Abstract
We have examined the relationship between nuclear localization and transcriptional activity of the endogenous murine β-globin locus during erythroid differentiation. Murine fetal liver cells were separated into distinct erythroid maturation stages by fluorescence-activated cell sorting, and the nuclear position of the locus was determined at each stage. We find that the β-globin locus progressively moves away from the nuclear periphery with increasing maturation. Contrary to the prevailing notion that the nuclear periphery is a repressive compartment in mammalian cells, βmajor-globin expression begins at the nuclear periphery prior to relocalization. However, relocation of the locus to the nuclear interior with maturation is accompanied by an increase in βmajor-globin transcription. The distribution of nuclear polymerase II (Pol II) foci also changes with erythroid differentiation: Transcription factories decrease in number and contract toward the nuclear interior. Moreover, both efficient relocalization of the β-globin locus from the periphery and its association with hyperphosphorylated Pol II transcription factories require the locus control region (LCR). These results suggest that the LCR-dependent association of the β-globin locus with transcriptionally engaged Pol II foci provides the driving force for relocalization of the locus toward the nuclear interior during erythroid maturation.
Keywords: β-globin locus, nuclear organization, nuclear periphery, erythroid differentiation, RNA polymerase II, fluorescence in situ hybridization
The eukaryotic nucleus is structurally and functionally compartmentalized. Chromatin is subdivided into compacted and repressive domains (heterochromatin) or relatively decondensed domains that are enriched with genes that are active or poised for expression (euchromatin). Similarly, local accumulations of specific proteins mark nuclear subcompartments and form a number of functionally distinct nuclear bodies. The dynamic organization of the nucleus may provide an essential level of gene regulation in multicellular eukaryotes during tissue-specific differentiation when subsets of genes are transcriptionally activated and others stably silenced. For example, in developing B and T cells, the transcription factor Ikaros recruits genes destined for silencing to pericentromeric heterochromatin (PCH) through direct binding of the target genes and heterochromatic repeats (Brown et al. 1997, 1999; Cobb et al. 2000). Euchromatic sequences that become integrated or relocated to PCH similarly become silenced following this association (Csink and Henikoff 1996; Francastel et al. 1999). In mammalian systems, the nuclear periphery has also been regarded as a repressive compartment where silenced genes are sequestered. For example, the CFTR gene and its nearest neighbors associate with the periphery in a tissue-specific fashion when inactive and relocate away from the periphery when active (Zink et al. 2004). The immunoglobulin heavy chain (IgH) locus is located at the nuclear periphery in lymphoid progenitors prior to its activation (Kosak et al. 2002). During B-cell development, as the locus becomes transcriptionally active and undergoes rearrangement, it is relocated to the nuclear interior (Kosak et al. 2002). In addition, the nuclear periphery preferentially harbors gene-poor chromosomes, as has been demonstrated in human nuclei, possibly reflecting an overall lower level of transcriptional activity from these domains (Croft et al. 1999; Boyle et al. 2001).
Gene localization with respect to chromosome territories (CTs; the most densely staining regions of interphase chromosomes) has also been correlated with different activation states. For example, during robust transcriptional activity, several multigene loci extend in loops from their CTs, whereas they remain associated with their CTs when inactive (Volpi et al. 2000; Williams et al. 2002). In the case of the Hoxb locus, extrusion from the CT appears to be developmentally regulated and is limited to the stage-specific active gene (Chambeyron and Bickmore 2004; Chambeyron et al. 2005). In contrast, CT looping of the β-globin locus can also reflect a poised state prior to transcriptional activation or even repression if the chromatin loop extends to a repressive compartment (Ragoczy et al. 2003). The molecular mechanisms leading to differentiation- or activation-state-specific subnuclear localization of genes remain to be elucidated. Similarly, it is not understood whether regulatory elements directly contribute to localization or what the sequence requirements might be.
We have sorted murine fetal liver cells into distinct erythroid maturation stages and examined the nuclear position and expression state of the β-globin locus during cellular differentiation. We find that the locus is relocated away from the nuclear periphery and PCH with progressive maturation. Importantly, significant expression of the α- and βmajor-globin genes begins prior to the most dramatic shift in positioning, suggesting that relocation may be a result of, rather than requirement for, transcriptional activity. In support of this possibility, the internalization of the β-globin locus during erythroid maturation is accompanied by a redistribution of RNA polymerase II (Pol II) foci. In differentiated cells, Pol II foci decrease in abundance and become more restricted to the nuclear interior, which may drive the relocation of the β-globin locus. Furthermore, in the absence of the locus control region (LCR), which is required for high-level expression of the β-globin genes, the locus does not efficiently relocate to the nuclear interior and fails to colocalize consistently with foci of engaged (highly phosphorylated) Pol II. Thus, the LCR increases the locus’ affinity for, or ability to nucleate, active transcription factories, resulting in activated transcription and nuclear relocalization.
Results
Separation of fetal liver cells into distinct erythroid maturation stages
The characterization of changes in nuclear organization during murine erythropoiesis requires the separation of erythroid cells into distinct maturation stages. While spleen or marrow has been used for isolating primary erythroid cells in distinct maturational compartments (Socolovsky et al. 2001), we have used day-13.5 post-conception (dpc) fetal liver as a source of material, as it is at least 90% erythroid (Trimborn et al. 1999). Fresh fetal liver cells were stained with a monoclonal antibody to the surface marker c-kit (CD117) and monoclonal antibodies ERY-1 (Bacon and Sytkowski 1987) and Ter-119, a lineage-specific antibody staining all erythroid cells after the CFU-E stage. While CD117 marks the most immature hematopoietic cells and is not erythroid specific, the morphology and gene expression profile of the vast majority of CD117+ cells in fetal liver is consistent with the erythroid lineage (M.A. Bender, unpubl.). The erythroid-specific ERY-1 was used as an intermediate maturation marker (Bacon and Sytkowski 1987), since its surface expression begins prior to the loss of CD117 staining, permitting the isolation of immature definitive erythroid cells. Furthermore, ERY-1 expression precedes the emergence of TER-119 during maturation, but while TER-119 expression is maintained, ERY-1 levels eventually fall. Figure 1A shows the fluorescence profiles of combinations of these antibodies, with the arrows indicating the changes in fluorescence observed during erythroid maturation. On the basis of these three surface markers, we sorted fetal liver cells into the following four fractions: CD117+, ERY-1−, TER-119− (stage 1); CD117+, ERY-1+, TER-119− (stage 2); CD117−, ERY-1+, TER-119+ (stage 3); and CD117−, and ERY-1−, TER-119+ (stage 4). Wright-Giemsa staining confirmed that these subpopulations represent progressive maturation stages (Fig. 1B): Cells in the more mature fractions exhibit increased chromatin condensation and an obvious decrease in nuclear size. The final fraction is a mix of mature cells, including polychromatic, pyknotic, and enucleated red cells. Thus, through Fluorescence-Activated Cell Sorting (FACS) of fetal liver stained with CD117, ERY-1, and TER-119, a spectrum of murine erythroid differentiation stages can be distinguished and isolated.
