Abstract
We discuss the use of a photoactivated polycarbonate (PPC) microfluidic chip for the solid-phase, reversible immobilization (SPRI) and purification of genomic DNA (gDNA) from whole cell lysates. The surface of polycarbonate was activated by UV radiation resulting in a photo-oxidation reaction, which produced a channel surface containing carboxylate groups. The gDNA was selectively captured on this photoactivated surface in an immobilization buffer, which consisted of 3% polyethylene glycol, 0.4 M NaCl and 70% ethanol. The methodology reported herein is similar to conventional SPRI in that surface-confined carboxylate groups are used for the selective immobilization of DNA; however, no magnetic beads or a magnetic field are required. As observed by UV spectroscopy, a load of ∼7.6 ± 1.6 µg/ml of gDNA was immobilized onto the PPC bed. The recovery of DNA following purification was estimated to be 85 ± 5%. The immobilization and purification assay using this PPC microchip could be performed within ∼25 min as follows: (i) DNA immobilization ∼6 min, (ii) chip washout with ethanol 10 min, and (iii) drying and gDNA desorption ∼6 min. The PPC microchip could also be used for subsequent assays with no substantial loss in recovery, no observable carryover and no need for ‘reactivation’ of the PC surface with UV light.
INTRODUCTION
The ability to analyze cells (mutational content, identification and the like) via signature sequences elucidated from their genomic DNA (gDNA) requires the ability to effectively recover and/or purify the gDNA from the whole cell lysate. Following cell lysis, it is often necessary to remove cellular debris, proteins and other intracellular components that may potentially interfere with the subsequent bioenzymatic reactions required to process the gDNA. For example, PCR processing of gDNA from whole cell lysates typically requires an extraction protocol to remove potential interferents with the Taq polymerization step of PCR (1). A typical isolation protocol of gDNA involves organic extractions, dialysis or precipitations using ethanol, phenol or 2-propanol. In many of these precipitation-type purification protocols, ultracentrifugation is required, which can be a coarse process that may damage the DNA due to shearing artifacts and is difficult to automate. In order to obtain pure and high molecular weight gDNA, very gentle handling during these processes is required (1).
An automated method designed for the purification of dye-terminator-based DNA sequencing products and employing magnetic beads was developed by Hawkins et al. (http://www.agencourt.com/technical/) (2–5). This DNA purification protocol, solid-phase reversible immobilization (SPRI), has been demonstrated to generate sequencing fragments that are free from template DNA, salts and excess dye-terminator products. In comparison to traditional ethanol precipitation, this method uses no centrifugation, is rapid and highly amenable to automation (http://www.agencourt.com/technical/). SPRI (2–5) is based on the selective immobilization of DNA onto carboxy-coated magnetic particles (5). Carboxylates and iron sites on the surface of these magnetic particles act as a bioaffinity adsorbent for the targeted nucleic acids. Under conditions of high concentrations of poly(ethylene glycol), PEG, or tetra(ethylene glycol), TEG, and salts, DNA binds selectively to the surface of these magnetic particles. Once bound, the DNA–bead complexes are magnetically captured and washed with ethanol to remove contaminants with the DNA subsequently released from the beads in water or low ionic strength buffer yielding highly purified DNA. The SPRI method provides high DNA-binding capacity and assay reproducibility (5). Also, modification of the binding buffer, as shown by Hawkins et al., has allowed for the purification of different DNA moieties, such as plasmids (4) and PCR products (3).
The incorporation of a purification platform for nucleic acids (NAs) into a microfluidic chip is very appealing because it offers the ability to handle and preconcentrate nucleic acids in a high-throughput, automated format and in a closed architecture to minimize contamination effects. In fact, most microfluidic chips require preconcentration of target species because of the ability of these devices to process only ultra-small volumes of sample (e.g. 10–1000 pl), making it difficult to detect low bioanalyte concentrations without a preconcentration step. Microfluidic chips are especially attractive for cellular-based assays because many of the sample processing steps can be integrated into a single module, minimizing the dilution of intracellular components following cell lysis. Also, sample handling in microfluidics can be gentle minimizing DNA shearing artifacts, which is important if high molecular weight DNA fragments, such as gDNA, are to be interrogated (1). Unfortunately, centrifugation processes, which are frequently employed in DNA purification strategies, are difficult to integrate into a microfluidic platform.
Several examples of the implementation of different purification strategies for nucleic acids configured into microfluidic chips can be found in the literature. For instance, Breadmore et al. (6) reported on a microchip-based purification method of DNA using silica beads packed into glass microchips and immobilized within a sol-gel. Using this procedure, gDNA from human whole blood was isolated. Northrup and co-workers (7) extracted DNA using a silicon fluidic microchip containing pillars within the microchannel. In these experiments, chaotropic salt solutions were used as the binding agents and the wash and elution solvents consisted of ethanol and water, respectively. Extraction efficiencies of ∼50% were reported using this chip. Tian et al. (8) evaluated the use of silica resins for the direct extraction of DNA from biological samples in a miniaturized microfluidic format. DNA was recovered from white blood cells with an efficiency of ∼70%, while ∼80% of cellular proteins were removed. Chung et al. (9) demonstrated the use of a microfluidic chip loaded with silica beads in a microchannel. They observed that the DNA capture efficiency was higher than that from free-flowing beads. Additionally, they improved DNA extraction efficiency by flowing the solution containing DNA repeatedly backward and forward within the microchannel. Xu et al. (10) demonstrated the purification of Sanger cycle sequencing fragments using SPRI technology carried out in a UV-modified polycarbonate microchip containing an array of microposts. The chip was fabricated in polycarbonate (PC) that was stamped from a metal mold master using simple hot embossing.
