Abstract
Nitric oxide (NO•) can stabilize mRNA by activating p38 mitogen-activated protein kinase (MAPK). Here, transcript stabilization by NO• was investigated in human THP-1 cells using microarrays. After LPS pre-stimulation, cells were treated with actinomycin D and then exposed to NO• without or with the p38 MAPK inhibitor SB202190 (SB). The decay of 220 mRNAs was affected; most were stabilized by NO•. Unexpectedly, SB often enhanced rather than antagonized transcript stability. NO• activated p38 MAPK and Erk1/2; SB blocked p38 MAPK, but further activated Erk1/2. RT–PCR confirmed that NO• and SB could additively stabilize certain mRNA transcripts, an effect abolished by Erk1/2 inhibition. In affected genes, these responses were associated with CU-rich elements (CURE) in 3′-untranslated regions (3′-UTR). NO• stabilized the mRNA of a CURE-containing reporter gene, while repressing translation. Dominant-negative Mek1, an Erk1/2 inhibitor, abolished this effect. NO• similarly stabilized, but blocked translation of MAP3K7IP2, a natural CURE-containing gene. NO• increased hnRNP translocation to the cytoplasm and binding to CURE. Over-expression of hnRNP K, like NO•, repressed translation of CURE-containing mRNA. These findings define a sequence-specific mechanism of NO•-triggered gene regulation that stabilizes mRNA, but represses translation.
INTRODUCTION
Gene expression in eukaryotic cells is a dynamic process that includes transcription, pre-mRNA splicing, nucleo-cytoplasmic transport, subcellular localization of mRNA and finally transcript translation or degradation. In addition to the many mechanisms that control gene transcription, the importance and complexity of post-transcriptional gene regulation has been increasingly recognized. Recent studies using microarrays have shown that regulation of mRNA stability accounts for about one-half of all changes in mRNA steady-state levels (1,2). Like the role of DNA sequence in regulating transcription, post-transcriptional events, in particular mRNA translation and degradation, have been linked to tightly regulated mechanisms that are dependent on specific cis-acting mRNA elements and trans-factors. Important examples of cis-acting sequences that control post-transcriptional mRNA regulation include AU-rich elements (ARE) and the less-well characterized differentiation control element (DICE), a CU-rich repetitive motif.
ARE consist of multiple, frequently overlapping copies of the AUUUA motif in the 3′-untranslated region (3′-UTR) of many cytokines, growth factors and proto-oncogenes (3,4). ARE induce rapid shortening of the poly(A) tail followed by exosomal degradation of the mRNA body (5,6). Several ARE-binding trans-factors, such as HuR, tristetraprolin, AUF1 and CUGBP2 have been identified and their various functions have become better understood. By interaction with ARE, HuR stabilizes (7), but tristetraprolin destabilizes mRNA (8,9). AUF1 can either stabilize or destabilize ARE-containing transcripts depending on the relative abundance of different AUF1 isoforms (10). CUGBP2 stabilizes ARE-bearing mRNA but silences its translation (11). DICE, originally described in the 3′-UTR of lipoxygenase mRNA (12), is characterized by repetitive, tandem-like CU-rich sequences with a (C/U)CCANX CCC(U/A) (C/U)y UC(C/U)CC consensus architecture (13,14). DICE binds heterogeneous nuclear ribonucleoprotein (hnRNP) K or E2/E1 to stabilize mRNA (14,15), and to either silence (13,16,17) or drive translation (18). These molecular details of cis-acting sequences and trans-factors have provided important tools for studying their interactions with major signal transduction networks, such as the stress kinase pathways that regulate mRNA stability and translation.
The connection between p38 mitogen-activated protein kinase (MAPK) signaling and ARE-binding trans-factors has recently been investigated for tristetraprolin. In vitro evidence shows that tristetraprolin can be phosphorylated by p38 MAPK, which inhibits its binding to ARE, thereby stabilizing target transcripts (19,20). Alternatively, as shown for IL-3 mRNA, p38 MAPK can also phosphorylate other ARE-stabilizing trans-factors, such as HuR and subsequently antagonize the effects of tristetraprolin (21). To date, the p38 MAPK signaling pathway has been implicated in stabilizing mRNA half-lives of more than 40 ARE genes (22), including cyclooxygenase 2 (23), TNFα (19), IL-3 (21), IL-8 (22,24), vascular endothelial growth factor (25) and p21/Waf1/Cip1 (26). Inhibitors of p38 MAPK or expression of a dominant-negative mutant of p38 MAPK activated protein kinase 2 abolish mRNA stabilization of these genes (19,23,24,27). Likewise, the Erk1/2 signaling pathway has been implicated in the regulation of DICE -containing transcripts. Through phosphorylation of hnRNP K, Erk1/2 increases hnRNP K cytoplasmic accumulation and thereby silences the translation of DICE-containing genes (16).
Nitric oxide (NO•) is an important signaling molecule that regulates a wide range of cellular activities including gene expression. It has been demonstrated that NO• regulates transcription through Sp1 (28,29), NF-kB (30), AP-1 (31), Egr-1 (32) and HIF-1 (33). Besides these defined effects on gene transcription, NO• has been further implicated in regulating the mRNA stability of a number of genes including heme oxygenase-1 (34), cytochrome C oxidase (35), flavin-containing monooxygenase (36), transforming growth factor-β3 (37), matrix metalloproteinase-9 (38), IL-8 (24) and p21/Waf1/Cip1 (26). NO• was found to destabilize matrix metalloproteinase-9 mRNA through the cGMP-dependent down-regulation of HuR (38). Conversely, NO• stabilized IL-8 and p21/Waf1/Cip1 mRNA through the cGMP-independent activation of p38 MAPK (24,26). For other genes, the mechanism by which NO• signaling regulates mRNA turnover has not yet been determined.
To more completely characterize transcript stabilization by NO• and to further explore the role of p38 MAPK in these events, we performed a large-scale analysis of mRNA decay using oligonucleotide microarrays in lipopolysaccharide (LPS)-stimulated human THP-1 cells, a monocytic line. In the presence of LPS, a very strong activator of p38 MAPK, NO• was found to increase the half-life of relatively few genes by further engaging this pathway. Unexpectedly, most genes stabilized by NO• were further stabilized by p38 MAPK inhibition. This result prompted a search of UTR databases for cis-acting elements that might explain this finding. Downstream experiments were then conducted to define possible mechanisms. NO• was shown to stabilize transcripts while suppressing their translation through DICE-like, CU-rich elements (CURE) in target transcripts. NO• activation of Erk1/2 was required, as was an associated increase in the binding of hnRNP proteins to mRNA. These findings suggest a novel mechanism of NO•-mediated post-transcriptional regulation that functions as both a mRNA stabilizer and a translation inhibitor.
MATERIALS AND METHODS
Reagents
Salmonella minnesota Re595 LPS was obtained from List Biologic (Campbell, CA). S-nitrosoglutathione (GSNO), SB202190 (SB) and PD98059 (PD) were purchased from Calbiochem (San Diego, CA). Actinomycin D (ActD), glutathione (GSH), β-mercaptoethanol and dimethyl sulfoxide (DMSO) were from Sigma-Aldrich (St. Louis, MO). DMSO was used to dissolve SB and PD and was similarly added to control cells (final concentration 0.0033%) in experiments that tested these reagents. Rabbit polyclonal antibodies detecting p38 MAPK, phospho-p38 MAPK (Thr180/Tyr182), Erk1/2 and phospho-Erk1/2 (Thr202/Tyr204) were obtained from Cell Signaling Technology, Inc. (Beverly, MA). Goat polyclonal antibodies against hnRNP K and hnRNP E2/E1, normal goat serum and mouse monoclonal antibody against α tubulin were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). MAP3K7IP2 (MAPK kinase kinase 7 interacting protein 2) antibody was obtained from Affinity BioReagents (Golden, CO).
Microarray experiments
THP-1 cells (2 × 107) were first stimulated with LPS (1 µg/ml) for 4 h to activate the cells and boost transcript levels. After 30 min treatment with ActD (2.5 µg/ml), a transcription inhibitor, in the absence or presence of p38 MAPK inhibitor SB (0.1 µM), cells were then further incubated for 0–180 min with 400 µM of GSNO or GSH control (N = 4). Total RNA at different time points (0, 45, 90 and 180 min) was extracted, labeled and hybridized to human U133A microarrays following standard Affymetrix procedures as described previously (26). After staining with streptavidin phycoerythrin (Molecular Probes), microarrays were scanned using Agilent GeneArray Scanner.