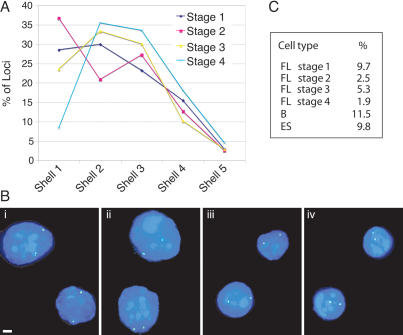
Figure 1.
Isolation of erythroid cells at progressive maturation stages. (A) FACS profiles of the fetal liver cells: pairwise comparison of ERY-and 1 CD117 staining (panel i), Ter-119 and CD117 (panel ii), and ERY-1 and Ter-119 (panel iii). The arrows indicate changes in the fluorescence observed with increasing erythroid maturation. (B) Wright-Giemsa staining of sorted fetal liver populations. The four fractions (stage 1: CD117+, ERY-1−, TER-119−; stage 2: CD117+, ERY-1+, TER-119−; stage 3: CD117−, ERY-1+ TER-119+; stage 4: CD117−, ERY-1−, TER-119+) exhibit a progressive decrease in cell size, increase in cytoplasm to nuclear volume ratio, nuclear compaction, and enucleation.
The β-globin locus relocates away from the nuclear periphery and PCH during erythroid maturation
In mammalian cells, the nuclear periphery and PCH are considered to be repressive nuclear compartments. To assess the position of the β-globin gene locus relative to these compartments during erythroid differentiation, we analyzed, sorted, and formaldehyde-fixed cell populations by DNA FISH (fluorescence in situ hybridization) with β-globin and γ-satellite probes. The latter marks all murine pericentromeres.
As shown in Figure 2A, the most dramatic effect of erythroid maturation on β-globin position is a steep drop in the number of peripheral loci between stages 2 and 4. While 28% and 36% of alleles are near the nuclear envelope in the immature cell fractions (stages 1 and 2, respectively), this peripheral fraction falls to 8% in the most mature cells. As in previous two-dimensional (2D) studies, loci within 20% of the nuclear radius from the envelope were considered to be peripheral (shell 1) (Kosak et al. 2002). The mean of the distances from the periphery of the most mature cell fraction (stage 4) was statistically significantly different from that of the more immature cells (for stages 1–3, P < 0.001). As the measurements are corrected for nuclear radius, the smaller dimensions of the mature cells cannot account for this shift in position. In addition, if the distribution of β-globin loci was random, we would expect 22% of the alleles to be located within the peripheral shell (Kosak et al. 2002). Thus, the fact that only 8% of βglobin loci are peripheral clearly demonstrates a nonrandom positioning toward the nuclear interior with the final stage of erythroid maturity. To further validate our 2D results, we also performed three-dimensional (3D) ImmunoFISH analyses of the position of the β-globin locus relative to lamin B staining on the four maturation stages. In immature cells (stages 1 and 2), 66% and 75%, respectively, of β-globin loci are in contact with the lamina and thus are perinuclear, whereas at stage 3, 47% of the loci are peripheral, and in the most mature cells (stage 4), only 27% of the loci are peripheral (Supplementary Fig. 1B). These data reinforce the shift in nuclear position of the β-globin locus with erythroid maturation.
Figure 2.
Nuclear location of the β-globin locus. (A) Distribution of the β-globin loci among five concentric shells at the different maturation stages as assayed by DNA FISH (each shell encompasses 20% of the nuclear radius, shell 1 representing the periphery and shell 5 the center of the nucleus). Image stacks of formaldehyde-fixed cells were collapsed onto a single plane and analyzed in 2D. Between 150 and 300 loci were scored for each cell population. (B, panels i–iv) Examples of β-globin locus position at the sorted maturation stages 1–4 by DNA FISH. The β-globin locus probe is stained green; nuclear DNA is counterstained blue with DAPI. Bars, 2 μm. (C) Association of the β-globin locus with PCH by DNA FISH. Nuclear volumes were reconstructed and rotated in 3D, and colocalization of the β-globin locus probe with γ-satellite repeat probe scored. All four fetal liver stages and splenic B cells and ES cells were analyzed.
Similarly, we observed a relocation of the β-globin locus away from PCH in maturing erythroid cells (Fig. 2C). Three-dimensional analysis reveals that the β-globin locus does not associate frequently with PCH in cells in which it is inactive, including the most immature erythroid cells, undifferentiated embryonic stem (ES) cells, and splenic B cells. The fraction of the β-globin loci associated with PCH did not significantly exceed 10% in any cell population analyzed. As fetal liver cells mature, the association with PCH is reduced even further, decreasing to 2% in cells at stage 4. In combination, these results indicate that the locus is relocated away from nuclear compartments that are conventionally considered repressive, particularly the nuclear periphery, during erythroid maturation.
Nuclear position is gene and cell type specific
To address the possibility that genes are nonspecifically relocated to the nuclear interior in terminally differentiating erythroid cells, we compared the positions of the β-globin and the IgH loci in sorted fetal liver cells. FISH analysis with an IgH-specific probe revealed that, similar to the β-globin genes, many of the IgH loci (33.3%) are peripheral in immature erythroid cells (Fig. 3A). However, the number of peripheral IgH loci did not fall as markedly as that of the β-globin loci with maturation. Instead, at the third maturation stage, 38.5% IgH alleles remained peripheral, and only the most mature cells (stage 4) exhibit reduced peripheral localization. Notably, the peripheral incidence of IgH at stage 4 (23.4%) is significantly higher than that of the β-globin locus (P < 0.05). Furthermore, the frequencies of peripheral localization for the IgH locus at the stages 1 and 3 (>30% by 2D analysis) strongly suggest a nonrandom enrichment at the periphery. Whereas the βglobin locus is depleted from the periphery by stage 4, the number of IgH loci in the peripheral shell reflects a random distribution (close to the calculated value of 22%).
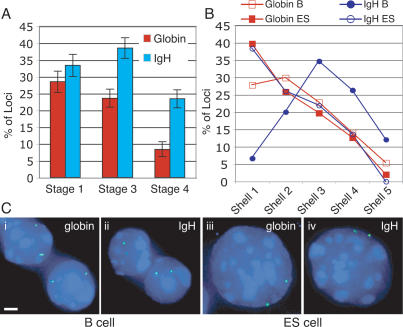
Figure 3.