Polymers are viewed as attractive materials for microfluidic chips because of their flexibility in micro-manufacturing the chip and the low cost of replicating chips from masters using either hot-embossing or injection molding techniques (11). Additionally, the wide choice of modification chemistries for polymeric surfaces (12,13) opens new opportunities for potential applications. For example, PC exposed to 254–300 nm light results in the formation of polymer phenyl salicylates and hydroxybenzophenones (14–16) and a wide range of carbonyl/carboxyl-containing groups. Such functionalities (2–5) can serve as a capture medium for a wide range of molecular weight-sized DNAs for the isolation, preconcentration or purification of DNA embedded in complex sample matrices.
In this manuscript, we report on the use of a photoactivated polycarbonate (PPC) microchip as a capture medium for gDNA isolated from whole cell lysates. The chip's ability to capture gDNA was demonstrated by immobilization of gDNA from Escherichia coli whole cell lysates. Purification of gDNA from whole blood spiked with E.coli will also be demonstrated. Cellular debris and proteins were successfully removed and purified gDNA was isolated and collected. A PCR performed without purification demonstrated inhibition of the PCR by constituents found in whole blood, but the PPC could effectively isolate the gDNA from PCR inhibitors making it readily amplifiable.
MATERIALS AND METHODS
Reagents
E.coli type B cells were obtained from Aldrich (Milwaukee, WI). Human red blood cells (RBC's, group AB) and serum were purchased from Sigma, St Louis, MO. Whole blood was purchased from Colorado Serum Company, Denver, CO. PEG, Mw = 8000), NaCl, ethanol and 2-propanol were all purchased from Sigma. All reagents were used as received unless stated otherwise.
Microchip fabrication
Polymer microchips were fabricated using procedures reported previously (11,17,18). Briefly, the procedure involved fabrication of a metal molding die by LiGA (German acronym for a three-step micro-manufacturing process consisting of lithography, electroplating and molding). The die consisted of raised Ni microstructures electroplated onto a stainless steel base plate. Replicates of the molding die were hot-embossed into polycarbonate (PC) using a PHI Precision Press model number TS-21-H-C (4A)-5 (City of Industry, CA). PC stock sheets of 5.0 mm thickness were purchased from Goodfellow (Berwyn, PA) and prior to embossing were cut into 133 mm diameter wafers. Polymer wafers were rinsed with 2-propanol (Sigma) and deionized (DI) water (>17.9 MΩ) from an E-pure water purification system (Barnstead, Dubuque, IA) and subsequently dried in an oven at 80°C overnight. Embossing in PC was performed at 1000 lbs pressure for 2–4 min at 190°C. Solution and buffer reservoirs (1 mm in diameter) were drilled into the 5 mm thick chips. Before final assembly (attachment of cover plate to enclose fluidic channels), the PC chips were washed with ∼0.5% Alconox solution, rinsed with DI water, 2-propanol and ultrasonicated for 10 min in DI water. The hot-embossed PC microchannels and cover plates (0.125 mm thick) were exposed to UV radiation (UV lamp from ABM, Inc., San Jose, CA; 254 nm, 15 mW/cm2) for 30 min prior to assembly. After illumination, the PC parts were rinsed with DI water, dried and assembled by heat annealing at 144°C for 20 min.
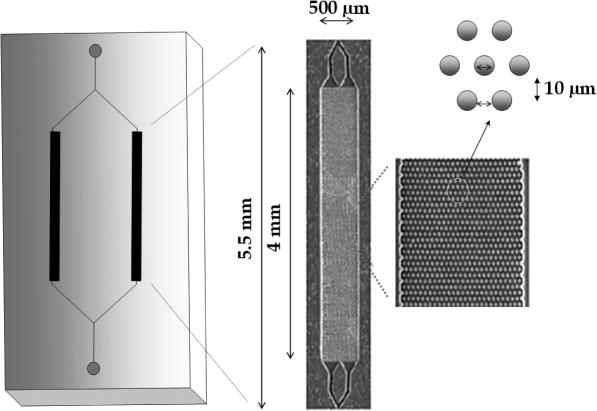
Figure 1 depicts the topographical layout of the PPC microchip and its details. Each of the two immobilization beds of the PPC chip was 500 µm wide, 50 µm deep and 4.0 mm long. The beds consisted of an ordered array of 5:1 aspect ratio—defined as the ratio of structure height to lateral dimension—microposts (d = 10 µm, spacing = 10 µm), which provided a higher surface area compared with an open channel of the same dimensions, allowing larger gDNA loading capacities. The immobilization bed possessed a total available surface area of 2.3 × 107 µm2 and a net volume of 1.6 × 108 µm3 (160 nl) (10). The device's inlet and outlet reservoirs were fitted with capillaries (41 µm i.d.; Polymicro Technologies, Inc., Phoenix, AZ) through which solutions were introduced into the device or collected after purification. A waste/sample collection container (200 µl PCR micro-tube, RNase and DNase free) was mounted at the capillary outlet.
Figure 1.
Schematic diagram and dimensions of the PPC microfluidic chip used for gDNA isolation and purification. The chip was hot-embossed into PC using a metal mold master. Also shown are SEMs of the capture bed and a high magnification view of the microposts configured within the capture bed used to increase the load of gDNA.
Cell lysis and immobilization of gDNA
Thermal E.coli cell lysis was performed at 94°C for 15 min. Lysed E.coli cells (10 µl of whole cell lysate) were suspended in 50 µl of immobilization buffer (IB) containing 3% PEG, 0.4 M NaCl and 70% ethanol, and was run through the PPC microchip at a volumetric flow rate of ∼10 µl/min. The chip was operated in the following manner: first, the chip was filled with the gDNA sample suspended in the IB. The cell suspensions were introduced into the PPC-SPRI bed using a syringe pump (Harvard 22, Harvard Apparatus; Holliston, MA) equipped with a 250 µl glass syringe (Hamilton, Reno, NV). The syringes were equipped with a luer lock to CE column adapter (InnovaQuartz, Inc., Phoenix, AZ). After sample immobilization was complete, the gDNA was washed with 200 µl of 85% ethanol (unless stated otherwise) at a volumetric flow rate of 20 µl/min. The microchannel was then dried with N2 (2 min) and the gDNA released from the PPC chip with ∼20 µl of DI water (5 µl/min). The DI water effluent was collected into the 200 µl PCR micro-tube mounted at the capillary outlet.
