, Moroder L. Bivalency as a principle for proteasome inhibition. Proc. Natl. Acad. Sci. U. S. A. 1999;96:5418–5422. doi: 10.1073/pnas.96.10.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loidl G, Musiol HJ, Groll M, Huber R, Moroder L. Synthesis of bivalent inhibitors of eucaryotic proteasomes. J. Pept. Sci. 2000;6:36–46. doi: 10.1002/(SICI)1099-1387(200001)6:1<36::AID-PSC232>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 38.Love JA, Morgan JP, Trnka TM, Grubbs RH. A practical and highly active ruthenium-based catalyst that effects the cross metathesis of acrylonitrile. Angew. Chem. Int. Ed. Engl. 2002;41:4035–4037. doi: 10.1002/1521-3773(20021104)41:21<4035::AID-ANIE4035>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.