Location of the β-globin and IgH loci is gene and tissue specific. (A) The nuclear position of the IgH and β-globin loci was compared at three different erythroid maturation stages by DNA FISH. Shown are only the frequencies of peripheral loci within 20% of the nuclear radius from the envelope (red bars indicate β-globin, blue bars indicate IgH locus, and error bars indicate SE). (B) Comparison of the nuclear positions of the β-globin and IgH loci in B and ES cells. The graph shows the distribution of the gene loci among five concentric shells in B- and ES-cell nuclei (each shell encompasses 20% of the nuclear radius, shell 1 representing the periphery and shell 5 the center of the nucleus; red open squares indicate β-globin locus in B cells, red closed squares indicate β-globin locus in ES cells, blue closed circles indicate IgH locus in B cells, and blue open circles indicate IgH locus in ES cells). (C) Examples of β-globin and IgH locus position in B and ES cells by DNA FISH. The locus probes are green; nuclear DNA is counterstained blue with DAPI. Bars, 2 μm.
The position of the IgH locus in fetal liver erythroid cells contrasts with its behavior in B cells. As reported previously, the IgH locus is relocated to the nuclear center during B-cell development (Kosak et al. 2002). We isolated B220+ B cells from mouse spleens (predominantly resting mature B lymphocytes) and confirmed the internal position of the IgH locus (Fig. 3B); the mean distance to the nuclear periphery is significantly higher than at any fetal liver maturation stage, and very few loci are near the periphery itself. On the other hand, 28% of globin loci were peripheral in B cells, similar to the proportion seen in immature fetal liver cells. We also compared the position of the IgH and β-globin loci in undifferentiated ES cells, a nonpermissive cell type for either locus. In this context, both loci are highly enriched at the nuclear periphery (38% and 40%, respectively) and their overall distributions are indistinguishable. These results strongly suggest a tissue-specific nuclear location for both the β-globin and IgH loci, with a preferred internal nuclear position in tissue where the particular gene is active or poised for transcriptional activity.
The LCR is required for the relocation of the β-globin locus to the nuclear interior in mature erythroid cells
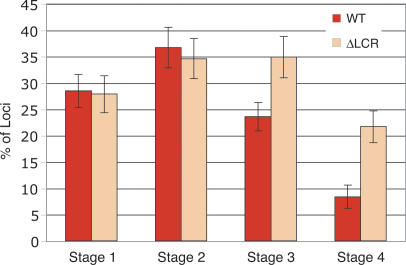
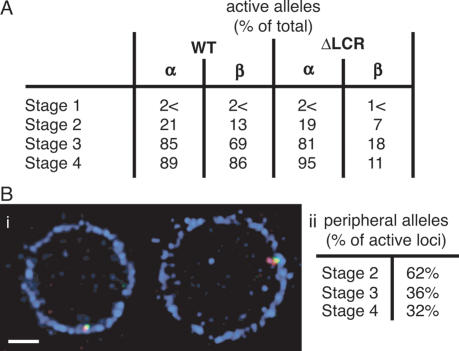
Deletion of the LCR from the native β-globin locus results in a 25- to 100-fold reduction in β-like globin gene expression throughout development (Bender et al. 2000). One possible mechanism by which the LCR regulates gene activity might be to direct the β-globin locus toward a transcriptionally favorable nuclear environment. To assess the potential contribution of the LCR to nuclear positioning of the β-globin locus, we isolated fetal liver cells from mice in which the LCR was deleted from its native position in both β-globin alleles by homologous recombination (Bender et al. 2000). The nuclear positions of the ΔLCR alleles and the wild-type β-globin loci were compared in each of the four erythroid maturation stages. Figure 4 reveals that the positions of the mutant and wild-type alleles relative to the nuclear periphery are indistinguishable in immature erythroid cells (stages 1 and 2). Both alleles exhibit a slightly higher prevalence near the periphery at stage 2 than at stage 1. However, with increasing maturation, the ΔLCR allele remains at the periphery at a significantly higher frequency than the wild-type locus, which progressively relocates to the nuclear interior at stages 3 and 4 (P < 0.05). The peripheral enrichment of the ΔLCR relative to the wild-type allele was also revealed in 3D ImmunoFISH: Of ΔLCR loci, 76% are in contact with the nuclear lamina in cells at stages 1 and 2, 58% at stage 3, and 53% at stage 4 (Supplementary Fig. 1C). The values for the latter two stages are significantly higher than are those observed for the wild-type allele (P < 0.05) and demonstrate the propensity of the ΔLCR allele to remain predominantly in a peripheral nuclear position.
Figure 4.
The LCR is required for relocation away from the periphery. The nuclear position of the ΔLCR globin locus during erythroid differentiation was compared with the wild-type allele by DNA FISH. Shown are only the frequencies of peripheral loci (within 20% of the nuclear radius from the envelope; red bars indicate wild type, orange bars indicate ΔLCR locus, and error bars indicate SE).
The deletion of the LCR also affected the position of the β-globin locus with respect to PCH with progressing differentiation (data not shown). In the most immature cell population, the mutant allele associated with PCH to a similar extent as the wild type (13% vs.10%). However, at stage 4, 9% of ΔLCR loci remain in contact with PCH, whereas only 2% of wild-type loci contact PCH at this stage (see above). This failure to completely dissociate from PCH underscores the potential role of the β-globin LCR in positioning the β-globin locus in an increasingly erythroid cell environment.
Relocation of the β-globin locus coincides with changes in RNA Pol II distribution during erythroid maturation
The nuclear position of certain gene loci has been linked to their transcriptional status (Francastel et al. 1999; Brown et al. 2001; Schubeler et al. 2001; Chambeyron and Bickmore 2004; Zink et al. 2004). In erythroid cells, where globin genes are highly expressed, the β-globin locus has been shown to consistently colocalize with nuclear staining of RNA Pol II (Osborne et al. 2004). Consequently, it is possible that our observed relocation of the β-globin locus may result from a change in the nuclear organization of the transcriptional machinery during terminal erythroid differentiation. To address this possibility, we analyzed the nuclear distribution of Pol II at successive erythroid maturation stages.
Indirect immunostaining shows Pol II exists in freely diffusible form and in distinct foci (transcription factories) that are the sites of the majority of RNA synthesis and contain multiple transcription factors (Grande et al. 1997). These transcription factories are attached to the underlying nucleoskeleton and show very little mobility and may thus represent one facet of nuclear architecture (Kimura et al. 1999).