Plasmid construction
The plasmid pGL3 containing a firefly luciferase (LUC) reporter gene driven by the SV40 promoter was purchased from Promega (Madsion, WI). Synthetic oligonulceotides containing the consensus CURE sequence (5′-CTTTCTCCCCCACCCTCTTCTCCCCCTTCCCCCTCCCC-3′; the core sequences are underlined) or its antisense form (5′-GGGGAGGGGGAAGGGGGAGAAGAGGGTGGGGGAGAAAG-3′) were cloned into pGL3 at the XbalI site residing in the 3′-UTR of LUC, generating the plasmids pGL3/CURE and pGL3/CUREmut, respectively. The plasmid pMEK1-DN encoding a dominant-negative mutant of Mek1 was a gift from Dr Ae-Kyung Yi at University of Tennessee Health Science Center, Memphis, Tennessee (39). The phnRNP-K, plasmid that expresses hnRNP K was kindly provided by Dr Ze'ev Ronai at Mount Sinai School of Medicine, New York (16). The parental plasmids, pUSEamp of pMEK1-DN and pcDNA3 of phnRNP-K, were obtained from Upstate (Charlottesville, VA) and Invitrogen (Carlsbad, CA), respectively.
Cell culture and transfection
THP-1 cells, a human monocytic line obtained from ATCC (Manassas, VA) were maintained in RPMI supplemented with 10% fetal calf serum (FCS) (Cellgro, Herndon, VA) and 50 µM β-mercaptoethanol as described previously (24). Transfections were performed using Nucleofector® Kit V (amaxa Inc. Gaithersburg, MD) according to the manufacture's instructions. For each transfection, 0.3 µg of pGL3, pGL3/CURE or pGL3/CUREmut and 0.1 µg of pRL-TK were cotransfected into 1.5 × 106 THP-1 cells. In some transfections, 0.2 µg of pMEK1-DN, pUSEamp, phnRNP-K or pcDNA3 were added as indicated. Cells were allowed to recover for 16 h post-transfection in fresh media before exposure to the various conditions tested in each experiment. LUC activities were subsequently measured using the Dual-Luciferase reporter assay system (Promega, Madison, WI) while reporter gene mRNA levels were quantified using real-time RT–PCR (see below). LUC activities and LUC mRNA levels were normalized to Renilla luciferase expressed by co-transfected pRL-TK (Promega, Madison, WI) to adjust transfection efficiency.
Real-time RT–PCR
TaqMan® real-time RT–PCR (ABI, Rockville, MD) was employed to quantify mRNA levels. Gene specific probes and PCR primers for GAPDH, MAP3K7IP2, MRPS18A (mitochondrial ribosomal protein S18A) and TP53BP2 (tumor protein p53 binding protein 2) were purchased from ABI (Foster City, CA). Probes and primers of LUC and Renilla luciferase were designed by us and synthesized through ABI. The sequences were as follows: LUC probe (5′-CATTTCGCAGCCTACCGTGGTGTTC-3′) and primers (5′-AACGTGAATTGCTCAACAGTATGG-3′ and 5′-TTTGCAACCCCTTTTTGGAA-3′); and Renilla luciferase probe (5′-CCTGATTTGCCCATACCAATAAGGTCTGG-3′) and primers (5′-AGCCAGTAGCGCGGTGTATT-3′ and 5′-TCAAGTAACCTATAAGAACCATTACCAGATT-3′). The High-capacity cDNA Archive kit (ABI, Foster City, CA) was employed to prepare cDNA from 2 µg of total RNA. Resulting cDNA was used for RT–PCR in triplicate according to the standard ABI protocol. The target mRNA of MAP3K7IP2, MRPS18A and TP53BP2 were normalized to GAPDH. The LUC mRNA were normalized to Renilla luciferase mRNA.
RNA electrophoretic mobility shift assays (REMSA)
Synthetic consensus CURE probe (5′-CUUUCUCCCCCACCCUCUUCUCCCCCUUCCCCCUCCCC-3′) and MAP3K7IP2 CURE (5′-AGACUCCGUCUCUACAGAAGGUUUUGAA-3′) were labeled with biotin-N4-CTP using Biotin 3′ end Labeling Kit (Pierce, Rockford, IL). Cytoplamic fractions were extracted using Nu-CLEAR™ extraction kit (Sigma–Aldrich. St. Louis, MO). Labeled CURE probe (0.5 pmol) was incubated with cytoplasmic protein (30 µg) in binding buffer [15 mM HEPES (pH 7.4), 10 mM KCl, 5 mM MgCl2, 5% glycerol and 1 mM DTT] for 20 min at room temperature. To prevent nonspecific binding, yeast tRNA (final concentration, 0.1 mg/ml) was added. RNA–protein complexes were separated in 6% polyacrylamide gel with 0.5× Tris/boric acid/EDTA buffer, transfered on to nylon membrane and detected using the LightShiftTM Chemiluminescent EMSA Kit (Pierce, Rockford, IL). In competition experiments, a 100-fold molar excess of unlabeled consensus CURE, or its mutant (5′-CUUUAGAGAAGACACAGAAGAAGAAGAAGACACAGACC-3′) or MAP3K7IP2 CURE, were added to the incubation mixture. For antibody supershift assays, 2 µg of specific polyclonal antibody or control goat serum were preincubated with cytoplasmic proteins for 20 min at room temperature prior to addition of labeled CURE probes.
Bioinformatics and data analysis
Affymetrix MAS5 signal values and present call results were stored in the NIHLIMS, a database for storage and retrieval of Affymetrix GeneChip data in use at the NIH. This entire microarray dataset were also submitted to GEO repository (GSE4228). Data were retrieved and analyzed using the MSCL Analyst's Toolbox (http://abs.cit.nih.gov/MSCLtoolbox/) and the JMP statistical software package (SAS, Inc, Cary, NC; http://www.jmp.com). Data were first normalized to the 97th percentile, a value corresponding to the expression level of the 678th most intense probeset on the array. This normalization strategy assumed that the most intense probesets corresponded to mRNA species which were most stable and were generally unaffected by the treatments studied here. Then logarithmically transformed normalized data were subject to linear regression with respect to the four time points studied (0, 45, 90, 180 min following the start of incubation with GSH or GSNO), to estimate a slope corresponding to a first-order decay rate. As expected, the distribution of slope values across probesets showed a long negative tail, corresponding to genes which decayed over the time period studied. The decay slope was calculated for each probeset, for each of the four conditions (GSH, GSNO, SB/GSH and SB/GSNO) using an Analysis of Covariance (ANCOVA), constraining the time 0 expression value to be identical for the pair of conditions without SB and the pair with SB, as necessitated by the design of the experiment. Further, since the experiment was replicated in four distinct batches, a blocked ANCOVA was utilized. A specialized Toolbox script, ANCOVAbatch, was written for this purpose. The analysis results were then used to select genes which decayed, and whose decay rate changed following treatment. The P-value for a one-way, four level ANCOVA was calculated and used to compute a false discovery rate (FDR) (40). A total of 238 probesets with the lowest P-values were selected, corresponding to a FDR of 10%. Probesets were annotated identifying 220 unique transcripts based on information presented by Affymetrix at the web-site http://www.affymetrix.com/analysis/index.affx as of April 12, 2004.
Mean signal intensities of the 220 identified transcripts across four independent experiments were computed and normalized using the 0 min values for all conditions and time points. These normalized mean signal intensities were hierarchically clustered (Figure 1A) using the complete linkage algorithm in JMP (SAS Institute, Cary, NC). For Figure 1B, mRNA decay slopes for every identified gene in different conditions were hierarchically clustered with an average linkage algorithm and the centered Pearson correlation coefficient as the similarity metric using HCE (Hierarchical Clustering Explorer 2.0 beta, available at http://www.cs.umd.edu/hcil/hce/) (41).
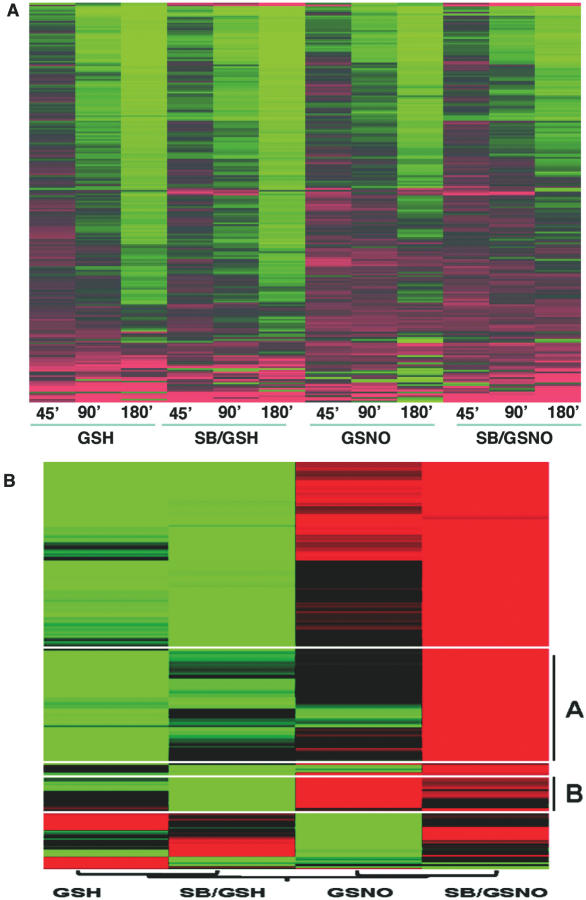
Figure 1.