PCR
PCR was performed on the isolated gDNA samples using 1 µl of the SPRI/PPC-purified material taken from the whole cell lysate. A GeneAmp® PCR reagent kit with AmpliTaq® DNA polymerase (Applied Biosystems, Foster City, CA) was used for the PCR. PCR cocktails consisted of 1 µl of each of the primers (final concentration 1 µM), 2 µl dNTP's (final concentration of each 0.2 mM), 10 µl of PCR buffer, 1 µl of Taq DNA Polymerase (0.5 U/µl) and 78 µl of DI water. PCR was carried out using a commercial thermal cycling machine (Techne, Burlington, NJ) with cycles consisting of an initial denaturation step at 94°C for 5 min followed by 30 cycles of the following: 94°C for 30 s, 69°C for 40 s, 72°C for 60 s. A final extension at 72°C for 7 min was followed by a cooling step at 4°C.
Information on the sequence of E.coli Type B was obtained from the National Center for Biotechnology Information (http://www.ncbi.nih.gov). The primers were designed for the nucleotide sequence of E.coli between argI and valS producing a 600 bp amplicon. The sequences of the primers were forward, 5′-ATG GCA AAC CCG GAA CAA CTG G-3′ (Tm = 62°C) and reverse 5′-CGC TGC TAT CTG GAA ACG ACC G-3′ (Tm = 61.3°C). The primers were obtained from Integrated DNA Technologies, Inc., Coralville, IA.
SDS–PAGE
Following cell lysis, the suspension (0.5 mg/ml) was centrifuged at 3000 r.p.m. (735 × g) for 10 min. Two 10 µl samples of supernatant were removed and the remaining solution was rigorously vortexed to homogeneity. Then, two 10 µl portions of the suspended cellular debris were removed. One set of supernatant and debris samples were suspended in IB and subjected to PPC-SPRI purification. Samples with and without PPC-SPRI purification taken from the whole cell lysates were separated on a 7.5% SDS–polyacrylamide gel using the Mini-PROTEAN 3 cell (Bio-Rad Laboratories, Inc., Hercules, CA). The separation was performed at 12 V/cm in 1× Tris/Glycine/SDS buffer, pH = 8.8. Proteins were indexed against a sizing ladder (37–250 kDa; Bio-Rad Laboratories, Inc.). After the separation, the gels were stained with Coomasie blue for 30 min and destained overnight. Images were collected using a Logic Gel imaging system (Eastman Kodak Company, Rochester, NY).
Slab gel electrophoresis
PCR products were electrophoresed on a 3% agarose gel (Bio-Rad Laboratories). Amplicons were indexed against a DNA sizing ladder (50–1000 bp; Molecular Probes, Eugene, OR) with the separation performed at 4.8 V/cm in 1× TBE (Tris–boric acid/EDTA) (Bio-Rad Laboratories). After separation, the gels were stained with SYBR Green I (∼0.3 µl/mL; Sigma) for 15 min and images were collected using a Logic Gel imaging system (Eastman Kodak Company).
UV-Vis spectroscopy
UV absorption was performed with an Ultraspec 4000 Spectrometer (Amersham Pharmacia Biotech, Piscataway, NJ). Collected whole cell lysate samples (10 µl) before and after purification in the PPC microchip were examined using UV-Vis spectroscopy. The total DNA concentration and its purity were estimated from the absorbance at 260 and 280 nm.
Fluorescence spectroscopy
Emission spectra of unpurified and purified DNA samples (20 µl) stained with YOPRO-1 (2 µM, YO-PRO-1 iodide, excitation 491 nm/emission 509 nm; Molecular Probes) (30 µl) were acquired using a FLUOROLOG®-3 spectrofluorometer (Jobin Yvon Inc., Edison, NJ) equipped with a 450 W xenon lamp and a cooled Hamamatsu R928 photomultiplier (Bridgewater, NJ) operated at 900 V in a photon-counting mode. The bandpass was set to 5 nm on both the excitation and emission spectrometers. Excitation of the sample was at 488 nm and emission was recorded between 495 and 535 nm. The scans were taken under ambient room conditions and DM300 software was used for analysis.
Fluorescence microscopy
Examination of the UV-modified PC surfaces and observations of DNA immobilization onto the PPC was accomplished using a Zeiss Axiovert 200M Inverted Microscope with a 40× fluorescence objective equipped with a 480 nm excitation filter. The microscope was fitted with a JAI CV 252 monochrome video camera. The images were captured directly to the hard drive of a Dell Precision 500M workstation via a Pinnacle Systems DV500 video capture card that was interfaced to a Pinnacle Systems Bluebox (Pinnacle Systems, Inc., Mountain View, CA). The Bluebox served as a conduit for the video transfer to the computer. Adobe Premiere 6.0 (Adobe Systems, Inc., San Jose, CA) was used for image acquisition and subsequent image processing.
The PPC surface for microscopic interrogation was prepared as follows. Precut pieces of 0.5 mm thick PC were rinsed with 2-propanol and DI water and dried with nitrogen. A 500-mesh Ni TEM grid was placed over the pristine PC and the masked pieces were placed 1 cm from the UV source and subsequently irradiated for 30 min in air. Upon removal from the UV source, the PPC pieces were rinsed with DI water. The cell lysate (∼5 µg/ml) was suspended in the IB. To visualize the DNA, YOPRO-1 (3 µM, YO-PRO-1 iodide, excitation 491 nm/emission 509 nm; Molecular Probes) was added to the whole cell lysate. These pieces were subsequently imaged using fluorescence microscopy following washing of the PC surface using 85% ethanol.