We performed immunostaining experiments on sorted immature and fully differentiated nucleated erythroid fetal liver cells using an antibody recognizing both hyperphosphorylated and nonphosphorylated Pol II (for review, see Dahmus 1996). In the immature c-kit-positive population (stage 1), Pol II is distributed over hundreds of foci closely packed throughout the nucleoplasm with the exclusion of PCH, nucleoli, and most of the immediate nuclear periphery (Fig. 5a–c). As cells differentiate along the erythroid lineage, Pol II foci gradually diminish in number and no longer fill out most of the nucleoplasm. In the last sorted maturation stage, Pol II staining frequently resembles a cage-like structure, centered mostly in the nuclear interior but also reaching out to the periphery (Fig. 5d–f). These results demonstrate a changing nuclear landscape with erythroid differentiation, reflecting an increasingly restricted compartment available for gene expression.
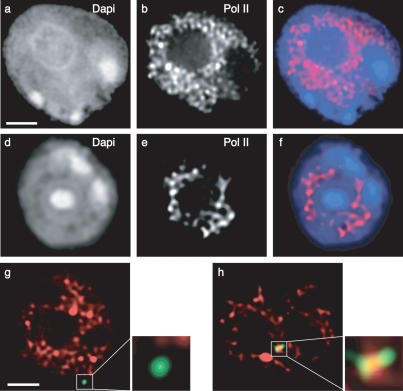
Figure 5.
Nuclear distribution of Pol II during erythroid differentiation and position of the β-globin locus with respect to Pol II. Sorted fetal liver cells (a–c, stage 1; d–f, stage 4) were immunostained for Pol II. (a,d) Nuclear counterstain DAPI. (b,e) Pol II. (c,f) Merged images of DAPI (blue) and Pol II (red). (g) In immature erythroid cells (stage 1), the predominantly peripheral β-globin locus (green) falls just outside Pol II staining (red). (h) In mature cells (stage 4), the locus lies most often in the nuclear interior and overlaps with strong foci of Pol II staining. Bars, 2 μm.
Next we examined the location of the wild-type β-globin locus relative to Pol II in immature and mature erythroid cells by ImmunoFISH. In the immature cell fraction, we found only limited direct overlap between the β-globin and Pol II signals. In particular, many of the peripheral β-globin loci remain just beyond the volume occupied by most Pol II foci (Fig. 5g). In contrast, in mature erythroid cells (stage 4), 94% of wild-type β-globin loci overlap with prominent Pol II staining (Fig. 5h). Thus, despite a decrease in available Pol II foci in maturing erythroid cells, enhanced overlap of the β-globin locus with Pol II is observed with maturation. This adherence to polymerase staining could explain the relocation of the β-globin locus toward the nuclear interior, as the Pol II foci are increasingly concentrated away from the periphery. However, analysis of the ΔLCR allele revealed that the mutant β-globin locus also coincides with Pol II foci in mature cells at a high frequency (87%; data not shown). This suggests that the ΔLCR allele is accessible to Pol II even though the locus tends to be located at the periphery (Fig. 4). This result is consistent with our previous finding that the LCR is not required for Pol II loading to the adult β-globin promoter (Sawado et al. 2003). Therefore, our results suggest that Pol II loading onto the locus is not sufficient for relocating the locus away from the periphery.
Only the wild-type β-globin locus consistently colocalizes with engaged Pol II
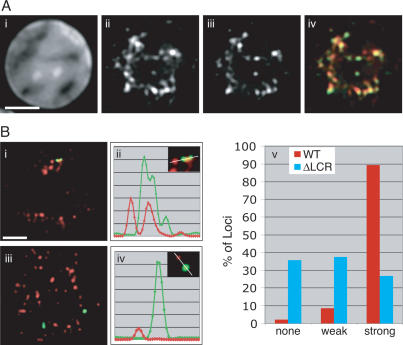
Although both the wild-type and ΔLCR β-globin loci frequently colocalize with Pol II in mature erythroid cells, transcription from the ΔLCR allele is much lower than wild-type levels (Bender et al. 2000). This suggests that the ΔLCR allele may not be contacting functionally equivalent Pol II foci. By extension, not all of the labeled Pol II foci may be actively transcribing (Kimura et al. 1999). Therefore, using an antibody directed specifically against the C-terminal domain (CTD) of Pol II phosphorylated at Ser 5 (Pol II-S5-P), we determined the association of both alleles with only the hyperphosphorylated form of Pol II, which represents the engaged population of the enzyme and true transcription factories, in erythroid cells. Immunostaining of cells at stage 4 revealed that Pol II hyperphosphorylated at the CTD marks a subpopulation of Pol II foci Fig. 6A). When analyzed by ImmunoFISH, nine out of 10 wild-type β-globin loci colocalize with or contact a strong Pol II-S5-P focus in these cells, including the peripheral loci (Fig. 6B; Supplementary Fig. 2). Association with high concentrations of transcriptionally active Pol II is consistent with the robust expression from the wild-type locus. In contrast, the ΔLCR allele does not show a preference for strong Pol II-S5-P foci (Fig. 6B). Instead, it appears evenly distributed among three categories: (1) no colocalization, (2) colocalization with weakly staining foci, and (3) colocalization with strong foci of hyperphosphorylated Pol II (Fig. 6B, panel v). We note that the signal strength of the majority of the weak Pol II-S5-P foci is <10% of the strong foci and likely represents background staining. Therefore, the difference in association of the wild-type and ΔLCR loci with strong Pol II-S5-P foci is highly significant. In particular, 80% of peripheral alleles do not contact strong foci of phosphorylated Pol II (Supplementary Fig. 2; data not shown), suggesting that the presence of the LCR is correlated with an increased probability of the β-globin locus to be associated with transcription factories containing engaged Pol II within the nuclear interior.
Figure 6.
The LCR is required for association with hyperphosphorylated Pol II. (A) Pol II hyperphosphorylated at the CTD (Pol II-Ser5-P) stains a subpopulation of Pol II foci. (Panel i) DAPI stain of a mature fetal liver cell. (Panel ii) Pol II stain. (Panel iii) Pol II-Ser5-P. (Panel iv) Merged image of panels ii and iii (Pol II, red; Pol II-Ser5-P, green). Bar, 2 μm. (B, panel i) In mature erythroid cells (stage 4), the wild-type β-globin locus (green) consistently colocalizes with Pol II-Ser5-P (red), unlike the ΔLCR alleles (in panel iii). Bar, 2 μm. (Panels ii,iv) Linescans of the images on the left, demonstrating the signal overlap between the β-globin locus and Pol II-Ser5-P for the wild-type locus (panel ii) and lack thereof for the mutant (panel iv). (Panel v) Frequency of colocalization of wild-type (red) and ΔLCR (blue) globin loci with Pol II-Ser5-P foci. “Weak” foci exhibit a signal intensity of <20% of the brightest foci detectable in a given nucleus.