Heat maps; effects of NO• and the p38 MAPK inhibitor SB202190 (SB) on mRNA degradation as determined by microarray. THP-1 cells (2 × 107) were stimulated with LPS (1 µg/ml) for 4 h. After 30 min treatment with ActD (2.5 µg/ml) in the absence or presence of SB (0.1 µM), cells were incubated with GSNO (400 µM) or control GSH (400 µM) for 0–180 min. At the indicated time-points, cells were harvested to extract total RNA for microarray analysis. The half-lives of 220 genes were found to be differentially regulated (see Materials and Methods). (A) Hierarchical clustering of normalized mean signal intensities from four independent experiments for all 220 genes at each time point and condition. (B) Same results as (A) after conversion of individual time point data into slopes based on a first order mRNA decay model.
MAP3K7IP2, MRPS18A and TP53BP2 mRNA levels over time were analyzed using a two-way ANOVA (the first factor was time, the second factor was treatment) followed by post-hoc tests. Luciferase mRNA level and activity were analyzed using paired t-tests to compare different reporter gene constructs and experimental treatments.
To find putative CURE cis-acting elements in 3′-UTR, the mRNA RefSeq for the 220 genes identified by microarray were downloaded from the NCBI nucleotide database (http://www.ncbi.nlm.nih.gov/), and then each sequence was scanned using the UTRScan database (http://www.ba.itb.cnr.it/BIG/UTRScan/) (42) or MacVector™. The ARE-containing transcripts of the 220 genes were found through searching the ARE database (http://rc.kfshrc.edu.sa/ared/) (4). Fisher's exact test for 2 × 2 contingency tables was used to compare proportions of ARE- and CURE-containing mRNAs between different gene clusters.
RESULTS
NO• regulation of mRNA stability
NO• regulates degradation of IL-8 and p21/Waf1/Cip1 mRNA through the activation of p38 MAPK (24,26). Here, NO• stabilization of mRNA was explored in LPS-stimulated human THP-1 cells using oligonucleotide microarrays (Affymetrx U133A Genechips®). As shown in the Supplementary Table, 220 genes were identified whose mRNA stabilities were significantly regulated by NO•, p38 MAPK inhibitor SB or NO• plus SB. A heat map of expression levels for these 220 genes, arranged by hierarchical clustering (Figure 1A), demonstrated degradation of mRNA over time within each condition (shift from red to green). Also visible in this figure, NO• generally increased overall mRNA stability. Note that the shift from red to green is less pronounced for the NO• and NO•/SB conditions. Unexpectedly, the addition of SB to NO• enhanced rather than antagonized NO•-mediated mRNA stabilization for many of these transcripts, while SB in the absence of NO• had no or only a modest effect. This effect is more obvious in a clustered heat map that condensed expression levels at the four time points into a single mRNA decay rate (Figure 1B). Here, compared to the GSH control without or with SB, GSNO generally stabilized mRNA (shift from green to red). Again, this effect of NO• was further augmented by SB for many of these transcripts, of which 60 genes (cluster A) were also stabilized by SB alone. Antagonism of NO•-mediated effects by SB as would be expected for p38 MAPK stabilized transcripts was seen for only 20 genes (cluster B). The rest of this investigation focused on identifying a mechanism of NO•-mediated mRNA stabilization that could be enhanced by p38 MAPK inhibition.
Activation of p38 and Erk1/2 by NO•
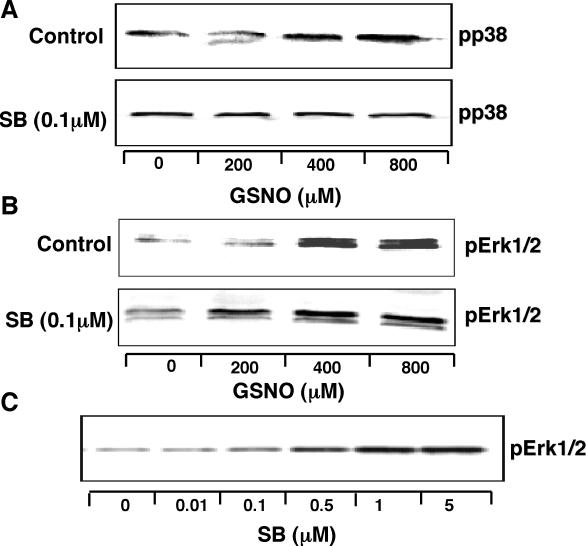
Consistent with our previous findings in THP-1 cells, GSNO activated p38 MAPK in dose-dependent manner (Figure 2A, upper panel) and the p38 inhibitor SB blocked this effect (Figure 2A, lower panel). GSNO also dose-dependently activated Erk1/2 as shown in the upper panel of Figure 2B. Interestingly, this effect of GSNO on Erk1/2 was further enhanced by the p38 MAPK inhibitor SB (Figure 2B, the lower panel). In addition, SB alone dose-dependently activated Erk1/2 in THP-1 cells, though this effect was barely detectable at the dose of SB used here in our microarray experiments (Figure 2C). As expected, neither GSNO nor SB changed total p38 MAPK or total Erk1/2 levels (data not shown).
Figure 2.
Effects of NO• and the p38 MAPK inhibitor SB202190 (SB) on MAPK phosphorylation. (A) NO• increases p38 MAPK phosphorylation, an effect blocked by SB (0.1 µM). (B) NO• increases Erk1/2 phosphorylation, an effect enhanced by SB (0.1 µM). (C) SB (0–5 µM) alone increases Erk1/2 phosphorylation. THP-1 cells (1 × 107) were stimulated with LPS (1 µg/ml) for 4 h. After 30 min treatment with ActD (2.5 µg/ml) in the absence (control) or presence of SB, cells were incubated without or with GSNO (0–800 µM) for another 30 min, as indicated and then lysed. Each experiment was repeated at least twice with similar results.
NO• stabilization of mRNA through Erk1/2
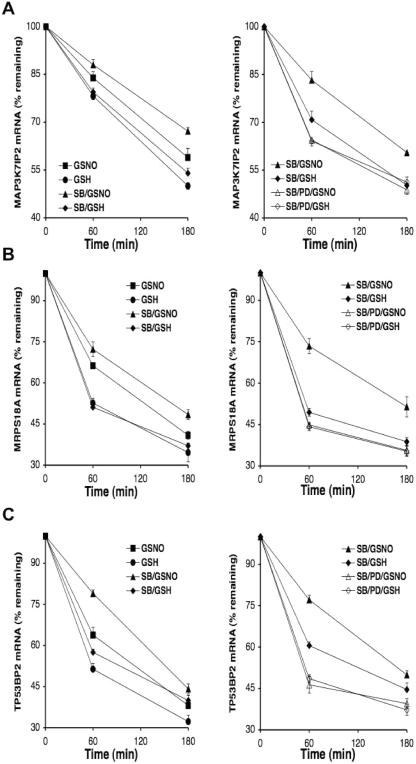
Enhanced NO• activation of Erk1/2 in the presence of SB (Figure 2B) might explain the additive effect of these agents in stabilizing the mRNA transcripts identified by microarray. In order to test whether Erk1/2 activation may be an important signaling pathway involved in NO•-induced mRNA stabilization, we investigated in detail MAP3K7IP2, MRPS18A and TP53BP2, three prototypic genes from cluster A, using real-time RT–PCR. In our microarray experiment, these transcripts were stabilized by NO• and the addition of SB further enhanced this effect. Consistent with these results, RT–PCR (Figure 3A–C; left panels) demonstrated that GSNO significantly stabilized the mRNA of MAP3K7IP2, MRPS18A and TP53BP2 compared with the GSH control (P < 0.005 for all); the p38 MAPK inhibitor SB further enhanced the effect of NO• (P < 0.009 for all). Treatment with the specific Erk1/2 inhibitor PD prior to GSNO incubation abolished the effects of NO• and SB on the mRNA stability of these three transcripts (Figure 3A–C; right panels). These results suggested that Erk1/2 activation is involved in both the NO•-mediated mRNA stabilization of MAP3K7IP2, MRPS18A and TP53BP2 and the enhancing effects of SB.
Figure 3.
NO• stabilizes (A) MAP3K7IP2, (B) MRPS18A and (C) TP53BP2 mRNA through Erk1/2 as determined by RT–PCR. Left panels show the effects of NO• and the p38 MAPK inhibitor, SB202190 (SB; 0.1 µM), on mRNA degradation. Right panels show the effects of the Erk1/2 inhibitor, PD98059 (PD; 30 µM), on mRNA degradation in the presence of SB. THP-1 cells (2 × 107) were stimulated with LPS (1 µg/ml) for 4 h. After 30 min treatment with transcription inhibitor ActD (2.5 µg/ml) in the absence or presence of indicated MAPK inhibitors, cells were incubated with GSNO (400 µM) or GSH control (400 µM) for 0–180 min. All mRNA levels were quantitated by TaqMan® RT–PCR and normalized to GADPH mRNA. Data, presented as percentage relative to mRNA levels at 0 min, are the mean ± SEM of three independent experiments. The respective mRNA half-lives of MAP3K7IP2, MRPS18A and TP53BP2 were as follows: 179, 98 and 91 min for control GSH; 236, 132 and 121 min for GSNO; 200, 103 and 119 min for SB/GSH; 314, 166 and 155 min for SB/GSNO; 171, 89 and 100 min for SB/PD/GSH; and 160, 90 and 103 min for SB/PD/GSNO.
