Abstract
Bacteriophage Mu uses non-replicative transposition for integration into the host's chromosome and replicative transposition for phage propagation. Biochemical and structural comparisons together with evolutionary considerations suggest that the Mu transposition machinery might share functional similarities with machineries of the systems that are known to employ a hairpin intermediate during the catalytic steps of transposition. Model transposon end DNA hairpin substrates were used in a minimal-component in vitro system to study their proficiency to promote Mu transpososome assembly and subsequent MuA-catalyzed chemical reactions leading to the strand transfer product. MuA indeed was able to assemble hairpin substrates into a catalytically competent transpososome, open the hairpin ends and accurately join the opened ends to the target DNA. The hairpin opening and transposon end cleavage reactions had identical metal ion preferences, indicating similar conformations within the catalytic center for these reactions. Hairpin length influenced transpososome assembly as well as catalysis: longer loops were more efficient in these respects. In general, MuA's proficiency to utilize different types of hairpin substrates indicates a certain degree of flexibility within the transposition machinery core. Overall, the results suggest that non-replicative and replicative transposition systems may structurally and evolutionarily be more closely linked than anticipated previously.
INTRODUCTION
A multitude of mobile genetic elements use the reactions of transpositional DNA recombination for movement within and between the genomes of their host organisms (1). Depending on small differences in the details of the reaction mechanisms, dictated primarily by the structure–function characteristics and regulation of the responsible machineries, the outcome of DNA transposition can be either replicative or non-replicative by nature (2). Of particular importance are the ways the element-encoded transposase or integrase proteins catalyze DNA cleavage and joining reactions (3,4).
Transposases constitute a group of structurally and functionally related proteins with distinct domains responsible for separate subfunctions (5,6). The most important units include a catalytic core and a DNA-binding domain responsible for transposon end recognition. Other domains may provide functions for interactions with auxiliary proteins and accessory DNA sites. Transposases function as a multimer within the context of a specific protein–DNA complex, a transpososome and catalyze two common reactions that unite transposition pathways: (i) a pair of initial cleavages at the transposon ends (typically at the 3′ ends but in some systems at the 5′ ends, e.g. see Hermes transposition below) and (ii) a pair of strand transfer reactions, in which the liberated transposon 3′ ends are joined to the target DNA (7). At least in the two known cases, Mu and Tn5, where assembly of the transpososome is a prerequisite for the chemical steps, the catalysis occurs in trans: i.e. a transposase protomer that is bound to one transposon end via its transposon end-binding domain catalyzes cleavage and strand transfer at the other end of the transposon (8–13). A majority of transposases belong to a family of DDE transposases and retroviral integrases, referring to an active site that contains a triad of catalytically important amino acids, supposedly involved in the coordination of divalent metal ions (6).
Transposing bacteriophage Mu is a widely studied model for DNA transposition in general (14). During the lytic phage propagation, Mu replicates via replicative transposition (15), and this pathway is relatively well characterized (14). Conversely, the outcome of the reaction pathway that leads to the initial Mu integration into the host's chromosome (following phage infection) is distinctively non-replicative (16–19), but details of this latter pathway remain to be elucidated.
In general, Mu transposition proceeds within a protein–DNA complex called the Mu transpososome (13,14,20,21). This functional unit forms via an elaborate assembly pathway and synapses the transposon ends. In its core, it contains a tetramer of the critical catalytic protein, MuA (75 kDa monomer), a well-characterized member of the DDE-transposase/integrase family. A number of studies [reviewed by (14)] indicate that the reactions catalyzed by MuA lead to a branched DNA structure, also known as the Shapiro intermediate, that subsequently is processed by host-encoded replication or repair factors, leading to a cointegrate formation or simple insertion, respectively. In both cases, a 5 bp target site duplication is generated. In addition to MuA, a number of cofactors and certain DNA topology are important for Mu propagation in vivo [reviewed by (14)]. The cofactors include specific MuA-binding sites within the phage genome as well as the Mu-encoded target selector protein, MuB. Important host-encoded functions are provided by DNA bending proteins, HU and IHF, as well as certain protein remodeling and DNA replication factors.
Biochemical properties of MuA can be studied effectively by a minimal-component in vitro transposition reaction that includes MuA and a short Mu end-specific DNA segment as the only macromolecular components (22). The reaction faithfully reproduces transpososome assembly, donor cleavage and strand transfer steps; and it has been used for detailed analyses of DNA substrates with variable configuration or sequence (22–27).
Studies of transposon Tn10 and Tn5 systems have revealed a mechanism that uses a transposon end DNA hairpin intermediate during non-replicative transposition (28,29). Between the 3′ end cleavage and strand transfer, the transposases of Tn10 and Tn5 catalyze two additional reaction steps: hairpin formation via the attack of the transposon end 3′-OH group on the opposite strand of the duplex and hairpin opening by hydrolysis that regenerates the 3′-OH residue. Hairpinning of DNA has also been described in related systems such as V(D)J recombination and Hermes transposition, but in these cases the hairpins are formed to seal the flanking DNA ends (30,31).
The evolutionary history of transposable elements must be reflected in their encoded transposition machineries. Yet, although distinct similarities unite a variety of DNA transposition systems, the structural and functional relationships among different elements that use different strategies are unclear. In particular, it is not known when and how different transposases have adapted or lost the catalytic potential for DNA hairpinning. In principle, it might be possible to detect transposases in transition, i.e. retained or potentially emerging activities might be revealed by a detailed analysis. We addressed these issues directly by assembling Mu transpososomes with preformed model hairpin substrates and analyzed the catalytic activities of the ensuing complexes. The Mu transpososome was proficient to accommodate and process a variety of different hairpin substrates, indicating a degree of flexibility within the catalytic machinery. Such flexibility within the active site may be a general determinant that influences or has influenced the choice between the transposition pathways among a variety of transposons.
MATERIALS AND METHODS
Bacteria, proteins and reagents
Escherichia coli strain DH5α (Invitrogen) was used as a cloning host and for plasmid propagation. Bacteria were grown in Luria–Bertani (LB) broth or on LB agar plates (32). To select for plasmid maintenance, the media were supplemented with appropriate antibiotics: ampicillin (Ap, 100 µg/ml) and chloramphenicol (Cm, 10 µg/ml). T4 DNA polymerase was from Promega. T4 polynucleotide kinase (PNK) and T4 DNA ligase were from New England Biolabs. Deoxynucleotides and Dynazyme EXT DNA polymerase preparation were from Finnzymes (Espoo, Finland). BSA was from Sigma, ATP from Roche, and [γ-33P]ATP (1000–3000 Ci/mmol) from GE Healthcare. The commercial enzymes were used under recommended reaction conditions. MuA protein variants were overexpressed and purified as described elsewhere (33). MuB protein was a gift from Dr Kiyoshi Mizuuchi (National Institutes of Health, Bethesda). This protein preparation was made according to Chaconas et al. (34) with the additional step to remove aggregated protein, as described by Adzuma and Mizuuchi (35).