Globin gene expression increases with erythroid maturation
Next we determined the expression status of the globin genes in the isolated erythroid maturation stages. We used primary transcript RNA FISH with labeled oligonucleotides directed against α-globin and βmajor-globin introns to assess globin transcriptional activity at the single allele level. The combined detection of α- and β-globin transcripts also permitted us to compare the temporal expression patterns of the two gene loci.
As shown in Figure 7A, an increasing fraction of cells show globin gene expression with progressing maturation. Detectable transcription foci were largely absent in the most immature cells (<2%). Both α (21%) and β-globin (13%) transcripts began to emerge with the appearance of the erythroid cell surface marker ERY-1 (stage 2). However, almost three-quarters of nuclei scoring positive for transcription foci exhibited only a single RNA FISH signal (either α-globin or βmajor-globin), suggesting that this cell population is only at the threshold of globin gene activation. By the intermediate stage (stage 3), most globin loci are actively transcribing. At this stage, only 69% of β-globin alleles appear active compared with 83% of α-globin alleles. Thus, α-globin may undergo longer or more frequent transcription bursts, as suggested by previous reports that α-globin expression slightly precedes that of β-globin (de Krom et al. 2002). This notion is supported by the observation that while 60% of nuclei express all α-globin and β-globin alleles (four RNA FISH spots), another 21% show two α-globin transcript signals but only one or no β-globin signal (data not shown). In contrast, in the most mature cell population, 89% of all α-globin and 86% of β-globin alleles exhibit primary transcript signals, and 79% of nuclei contain signals from all four loci. However, we detected a small percentage (∼7%) of nuclei without active α-globin or βmajor-globin transcription. These nuclei were frequently also the smallest and most compact; the absence of a visible RNA FISH signal may thus signal the final shutdown of globin transcription prior to enucleation. In support of this finding, in a human transcriptional analysis, Brown et al. (2006) observed a decrease in globin gene expression in their most mature isolated erythroid population.
Figure 7.
Expression of α- and β-globin during erythroid differentiation by primary transcript RNA FISH. (A) Sorted fetal liver cell populations were fixed and probed with oligonucleotides to introns of α and β-major transcripts. Seventy to 100 nuclei were analyzed for each cell fraction. (B) β-globin expression at the nuclear periphery. (Panel i) Examples of transcriptionally active peripheral β-globin loci. Fetal liver cells were probed for the β-globin locus (green) and β-major primary transcripts (red) by RNA/DNA FISH followed by immunostaining of lamin B (blue). Bar, 2 μm. Especially at early maturation stages, the β-globin locus colocalizes with the nuclear lamina, even when the locus is transcriptionally active. (Panel ii) Percentage of active β-globin loci at the nuclear periphery by 3D analysis at all three erythroid maturation stages that exhibit globin transcription. At least 50–60 active loci were analyzed for each stage.
In the ΔLCR cells, the expression pattern of α-globin was very similar to wild-type fetal liver (Fig. 7A), with transcriptional activity beginning at stage 2 (19% of alleles) and peaking at the most mature stage (95% of alleles). Given the requirement of the LCR for robust β-globin expression, only a small percentage (∼7%) of βmajor-globin alleles exhibited weak transcription signals at stage 2, and the proportion of visibly active alleles fluctuated between 11% and 18% in the more mature cells.
Transcriptional activity of the β-globin locus does not depend on an internal nuclear position
The above localization experiments could not determine whether the relocation of the β-globin locus during erythroid differentiation is a cause or consequence of transcriptional activity or whether the LCR directly contributes to the process. However, our RNA FISH data reveal an apparent disconnect between transcriptional activity and nuclear localization: Primary transcript foci become visible (stage 2) before significant relocation of the β-globin locus from the peripheral shell (stages 3 and 4). To locate the transcriptionally active loci, we performed RNA/DNA FISH experiments to determine position and expression status concomitantly. We included a lamin B stain to accurately assess peripheral location in 3D. Contrary to expectation, we find that expressing loci are frequently located at the nuclear periphery, especially in the more immature erythroid cells (Fig. 7B, panel i). While activated transcription has been reported at the nuclear periphery in yeast (Ishii et al. 2002; Brickner and Walter 2004; Casolari et al. 2004, 2005), this compartment is predominantly regarded as repressive in mammalian systems. However, 3D analysis of stage 2 cells, which are at the threshold of robust β-globin gene expression, reveals that 62% of transcriptionally active loci are in contact with lamin B and thus peripheral (Fig. 7B, panel ii). This proportion subsequently decreases to 36% and 32% at the more mature stages 3 and 4, respectively, when the majority of loci shows strong primary transcript foci. These results suggest that nuclear relocation of the loci at later maturation stages is likely to be a consequence of continued transcription rather than a requirement for transcription. Furthermore, we show that robust gene expression can also occur at the nuclear periphery in mammalian cells.
Discussion
Recent work from a number of laboratories has revealed that the interphase nucleus is organized nonrandomly and in a tissue-specific manner (for reviews, see Cremer and Cremer 2001; Kosak and Groudine 2004; Parada et al. 2004). Erythropoiesis represents a well-characterized example of differentiation marked by increasingly restrictive gene expression (Bruno et al. 2004), nuclear compaction, progressive heterochromatinization, and eventual enucleation. We have examined the spatial organization associated with the activation of the β-globin locus during erythropoiesis.
Nuclear position and expression of the β-globin locus
On the basis of differential expression of three surface markers, we isolated murine fetal liver cells from four stages of erythroid maturation and characterized these cells with respect to morphology, β-globin locus position, and β-globin gene expression (Figs. 1, 2, 7A; Supplementary Fig. 1). In contrast to mature erythroid cells, in early progenitors and immature erythroblasts (stages 1 and 2) few β-globin loci are transcriptionally active, and the β-globin locus is more often found at the nuclear periphery. The β-globin locus is similarly biased toward the nuclear envelope in B lymphocytes and undifferentiated ES cells in which the globin genes are silent, consistent with reports that proximity to perinuclear heterochromatin correlates inversely with transcriptional activity (Kosak et al. 2002; Zink et al. 2004). With erythroid maturation, the β-globin locus is less likely to be located at the nuclear periphery while expression increases, peaking in the most mature fraction. This behavior contrasts with that of the IgH locus, which remains near the periphery in a high percentage of mature erythroid cells, but relocates to the nuclear interior in splenic B lymphocytes. Thus, both loci exhibit cell type- and activation-dependent nuclear localization associated with differentiation.