Cis-acting elements mediating NO•-MAPK regulation of mRNA stability
3′-UTR elements play pivotal roles in mRNA stabilization (3,5,6). The regulation of mRNA decay by p38 MAPK has been reported to be dependent on ARE within 3′-UTR (19–23,27). We recently found that mRNAs containing ARE are over-represented among NO•-regulated genes in differentiated U937 cells (26). Consistent with these results, 65% (13/20) of transcripts that were NO• stabilized and p38 MAPK inhibitor destabilize (cluster B; Figure 1B and Table 1) contained ARE compared to only 31 of the remaining 200 transcripts (15.5%, P < 0.001) identified by microarray. An in depth investigation of ARE functionality in mRNA decay regulated by NO•-MAPK signaling is being conducted in a separate study.
Table 1.
Cluster B: mRNA transcripts stabilized by NO• through p38 MAPK activation
| Probe set ID | RefSeq | Symbol | Gene name |
|---|---|---|---|
| 201281_at | NM_007002 | ADRM1 | Adhesion regulating molecule 1 |
| 202518_at | NM_001707 | BCL7B | B-cell CLL/lymphoma 7B |
| 205780_at | NM_001197 | BIKa | BCL2-interacting killer (apoptosis-inducing) |
| 205114_s_at | NM_002983 | CCL3a | Chemokine (C–C motif) ligand 3-like, centromeric |
| 204103_at | NM_002984 | CCL4a | Chemokine (C–C motif) ligand 4 |
| 210046_s_at | NM_002168 | IDH2a | Isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| 203064_s_at | NM_004514 | ILF1 | Forkhead box K2 |
| 201627_s_at | NM_005542 | INSIG1a | Insulin induced gene 1 |
| 201285_at | NM_013446 | MKRN1a | Makorin, ring finger protein, 1 |
| 208620_at | NM_006196 | PCBP1a | Poly(rC) binding protein 1 |
| 212100_s_at | NM_032311 | PDIP46 | Polymerase delta interacting protein 3 |
| 209533_s_at | NM_004253 | PLAAa | Phospholipase A2-activating protein |
| 204958_at | NM_004073 | PLK3a | Polo-like kinase 3 (Drosophila) |
| 208361_s_at | NM_001722 | POLR3Da | Pol III (DNA directed) polypeptide D, 44 kDa |
| 209158_s_at | NM_004228 | PSCD2a | Pleckstrin-like, Sec7 and coiled-coil domains 2 |
| 210573_s_at | NM_006468 | RPC62 | Pol III (DNA directed) polypeptide C (62 kDa) |
| 58696_at | NM_019037 | RRP41 | Exosome component 4 |
| 213330_s_at | NM_006819 | STIP1a | Stress-induced-phosphoprotein 1 |
| 203112_s_at | NM_005663 | WHSC2a | Wolf–Hirschhorn syndrome candidate 2 |
| 209428_s_at | NM_006782 | ZFPL1 | Zinc finger protein-like 1 |
aWith ARE in 3′-UTR.
Since the Erk1/2 signaling pathway has been shown to regulate DICE-containing mRNA transcripts (16), we specifically looked for DICE-like sequences in the 3′-UTRs of all 220 genes identified by micorarray using UTRScan database (42). As shown in the Supplementary Table, 64 of 220 genes were found to have identifiable DICE-like sequences, that here, we call CURE to distinguish them from DICE which has multiple CURE repeats. Of these 64 putative CURE-containing genes, 28 were among the 60 genes in cluster A (46.7%, Figure 1B and Table 2) that were additively stabilized by NO• and SB. In contrast, only 36 of the remaining genes (22.5%, P < 0.001) similarly contained potential CURE sequences. MAP3K7IP2, MRPS18A and TP53BP2 mRNA, shown here by RT–PCR to be stabilized by NO• through Erk1/2 activation, were among the 28 CURE-containing mRNAs in cluster A. The CURE sequence found within the 3′-UTR of MAP3K7IP2, MRPS18A and TP53BP2 through the UTRScan database is CUCCGUCUCUACAGAAG, UCCCAUCCUCUUCATG and CCAGUCCUCCUGCCAGAAAG, respectively.
Table 2.
Cluster A: mRNA transcripts stabilized by NO• through Erk1/2 activation
| Probe set ID | RefSeq | Symbol | Gene name |
|---|---|---|---|
| 221492_s_at | NM_022488 | APG3a | APG3 autophagy 3-like (Saccharomyces cerevisiae) |
| 202511_s_at | AK001899 | APG5Lb | APG5 autophagy 5-like (S.cerevisiae) |
| 215411_s_at | AL008730 | C6orf4 | Chromosome 6 open reading frame 4 |
| 212711_at | NM_015447 | CAMSAP1 | Calmodulin regulated spectrin-associated protein 1 |
| 205379_at | NM_001236 | CBR3 | Carbonyl reductase 3 |
| 209056_s_at | NM_001253 | CDC5L | CDC5 cell division cycle 5-like (Schizosaccharomyces pombe) |
| 203721_s_at | NM_016001 | CGI-48 | CGI-48 protein |
| 203044_at | NM_014918 | CHSY1a,b | Carbohydrate (chondroitin) synthase 1 |
| 212180_at | NM_005207 | CRKLb | V-crk sarcoma virus CT10 oncogene avian-like |
| 218648_at | NM_022769 | CRTC3 | CREB regulated transcription coactivator 3 |
| 207614_s_at | NM_003592 | CUL1b | Cullin 1 |
| 201371_s_at | NM_003590 | CUL3a | Cullin 3 |
| 202703_at | NM_003584 | DUSP11b | Dual specificity phosphatase 11 |
| 213848_at | NM_001947 | DUSP7b | Dual specificity phosphatase 7 |
| 202776_at | NM_014597 | ERBP | Estrogen receptor binding protein |
| 202949_s_at | NM_001450 | FHL2 | Four and a half LIM domains 2 |
| 219083_at | NM_018130 | FLJ10539b | Hypothetical protein FLJ10539 |
| 219933_at | NM_016066 | GLRX2 | Glutaredoxin 2 |
| 217957_at | NM_013242 | GTL3b | Likely ortholog of mouse gene trap locus 3 |
| 219484_at | NM_013320 | HCF-2 | Host cell factor C2 |
| 217965_s_at | NM_013260 | HCNGPb | Transcriptional regulator protein |
| 218603_at | NM_016217 | HECAa,b | Headcase homolog (Drosophila) |
| 218946_at | NM_015700 | HIRIP5 | HIRA interacting protein 5 |
| 205526_s_at | NM_007044 | KATNA1 | Katanin p60 (ATPase-containing) subunit A 1 |
| 202417_at | NM_012289 | KEAP1b | Kelch-like ECH-associated protein 1 |
| 203702_s_at | NM_014640 | KIAA0173 | Tubulin tyrosine ligase-like family, member 4 |
| 212846_at | NM_015056 | KIAA0179b | KIAA0179 |
| 203322_at | NM_014913 | KIAA0863b | KIAA0863 protein |
| 200650_s_at | NM_005566 | LDHAb | Lactate dehydrogenase A |
| 219631_at | NM_024937 | LRP12 | Low density lipoprotein-related protein 12 |
| 212184_s_at | NM_145342 | MAP3K7IP2b | MAPK kinase kinase 7 interacting protein 2 |
| 202484_s_at | NM_015832 | MBD2b | Methyl-CpG binding domain protein 2 |
| 219348_at | NM_018467 | MDS032 | Hematopoietic stem cells protein MDS032 |
| 219406_at | NM_024097 | MGC955b | Hypothetical protein MGC955 |
| 218385_at | NM_018135 | MRPS18Ab | Mitochondrial ribosomal protein S18A |
| 201829_at | NM_005863 | NET1a | Neuroepithelial cell transforming gene 1 |
| 218889_at | NM_022451 | NOC3Lb | Nucleolar complex associated 3 homolog |
| 204441_s_at | NM_002689 | POLA2b | Polymerase (DNA-directed), alpha (70 kDa) |
| 207830_s_at | NM_002713 | PPP1R8b | Protein phosphatase 1, regulatory subunit 8 |
| 201934_at | NM_025222 | PRO2730b | Hypothetical protein PRO2730 |
| 203401_at | NM_002765 | PRPS2b | Phosphoribosyl pyrophosphate synthetase 2 |
| 212296_at | NM_005805 | PSMD14 | Proteasome 26S subunit, non-ATPase, 14 |
| 202990_at | NM_002863 | PYGL | Phosphorylase, glycogen |
| 200833_s_at | NM_015646 | RAP1B | RAP1B, member of RAS oncogene family |
| 218535_s_at | NM_018343 | RIOK2a | RIO kinase 2 (yeast) |
| 218016_s_at | NM_018119 | RPC5 | Pol III (DNA directed) polypeptide E (80 kDa) |
| 212018_s_at | NM_015659 | RSL1D1 | Homo sapiens ribosomal L1 domain containing 1 |
| 218137_s_at | NM_021940 | SMAP1b | Stromal membrane-associated protein 1 |
| 210053_at | NM_006951 | TAF5 | TAF5 Pol II, TBP-associated factor, 100 kDa |
| 213301_x_at | NM_015905 | TIF1 | Transcriptional intermediary factor 1 |
| 218118_s_at | NM_006327 | TIMM23 | Translocase of mitochondrial membrane 23 homolog |
| 202633_at | NM_007027 | TOPBP1a | Topoisomerase (DNA) II binding protein 1 |
| 203120_at | NM_005426 | TP53BP2b | Tumor protein p53 binding protein, 2 |
| 218855_at | NM_016372 | TPRA40b | Seven transmembrane domain orphan receptor |
| 212544_at | NM_004773 | TRIP3 | Thyroid hormone receptor interactor 3 |
| 202413_s_at | NM_003368 | USP1 | Ubiquitin specific protease 1 |
| 218806_s_at | NM_006113 | VAV3b | Vav 3 oncogene |
| 210275_s_at | NM_006007 | ZNF216 | Zinc finger, A20 domain containing 2 |
| 209944_at | NM_021188 | ZNF410b | Zinc finger protein 410 |
| 213097_s_at | AI338837 | ZRF1 | Zuotin related factor 1 |
aWith ARE in 3′-UTR.