RESULTS AND DISCUSSION
Immobilization buffer and the purification protocol
Our group has demonstrated previously the ability to purify single-stranded DNA (ssDNA) sequencing ladders using SPRI technology configured into a microchip fabricated from PC and containing microposts that provided high surface area (10). An immobilization bed was prepared by exposing pristine PC surfaces to UV radiation that resulted in the formation of surface carboxylate groups through a photo-oxidation reaction. DNA was precipitated onto the activated PC in TEG/ethanol binding buffer, washed with ethanol and subsequently released from the PC surface with DI water. The loading density of DNA on the activated surface was found to be 3.9 pmol/cm2 as estimated by scintillation counting (10). The method was shown to effectively remove excess dye-terminator, as observed in the capillary gel electrophoresis tract. Separation of purified ssDNA sequencing fragments produced a read length comparable with the conventional SPRI format (5), which utilized carboxylated magnetic beads and a magnetic field (10).
In the studies presented herein, the PPC microchip was investigated for the purification and preconcentration of gDNA from crude, whole cell lysates. Precipitation of biological moieties using PEG is a well-known method. It has been shown by Lis (19) that low concentrations of PEG tend to precipitate larger and more anisometric structures. For example, bacterial cells require 1% PEG for quantitative precipitation of their gDNA, while proteins require PEG concentrations of 20% or more (19). Precipitation of relatively small DNA fragments shows an inverse relationship with PEG concentration: a 5% PEG solution will precipitate DNA fragments larger than ∼1.6 kb, while smaller DNA fragments remain in the supernatant or can be precipitated by 11% PEG concentrations. The precipitation of DNA fragments longer than 2 kb, however, with high efficiency has been noted to be rather poor (19). In a subsequent report by Hawkins et al. (5), it was shown that binding and immobilization of bacterial DNA to carboxyl-coated magnetic particles is possible and occurs in buffers containing 10% PEG and 1.25 M NaCl. In that report, however, the purification and isolation procedure was rather lengthy, involving the preparation of a cleared cell lysate prior to incubation with the SPRI beads. Therefore, we sought to investigate the potential for isolating gDNA from crude cell lysates using a variant of SPRI, in which the immobilization bed was created in a microfluidic chip fabricated via simple hot-embossing from a metal master into PC. The PC could subsequently be photoactivated to produce a surface that showed high propensity to immobilize nucleic acids.
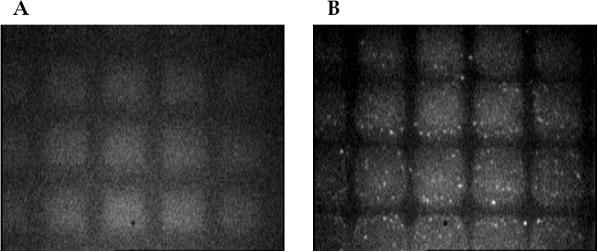
The IB used in these experiments consisted of 0.4 M NaCl, 3% PEG and 70% ethanol. The performance of this IB cocktail was first evaluated with YOPRO-labeled gDNA from E.coli under a fluorescence microscope for its immobilization onto PPC. The PC surface was selectively UV-modified using a TEM grid (500 mesh) serving as a shadow mask to create activated and non-activated regions on the PC. The resulting fluorescence images of the PPC before and after incubation with gDNA in the IB are shown in Figure 2. As can be seen, those areas exposed to the UV radiation through the TEM grid produced a higher level of autofluorescence compared with those areas not exposed to the UV radiation (Figure 2A). This observation is consistent with our previous results and is a consequence of photo-oxidation reactions generating endogenous fluorescent species (20,21). Figure 2B shows the fluorescence image of the masked PPC following incubation with fluorescently-labeled gDNA. It can be seen that the gDNA (bright small spots) are present predominantly only within those areas exposed to the UV radiation with negligible amounts of gDNA found in areas that were not exposed to the UV radiation.
Figure 2.
Fluorescence microscopic images of the PC surfaces exposed to 254 nm UV radiation through a TEM grid incubated with no gDNA (A) and then, incubated with gDNA labeled with YOPRO-1 (B) in the IB, which consisted of 3% PEG, 0.4 M NaCl and 70% ethanol. The fluorescence images were acquired with a 40× objective using 480 nm excitation and the emission monitored at ∼520 nm. The PC sheets were exposed to UV radiation (15 mW/cm2) for 30 min through a TEM grid.
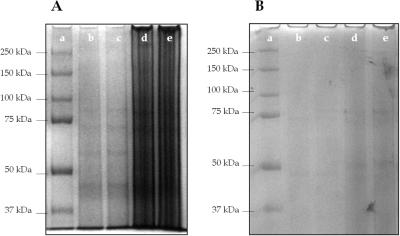
A thermally lysed E.coli cell suspension (0.5 mg/ml) was purified using a PPC chip according to the procedure described in Materials and Methods. The effluent containing purified gDNA was collected and SDS–PAGE was performed in order to evaluate the ability of the chip to remove cellular proteins. Also, the collected samples were subjected to PCR in order to verify the presence and quantity of the gDNA isolated. PCR products were separated on an agarose gel to confirm the presence of a 600 bp amplicon generated from E.coli gDNA. Figure 3 shows the SDS–PAGE protein separation results performed before (A) and after (B) purification of the whole cell lysate using the PPC microchip. The cell lysates not subjected to PPC-SPRI contained high levels of proteins ranging in size between 37 and 250 kDa. After transport of the cell lysate through the PPC microchip, the intensity of the protein bands decreased substantially. These results clearly demonstrate the ability of the PPC to remove cellular proteins from the raw cell lysate by showing minimal amounts of adsorption of these proteins to the photoactivated PC surface. As judged by UV spectrophotometry, the ratio of A260/A280 calculated before purification of the whole cell lysate was 1.47 ± 0.13 (n = 7) and after purification this ratio was found to be 1.89 ± 0.10 (n = 7). Based on the values of absorbance at 280 nm measured before and after purification, ∼80% of the protein content was removed during the purification process using this PPC chip.