Both a peripheral position or proximity to PCH has been correlated with transcriptional repression during differentiation (Brown et al. 1997; Brown et al. 1999; Francastel et al. 1999; Su et al. 2004). For example, we demonstrated that up to 60% of β-globin loci colocalized with PCH in undifferentiated murine erythroleukemia (MEL) cells in which the β-globin genes are transcriptionally inactive, but only 8% of β-globin alleles contacted PCH after induction of differentiation (Francastel et al. 2001). In contrast, the β-globin locus infrequently colocalizes with PCH in primary fetal liver cells, even in immature cells prior to transcriptional activation. Instead, the locus is often at the nuclear periphery. This discrepancy may reflect a biological difference between the nuclear organization of transformed cell lines and primary cells and/or differences revealed by the current 3D and former 2D analyses. Regardless, the bias of the β-globin locus for the nuclear periphery rather than PCH when inactive in immature fetal liver (i.e., stages 1 and 2) and ES cells might reflect functional differences between these two repressive compartments. In fact, several reports suggest that any particular silenced gene associates either with PCH or the periphery, but rarely both (Skok et al. 2001; Kosak et al. 2002; Zink et al. 2004; Williams et al. 2006). PCH consists of constitutive heterochromatin that replicates late in S phase, is marked by high levels of trimethyl-H3K9 (Dimitrova and Gilbert 1999; Wu et al. 2005), and represents a compartment for stable epigenetic gene repression (as observed in T- and B-cell differentiation) (Brown et al. 1997, 1999; Su et al. 2004). In contrast, the bulk of the chromatin at the nuclear periphery replicates in mid-S phase and is enriched in dimethyl-H3K9, consistent with facultative heterochromatin (Wu et al. 2005). Furthermore, experiments by Hutchison and Weintraub (1985) demonstrated that, unlike PCH, the nuclear periphery is enriched in DNase I-sensitive chromatin, suggesting the presence of poised or active genes. As such, the periphery may harbor gene loci that are silent but responsive to developmental cues to become activated or repressed. In ES cells, in which the globin locus is peripheral, differentiation along the hematopoietic and then erythroid lineages is one possible fate. Therefore, the locus may be competent for activation in the nuclear periphery of ES cells, while in immature fetal liver cells it is poised to activate the globin genes. This is in contrast to activated T cells where the β-globin locus is stably silenced by recruitment to PCH (Brown et al. 2001). We find that the β-globin locus associates with PCH only at a very low frequency in splenic B cells. However, most of these B cells are in a quiescent state, and recruitment of heritably silenced genes to PCH only occurs in cycling but not resting lymphoid cells (Brown et al. 1997, 1999).
One striking finding is that robust β-globin expression (commencing at stage 2) precedes the most dramatic relocation of the β-globin locus away from the nuclear periphery (stages 3 and 4). In fact, many β-globin loci that associate with strong transcription foci colocalize with the nuclear lamina. Thus, our results provide further evidence that robust transcription is possible in the immediate vicinity of the nuclear periphery in mammalian systems, and that the relocation of the β-globin locus to the nuclear interior is more likely a consequence of transcription rather than the basis for transcription.
Although the nuclear periphery has been described traditionally as a repressive compartment and repository for gene-poor chromosome regions (Gottschling et al. 1990; Gotta and Gasser 1996; Andrulis et al. 1998, 2002; Laroche et al. 1998; Croft et al. 1999; Galy et al. 2000; Boyle et al. 2001), several recent studies in yeast have demonstrated that repressive and activating domains can coexist at the periphery. A genome-wide analysis revealed that a subset of highly active genes (e.g., induced GAL genes) associate with the nuclear export machinery and are relocated to the periphery (Casolari et al. 2004). In addition, the majority of α-factor-induced genes, involving loci from multiple chromosomes, associate with components of the nuclear envelope upon activation, suggesting a global nuclear reorganization (Casolari et al. 2005). INO1, a nucleoplasmic gene locus when repressed, relocates to the periphery when activated by the unfolded protein response (Brickner and Walter 2004). Furthermore, Ishii et al. (2002) demonstrated that genes can be tethered to the periphery and still be transcriptionally active when protected by boundary activities conferred by components of the nuclear transport machinery. More recently, Schmid et al. (2006) reported that a transient association of genes with the nuclear pore basket may represent an integral part of the activation process for many genes.
In mammalian systems, the periphery is largely considered to be a transcriptionally inactive compartment, particularly in undifferentiated or progenitor cells (Imai et al. 1997; Croft et al. 1999; Boyle et al. 2001; Kosak et al. 2002; Zhou et al. 2002; Zink et al. 2004; Williams et al. 2006). Moreover, movement away from the periphery has in several instances been correlated with transcriptional activity. For example, in a Chinese hamster ovary (CHO) cell clone, a lacO array relocates away from the periphery when targeted by VP16 and induced to express a reporter gene (Tumbar and Belmont 2001). Rudolph and colleagues (Zink et al. 2004) showed that genes adjacent to CFTR independently adopt tissue-specific positions relative to the nuclear periphery that reflect their individual transcriptional state with interior location correlating with activity. The Mash1 locus is positioned at the periphery when repressed in ES cells but is relocated to the interior when it becomes transcriptionally active during neuronal differentiation (Williams et al. 2006). However, Fisher’s group (Hewitt et al. 2004) has also shown that the mammalian nuclear periphery is not universally repressive, as the IFNγ gene remains peripheral in developing T-helper cells (as well as B cells and fibroblasts) even when transcriptionally active.
It is tempting to invoke the gene gating hypothesis, first proposed by Blobel (1985), to explain the significant number of β-globin loci we observe expressing at the nuclear periphery (e.g., >60% of active loci at stage 2 are in contact with the nuclear lamina). Placement of active genes in the proximity of nuclear pore complexes (NPCs) would ensure the rapid export of mature transcripts from the nucleoplasm and thus promote efficient downstream processing. This proximity could be particularly advantageous during the earliest bursts of β-globin gene expression, when it may be imperative to quickly reach a threshold of β-globin protein synthesis to balance α-globin levels. In fact, as assayed by ImmunoFISH, peripheral loci also colocalize with foci of NPC components (data not shown). While this observation is in accord with the gene gating model, NPCs are embedded within the lamina and are present at high density in the erythroid cells. Thus, at this resolution, it is not possible to distinguish association of the β-globin locus with the lamina versus with NPCs. Furthermore, the fraction of transcriptionally active β-globin loci at the periphery decreases with erythroid maturation and higher frequency of expression. This result is more consistent with a model of stochastic initiation of globin gene activation (de Krom et al. 2002), where a fraction of loci may be triggered while still at the periphery and subsequently relocate to the interior. On the other hand, the β-globin locus may not need to be near the nuclear periphery in the more mature erythroblasts with their high levels of globin transcripts and steady flow of newly synthesized RNA to the cytoplasm.