bWith CURE in 3′-UTR.
NO•-Erk1/2-CURE regulation of mRNA stability and translation using a reporter gene-CURE construct
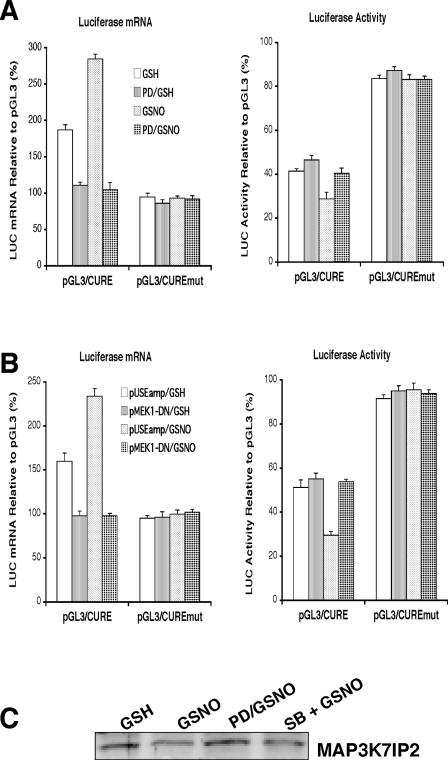
Since CURE-containing mRNAs were over represented among transcripts that were additively stabilized by NO• and SB, the possible role of CURE in transducing this response was investigated. Inserting a 38 nt consensus CURE, identified from DICE-containing mRNAs (14) or its antisense form into the 3′-UTR of a LUC reporter gene, we generated pGL3/CURE and its mutant pGL3/CUREmut (See Materials and Methods). The responsiveness of these constructs to NO• was then examined after transfection into THP-1 cells. As shown in Figure 4A, wild-type pGL3/CURE, but not pGL3/CUREmut, increased LUC mRNA levels to 180% of control pGL3 values. The NO• donor GSNO further elevated pGL3/CURE mRNA to 280% of control values, but had no effect on pGL3/CUREmut (Figure 4A). NO•-induced LUC mRNA elevations seen with pGL3/CURE were abolished by the Erk1/2 inhibitor PD (Figure 4A; P < 0.005). This result suggested that Erk1/2 mediated NO• stabilization of CURE-containing mRNA transcripts.
Figure 4.
NO• stabilizes CURE-containing mRNA but inhibits its translation through an Erk1/2-dependent mechanism. (A) Effect of the Erk1/2 inhibitor, PD98059 (PD; 30 µM), on LUC mRNA levels and LUC activity, respectively. THP-1 cells, transfected with pGL3/CURE, mutant pGL3/CUREmut or control pGL3, were treated with ActD (2.5 µg/ml) for 30 min (for mRNA determinations only) and then incubated with GSH (400 µM) or GSNO (400 µM) for 5 h to measure LUC mRNA by TaqMan® RT–PCR or for 20 h to measure LUC activity. (B) Effect of a Mek1 dominant-negative mutant on LUC mRNA levels and LUC activity, respectively. THP-1 cells, co-transfected with pGL3/CURE or mutant pGL3/CUREmut or control pGL3 plus either pUSEamp (empty vector) or pMEK1-DN (dominant-negative Mek1), were similarly treated as in A for measurement of LUC mRNA levels and LUC activity. Data, presented as percentage relative to LUC mRNA level or LUC activity of pGL3, are the mean ± SEM of three to six independent experiments. (C) Effect of NO• on the expression of MAP3K7IP2, a naturally-occurring, CURE-containing gene. THP-1 cells (1 × 107) were pretreated with SB (0.1 µM) or PD (30 µM) for 30 min. After 20 h incubation of GSH (400 µM) or GSNO (400 µM), cells were then lysed for western blotting. Each experiment was repeated twice with similar results.
Despite NO•-induced stabilization of LUC mRNA containing CURE sequence, LUC expression as measured by its activity was significantly decreased by CURE (pGL3/CURE versus pGL3/CUREmut: P < 0.005) and further reduced by NO• (GSNO versus GSH for pGL3/CURE: P < 0.04; Figure 4A). This inhibitory effect of NO• on LUC translation in pGL3/CURE transfected cells was blocked by the Erk1/2 inhibitor PD (P < 0.01; Figure 4A). Although pGL3/CUREmut also demonstrated a slightly lower LUC activity relative to pGL3 control, neither NO• nor PD altered it (Figure 4A). These data suggested that the NO•-Erk1/2 signaling stabilizes mRNA, but represses translation of genes that harbor these DICE-like, CURE sequences. In support of this conclusion, co-transfection of the plasmid pMEK1-DN, encoding a dominant-negative mutant of Mek1, but not its control empty vector pUSEamp, functioned like the Erk1/2 inhibitor PD, abrogating the ability of NO• to stabilize LUC mRNA or repress its translation (Figure 4B) in cells transfected with the pGL3/CURE construct. Mek1 is the upstream kinase that activates Erk1/2 (39).
Finally, we tested whether NO•-Erk1/2 signaling represses protein expression of a natural CURE-containing mRNA, MAP3K7IP2. This transcript was stabilized by NO• plus SB in our microarray study, a finding confirmed by RT–PCR. Importantly, RT–PCR also demonstrated that PD, an Erk1/2 inhibitor, abolished the effects of NO• and SB on the stability of MAP3K7IP2 mRNA. As shown in Figure 4C, GSNO decreased the expression of MAP3K7IP2 protein; this effect of NO• was enhanced by SB but inhibited by the Erk1/2 inhibitor PD.
Trans-factors associated with the regulation of mRNA stabilization and translation by NO•-Erk1/2-CURE
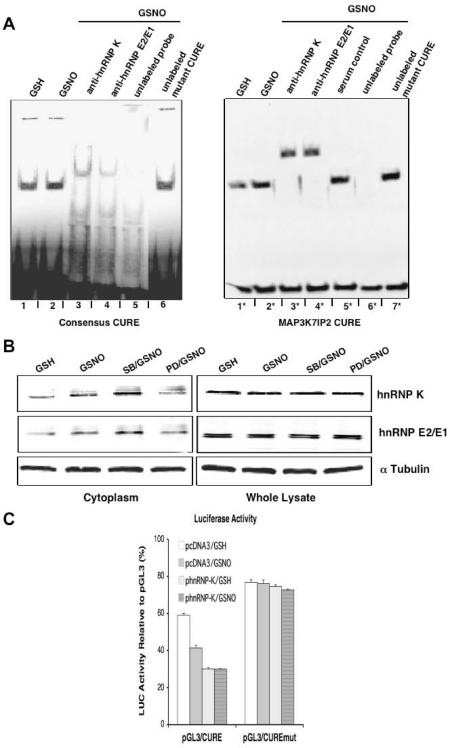
Trans-factors hnRNP K and hnRNP E2/E1 are major DICE-binding proteins that have recently been implicated in mRNA stabilization (14,15) or translation silencing (13,16,17). To assess the role of these proteins in regulating the stabilization and translation of CURE-containing mRNA by NO•-Erk1/2 signaling, we performed REMSA using two biotin-labeled riboprobes. One probe contained the consensus CURE sequence, and another contained the 3′ MAP3K7IP2 CURE sequence (Figure 5A). In Figure 5A, one major RNA–protein complex was formed by incubating THP-1 cytoplasmic extracts with either of the labeled riboprobes. The specificity of this complex was confirmed by competition in which unlabeled consensus CURE (Figure 5A, lane 5) or MAP3K7IP2 CURE (Figure 5A, lane 6*) prevented formation of this complex. Mutated consensus CURE (Figure 5A, lanes 6 and 7*) showed no effect on the complex. Antibodies against hnRNP K (Figure 5A, lanes 3 and 3*) and hnRNP E2/E1 (Figure 5A, lanes 4 and 4*), but not control serum (Figure 5A, lane 5*), super-shifted the CURE–protein complex, suggesting that both hnRNP K and E2/E1 proteins are present and specifically bind to CURE mRNA in THP-1 cells. NO• was found to induce the formation of this complex (Figure 5A, lane 2 versus 1 and lane 2* versus 1*) and also to increase the cytoplasmic accumulation of both hnRNP K and hnRNP E2/E1 without changing their overall expression (Figure 5B). Like effects seen on gene regulation, NO•-induced cytoplasmic accumulation of these proteins was enhanced by the p38 MAPK inhibitor SB, but inhibited by the Erk1/2 inhibitor PD (Figure 5B). These results suggest that hnRNP K and hnRNP E2/E1 respond to NO•-Erk1/2 signal transduction by accumulating in the cytoplasm where they differentially regulate the stability and translation of transcripts containing CURE. In support of this concept, cytoplasmic over-expression of hnRNP K was found to repress the translation of LUC linked to a 3′-UTR containing CURE (phnRNP-K/GSH versus pcDNA3/GSH in pGL3/CURE transfected cells; P < 0.001) and to mitigate the repressive effect of NO• (Figure 5C).