Figure 3.
An SDS–PAGE electrophoretic separation of proteins from an E.coli crude whole cell lysate before (A) and after (B) purification in the PPC microchip. Following thermal lysis, the cellular suspension was centrifuged at 3000 r.p.m. (735 × g) for 10 min. The supernatant (2 × 10 µl samples) was removed from the centrifuge tube and the remaining debris was rigorously vortexed and another two 10 µl samples were removed. The lanes in this figure represent standard sizing ladder (a); thermally lysed cells, supernatant (b and c); and thermally lysed cells, debris (d and e). Proteins were separated using a 7.5% acrylamide gel at 12 V/cm in 1× Tris–Glycine–SDS buffer, pH 8.8. Proteins were indexed against a sizing ladder, which ranged from 37 to 250 kDa in size.
PCR of PPC-purified samples
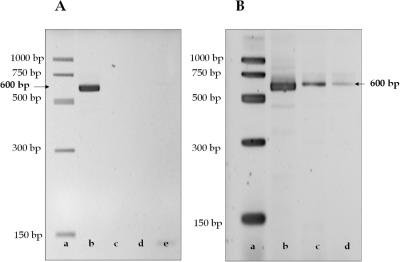
To demonstrate the ability of the PPC-SPRI format to immobilize gDNA, lysed 0.05 mg/ml E.coli cell suspensions in IB were hydrodynamically introduced through the PPC-SPRI chip. After release of the gDNA from the PPC surface using DI water, PCR was performed on the collected sample in order to verify the presence of the genetic material. When the chip was rinsed with 150–200 µl of 85% ethanol at 20 µl/min followed by 20 µl of DI water, PCR resulted in the production of a 600 bp amplicon (Figure 4A, lane b). However, when only 100 µl or less of ethanol was used for the washing step, no PCR products were observed (Figure 4A, lane c). Interestingly, the UV spectrum confirmed the presence of gDNA in the water wash (data not shown). The failure of the PCR was most likely due to the presence of >50 mM salt concentrations and/or traces of ethanol in the eluted DNA that may inhibit Taq polymerase activity. To test this hypothesis, we performed PCR on a positive control sample with the addition of 0.5 µl of IB but the PCR failed (Figure 4A, lane d). When present in the sample, NaCl either caused deactivation of the Taq polymerase or changed the Tm's of the primer/target DNA duplex (1). We have not established what the concentration of NaCl was after rinsing; however, control PCRs with NaCl concentrations between 20 and 45 mM showed low efficiency PCR amplifications. Control samples with a NaCl concentration >50 mM inhibited the reaction completely. Therefore, it was imperative that a sufficient amount of ethanol be used to efficiently remove salts prior to PCR. We also observed that when the PPC-SPRI microchip was not dried following washing with ethanol, the PCR was also unsuccessful. We performed control PCRs containing gDNA isolated from E.coli containing 1.7% ethanol, which also produced a failed PCR (Figure 4A, lane e).
Figure 4.
(A) Fluorescence images of agarose gels showing the presence of the 600 bp amplicon generated from PCR performed on the PPC-purified E.coli gDNA obtained from purifications run on the same chip. After gDNA immobilization, DNA sizing ladder (lane a); the microchip was rinsed with 200 µl of 85% ethanol followed by 20 µl of DI water (lane b); the chip was rinsed with 50 µl of 85% ethanol followed by 20 µl of DI water (lane c); control PCR containing gDNA isolated from E.coli containing 1.7% ethanol (lanes d and e). (B) Gel images of the 600 bp amplicons obtained from PCR performed with the PPC-purified E.coli at different aliquot numbers of water (20 µl each) used to elute the DNA from the immobilization bed. Lane a, DNA sizing ladder; lane b, fraction 1 (20 µl); lane c, fraction 2 (20 µl); and lane d, fraction 3 (20 µl). The samples were immobilized in buffer containing 3% PEG, 0.4 M NaCl and 70% ethanol. The separation was performed on a 3% agarose gel using a field strength of 4.8 V/cm in 1× TBE. The sample volume loaded onto the gel was 10 µl, which also contained 2 µl of the loading dye. The gels were stained with SYBR Green I prior to imaging.
To investigate the volume of DI water required to remove the DNA from the carboxylated PC surface, we collected three consecutive fractions of gDNA with each fraction's volume set at 20 µl. Then, we performed a PCR on each of these fractions. Figure 4B shows a fluorescence image of the gel electropherograms of the PCR products obtained from this experiment. Lanes b–d correspond to the first, second and third DI water fractions collected, respectively, when the sample was immobilized in buffer containing 3% PEG, 0.4 M NaCl and 70% ethanol. The highest intensity amplicon band was observed in lane b—the very first fraction of the DNA collected. Band intensities in lanes c and d were much weaker, suggesting that most of the gDNA was eluted from the microchip within the first 20 µl of DI water addition. Additional 20 µl washings of the PPC chip with DI water indicated the absence of any detectable gDNA, which was analyzed by PCR and gel electrophoresis (data not shown).