Oligonucleotides, DNA and DNA techniques
The oligonucleotides (Supplementary Data) were obtained from commercial sources and purified by urea–PAGE (32) prior to use. When required, the purified oligonucleotides were labeled at their 5′ end with T4 PNK and [γ-33P]ATP (32). Unincorporated nucleotides were removed by sequential phenol and chloroform extractions, passage through a spin column (Bio-Spin 30; Bio-Rad), and ethanol precipitation. To generate double-stranded donor DNA substrates for in vitro transposition reactions, appropriate oligonucleotides were annealed (intramolecularly and/or intermolecularly, see Figure 1) in TEN-buffer (10 mM Tris–HCl, pH 7.5, 0.5 mM EDTA and 50 mM NaCl) as described elsewhere (22). The specific activities of donor DNA substrates were scaled to an equal level of radioactivity by adjustment with corresponding unlabeled substrates. Plasmid DNA was prepared and PCR products were purified using appropriate Qiagen kits. The transposition target plasmid pUC19 (New England Biolabs) was additionally purified by CsCl gradient centrifugation (32) to enrich supercoiled plasmid forms. Standard DNA techniques were performed as described elsewhere (32). DNA sequencing was done at the sequencing service unit of the Institute of Biotechnology, University of Helsinki.
Figure 1.
Donor DNA substrates. (A) Substrate schematic representation. All the Mu-specific substrates contain ∼50 bp, starting from the Mu R-end and including the MuA-binding sites R1 and R2 (22). These substrates also include a flanking DNA region, which can be in duplex DNA, linear single strand or in a hairpin loop configuration. Some substrates contain nucleotide substitutions close to the transposon end or within the loop region [see (B)]. The depicted 16 bp flanking sequence has been shown earlier to support efficient catalysis by MuA in an in vitro model substrate assay (22), and the standard loop sequences are derived from that same sequence by joining the endmost Mu-specific 3′-adenine to various nucleotide positions in the opposite strand of the duplex. The Mu end cleavage point is indicated by a solid arrow and the position of the radioactive label by an asterisk. The open arrow shows the position of the nick present in most substrates [see (B)]. (B) Substrate characteristics. The donor substrates were made by annealing one or two oligonucleotides to form double-stranded species as indicated. Most of the hairpin substrates were generated from two oligonucleotides, and these substrates therefore contain a nick [within the R1 MuA-binding site, see (A)]. This nick does not interfere with in vitro transposition reactions as indicated by a direct comparison of reactions with the nicked and corresponding unnicked substrate (compare lanes 4 and 12 in Figure 5). The endmost nucleotides of the transposon DNA are shown in bold, and flanking nucleotides are indicated in regular font. The rHP4 substrate includes a randomly chosen non-Mu sequence with two conserved base pairs mimicking the transposon end.
In vitro transposition reaction and analysis of reaction products
The standard in vitro transposition reaction was performed in three stages. Initially, transpososomes were assembled for 1 h at 30°C in the absence of target DNA and divalent metal ions (a sample was withdrawn at this stage for the analysis of protein–DNA complexes, see below). Next, target DNA (1 µl of 220 ng/µl stock) was added to the reaction mixture (23 µl) and the incubation was continued for 10 min to allow target capture by transpososomes. To activate catalysis, MgCl2 (1 µl of 250 mM stock) was then added, and the incubation was continued for the times indicated. At this final incubation stage, a standard (1-fold, 1×) reaction (25 µl) contained 50 nM 33P-labeled donor DNA fragment, 116 nM transposase (MuA or MuAE392Q as indicated), 25 mM Tris–HCl, pH 8.0, 100 µg/ml BSA, 15% (w/v) glycerol, 15% (v/v) dimethyl sulfoxide, 0.05% (w/v) Triton X-100, and 126 mM NaCl, 220 ng pUC19 DNA (target DNA) and 10 mM MgCl2. The reactions were stopped by freezing in liquid nitrogen prior to the analysis of reaction products (see below). For some reactions (3-fold scaled up), three times more donor DNA (150 nM) as well as transposase (348 nM) were used. When indicated, reactions also contained 0.3 µg MuB and 2 mM ATP, both added following a 60 min assembly step, 10 min prior to target addition. In metal ion analysis, MgCl2 was substituted in certain reactions with an equivalent concentration of CaCl2 or MnCl2.
The detection of protein–DNA complexes has been described in detail elsewhere (22). Briefly, competitor DNA to trap loosely bound MuA and 0.2 vol of Ficoll (25%, Pharmacia) were added to the samples prior to gel loading and analysis on a native 2% Metaphor agarose (Cambrex) gel. To detect strand transfer products involving a plasmid target, 0.2 vol of loading dye (0.1% bromophenol blue, 2.5% SDS, 50 mM EDTA and 25% Ficoll 400) was added to the samples prior to gel loading and analysis on a 1.5% LE agarose gel (Promega) in 1× TAE buffer (32) at 5.3 V/cm for 2 h. Agarose gels were dried onto DEAE (Whatman DE-81) paper for autoradiography. Hairpin opening products were analyzed by denaturing 7 M urea–10% PAGE and autoradiography as described elsewhere (36).
Analysis of donor–target junction sequences of transposition reaction products
A 3-fold scaled up transposition reaction was performed with a 3 h final incubation time for catalysis, using HP7 as a donor DNA fragment and pUC19 as a target DNA. The reaction was diluted 1:20 with water, and 1 µl of the diluted mixture was then used as a template for PCR amplification. In the PCR strategy used, the exposed target plasmid 3′-OH groups within the transposition reaction product are initially used for elongation towards the transposon DNA. Following this initial phase, a single transposon-specific primer amplifies a duplex that contains a stretch of transposon DNA attached to each end of the linearized pUC19 target. Thirty cycles of PCR were performed with Dynazyme EXT DNA polymerase as specified by the supplier, using the Mu end-specific primer HSP6. The resulting 2.8. kb PCR product was purified, treated with T4 DNA polymerase in the presence of dNTPs to generate blunt ends (32), and ligated into a DNA fragment containing a selectable marker gene for chloramphenicol resistance. This fragment, the 1.1 kb BamHI fragment of cat-Mu artificial transposon (37), was also treated with T4 DNA polymerase and dNTPs prior to ligation. Ligation products were electrotransformed (38) into DH5α cells, and bacteria were selected for antibiotic resistance on LB-Ap-Cm plates. Plasmid DNA was isolated from 11 clones, and the target sequences flanking transposon DNA were determined using the marker fragment-specific primers HSP350 and HSP349.