Figure 5.
Role of hnRNP K and hnRNP E2/E1 in NO•-Erk1/2-CURE signaling. (A) RNA REMSAs with either a consensus (left panel) or MAP3K7IP2 (right panel) CURE riboprobes. GSNO (400 µM) treatment for 3 h increased complex formation compared to control GSH; anti-hnRNP K and anti-hnRNP E2/E1 both super-shift the complex; the unlabeled CURE riboprobes, but not the mutant of consensus CURE (mutant CURE) compete with the labeled CURE riboprobes. (B) Translocation of hnRNP K and hnRNP E2/E1 to the cytoplasm by western blotting. GSNO (400 µM) treatment for 3 h increased the presence of hnRNP K and hnRNP E2/E1 in the cytoplasm but not in whole-cell lysates compared to control GSH. This effect was further enhanced by the p38 MAPK inhibitor SB202190 (SB; 0.1 µM), but blocked by the Erk1/2 inhibitor PD98059 (PD; 30 µM). A control protein α tubulin is shown for comparison. Experiments in (A and B) were repeated at least twice with similar results. (C) Overexpression of hnRNP K mimicked the effect of NO•, repressing the expression of a chimeric LUC-CURE reporter gene. THP-1 cells were co-transfected with pGL3/CURE, pGL3/CUREmut or control pGL3 and pcDNA3 (empty vector) or phnRNP-K (hnRNP K expression plasmid). After treatment with GSH (400 µM) or GSNO (400 µM) for 20 h, LUC activities were measured. Data, presented as percentage relative to LUC activity with pGL3, are the mean ± SEM of three independent experiments.
DISSCUSION
Transcripts containing ARE sites in their 3′-UTR have been shown to be stabilized by activation of p38 MAPK (19,22–24,27). Our previous work demonstrated that NO• stabilizes IL-8 and p21/Waf1/Cip1 mRNA through this mechanism (24,26). Likewise here, NO• was again shown to activate p38 MAPK and to stabilize 20 transcripts that were then destabilized by SB, a p38 MAPK inhibitor. Consistent with our previous findings in differentiated U937 cells (26), ARE-containing mRNAs were over-represented among these NO• up-regulated transcripts; 13 of 20 (65%) have ARE in their 3′-UTR. Three of these, CCL3, CCL4 (chemokine ligand 3 and 4) and PLAA (phospholipase A2-activating protein), were previously reported to be stabilized through p38 MAPK activation by other investigators (22).
However, in human THP-1 cells pre-stimulated with LPS, a strong activator of p38 MAPK, most NO•-stabilized mRNAs had their half-lives further extended by SB, an unexpected finding that we sought to explain. Associated with these microarray results, NO• was found to activate Erk1/2 in addition to p38 MAPK, an effect that was enhanced by SB. This pattern of response and previous reports linking Erk1/2 signaling with post-transcriptional regulatory events (16,43–45) suggested a candidate mechanism. First, three genes that strongly displayed the characteristics of interest, MAP3K7IP2,MRPS18A and TP53BP2, were chosen to confirm the microarray results and to test for any potential connection with Erk1/2 signaling. By RT–PCR, SB enhanced, while PD, a specific Erk1/2 inhibitor, was shown to block NO• effects on the mRNA stability of MAP3K7IP2, MRPS18A and TP53BP2. NO• activation of both p38 MAPK and Erk1/2, observed here in THP-1 cells, has also been reported in Jurkat T cells (46,47), but the underlying molecular mechanisms still need to be defined. Notably, previous work has shown that p38 MAPK activation can negatively regulated Erk1/2 in other cell types (48,49). Therefore, the p38 MAPK inhibitor SB potentially may enhance NO• activation of Erk1/2 by blocking this inhibitory effect. Alternatively, SB could activate Erk1/2 through effects on up-stream c-Raf kinase (50), though p38 MAPK modulation of Erk1/2 signaling seems more likely.
In agreement with our data, several groups have demonstrated a major role for Erk1/2 in stabilizing the mRNA of amyloid precursor protein (51), granulocyte–macrophage colony-stimulating factor (44), tristetraprolin (43), nucleolin (52) and transforming growth factor β 1 (45). ARE sites and the RNA-binding proteins HuR and tristetraprolin (44), or a short C/U region and the RNA-binding proteins nucleolin and hnRNP C (51) were variably proposed to mediate these Erk1/2 effects. Of the 60 genes in cluster A that were stabilized by NO• and SB here, only 7 (11.7%) have ARE, but 28 (46.7%) have identifiable CURE in their 3′-UTRs. Of the remaining 160 genes, 37 (23.1%) have ARE, but only 36 (22.5%) have CURE, indicating CUREs but not AREs are over-represented in cluster A. This suggests that CURE may be the main cis-element that transduces the mRNA stabilizing effect of Erk1/2. In further support of this, insertion of a consensus CURE sequence, identified from several DICE-containing mRNAs (14), but not its antisense form into 3′-UTR of a LUC reporter gene significantly stabilized its mRNA. NO• further increased the mRNA half-life of this hybrid transcript and the Erk1/2 inhibitor PD or expression of Mek1 dominant-negative mutant both abolished this NO• stabilizing effect.
In concurrent experiments that measured LUC activities instead of mRNA levels, NO•-Erk1/2 signaling was demonstrated to inhibit, rather than enhance, the translation of stabilized, chimeric CURE-containing mRNA. Consistently, NO•-Erk1/2 signaling was further shown to repress protein expression of a natural CURE-containing mRNA, MAP3K7IP2, as measured by western blotting. As discussed earlier, MAP3K7IP2 mRNA was stabilized by NO• through Erk1/2 activation, as demonstrated by RT–PCR. So, NO• activation of Erk1/2 in THP-1 cells stabilized CURE-containing mRNAs but inhibited their translation. These patterns of response, either mRNA stabilization or decreased translation, have been previously reported separately for several DICE-containing genes including α-globin, α-collagen and lipoxygenase (13,14,16,53).
Trans-factors binding to consensus CURE and MAP3K7IP2 CURE were identified by REMSAs performed with cell cytoplasm. NO•-Erk1/2 signaling was shown to increase the cytoplasmic accumulation of hnRNP K and hnRNP E2/E1 and to enhance the formation of a major complex between these proteins and CURE probes. The role of hnRNP K was then confirmed by hnRNP K over-expression which repressed the translation of CURE-containing mRNA, thereby mimicking the effects of NO•. Phosphorylation of hnRNP K by Erk1/2 is known to prompt its cytoplasmic accumulation (16).
Ubiquitously expressed, hnRNP K and hnRNP E2/E1 have triple KH (hnRNP K homology) domains and CURE-binding specificity (54). By binding to CURE, interacting with poly(A)-binding protein, and then blocking deadenylation, these KH domain proteins have been shown to stabilize the mRNA of α-globin (14,54,55), renin (15) and α1 collagen (53). In contrast to their well-characterized ability to stabilize mRNA, hnRNP K and hnRNP E2/E1 have also been shown to repress translation of 15-lipoxygenase (12,13,18) and C/EBPα (17) by blocking 80S ribosome assembly at the AUG initiation site (12,13,17,18). Phorsphorylation of hnRNP K by Erk1/2 increases (16), while phosphorylation by c-Src kinase decreases its repressive effect on 15-lipoxygenase translation (18). Prior data demonstrated that the KH domain complex contributes to the stability of translationally active as well as translationally blocked α-globin mRNA (56,57).