Efficiency of PPC-SPRI
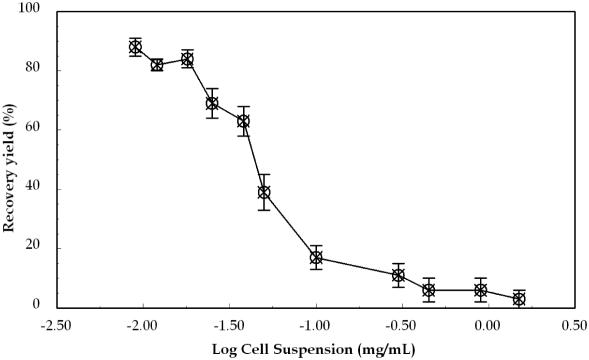
Recovery of gDNA after PPC-SPRI purification was calculated by examining the mass of gDNA in a 10 µl E.coli whole cell lysate before and the mass of gDNA in 20 µl after purification in the PPC microchip. A 10 µl whole cell lysate was suspended in 50 µl of IB and the entire volume was loaded and run through the PPC microchip at a volumetric flow rate of ∼10 µl/min. Samples containing E.coli cells in a concentration range of 0.02–1.5 mg/ml were examined. After purification and elution, as judged by UV spectrophotometry at 260 nm and fluorescence, the average concentration of gDNA immobilized onto the PPC-SPRI bed was 7.6 ± 1.6 µg/ml (∼150 ± 30 ng; n = 5). For a 0.05 mg/ml E.coli cell suspension, a capture efficiency of gDNA was estimated to be 39%, while it increased to 85% when the cell suspension concentration was set below 0.02 mg/ml (Figure 5). The lower recoveries observed with higher concentrations of E.coli cells suggests that the amount of DNA introduced exceeded the load capacity of the capture bed (i.e. available –COOH sites). The saturation point for gDNA capture was estimated from Figure 5 to be ∼9.6 ± 2.0 µg/ml (190 ± 40 ng of gDNA; n = 7), in fair agreement to the surface load calculated using UV and fluorescence spectroscopies. Based on the mass saturation point and the available surface area of the capture bed, the surface loading density of E.coli gDNA (4.6 Mb) was determined to be 0.26 fmol/cm2 or 0.79 µg/cm2, as compared with 3.9 pmol/cm2 (0.65 µg/cm2 assuming an average DNA length of 500 bp) reported for DNA sequencing fragments (10). The lower molar surface loading density of gDNA as compared with the ssDNAs strongly suggests that there are multiple carboxylate sites involved in the capture of DNA molecules, with larger DNAs showing a lower number density on the surface due to the fact that they occupy more potential surface binding sites. The binding capacity of the PPC chip could be improved by increasing the active surface area of the immobilization bed by incorporating a higher density of posts (i.e. use of smaller posts diameters and reduced inter-post spacing) or a larger PPC bed.
Figure 5.
The recovery of gDNA loaded onto the microchip using the PPC-SPRI purification assay for E.coli gDNA, which was immobilized to the activated surface using 3% PEG, 0.4 M NaCl and 70% ethanol. The total DNA mass before and after PPC-SPRI was estimated from absorbance values at 260 nm and fluorescence data. Error bars in the graph represent SDs from three replicate runs.
Reproducibility of the PPC-SPRI Microchip
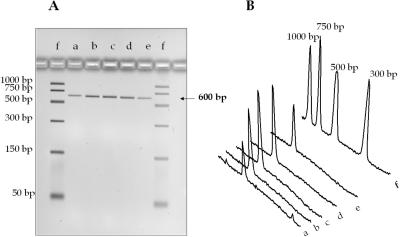
The reproducibility of gDNA immobilization with the PPC microchip was investigated using several chips. An E.coli cell suspension (0.05 mg/ml) prepared in IB was thermally lysed and introduced into the chip. To test intrachip reproducibility, following release of the captured gDNA, the channel was washed with ∼60 µl of DI water, 100 µl of 85% ethanol, dried with nitrogen, and the next sample was then introduced into the chip. To verify the lack of carryover contamination, a 20 µl aliquot of water was run through the PPC microchip with the effluent checked for the presence of nucleic acids using fluorescence and UV spectrophotometry. The capture bed was not reactivated between purifications. After the purification, the amount of recovered gDNA was analyzed using UV spectrophotometry and fluorescence spectroscopy from which the gDNA concentration was estimated. The collected samples were also PCR amplified and using the integrated intensity of the amplicon bands, the performance of the assay was determined. Figure 6 shows the intensities of the 600 bp amplicon bands obtained from a gel image following PCR. The data were obtained using five different PPC-SPRI chips to test inter-chip reproducibility. Based on the spectroscopic data, the chip-to-chip RSD was ∼15% (n = 7), while the intrachip RSD was 7%.
Figure 6.
(A) Agarose gel electrophoresis images of 600 bp PCR products generated from purified E.coli gDNA using the PPC microchip, and (B) 2D traces of the electropherograms in the accompanying image. Samples were immobilized to the capture bed in 3% PEG, 0.4 M NaCl and 70% ethanol. Lanes a–e, chip #1–5 and lane (f) DNA sizing ladder. The sample volume loaded onto the gel was 5 µl with 1 µl of the loading dye. Separation parameters are described in Figure 4.
Immobilization of E.coli gDNA from whole blood
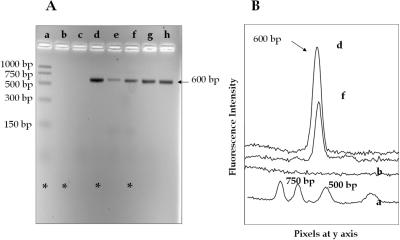
The critical limitation of PCR is the potential inhibition of polymerase activity produced by interferents contained within the sample matrix. Many clinical samples contain polymerase inhibitors, such as hemoglobin, urea, heparin, organic and phenolic compounds as well as intracellular inhibitors found in bacterial cells (e.g. bacterial proteases) (1). The application of PCR to identify microbial DNA in blood is used for a number of diagnostic purposes, which are among the many important tests used for the diagnosis of infectious diseases. For example, detection of hepatitis B virus and a number of bacterial strains by PCR is widely performed in clinical laboratories (22–26). However, prior to PCR amplification of nucleic acids found in cell lysates, it is advisable to purify and preconcentrate the DNA to prevent polymerase inactivation. We therefore set out to test the ability of the PPC-SPRI microchip to purify bacterial gDNA from whole blood. An E.coli cell suspension (1.0 mg/ml) was prepared in whole blood and the sample was lysed. Of this lysate 10 µl was mixed with 50 µl of IB after which the sample was passed through the PPC-SPRI microchip. After immobilization and washing with ethanol, the DNA was collected in DI water and PCR was performed on the collected sample. Electrophoretic analysis indicated a lack of amplicon bands for four different whole blood samples spiked with E.coli (data not shown). Therefore, we modified the purification procedure by eliminating ethanol in the IB. After immobilization and washing with ethanol, the DNA isolated on the PPC microchip was collected in DI water and subjected to PCR. The electropherogram obtained following PCR of unpurified and purified E.coli spiked into whole blood is shown in Figure 7A and B.As can be seen, no 600 bp amplicon was observed using unpurified E.coli from the blood sample (lanes b and c in Figure 7A). The inhibition of Taq polymerase most likely occurred due to the presence of high concentrations of hemoglobin. Purified bacterial DNA from whole blood produced a successful PCR, as the fluorescence signal for the 600 bp amplicon was clearly evident in lanes e–h in Figure 7A. After the DNA had been eluted from the device, the immobilization bed was washed again with 50 µl of DI water, 10 µl of eluent was collected and again subjected to PCR. No 600 bp amplicons were detected, suggesting complete removal of the gDNA and a lack of potential carryover contamination into subsequent analyses. However, purification of blood samples containing E.coli gDNA resulted in lower total recoveries of the target gDNA compared with a sample containing bacterial cells only (Figure 7, lane d), most likely due to potential interferents in the whole blood sample competing for effective adsorption sites on the activated PC.