RESULTS
MuA catalyzes hairpin processing
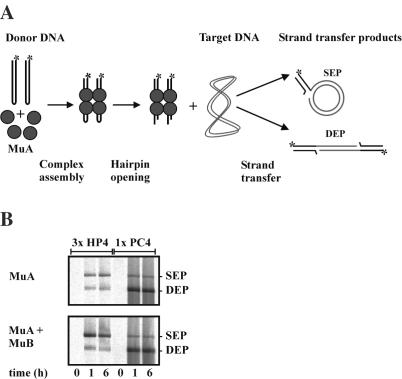
DNA hairpin processing by a transposase can be studied in vitro by assembling transpososomes with synthetic DNA model substrates that contain hairpin ends and critical transposase binding sites, as shown for Tn10 and Tn5 (28,29). To elucidate whether the Mu transposition machinery is able to accommodate and process analogous hairpin molecules, we generated a number of ∼50 bp Mu R-end model substrates (Figure 1) and used them as a radiolabeled donor DNA in an in vitro transposition assay, the plasmid pUC19 serving as a target DNA (Materials and Methods). Following dissociation of transpososomes by SDS-containing loading buffer, products of this coupled donor cleavage and strand transfer reaction were analyzed by agarose gel electrophoresis and visualized by autoradiography (Figure 2). Two species of reaction products involving the target plasmid are expected in this assay (Figure 2A): the double-ended reaction product (DEP) and the single-ended reaction product (SEP), resulting from utilization of one or two donor molecules, respectively (23–27).
Figure 2.
MuA transposase catalyzes the processing of model DNA hairpin substrates. (A) Reaction schematic representation. As with standard uncleaved and precleaved model substrates (22), model hairpin substrates and MuA protein assemble the tetrameric Mu transpososome that catalyzes the subsequent reactions. Upon addition of MgCl2, MuA first catalyzes a hydrolytic strand cleavage reaction (hairpin opening) and then executes a strand transfer reaction, in which the target becomes joined to transposon DNA. The end products include circular or linear target molecules, depending on whether one or two transposon ends become joined to the target, respectively. The radiolabel included in the donor DNA fragment will be incorporated in the DNA product, providing an easy assay for catalysis (23–27). (B) Coupled cleavage and strand transfer reaction. HP4 substrate (and PC4 for control) was incubated in the presence of MuA (upper panel), or MuA and MuB (lower panel) for 1–6 h. The strand transfer reaction products (SEPs and DEPs) were analyzed by agarose gel electrophoresis and autoradiography. The unreacted donor substrates that migrate close to the bottom of the gel are not shown. To increase the yield of reaction products, 3-fold scaled up (3×) reactions were used with the HP4 donor, whereas PC4 reactions were 1-fold (1×) for convenient identification of reaction products (Materials and Methods).
Initially, we analyzed MuA-catalyzed hairpin processing using the substrate HP4, which contains four unpaired hairpin nucleotides (Figure 2B). The precut substrate PC4 served as a positive control for identifiable reaction products. We also included MuB in this experiment due to its stimulatory effect(s) in various types of in vitro assays [(14,23,26,39) and references therein]. The hairpin substrate generated the expected reaction product species (SEPs and DEPs) but much less efficiently than the precut control substrate. In addition, the control substrate yielded predominantly DEPs, whereas the hairpin donor produced more SEPs than DEPs. The amount of reaction products did not increase after 1 h incubation, suggesting that the assembly of complexes probably was a limiting factor in the reaction. Addition of MuB to the hairpin substrate reaction increased formation of reaction products to some extent. With the hairpin substrate (data not shown), no reaction products were observed in the absence of MuA, or if the wild-type MuA was replaced by an active-site mutant MuAE392Q (33) defective in catalysis. These data indicate that MuA can catalyze a DNA hairpin cleavage reaction and the subsequent strand transfer step of transposition.
Influence of divalent metal ions
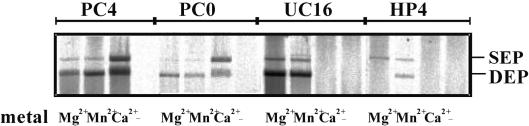
In Mu transposition, donor DNA cleavage and strand transfer require the presence of an appropriate divalent metal ion. While Mg2+ and Mn2+ can support both the cleavage and strand transfer reactions, Ca2+ can support only strand transfer (22). To investigate the divalent metal ion requirements of MuA-catalyzed hairpin processing, we performed transposition reactions with the hairpin donor DNA substrate HP4 in the presence of Mg2+, Mn2+ or Ca2+ (Figure 3). Control reactions included an uncleaved substrate (UC16) as well as two precleaved substrates with different transposon end configurations: a 4 nt 5′-overhang (PC4) and a blunt end (PC0). All three divalent metal ions supported strand transfer with the precut donors, and the uncleaved donor generated reaction products in the presence of Mg2+ and Mn2+. With the hairpin substrate, detectable amounts of reaction products were generated with Mg2+ and Mn2+ but, similar to uncleaved donor, no products were generated with Ca2+, suggesting that the hairpin opening stage of hairpin processing probably mimics the donor cleavage step. With the hairpin substrate, Mn2+ generated more DEPs than SEPs, whereas Mg2+ produced mainly SEPs (also see Discussion).
Figure 3.
Utilization of various metal ions for the catalysis of hairpin processing. In vitro transposition reactions were performed with the indicated substrates by first assembling complexes in the absence of metal ions and then initiating reaction chemistry by adding MgCl2, MnCl2 or CaCl2 (Materials and Methods). The reaction products, SEPs and DEPs, were analyzed by agarose gel electrophoresis and autoradiography following a 3 h incubation in the presence of metal ion.
Hairpin processing takes place within the Mu transpososome
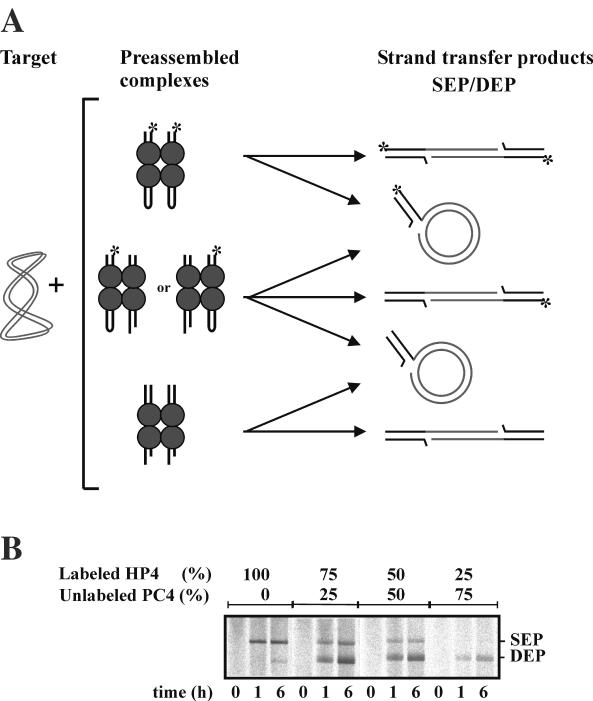
To study whether hairpin processing takes place within the Mu transpososome, we generated mixed transpososomes (Figure 4A) that contained a labeled hairpin donor DNA (HP4) in combination with unlabeled precut donor DNA (PC4). In these mixed complexes, the precut transposon end is readily available for strand transfer, whereas the hairpin end needs to be opened prior to strand transfer. If both precut and hairpin ends are utilized within a context of a single mixed complex, we should see the relative increase in the generation of labeled DEPs over SEPs in an experiment where increasingly more unlabeled precleaved substrate is used to substitute the labeled hairpin substrate. Analysis of reaction products from such an experiment verified this prediction: the relative amount of labeled DEPs increased upon substitution of increasing amounts of unlabeled precut donor DNA (Figure 4B), indicating that critical reactions involved in hairpin processing must take place within the Mu transpososome.