In the present investigation, we have shown that NO• activates Erk1/2 in LPS-stimulated THP-1 cells and subsequently affects hnRNP K and hnRNP E2/E1 stabilization of CURE-containing mRNA transcripts while also suppressing their translation. Although hnRNP K-induced translational repression of 15-lipoxygenase was thought to be dependent on a highly repetitive CU-rich motif previously referred to as DICE (58), the cis-element that transduces NO•-Erk1/2 signaling here appears to correspond to a much shorter CURE sequence. CURE is related to a CU-rich consensus sequence (CUUUCUCCCCCACCCUCUUCUCCCCCUUCCCCCUCCCC; the underlined nucleotides represent core sequences) that was derived from four DICE-containing mRNAs, namely α-globin, α1 collagen, 15-lipoxygenase and tyrosine hydroxylase (14). However, consistent with the short CURE concept, hnRNP E2/E1-induced C/EBPα repression has been ascribed to a more simple CUCCCCC sequence (17), and the 3′-UTR CURE sequence of MAP3K7IP2 (CUCCGUCUCUACAGAAG), MRPS18A (UCCCAUCCUCUUCATG) and TP53BP2 (CCAGUCCUCCUGCCAGAAAG) described here are also relatively short. Whether NO•-Erk1/2 signaling uniformly stabilizes CURE-containing mRNAs while suppressing their translation cannot be determined from the current study. More likely, under other cellular conditions and through interactions with additional cis-acting elements and their binding proteins (such as ARE and HuR or CUGBP2), a variety of responses may be possible.
Accumulating evidence shows that mRNA turnover is linked to translation and these two post-transcriptional processes may be regulated in the same or opposite directions (2,11,24,26,56,57). For rapidly degraded ARE-containing transcripts, including cytokines like IL-8 and cell cycle genes like p21/Waf1/Cip1, mRNA stabilization by NO•-p38 MAPK signaling appears to result in increased protein production (24,26). Conversely, stress-related stabilization of other ARE-containing mRNAs by specific RNA-binding proteins, such as TIAR, HuR and CUGBP2 is associated with translation inhibition (11,59,60). Likewise, a recent microarray study of the endoplasmic reticulum stress response linked global mRNA stabilization to widespread translational repression (2). Adding to these previous reports, our data provide another example of mRNA stabilization coupled to translational inhibition, implicating a CURE cis-acting sequence and the RNA-binding proteins hnRNP K and hnRNP E2/E1. NO• regulates this process by activating Erk1/2, which stabilizes mRNA and represses translation by inducing the formation of a mRNA–protein complex. NO•-Erk1/2-hnRNP-CURE represents a novel mechanism for post-transcriptional regulation that can function as both a mRNA stabilizer and translation inhibitor.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
This research was supported by the Intramural Research Program of the National Institutes of Health, Clinical Center. Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health, Clinical Center.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fan J., Yang X., Wang W., Wood W.H., IIIrd, Becker K.G., Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc. Natl Acad. Sci. USA. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawai T., Fan J., Mazan-Mamczarz K., Gorospe M. Global mRNA stabilization preferentially linked to translational repression during the endoplasmic reticulum stress response. Mol. Cell Biol. 2004;24:6773–6787. doi: 10.1128/MCB.24.15.6773-6787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen C.Y., Shyu A.B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 4.Bakheet T., Williams B.R., Khabar K.S. ARED 2.0: an update of AU-rich element mRNA database. Nucleic Acids Res. 2003;31:421–423. doi: 10.1093/nar/gkg023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shyu A.B., Belasco J.G., Greenberg M.E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 6.Chen C.Y., Gherzi R., Ong S.E., Chan E.L., Raijmakers R., Pruijn G.J., Stoecklin G., Moroni C., Mann M., Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 7.Fan X.C., Steitz J.A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carballo E., Lai W.S., Blackshear P.J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 9.Lai W.S., Carballo E., Strum J.R., Kennington E.A., Phillips R.S., Blackshear P.J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raineri I., Wegmueller D., Gross B., Certa U., Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukhopadhyay D., Houchen C.W., Kennedy S., Dieckgraefe B.K., Anant S. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 12.Ostareck D.H., Ostareck-Lederer A., Shatsky I.N., Hentze M.W. Lipoxygenase mRNA silencing in erythroid differentiation: the 3′-UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- 13.Ostareck D.H., Ostareck-Lederer A., Wilm M., Thiele B.J., Mann M., Hentze M.W. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- 14.Holcik M., Liebhaber S.A. Four highly stable eukaryotic mRNAs assemble 3′-untranslated region RNA–protein complexes sharing cis and trans components. Proc. Natl Acad. Sci. USA. 1997;94:2410–2414. doi: 10.1073/pnas.94.6.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skalweit A., Doller A., Huth A., Kahne T., Persson P.B., Thiele B.J. Posttranscriptional control of renin synthesis: identification of proteins interacting with renin mRNA 3′-untranslated region. Circ. Res. 2003;92:419–427. doi: 10.1161/01.RES.0000059300.67152.4E. [DOI] [PubMed] [Google Scholar]
- 16.Habelhah H., Shah K., Huang L., Ostareck-Lederer A., Burlingame A.L., Shokat K.M., Hentze M.W., Ronai Z. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nature Cell Biol. 2001;3:325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 17.Perrotti D., Cesi V., Trotta R., Guerzoni C., Santilli G., Campbell K., Iervolino A., Condorelli F., Gambacorti-Passerini C., Caligiuri M.A., et al. BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nature Genet. 2002;30:48–58. doi: 10.1038/ng791. [DOI] [PubMed] [Google Scholar]
- 18.Ostareck-Lederer A., Ostareck D.H., Cans C., Neubauer G., Bomsztyk K., Superti-Furga G., Hentze M.W. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell Biol. 2002;22:4535–4543. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahtani K.R., Brook M., Dean J.L., Sully G., Saklatvala J., Clark A.R. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell Biol. 2001;21:6461–6469. doi: 10.1128/MCB.21.9.6461-6469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carballo E., Cao H., Lai W.S., Kennington E.A., Campbell D., Blackshear P.J. Decreased sensitivity of tristetraprolin-deficient cells to p38 inhibitors suggests the involvement of tristetraprolin in the p38 signaling pathway. J. Biol. Chem. 2001;276:42580–42587. doi: 10.1074/jbc.M104953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ming X.F., Stoecklin G., Lu M., Looser R., Moroni C. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol. Cell Biol. 2001;21:5778–5789. doi: 10.1128/MCB.21.17.5778-5789.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frevel M.A., Bakheet T., Silva A.M., Hissong J.G., Khabar K.S., Williams B.R. p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol. Cell Biol. 2003;23:425–436. doi: 10.1128/MCB.23.2.425-436.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lasa M., Mahtani K.R., Finch A., Brewer G., Saklatvala J., Clark A.R. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol. Cell Biol. 2000;20:4265–4274. doi: 10.1128/mcb.20.12.4265-4274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma P., Cui X., Wang S., Zhang J., Nishanian E.V., Wang W., Wesley R.A., Danner R.L. Nitric oxide post-transcriptionally up-regulates LPS-induced IL-8 expression through p38 MAPK activation. J. Leukoc. Biol. 2004;76:278–287. doi: 10.1189/jlb.1203653. [DOI] [PubMed] [Google Scholar]
- 25.Pages G., Berra E., Milanini J., Levy A.P., Pouyssegur J. Stress-activated protein kinases (JNK and p38/HOG) are essential for vascular endothelial growth factor mRNA stability. J. Biol. Chem. 2000;275:26484–26491. doi: 10.1074/jbc.M002104200. [DOI] [PubMed] [Google Scholar]
- 26.Cui X., Zhang J., Ma P., Myers D., Goldberg I.G., Sittler K.J., Barb J.J., Munson P.J., Del Pilar Cintron A., McCoy J.P., et al. cGMP-independent nitric oxide signaling and regulation of the cell cycle. BMC Genomics. 2005;6:151. doi: 10.1186/1471-2164-6-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winzen R., Kracht M., Ritter B., Wilhelm A., Chen C.Y., Shyu A.B., Muller M., Gaestel M., Resch K., Holtmann H. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999;18:4969–4980. doi: 10.1093/emboj/18.18.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang J., Wang S., Wesley R.A., Danner R.L. Adjacent sequence controls the response polarity of nitric oxide-sensitive Sp factor binding sites. J. Biol. Chem. 2003;278:29192–29200. doi: 10.1074/jbc.M213043200. [DOI] [PubMed] [Google Scholar]