Figure 7.
Agarose gel electrophoresis images of the 600 bp PCR product (A) and 2D gel electrophoresis traces of the same products (B) for several lanes depicted in (A). The amplicons were generated via PCR performed with template from unpurified E.coli/blood sample (lanes f and g), purified E.coli/blood sample (lanes a–d), and purified gDNA from E.coli cells only (lane e). Lane h shows the DNA sizing ladder. The samples were immobilized in buffer containing PEG and NaCl only. The sample volume loaded onto the gel was 5 µl with 1 µl of the loading dye. Gel electrophoresis parameters are described in Figure 4.
CONCLUSIONS
The SPRI-variant used herein was configured in a microfluidic chip fabricated in PC that was subsequently exposed to UV radiation, inducing a photo-oxidation reaction creating surface-confined carboxylates, which produce an affinity bed for isolating nucleic acids using a very simple, inexpensive and robust method. A protocol for the immobilization of gDNA from E.coli as the model system using PPC-SPRI and the subsequent amplification of the DNA was established. Purification of the gDNA in the PPC-SPRI microchip was performed with a capture efficiency of 85 ± 5%. In addition, the PPC-SPRI chip could be reused for subsequent assays without significant loss in capture efficiency if desired. However, in cases where cross contamination is unacceptable, the low cost of the fabrication of these chips and the simple procedure for generating the affinity surface allows it to be configured in a one-time use, disposable format. The PPC-SPRI assay enables purification of DNA sequencing fragments (10) as well as the isolation of bacterial gDNA from whole cell lysates (this study). The process is simple and not labor-intensive. The entire purification assay using this PPC-SPRI chip required <25 min: (i) DNA immobilization ∼6 min; (ii) microchip washout with ethanol ∼10 min; and (iii) drying and gDNA desorption ∼6 min.
The advantages of the PPC-SPRI can be easily recognized. The simplicity of its fabrication is a major advantage. The immobilization bed is fabricated during embossing of the microfluidic network and activation of the PC surface requires only exposure to broadband UV radiation. In addition, with proper selection of components contained within the IB, the selective isolation of intermediate sized nucleic acids can also be envisioned, such as PCR products, mtDNA, clones and RNAs (2–5), making the approach flexible enough to be used in a variety of different applications where the isolation and preconcentration of nucleic acids is necessary prior to downstream processing. Another advantage of PPC-SPRI stems from the fact that these are simple flow-through type reactors. They can be easily implemented into the design of more complex systems performing multi-step bioassays. The fabrication of PPC-SPRI reactors opens the direct route toward low cost, disposable DNA sample purification units. For example, the cost of each device in material only is a few cents and can be mass produced using injection molding. To further lower the ‘per purification’ cost, SPRI reactors can be fabricated into multi-reactor arrays for parallel preparation of multiple samples. Our group is currently developing a 96-well SPRI card that can be used in conjunction with standard 96-well microtiter plates for high-throughput applications.