Figure 4.
Reactions of mixed transpososomes. (A) Schematic representation of mixed complex analysis. When unlabeled precleaved substrate (PC4) and 5′-labeled hairpin substrate (HP4) are mixed, four types of transpososomes will be assembled to generate strand transfer products. Only those reaction products containing the label originating from the use of the HP4 donor will be detectable by autoradiography. (B) Analysis of reaction products of mixed HP4 and PC4 donor complexes. Labeled HP4 and unlabeled PC4 donor substrates were used in the indicated ratios to assemble mixed complexes in standard 1-fold in vitro transposition reactions. Transpososomes were first assembled without divalent metal ions for 1 h, after which MgCl2 was added (Materials and Methods), and samples were withdrawn after incubation for 0, 1 and 6 h. Strand transfer products (DEPs and SEPs) were analyzed by agarose gel electrophoresis and autoradiography.
Effects of hairpin loop length
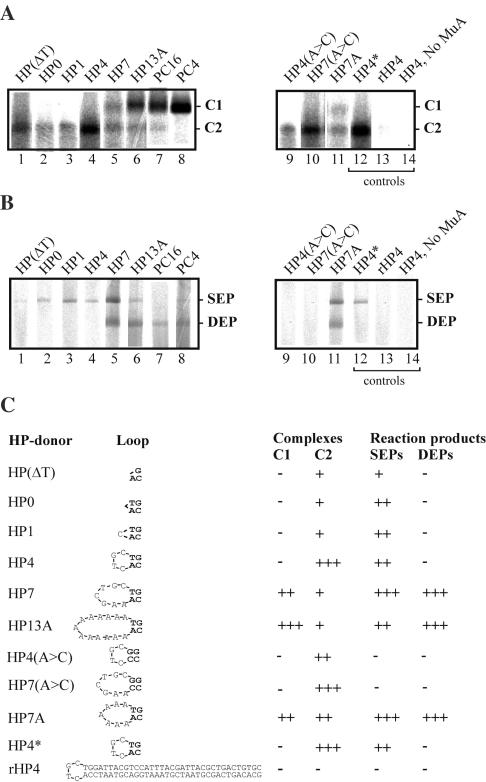
The length of the transposon flanking DNA within the donor DNA segment, as well as its end configuration, are critical variables determining the assembly, stability and activity characteristics of Mu transpososomes (22). The effect of hairpin loop length and sequence on hairpin processing was therefore investigated (Figure 5). Initially, formation of protein–DNA complexes was analyzed by agarose gel electrophoresis and autoradiography following a 1 h incubation in the absence of divalent metal ions. Under the electrophoretic conditions used, Mu transpososomes can be visualized as stable complexes that withstand a challenge by competitor DNA included in the loading buffer (22). However, depending on the exact reaction conditions and transposon end configuration, certain transpososomes are characteristically less stable and may not be detected by this assay (22). In the latter cases, other types of complexes may be revealed, and they may represent assembly or disassembly intermediates (22). Those hairpin substrates that contained the longest loops primarily yielded complexes that migrated in the gel similarly as the control transpososomes made with precut substrates (Figure 5A, indicated by C1). Upon shortening the loop length, this complex gradually disappeared and there was a simultaneous appearance of another complex having faster gel mobility (indicated by C2). With the hairpin loop lengths below 4 nt, essentially all of the observed complexes were of the latter type. Mutated nucleotides in the transposon end somewhat influenced the distribution of the complexes detected (compare lanes 4 and 9 as well as lanes 5 and 10), and the non-Mu hairpin substrate (lane 13) yielded no detectable complexes. The effect of loop sequence was studied with 7 nt loop substrates, and no influence on the complex formation was observed (compare lanes 5 and 11).
Figure 5.
Effect of hairpin loop length and sequence on stable complex formation and catalysis. (A) Complex formation. Complexes with hairpin (3-fold scaled up reactions) and precleaved (1-fold reactions) substrates were assembled for 1 h in the absence of metal ions, and the reaction products were analyzed by native agarose gel electrophoresis and autoradiography. Primarily, the assay detects transpososomes but may also detect assembly and/or disassembly intermediates. The slower migrating complexes (indicated by C1) include transpososomes. (B) Analysis of strand transfer products. Target DNA (pUC19) and MgCl2 were added to the assembly reactions to allow catalysis (Materials and Methods), and the reactions were incubated for 6 h prior to analysis of strand transfer products (as in Figure 4). (C) Tabulation of data from (A) and (B). Only a minor portion of the substrates were converted to reaction products (in most cases <1%). The levels of the detected products are based on visual inspection and indicated by a scale of four categories: (−), no products; (+), low level; (++), medium level; (+++), high level. Note that the HP4 substrate did not produce appreciable amounts of DEPs in this experiment, whereas in a similar experiment seen in Figure 2B it did. This apparent discrepancy is due to lower level of radioactivity in the substrate.
Next, to complete the coupled cleavage and strand transfer assay, pUC19 plasmid and MgCl2 were added to the above reactions that were further incubated for 1 h. The reaction products were analyzed by agarose gel electrophoresis and autoradiography (Figure 5B). Overall, the ability of each substrate to generate reaction products correlated well with the substrate's ability to yield C1 complexes. Those hairpin donors that contained the longest loops yielded both DEPs and SEPs. When the hairpin loop length was shortened to fewer than 4 nt, the DEPs disappeared and only SEPs were detected. No strand transfer products were detected with Mu hairpin donors containing mutated nucleotides (Figure 5B, lanes 9 and 10) or with non-Mu hairpin (lane 13), indicating that MuA processes only hairpins that contain the Mu end sequence. The loop sequence did not influence hairpin processing or strand transfer (Figure 5B, compare lanes 5 and 11).