It is possible that the shift in location of the β-globin locus is a consequence of general nuclear reorganization during erythroid differentiation. With the progression of an increasingly restricted tissue-specific expression program, numerous genes must be positioned in transcriptionally competent environments, while others are stably silenced. Colocalization of the β-globin locus with active Pol II factories would explain the relocation, since the Pol II distribution changes with maturation. While transcription factories fill the nucleoplasm in early progenitors, their abundance diminishes with differentiation, and they become concentrated in ridges in the nuclear interior. This change is accompanied by a decrease in the number of active genes in the terminally differentiating cells (Bruno et al. 2004) and an increase in facultative heterochromatin that renders whole chromatin domains inaccessible to the transcriptional machinery. Particularly in late erythroblasts, the number of transcription factories is likely to be lower than the number of genes that need to be expressed, suggesting that the Pol II foci are hubs of transcriptional activity, each shared by several genes (Iborra et al. 1996; Osborne et al. 2004; Chakalova et al. 2005). Evidence suggests that the transcription factories are firmly attached to the underlying nucleoskeleton and thus rather immobile (which does not preclude dynamic protein exchange within or between foci) (Kimura et al. 1999). Our own results are consistent with the hypothesis that once transcriptionally engaged, the β-globin locus remains associated with or in the immediate vicinity of a factory, which would gradually pull it toward the nuclear interior as cells differentiate.
Genetic analysis of globin gene positioning
We previously showed that the human β-globin LCR is required for a high incidence of looping of the locus away from its CT (Ragoczy et al. 2003). Our current results demonstrate that the murine β-globin LCR is necessary for efficient relocation of the locus from the periphery toward the nuclear interior. Our results suggest that the LCR-dependent relocation from the periphery follows, rather than precedes, robust globin gene activation.
We also observed that the ΔLCR allele contacts Pol II foci at a frequency similar to the wild-type allele in vivo. This result is consistent with our previous demonstration by chromatin immunoprecipitation (ChIP) that the ΔLCR and wild-type alleles similarly associate with Pol II and other components of the preinitiation complex (Sawado et al. 2003). The LCR is therefore not required for association of the locus with transcription factories. However, we demonstrate that association with Pol II foci is required, but not sufficient, for transcription. All active wild-type and ΔLCR loci are associated with Pol II foci, but only a small fraction of ΔLCR loci exhibit primary transcript signals. Our ChIP results revealed that the wild-type allele is much more highly associated with the engaged form of Pol II (hyperphosphorylated at the CTD [S5-P]) than is the ΔLCR allele (Sawado et al. 2003). By ImmunoFISH, we now find that the wild type, but not the ΔLCR allele, associates consistently with foci of Pol II S5-P in the most mature erythroid cells. Consequently, the LCR might either nucleate foci of active transcription factories or enhance the affinity of the locus for such foci. We cannot distinguish between these two possibilities. Nevertheless, association with hyperphosphorylated Pol II is insufficient to ensure transcription, since only ∼10% of ΔLCR alleles are transcriptionally active, yet ∼30% of loci colocalize with strong foci of engaged Pol II. The LCR therefore most likely contributes to the recruitment of additional erythroid-specific transcription factors to assemble fully competent transcription factories at the β-globin locus.
Materials and methods
Cells and cell sorting
Undifferentiated ES cells (AK7) were maintained in Dubelco’s modified media supplemented with 20% fetal bovine serum and leukemia inhibitory factor (LIF). CD45 (B220+) B cells were isolated from adult wild-type murine spleens by two rounds of positive AutoMACS selection and immediately fixed onto polylysine-coated microscope slides. Fetal liver cells were obtained from dpc-13.5 wild-type and homozygous ΔLCR mouse embryos. Livers were disrupted in PBS and cells were fluorescently labeled with an APC-conjugated antibody against CD-117 (c-kit; Pharmingen), PE-conjugated Ter-119 (Pharmingen), and antibody ERY-1 (Bacon and Sytkowski 1987), which was followed by a FITC-conjugated anti-rat polyclonal antiserum (Pharmingen). Cells were then separated into the CD117+/ERY-1−/Ter-119− (stage 1), CD117+/ERY-1+/Ter-119− (stage 2), CD117−/ERY-1+/Ter-119+ (stage 3), and CD117−/ERY-1−/Ter-119+ (stage 4) stages on a Vantage FACS sorter. Following separation, the cells were incubated for 1 h at 37°C in erythroid progenitor culture media (Carotta et al. 2004) prior to fixation onto microscope slides.
DNA, RNA FISH, and ImmunoFISH
DNA and RNA FISH were performed essentially as described (Wijgerde et al. 1995; Croft et al. 1999). For details on modifications to the protocols, probes, and antibodies used for ImmunoFISH, see the Supplemental Material.
Image Analysis
Image stacks (Z sections spaced 0.25 μm apart) were captured on a Zeiss Axiovert 100 TV microscope equipped with a cooled CCD camera and subsequently deconvolved using Deltavision SoftWorx software (Applied Precision). For 2D distance measurements, the image stacks were projected onto a single plane and analyzed in Photoshop CS (Adobe). The shortest 2D distance from the centers of signal intensity of a FISH spot (gene locus) to the nuclear periphery (defined by a sharp drop in DAPI stain) was measured in pixels. Measurements were standardized to nuclear size by dividing each distance by the average radius of its nucleus. Seventy to 140 nuclei for each cell type/maturation stage were scored. Experiments were performed up to three times to ensure consistency and until statistical significance was achieved. Statistical differences between observation groups were determined by the Kolmogorov-Smirnov test. To assess the position of the β-globin locus relative to PCH or protein staining (Pol II, lamin B), colocalization or contact of the locus’ FISH signal with the relevant compartment was analyzed in 3D by reconstruction of the nuclear volume using Softworx (Applied Precision) and Imaris (Bitplane) software. In RNA FISH experiments the presence of hybridization signals for α- and β-globin was determined in 2D projections of the Z stacks.