- 29.Wang S., Wang W., Wesley R.A., Danner R.L. A Sp1 binding site of the tumor necrosis factor alpha promoter functions as a nitric oxide response element. J. Biol. Chem. 1999;274:33190–33193. doi: 10.1074/jbc.274.47.33190. [DOI] [PubMed] [Google Scholar]
- 30.Spiecker M., Peng H.B., Liao J.K. Inhibition of endothelial vascular cell adhesion molecule-1 expression by nitric oxide involves the induction and nuclear translocation of IkappaBalpha. J. Biol. Chem. 1997;272:30969–30974. doi: 10.1074/jbc.272.49.30969. [DOI] [PubMed] [Google Scholar]
- 31.Pilz R.B., Suhasini M., Idriss S., Meinkoth J.L., Boss G.R. Nitric oxide and cGMP analogs activate transcription from AP-1-responsive promoters in mammalian cells. FASEB J. 1995;9:552–558. doi: 10.1096/fasebj.9.7.7737465. [DOI] [PubMed] [Google Scholar]
- 32.Berendji D., Kolb-Bachofen V., Zipfel P.F., Skerka C., Carlberg C., Kroncke K.D. Zinc finger transcription factors as molecular targets for nitric oxide-mediated immunosuppression: inhibition of IL-2 gene expression in murine lymphocytes. Mol. Med. 1999;5:721–730. [PMC free article] [PubMed] [Google Scholar]
- 33.Kimura H., Weisz A., Kurashima Y., Hashimoto K., Ogura T., D'Acquisto F., Addeo R., Makuuchi M., Esumi H. Hypoxia response element of the human vascular endothelial growth factor gene mediates transcriptional regulation by nitric oxide: control of hypoxia-inducible factor-1 activity by nitric oxide. Blood. 2000;95:189–197. [PubMed] [Google Scholar]
- 34.Bouton C., Demple B. Nitric oxide-inducible expression of heme oxygenase-1 in human cells. Translation-independent stabilization of the mRNA and evidence for direct action of nitric oxide. J. Biol. Chem. 2000;275:32688–32693. doi: 10.1074/jbc.275.42.32688. [DOI] [PubMed] [Google Scholar]
- 35.Wei J., Guo H., Kuo P.C. Endotoxin-stimulated nitric oxide production inhibits expression of cytochrome c oxidase in ANA-1 murine macrophages. J. Immunol. 2002;168:4721–4727. doi: 10.4049/jimmunol.168.9.4721. [DOI] [PubMed] [Google Scholar]
- 36.Ryu S.D., Kang J.H., Yi H.G., Nahm C.H., Park C.S. Hepatic flavin-containing monooxygenase activity attenuated by cGMP-independent nitric oxide-mediated mRNA destabilization. Biochem. Biophys. Res. Commun. 2004;324:409–416. doi: 10.1016/j.bbrc.2004.09.065. [DOI] [PubMed] [Google Scholar]
- 37.Abdelaziz N., Colombo F., Mercier I., Calderone A. Nitric oxide attenuates the expression of transforming growth factor-beta(3) mRNA in rat cardiac fibroblasts via destabilization. Hypertension. 2001;38:261–266. doi: 10.1161/01.hyp.38.2.261. [DOI] [PubMed] [Google Scholar]
- 38.Akool el,S., Kleinert H., Hamada F.M., Abdelwahab M.H., Forstermann U., Pfeilschifter J., Eberhardt W. Nitric oxide increases the decay of matrix metalloproteinase 9 mRNA by inhibiting the expression of mRNA-stabilizing factor HuR. Mol. Cell Biol. 2003;23:4901–4916. doi: 10.1128/MCB.23.14.4901-4916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeo S.J., Gravis D., Yoon J.G., Yi A.K. Myeloid differentiation factor 88-dependent transcriptional regulation of cyclooxygenase-2 expression by CpG DNA: role of NF-kappaB and p38. J. Biol. Chem. 2003;278:22563–22573. doi: 10.1074/jbc.M302076200. [DOI] [PubMed] [Google Scholar]
- 40.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 41.Seo J., Shneiderman B. Understanding hierarchical clustering results by interactive exploration of dendrograms: a case study with genomic microarray data. IEEE Comput. 2002;35:80–86. [Google Scholar]
- 42.Mignone F., Grillo G., Licciulli F., Iacono M., Liuni S., Kersey P.J., Duarte J., Saccone C., Pesole G. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brooks S.A., Connolly J.E., Rigby W.F. The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. J. Immunol. 2004;172:7263–7271. doi: 10.4049/jimmunol.172.12.7263. [DOI] [PubMed] [Google Scholar]
- 44.Esnault S., Malter J.S. Extracellular signal-regulated kinase mediates granulocyte-macrophage colony-stimulating factor messenger RNA stabilization in tumor necrosis factor-alpha plus fibronectin-activated peripheral blood eosinophils. Blood. 2002;99:4048–4052. doi: 10.1182/blood.v99.11.4048. [DOI] [PubMed] [Google Scholar]
- 45.Sullivan D.E., Ferris M., Pociask D., Brody A.R. Tumor necrosis factor-alpha induces transforming growth factor-beta1 expression in lung fibroblasts through the extracellular signal-regulated kinase pathway. Am. J. Respir. Cell. Mol. Biol. 2005;32:342–349. doi: 10.1165/rcmb.2004-0288OC. [DOI] [PubMed] [Google Scholar]
- 46.Lander H.M., Hajjar D.P., Hempstead B.L., Mirza U.A., Chait B.T., Campbell S., Quilliam L.A. A molecular redox switch on p21(ras). Structural basis for the nitric oxide-p21(ras) interaction. J. Biol. Chem. 1997;272:4323–4326. doi: 10.1074/jbc.272.7.4323. [DOI] [PubMed] [Google Scholar]
- 47.Lander H.M., Jacovina A.T., Davis R.J., Tauras J.M. Differential activation of mitogen-activated protein kinases by nitric oxide-related species. J. Biol. Chem. 1996;271:19705–19709. doi: 10.1074/jbc.271.33.19705. [DOI] [PubMed] [Google Scholar]
- 48.Singh R.P., Dhawan P., Golden C., Kapoor G.S., Mehta K.D. One-way cross-talk between p38(MAPK) and p42/44(MAPK). Inhibition of p38(MAPK) induces low density lipoprotein receptor expression through activation of the p42/44(MAPK) cascade. J. Biol. Chem. 1999;274:19593–19600. doi: 10.1074/jbc.274.28.19593. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H., Shi X., Hampong M., Blanis L., Pelech S. Stress-induced inhibition of ERK1 and ERK2 by direct interaction with p38 MAP kinase. J. Biol. Chem. 2001;276:6905–6908. doi: 10.1074/jbc.C000917200. [DOI] [PubMed] [Google Scholar]
- 50.Numazawa S., Watabe M., Nishimura S., Kurosawa M., Izuno M., Yoshida T. Regulation of ERK-mediated signal transduction by p38 MAP kinase in human monocytic THP-1 cells. J. Biochem. (Tokyo) 2003;133:599–605. doi: 10.1093/jb/mvg077. [DOI] [PubMed] [Google Scholar]
- 51.Westmark C.J., Malter J.S. Extracellular-regulated kinase controls beta-amyloid precursor protein mRNA decay. Brain Res. Mol. Brain Res. 2001;90:193–201. doi: 10.1016/s0169-328x(01)00112-7. [DOI] [PubMed] [Google Scholar]
- 52.Westmark C.J., Malter J.S. Up-regulation of nucleolin mRNA and protein in peripheral blood mononuclear cells by extracellular-regulated kinase. J. Biol. Chem. 2001;276:1119–1126. doi: 10.1074/jbc.M009435200. [DOI] [PubMed] [Google Scholar]
- 53.Lindquist J.N., Parsons C.J., Stefanovic B., Brenner D.A. Regulation of alpha1(I) collagen messenger RNA decay by interactions with alphaCP at the 3′-untranslated region. J. Biol. Chem. 2004;279:23822–23829. doi: 10.1074/jbc.M314060200. [DOI] [PubMed] [Google Scholar]
- 54.Kong J., Ji X., Liebhaber S.A. The KH-domain protein alpha CP has a direct role in mRNA stabilization independent of its cognate binding site. Mol. Cell Biol. 2003;23:1125–1134. doi: 10.1128/MCB.23.4.1125-1134.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z., Day N., Trifillis P., Kiledjian M. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ji X., Kong J., Liebhaber S.A. In vivo association of the stability control protein alphaCP with actively translating mRNAs. Mol. Cell Biol. 2003;23:899–907. doi: 10.1128/MCB.23.3.899-907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiss I.M., Liebhaber S.A. Erythroid cell-specific mRNA stability elements in the alpha 2-globin 3′ nontranslated region. Mol. Cell Biol. 1995;15:2457–2465. doi: 10.1128/mcb.15.5.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reimann I., Huth A., Thiele H., Thiele B.J. Suppression of 15-lipoxygenase synthesis by hnRNP E1 is dependent on repetitive nature of LOX mRNA 3′-UTR control element DICE. J. Mol. Biol. 2002;315:965–974. doi: 10.1006/jmbi.2001.5315. [DOI] [PubMed] [Google Scholar]
- 59.Kedersha N., Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 2002;30:963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 60.Katsanou V., Papadaki O., Milatos S., Blackshear P.J., Anderson P., Kollias G., Kontoyiannis D.L. HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell. 2005;19:777–789. doi: 10.1016/j.molcel.2005.08.007. [DOI] [PubMed] [Google Scholar]