Hairpin is cleaved at the transposon 3′ end
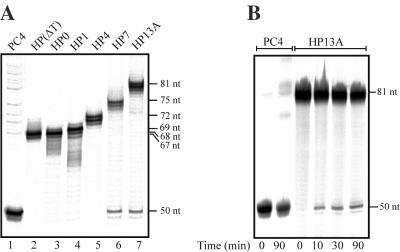
A hairpin opening step must precede strand transfer in the Mu hairpin processing reaction. We searched for a cleaved intermediate (i.e. opened hairpin molecule) by analyzing radioactively labeled reaction products in denaturing urea–PAGE. In our analysis (Figure 6A), a diagnostic 50 nt cleavage product would indicate hairpin opening at the transposon 3′ end DNA. Indeed, this product was detected with those hairpin substrates that contained a relatively large unpaired loop region (HP7 and HP13A), indicating cleavage at the exact 3′ end of the transposon. Notably, the same two substrates generated C1-type protein–DNA complexes (Figure 5A) and yielded reasonable amounts of DEPs (Figure 5B). The cleavage product was detectable already after 10 min of incubation, and the yield did not increase appreciably with longer incubation (Figure 6B). Thus, these data are consistent with the scenario where the DNA hairpin is opened at the transposon 3′ end and the end is then used for strand transfer.
Figure 6.
Analysis of hairpin opening products. (A) Effect of hairpin loop length. Transposition reactions were performed as in Figure 5. Hairpin opening products of hairpin donors having variable loop length were analyzed by denaturing urea–PAGE and visualized by autoradiography. The length of the expected opening product is 50 nt. The secondary bands seen in several lanes probably represent DNA synthesis impurities or they (mutually not exclusively) may represent DNA migrating in several major conformations, possibly influenced by the presence of hairpin ends. Minor products seen below 50 nt products (lanes 6 and 7) most probably represent cleavage from the (n − 1) synthesis impurities. Several bands seen in the top part of lane 1 represent strand transfer products. (B) Kinetics of hairpin opening reaction. Transposition reaction with HP13A donor was performed, and samples (taken after incubation of 0, 10, 30 and 90 min) were analyzed by urea–PAGE and autoradiography.
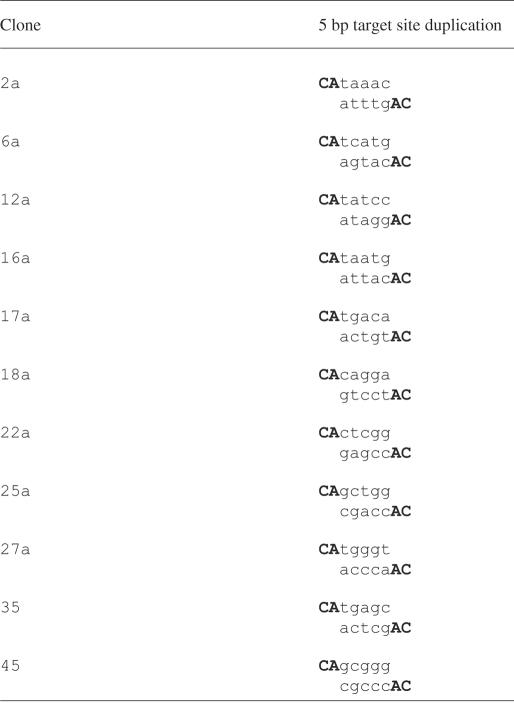
To verify that the hairpins are joined to the target DNA exactly at the 3′-OH end of the transposon DNA and that the target site duplication is generated by MuA during the two reactions involved in hairpin processing, we sequenced transposon–target junctions from several independent molecules representing transposition reaction products. For this experiment, we used a 3-fold scaled up transposition reaction with HP7 donor, pUC19 serving as the target, because such a reaction generated appreciable amounts of DEPs (target site duplication can be analyzed from DEPs only). The reaction products were amplified with a transposon-specific primer and individual reaction products were cloned for sequence analysis (Materials and Methods). All 11 clones sequenced (Table 1) contained a 5 bp target site duplication and, in each case, the ends of the transposon DNA were accurately joined to the pUC19 target plasmid. Accordingly, the data verified genuine two-ended integration upon transposition as well as utilization of authentic Mu transposition chemistry with the hallmark 5 bp target site duplication.
Table 1.
Transposon-target DNA junctions
DNA sequences of the transposon–target DNA junctions from 11 insertion events were determined from both transposon ends (Materials and Methods). The DNA regions flanking the transposon DNA are aligned appropriately to highlight in each case the 5 bp target site duplication (standard letters). The endmost nucleotides of the transposon are shown for both transposon ends (bold uppercase letters).
DISCUSSION
The transposition reaction mechanisms of non-replicative transposons Tn10 and Tn5 involve a DNA hairpin (28,29), which evidently is reflected on the proficiency of their respective transposition machineries to accommodate and process model hairpin substrates. A high degree of unity in DNA transposition reactions in general (3), the fact that Mu can transpose non-replicatively (19), and the previously discovered flexibility in MuA-catalyzed reactions (24,25,27) prompted us to investigate whether the Mu machinery also could accommodate and process transposon DNA end hairpins. We used a minimal-component in vitro assay system that enabled us to monitor the successful assembly of stable protein–DNA complexes and detect identifiable hairpin processing products.
Most of the hairpins detected in the Tn10 and Tn5 systems form via an accurate joining of the exact 3′ and 5′ ends of the transposon DNA (28,29). In the Tn5 system, however, a fraction of hairpins were imprecise [i.e. a connection from the 3′ end into the −1 position in the non-transferred strand (26)], indicating that the hairpin configuration within a particular system may vary. As even more extended variability may exist among different systems, it was difficult to make a priori predictions of potentially suitable hairpin substrates for the Mu machinery. Therefore, we generated several types (i.e. variable loop length and sequence) of substrates for the studies. Our initial experiments utilized an imprecise substrate with a 4 nt loop because: (i) such a loop resembles non-complementary nucleotides in a frayed substrate shown to be efficient in promoting the assembly of active transpososomes (22); and (ii) several unpaired loop nucleotides should provide conformational flexibility in DNA close to the active site, a feature that is important for the assembly, stability and activity of the Mu transpososome (22,23,26,40).
Our results show that MuA can open DNA hairpins and that the opened ends are subsequently used for strand transfer. No reaction products were generated without MuA or in the presence of an active-site mutant MuAE392Q, indicating that the appearance of SEPs and DEPs is dependent on the presence of a catalytically active MuA and suggesting that the hairpin processing reactions utilize the same active site that is used for the donor DNA cleavage and strand transfer steps. MuB was not required for hairpin processing, but its presence increased the amount of reaction products generated. The reason for this effect is currently unknown but may involve enhancement of transpososome assembly, increased stability of the transpososome, or stimulation of catalysis.
Products of hairpin processing reactions were generated in the presence of Mg2+ or Mn2+, but not with Ca2+. As the strand transfer step with precleaved substrates proceeds under all three conditions, the difference must reflect the characteristics of the hairpin opening reaction. At least with respect to ion utilization, the hairpin opening by MuA mimics the donor cleavage reaction. As Mn2+ rescues certain defects in MuA transposase (33,41,42) and relaxes requirements for donor substrates (39), it was of interest to study whether Mn2+ would have a similar influence on hairpin processing. In the presence of Mg2+, the strand transfer products generated were predominantly SEPs, and a minor fraction of DEPs could only be detected in experiments that utilized substrates with a high specific radioactivity (e.g. Figure 2). Reactions with Mn2+ yielded mainly DEPs, which suggests that Mn2+ stimulates coordination between the reactions at each transposon end.