Acknowledgments
The authors would like to thank the National Science Foundation (DBI-0138048) for financial support of this work. This work is also supported in part by the National Science Foundation under Grant Number EPS-0346411 and the State of Louisiana Board of Regents Support Fund. Funding to pay the Open Access publication charges for this article was provided by EPS-0346411.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wilson I.G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elkin C., Kapur H., Smith T., Humphries D., Pollard M., Hammon N., Hawkins T. Magnetic bead purification of labeled DNA fragments for high-throughput capillary electrophoresis sequencing. BioTechniques. 2002;32:1296. doi: 10.2144/02326st05. 1298–1300, 1302. [DOI] [PubMed] [Google Scholar]
- 3.DeAngelis M.M., Wang D.G., Hawkins T.L. Solid-phase reversible immobilization for the isolation of PCR products. Nucleic Acids Res. 1995;23:4742–4743. doi: 10.1093/nar/23.22.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elkin C.J., Richardson P.M., Fourcade H.M., Hammon N.M., Pollard M.J., Predki P.F., Glavina T., Hawkins T.L. High-throughput plasmid purification for capillary sequencing. Genome Res. 2001;11:1269–1274. doi: 10.1101/gr.167801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawkins T.L., O'Connor-Morin T., Roy A., Santillan C. DNA purification and isolation using a solid-phase. Nucleic Acids Res. 1994;22:4543–4544. doi: 10.1093/nar/22.21.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breadmore M.C., Wolfe K.A., Arcibal I.G., Leung W.K., Dickson D., Giordano B.C., Power M.E., Ferrance J.P., Feldman S.H., Norris P.M., et al. Microchip-based purification of DNA from biological samples. Anal. Chem. 2003;75:1880–1886. doi: 10.1021/ac0204855. [DOI] [PubMed] [Google Scholar]
- 7.Christel L.A., Petersen K., McMillan W., Northrup M.A. Rapid, automated nucleic acid probe assays using silicon microstructures for nucleic acid concentration. J. Biomech. Eng. 1999;121:22–27. doi: 10.1115/1.2798037. [DOI] [PubMed] [Google Scholar]
- 8.Tian H., Huhmer A.F.R., Landers J.P. Evaluation of silica resins for direct and efficient extraction of DNA from complex biological matrices in a miniaturized format. Anal. Biochem. 2000;283:175–191. doi: 10.1006/abio.2000.4577. [DOI] [PubMed] [Google Scholar]
- 9.Chung Y.-C., Jan M.-S., Lin Y.-C., Lin J.-H., Cheng W.-C., Fan C.-Y. Microfluidic chip for high efficiency DNA extraction. Lab. Chip. 2004;4:141–147. doi: 10.1039/b310849j. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y., Vaidya B., Patel A.B., Ford S.M., McCarley R.L., Soper S.A. Solid-phase reversible immobilization in microfluidic chips for the purification of dye-labeled DNA sequencing fragments. Anal. Chem. 2003;75:2975–2984. doi: 10.1021/ac030031n. [DOI] [PubMed] [Google Scholar]
- 11.Soper S.A., Ford S.M., Qi S., McCarley R.L., Kelly K., Murphy M.C. Polymeric microelectromechanical systems. Anal. Chem. 2000;72:643A–651A. doi: 10.1021/ac0029511. [DOI] [PubMed] [Google Scholar]
- 12.Soper S.A., Henry A.C., Vaidya B., Galloway M., Wabuyele M., McCarley R.L. Surface modification of polymer-based microfluidic devices. Anal. Chim. Acta. 2002;470:87–99. [Google Scholar]
- 13.Vaidya B., Soper S.A., McCarley R.L. Surface modification and characterization of microfabricated poly(carbonate) devices: manipulation of electroosmotic flow. Analyst. 2002;127:1289–1292. doi: 10.1039/b205801b. [DOI] [PubMed] [Google Scholar]
- 14.Adams M.R., Garton A. Surface modification of bisphenol-A-polycarbonate by far-UV radiation. Part I. In vacuum. Polym. Degrad. Stab. 1993;41:265–273. [Google Scholar]
- 15.Adams M.R., Garton A. Surface modification of bisphenol A-polycarbonate by far-UV radiation. Part II. In air. Polym. Degrad. Stab. 1993;42:145–151. [Google Scholar]
- 16.Johnson T.J., Ross D., Gaitan M., Locascio L.E. Laser modification of preformed polymer microchannels: application to reduce band broadening around turns subject to electrokinetic flow. Anal. Chem. 2001;73:3656–3661. doi: 10.1021/ac010269g. [DOI] [PubMed] [Google Scholar]
- 17.Ford S.M., McCandless A.B., Liu X., Soper S.A. Rapid fabrication of embossing tools for the production of polymeric microfluidic devices for bioanalytical applications. Proc. SPIE. 2001;4560:207–216. [Google Scholar]
- 18.Qi S., Liu X., Ford S., Barrows J., Thomas G., Kelly K., McCandless A., Lian K., Goettert J., Soper S.A. Microfluidic devices fabricated in poly(methyl methacrylate) using hot-embossing with integrated sampling capillary and fiber optics for fluorescence detection. Lab. Chip. 2002;2:88–95. doi: 10.1039/b200370h. [DOI] [PubMed] [Google Scholar]
- 19.Lis J.T. Fractionation of DNA fragments by polyethylene glycol induced precipitation. Methods Enzymol. 1980;65:347–353. doi: 10.1016/s0076-6879(80)65044-7. [DOI] [PubMed] [Google Scholar]
- 20.Witek M.A., Wei S., Vaidya B., Adams A.A., Zhu L., Stryjewski W., McCarley R.L., Soper S.A. Cell transport via electromigration in polymer-based microfluidic devices. Lab. Chip. 2004;4:464–472. doi: 10.1039/b317093d. [DOI] [PubMed] [Google Scholar]
- 21.McCarley R.L., Vaidya B., Wei S., Smith A.F., Patel A.B., Feng J., Murphy M.C., Soper S.A. Resist-free patterning of surface architectures in polymer-based microanalytical devices. J. Am. Chem. Soc. 2005;127:842–843. doi: 10.1021/ja0454135. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko S., Miller R.H., Feinstone S.M., Unoura M., Kobayashi K., Hattori N., Purcell R.H. Detection of serum hepatitis B virus DNA in patients with chronic hepatitis using the polymerase chain reaction assay. Proc. Natl Acad. Sci. USA. 1989;86:312–316. doi: 10.1073/pnas.86.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juvonen R., Satokari R., Mallison K., Haikara A. Detection of spoilage bacteria in beer by polymerase chain reaction. J. Am. Soc. Brew. Chem. 1999;57:99–103. [Google Scholar]
- 24.Belgrader P., Benett W., Hadley D., Richards J., Stratton P., Mariella R., Jr, , Milanovich F. PCR detection of bacteria in seven minutes. Science. 1999;284:449–450. doi: 10.1126/science.284.5413.449. [DOI] [PubMed] [Google Scholar]
- 25.Wang R.F., Cao W.W., Cerniglia C.E. A universal protocol for PCR detection of 13 species of foodborne pathogens in foods. J. Appl. Microbiol. 1997;83:727–736. doi: 10.1046/j.1365-2672.1997.00300.x. [DOI] [PubMed] [Google Scholar]
- 26.Holland J.L., Louie L., Simor A.E., Louie M. PCR detection of Escherichia coli O157:H7 directly from stools: evaluation of commercial extraction methods for purifying fecal DNA. J. Clin. Microbiol. 2000;38:4108–4113. doi: 10.1128/jcm.38.11.4108-4113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]