The results with mixed complexes indicate that chemical reactions involved in hairpin processing take place within the Mu transpososome. These data are consistent with the known behavior of MuA: the protein monomers are inert prior to tetramerization into an active transpososome (43), and the active sites within the Mu transpososome are formed by combining structural elements from two different MuA protomers (13). Non-Mu hairpins did not generate stable complexes, and these substrates were not processed, thus providing additional evidence for catalysis within the transpososome. Furthermore, formation of complexes that migrated similarly to known transpososomes in control experiments correlated with the generation of strand transfer products. The length of the hairpin loop influenced the formation of catalytically competent complexes, with longer loops being more productive than those containing only a few unpaired nucleotides. Supposedly, several of the critical contacts required for the transpososome assembly and stability are more optimally positioned with the longer loops. In addition, longer loops may allow more flexibility in the DNA at the vicinity of the endmost transposon nucleotides, thereby facilitating the proper conformation of the active site. The catalytically productive conformation in the Mu transpososome entails a criss-crossed architecture (13), within which the catalysis of both cleavage and strand transfer occurs in trans (8–10). For structural reasons, and because the strand transfer step is included, it is reasonable to assume that catalysis of the hairpin processing reactions detected in this study also must occur in trans.
The general requirements of the transposase active site for the hairpin formation and opening reactions are not known, but the model of the Tn5 transposase–DNA complex (12) has provided information regarding how the transpososome may engage its substrates and how the active site might function in catalysis. In the Tn5 transpososome, the hairpin forms via dramatic bending of the DNA backbone with concomitant flipping of a thymidine base. Interactions from the phylogenetically conserved motif known as the YREK signature (44) appear to be directly involved in the process (12, 45–47), and base-flipping may be a general feature of hairpinning mechanisms. As we were unable to identify such a motif in MuA, it is difficult to reconcile the mechanism by which the Mu transpososome accommodates hairpins. At least for longer loops, the active site might actually function in a similar fashion and conformation as it does for donor cleavage, and no additional conformational changes in the protein and DNA structures need to be postulated, a scenario also consistent with the metal ion analysis. However, to engage shorter DNA loops in a catalytically competent conformation, structural flexibility in the protein partner within the transpososome may be important.
The Mu transpososome was able to accommodate a wide range of hairpin substrates and engage them in the active site for productive catalysis, although different levels of efficiency were observed with regard to the loop length. MuA appears to process longer hairpins preferentially, probably reflecting more suitable protein–DNA contacts between MuA and a few critical nucleotides in the transposon end. Although shorter loops were still applicable substrates for catalysis, they appeared to uncouple the coordinated action of MuA within the transpososome. Possibly, the accommodation of such suboptimal substrates changes the structure and functioning of the Mu transpososome so that productive reactions in both transposon ends become less likely or impossible to execute. Based on information from the crystal structure of the Tn5 transpososome, in which the active site appears relatively crowded (12,48), the observed degree of flexibility within the Mu transpososome active site is somewhat surprising. However, it is notable that despite the apparent crowding within the active site, the Tn5 transpososome can not only accommodate but also generate two types of hairpins, precise without non-complementary nucleotides and imprecise with one such nucleotide (29).
In vitro studies have shown unequivocally that some transposons such as Tn5 and Tn10 utilize hairpinning to seal transposon ends (28,29), and systems like hAt and V(D)J recombination generate hairpinned flanking DNA ends (30,31). Transposases of some other elements may have a potential to use hairpinning or related strategies but only under certain circumstances. In fact, variation within the hairpinning theme exists, and some mechanisms have been revealed under highly artificial conditions. For example, a mechanism similar to hairpinning is used to form circular transposon intermediates of IS911 (49) and related products have been described for Tn7 (50) as well as Tn10 (51) and suggested for IS903 (52).
MuA was able to process preformed model DNA hairpins in vitro, but can Mu use hairpinning mechanism in vivo? The non-replicative initial integration of Mu has been explained by a model that involves a repair step following the formation of the Shapiro intermediate (53). A very recent study supports this model, as the flanking host DNA appears to remain attached to the Mu DNA upon integration (54). Theoretically, hairpinning could provide an alternate route: following the initial single-strand cleavage, MuA-catalyzed hairpinning would liberate the Mu genome from the flanking DNA, and hairpin opening followed by strand transfer would result in simple insertion. Although the above scenario is plausible, it is important to note that our current study does not directly address the question of hairpin formation by MuA, and we do not know whether MuA can form hairpins. Despite our attempts to characterize hairpin intermediates along the Mu transposition reaction pathway in vitro, such intermediates have been elusive, and no conclusive evidence has been gained in favor of their existence (unpublished data). Nevertheless, the formal possibility remains that MuA might use hairpinning as an alternate catalytic mechanism, in yet uncharacterized situations in vivo or under special reaction conditions in vitro. Interestingly, MuA can produce DNA hairpins under certain circumstances in vitro [P. A. Rice, H. Savilahti and K. Mizuuchi, unpublished data; (55)]. However, these hairpins differ from the transposon end DNA hairpins, as they are formed at the target DNA and represent reverse reaction products of the strand transfer.
It is possible that MuA might not be able to form hairpins but only process them, perhaps representing an evolutionary remnant of the hairpinning mechanism. Another possibility is that MuA has evolved its current, although imperfect, ability to process hairpins but not to form them. This latter scenario highlights the observation that both donor cleavage and hairpin opening are characteristically very similar reactions. Accordingly, at least with longer hairpin loops, the distinction between those two reactions is arbitrary, and hairpin opening in these cases can be regarded as a form of donor cleavage. The fact that MuA can process short hairpin loops, although relatively poorly, suggests that the evolutionary path from the non-hairpinning to hairpinning mechanism, or vice versa, may be shorter than anticipated previously. Another view is that, similar to what has been observed with protein–DNA complexes of several DNA-modifying enzymes (56–59), distortion of DNA might facilitate the initial cleavage in the Mu system. If this were the case, any hairpin substrate that could accommodate an appropriately distorted structure should be able to promote a productive cleavage reaction. Following cleavage, the catalytic machinery should then be proficient to adopt its normal configuration for the strand transfer step.
Acknowledgments
We thank Maria Pajunen and Anja Paatero for critical reading of the manuscript. Auli Saarinen and Pirjo Rahkola are acknowledged for excellent technical assistance. This research was funded by a grant from The Viikki Graduate School in Biosciences (to A.-H.S.) as well as grants from the Academy of Finland and Finnish National Technology Agency TEKES (to H.S.). Funding to pay the Open Access publication charges for this article was provided by TEKES.
Conflict of interest statement. None declared.
REFERENCES
- 1.Craig N.L., Craigie R., Gellert M., Lambowitz A.M. (eds) Mobile DNA II. Washington DC: American Society for Microbiology; 2002. [Google Scholar]
- 2.Curcio J.M., Derbyshire K.M. The outs and ins of transposition: from Mu to kangaroo. Mol. Cell. Biol. 2003;4:1–13. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 3.Craig N. Unity in transposition reactions. Science. 1995;270:253–254. doi: 10.1126/science.270.5234.253. [DOI] [PubMed] [Google Scholar]
- 4.Gueguen E., Rousseau P., Duval-Valentin G., Chandler M. The transpososome: control of transposition at the level of catalysis. Trends Microbiol. 2005;13:543–549. doi: 10.1016/j.tim.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Rice P., Mizuuchi K. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition. Cell. 1995;82:209–220. doi: 10.1016/0092-8674(95)90308-9. [DOI] [PubMed] [Google Scholar]
- 6.Haren L., Ton-Hoang B., Chandler M. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 1999;53:245–281. doi: 10.1146/annurev.micro.53.1.245. [DOI] [PubMed] [Google Scholar]
- 7.Mizuuchi K., Baker T.A. Chemical mechanisms for mobilizing DNA. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington DC: American Society for Microbiology; 2002. pp. 12–23. [Google Scholar]
- 8.Savilahti H., Mizuuchi K. Mu transpositional recombination: donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell. 1996;85:271–280. doi: 10.1016/s0092-8674(00)81103-4. [DOI] [PubMed] [Google Scholar]
- 9.Namgoong S.Y., Harshey R.M. The same two monomers within a MuA tetramer provide the DDE domains for the strand cleavage and strand transfer steps of transposition. EMBO J. 1998;17:3775–3785. doi: 10.1093/emboj/17.13.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams T.L., Jackson E.L., Carritte A., Baker T.A. Organization and dynamics of the Mu transpososome: recombination by communication between two active sites. Genes Dev. 1999;13:2725–2737. doi: 10.1101/gad.13.20.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naumann T.A., Reznikoff W.S. Trans catalysis in Tn5 transposition. Proc. Natl Acad. Sci. USA. 2000;97:8944–8949. doi: 10.1073/pnas.160107997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies D.R., Goryshin I.Y., Reznikoff W.S., Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 13.Yuan J.F., Beniac D.R., Chaconas G., Ottensmeyer F.P. 3D reconstruction of the Mu transposase and the Type 1 transpososome: a structural framework for Mu DNA transposition. Genes Dev. 2005;19:840–852. doi: 10.1101/gad.1291405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaconas G., Harshey R.M. Transposition of phage Mu DNA. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington DC: American Society for Microbiology; 2002. pp. 384–402. [Google Scholar]
- 15.Chaconas G., Harshey R.M., Sarvetnick N., Bukhari A.I. Predominant end-products of prophage Mu DNA transposition during the lytic cycle are replicon fusions. J. Mol. Biol. 1981;150:341–359. doi: 10.1016/0022-2836(81)90551-9. [DOI] [PubMed] [Google Scholar]
- 16.Liebart J.C., Ghelardini P., Paolozzi L. Conservative integration of bacteriophage Mu DNA into pBR322 plasmid. Proc. Natl Acad. Sci. USA. 1982;79:4362–4366. doi: 10.1073/pnas.79.14.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akroyd J.E., Symonds N. Evidence for a conservative pathway of transposition of bacteriophage Mu. Nature. 1983;303:84–86. doi: 10.1038/303084a0. [DOI] [PubMed] [Google Scholar]
- 18.Chaconas G., Kennedy D.L., Evans D. Predominant integration end products of infecting bacteriophage Mu DNA are simple insertions with no preference for integration of either Mu DNA strand. Virology. 1983;128:48–59. doi: 10.1016/0042-6822(83)90317-3. [DOI] [PubMed] [Google Scholar]
- 19.Harshey R.M. Transposition without duplication of infecting bacteriophage Mu DNA. Nature. 1984;311:580–581. doi: 10.1038/311580a0. [DOI] [PubMed] [Google Scholar]
- 20.Surette M.G., Buch S.J., Chaconas G. Transpososomes: stable protein–DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987;49:253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- 21.Craigie R., Mizuuchi K. Transposition of Mu DNA: joining of Mu to target DNA can be uncoupled from cleavage at the ends of Mu. Cell. 1987;51:493–501. doi: 10.1016/0092-8674(87)90645-3. [DOI] [PubMed] [Google Scholar]
- 22.Savilahti H., Rice P.A., Mizuuchi K. The phage Mu transpososome core: DNA requirements for assembly and function. EMBO J. 1995;14:4893–4903. doi: 10.1002/j.1460-2075.1995.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee I., Harshey R.M. Importance of the conserved CA dinucleotide at Mu termini. J. Mol. Biol. 2001;314:433–444. doi: 10.1006/jmbi.2001.5177. [DOI] [PubMed] [Google Scholar]
- 24.Goldhaber-Gordon I., Williams T.L., Baker T.A. DNA recognition sites activate MuA transposase to perform transposition of non-Mu DNA. J. Biol. Chem. 2002b;277:7694–7702. doi: 10.1074/jbc.M110341200. [DOI] [PubMed] [Google Scholar]
- 25.Goldhaber-Gordon I., Early M.H., Gray M.K., Baker T.A. Sequence and positional requirements for DNA sites in a Mu transpososome. J. Biol. Chem. 2002a;277:7703–7712. doi: 10.1074/jbc.M110342200. [DOI] [PubMed] [Google Scholar]
- 26.Lee I., Harshey R M. The conserved CA/TG motif at Mu termini: T specifies stable transpososome assembly. J. Mol. Biol. 2003;330:261–275. doi: 10.1016/s0022-2836(03)00574-6. [DOI] [PubMed] [Google Scholar]
- 27.Saariaho A.-H., Lamberg A., Elo S., Savilahti H. Functional comparison of the transposition core machineries of phage Mu and Haemophilus influenzae Mu like prophage Hin-Mu reveals interchangeable components. Virology. 2005;331:6–19. doi: 10.1016/j.virol.2004.09.041. [DOI] [PubMed] [Google Scholar]
- 28.Kennedy A.K., Guhathakurta A., Kleckner N., Haniford D.B. Tn10 transposition via a DNA hairpin intermediate. Cell. 1998;95:125–134. doi: 10.1016/s0092-8674(00)81788-2. [DOI] [PubMed] [Google Scholar]
- 29.Bhasin A., Goryshin I.Y., Reznikoff W.S. Hairpin formation in Tn5 transposition. J. Biol. Chem. 1999;52:37021–37029. doi: 10.1074/jbc.274.52.37021. [DOI] [PubMed] [Google Scholar]
- 30.McBlane J.F., van Gent D.C., Ramsden D.A., Romeo C., Cuomo C.A., Gellert M., Oettinger M.A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 31.Zhou L., Mitra R., Atkinson P.W., Hickman A.B., Dyda F., Craig N.L. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A laboratory manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Baker T.A., Luo L. Identification of residues in the Mu transposase essential for catalysis. Proc. Natl Acad. Sci. USA. 1994;91:6654–6658. doi: 10.1073/pnas.91.14.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaconas G., Gloor G., Miller J.L. Amplification and purification of the bacteriophage Mu encoded B transposition protein. J. Biol. Chem. 1985;260:2662–2669. [PubMed] [Google Scholar]
- 35.Adzuma K., Mizuuchi K. Steady-state kinetic analysis of ATP hydrolysis by the B protein of bacteriophage Mu. Involvement of protein oligomerization in the ATPase cycle. J. Biol. Chem. 1991;266:6159–6167. [PubMed] [Google Scholar]
- 36.Haapa-Paananen S, Rita H., Savilahti H. DNA transposition of bacteriophage Mu. A quantitative analysis of target site selection in vitro. J. Biol. Chem. 2002;277:2843–2851. doi: 10.1074/jbc.M108044200. [DOI] [PubMed] [Google Scholar]
- 37.Haapa S., Taira S., Heikkinen E., Savilahti H. An efficient and accurate integration of mini-Mu transposons in vitro: a general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res. 1999;27:2777–2784. doi: 10.1093/nar/27.13.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamberg A., Nieminen S., Qiao M., Savilahti H. Efficient insertion mutagenesis for bacterial genomes involving electroporation of in vitro-assembled DNA transposition complexes of bacteriophage Mu. Appl. Environ. Microbiol. 2002;68:705–712. doi: 10.1128/AEM.68.2.705-712.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coros C.J., Chaconas G. Effect of mutations in the Mu-host junction region on transpososome assembly. J. Mol. Biol. 2001;310:299–309. doi: 10.1006/jmbi.2001.4772. [DOI] [PubMed] [Google Scholar]
- 40.Lavoie B.D., Chan B.S., Allison R.G., Chaconas G. Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. EMBO J. 1991;10:3051–3059. doi: 10.1002/j.1460-2075.1991.tb07856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim K., Namgoong S.Y., Jayaram M., Harshey R.M. Step-arrest mutants of phage Mu transposase. Implications in DNA–protein assembly, Mu end cleavage, and strand transfer. J. Biol. Chem. 1995;270:1472–1479. doi: 10.1074/jbc.270.3.1472. [DOI] [PubMed] [Google Scholar]
- 42.Namgoong S.Y., Kim K., Saxena P., Yang J.Y., Jayaram M., Giedroc D.P., Harshey R.M. Mutational analysis of domain II beta of bacteriophage Mu transposase: domains II alpha and II beta belong to different catalytic complementation groups. J. Mol. Biol. 1998;275:221–232. doi: 10.1006/jmbi.1997.1466. [DOI] [PubMed] [Google Scholar]
- 43.Baker T.A., Mizuuchi K. DNA-promoted assembly of the active tetramer of the Mu transposase. Genes Dev. 1992;6:2221–2232. doi: 10.1101/gad.6.11.2221. [DOI] [PubMed] [Google Scholar]
- 44.Rezsohazy R., Hallet B., Delcour J., Mahillon J. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol. Microbiol. 1993;9:1283–1295. doi: 10.1111/j.1365-2958.1993.tb01258.x. [DOI] [PubMed] [Google Scholar]
- 45.Allingham J.S., Wardle S.J., Haniford D.B. Determinants for hairpin formation in Tn10 transposition. EMBO J. 2001;20:2931–2942. doi: 10.1093/emboj/20.11.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ason B., and Reznikoff W.S. Mutational analysis of the base flipping event found in Tn5 transposition. J. Biol. Chem. 2002;277:11284–11291. doi: 10.1074/jbc.M111119200. [DOI] [PubMed] [Google Scholar]
- 47.Naumann T.A., Reznikoff W.S. Tn5 transposase active site mutants. J. Biol. Chem. 2002;277:17623–17629. doi: 10.1074/jbc.M200742200. [DOI] [PubMed] [Google Scholar]
- 48.Reznikoff W.S. Tn5 as a model for understanding DNA transposition. Mol. Microbiol. 2003;47:1199–1206. doi: 10.1046/j.1365-2958.2003.03382.x. [DOI] [PubMed] [Google Scholar]
- 49.Polard P., Prere M.F., Fayet O., Chandler M. Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J. 1992;11:5079–5090. doi: 10.1002/j.1460-2075.1992.tb05615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biery M.C., Lopata M., Craig N.L. A minimal system for Tn7 transposition: the transposon-encoded proteins TnsA and TnsB can execute DNA breakage and joining reactions that generate circularized Tn7 species. J. Mol. Biol. 2000;297:25–37. doi: 10.1006/jmbi.2000.3558. [DOI] [PubMed] [Google Scholar]
- 51.Morisato D., Kleckner N. Transposase promotes double strand breaks and single strand joints at Tn10 termini in vivo. Cell. 1984;39:181–190. doi: 10.1016/0092-8674(84)90204-6. [DOI] [PubMed] [Google Scholar]
- 52.Tavakoli N.P., Derbyshire K.M. Tipping the balance between replicative and simple transposition. EMBO J. 2001;20:2923–2930. doi: 10.1093/emboj/20.11.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Craigie R., Mizuuchi K. Mechanism of transposition of bacteriophage Mu: structure of a transposition intermediate. Cell. 1985;41:867–876. doi: 10.1016/s0092-8674(85)80067-2. [DOI] [PubMed] [Google Scholar]
- 54.Au T.K., Agrawal P., Harshey R.M. Chromosomal integration mechanism of infecting Mu virion DNA. J. Bacteriol. 2006;188:1828–1834. doi: 10.1128/JB.188.5.1829-1834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Au T.K., Pathania S., Harshey R.M. True reversal of Mu integration. EMBO J. 2004;23:3408–3420. doi: 10.1038/sj.emboj.7600344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vipond I.B., Halford S.E. Structure-function correlation for the EcoRV restriction enzyme: from non-specific binding to specific DNA cleavage. Mol. Microbiol. 1993;9:225–231. doi: 10.1111/j.1365-2958.1993.tb01685.x. [DOI] [PubMed] [Google Scholar]
- 57.Gimble F.S., Wang J. Substrate recognition and induced DNA distortion by the PI-SceI endonuclease, an enzyme generated by protein splicing. J. Mol. Biol. 1996;263:163–180. doi: 10.1006/jmbi.1996.0567. [DOI] [PubMed] [Google Scholar]
- 58.Mueller J.E., Smith D., Bryk M., Belfort M. Intron-encoded endonuclease I-TevI binds as a monomer to effect sequential cleavage via conformational changes in the td homing site. EMBO J. 1995;14:5724–5735. doi: 10.1002/j.1460-2075.1995.tb00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luscombe N.M., Austin S.E., Berman H.M., Thornton J.M. An overview of the structures of protein–DNA complexes. Genome Biol. 2000;1:1–37. doi: 10.1186/gb-2000-1-1-reviews001. [DOI] [PMC free article] [PubMed] [Google Scholar]