Acknowledgments
We thank Arthur Sytkowski for the generous gift of the ERY-1 antibody, David Scalzo for discussions on and assistance with statistical analyses, and Barbara Trask, Steven Henikoff, Tomoyuki Sawado, and Steven Kosak for critical reading of the manuscript. This work was supported by American Society of Hematology Scholar and Cooley’s Anemia Foundation Awards (to M.A.B.) and NIH Grants DK 44746 and HL57620 (to M.G.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1419506
References
- Andrulis E.D., Neiman A.M., Zappulla D.C., Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- Andrulis E.D., Zappulla D.C., Ansari A., Perrod S., Laiosa C.V., Gartenberg M.R., Sternglanz R. Esc1, a nuclear periphery protein required for Sir4-based plasmid anchoring and partitioning. Mol. Cell. Biol. 2002;22:8292–8301. doi: 10.1128/MCB.22.23.8292-8301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon E.R., Sytkowski A.J. Identification and characterization of a differentiation-specific antigen on normal and malignant murine erythroid cells. Blood. 1987;69:103–108. [PubMed] [Google Scholar]
- Bender M.A., Bulger M., Close J., Groudine M. β-Globin gene switching and DNase I sensitivity of the endogenous β-globin locus in mice do not require the locus control region. Mol. Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- Blobel G. Gene gating: A hypothesis. Proc. Natl. Acad. Sci. 1985;82:8527–8529. doi: 10.1073/pnas.82.24.8527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S., Gilchrist S., Bridger J.M., Mahy N.L., Ellis J.A., Bickmore W.A. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 2001;10:211–219. doi: 10.1093/hmg/10.3.211. [DOI] [PubMed] [Google Scholar]
- Brickner J.H., Walter P. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2004;2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K.E., Guest S.S., Smale S.T., Hahm K., Merkenschlager M., Fisher A.G. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Brown K.E., Baxter J., Graf D., Merkenschlager M., Fisher A.G. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- Brown K.E., Amoils S., Horn J.M., Buckle V.J., Higgs D.R., Merkenschlager M., Fisher A.G. Expression of α- and β-globin genes occurs within different nuclear domains in haemopoietic cells. Nat. Cell Biol. 2001;3:602–606. doi: 10.1038/35078577. [DOI] [PubMed] [Google Scholar]
- Brown J.M., Leach J., Reittie J.E., Atzberger A., Lee-Prudhoe J., Wood W.G., Higgs D.R., Iborra F.J., Buckle V.J. Coregulated human globin genes are frequently in spatial proximity when active. J. Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno L., Hoffmann R., McBlane F., Brown J., Gupta R., Joshi C., Pearson S., Seidl T., Heyworth C., Enver T. Molecular signatures of self-renewal, differentiation, and lineage choice in multipotential hemopoietic progenitor cells in vitro. Mol. Cell. Biol. 2004;24:741–756. doi: 10.1128/MCB.24.2.741-756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotta S., Pilat S., Mairhofer A., Schmidt U., Dolznig H., Steinlein P., Beug H. Directed differentiation and mass cultivation of pure erythroid progenitors from mouse embryonic stem cells. Blood. 2004;104:1873–1880. doi: 10.1182/blood-2004-02-0570. [DOI] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Komili S., West J., Hieronymus H., Silver P.A. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Drubin D.A., Rando O.J., Silver P.A. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes & Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakalova L., Debrand E., Mitchell J.A., Osborne C.S., Fraser P. Replication and transcription: Shaping the landscape of the genome. Nat. Rev. Genet. 2005;6:669–677. doi: 10.1038/nrg1673. [DOI] [PubMed] [Google Scholar]
- Chambeyron S., Bickmore W.A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes & Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S., Da Silva N.R., Lawson K.A., Bickmore W.A. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- Cobb B.S., Morales-Alcelay S., Kleiger G., Brown K.E., Fisher A.G., Smale S.T. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes & Dev. 2000;14:2146–2160. doi: 10.1101/gad.816400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Croft J.A., Bridger J.M., Boyle S., Perry P., Teague P., Bickmore W.A. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csink A.K., Henikoff S. Genetic modification of heterochromatic association and nuclear organization in Drosophila. Nature. 1996;381:529–531. doi: 10.1038/381529a0. [DOI] [PubMed] [Google Scholar]
- Dahmus M.E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- de Krom M., de van Corput M., von Lindern M., Grosveld F., Strouboulis J. Stochastic patterns in globin gene expression are established prior to transcriptional activation and are clonally inherited. Mol. Cell. 2002;9:1319–1326. doi: 10.1016/s1097-2765(02)00558-0. [DOI] [PubMed] [Google Scholar]
- Dimitrova D.S., Gilbert D.M. The spatial position and replication timing of chromosomal domains are both established in early G1 phase. Mol. Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- Francastel C., Walters M.C., Groudine M., Martin D.I. A functional enhancer suppresses silencing of a transgene and prevents its localization close to centrometric heterochromatin. Cell. 1999;99:259–269. doi: 10.1016/s0092-8674(00)81657-8. [DOI] [PubMed] [Google Scholar]
- Francastel C., Magis W., Groudine M. Nuclear relocation of a transactivator subunit precedes target gene activation. Proc. Natl. Acad. Sci. 2001;98:12120–12125. doi: 10.1073/pnas.211444898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V., Olivo-Marin J.C., Scherthan H., Doye V., Rascalou N., Nehrbass U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 2000;403:108–112. doi: 10.1038/47528. [DOI] [PubMed] [Google Scholar]
- Gotta M., Gasser S.M. Nuclear organization and transcriptional silencing in yeast. Experientia. 1996;52:1136–1147. doi: 10.1007/BF01952113. [DOI] [PubMed] [Google Scholar]
- Gottschling D.E., Aparicio O.M., Billington B.L., Zakian V.A. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Grande M.A., van der Kraan I., de Jong L., van Driel R. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J. Cell Sci. 1997;110:1781–1791. doi: 10.1242/jcs.110.15.1781. [DOI] [PubMed] [Google Scholar]
- Hewitt S.L., High F.A., Reiner S.L., Fisher A.G., Merkenschlager M. Nuclear repositioning marks the selective exclusion of lineage-inappropriate transcription factor loci during T helper cell differentiation. Eur. J. Immunol. 2004;34:3604–3613. doi: 10.1002/eji.200425469. [DOI] [PubMed] [Google Scholar]
- Hutchison N., Weintraub H. Localization of DNase I-sensitive sequences to specific regions of interphase nuclei. Cell. 1985;43:471–482. doi: 10.1016/0092-8674(85)90177-1. [DOI] [PubMed] [Google Scholar]
- Iborra F.J., Pombo A., Jackson D.A., Cook P.R. Active RNA polymerases are localized within discrete transcription ‘factories’ in human nuclei. J. Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- Imai S., Nishibayashi S., Takao K., Tomifuji M., Fujino T., Hasegawa M., Takano T. Dissociation of Oct-1 from the nuclear peripheral structure induces the cellular aging-associated collagenase gene expression. Mol. Biol. Cell. 1997;8:2407–2419. doi: 10.1091/mbc.8.12.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Arib G., Lin C., Van Houwe G., Laemmli U.K. Chromatin boundaries in budding yeast: The nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- Kimura H., Tao Y., Roeder R.G., Cook P.R. Quantitation of RNA polymerase II and its transcription factors in an HeLa cell: Little soluble holoenzyme but significant amounts of polymerases attached to the nuclear substructure. Mol. Cell. Biol. 1999;19:5383–5392. doi: 10.1128/mcb.19.8.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak S.T., Groudine M. Form follows function: The genomic organization of cellular differentiation. Genes & Dev. 2004;18:1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- Kosak S.T., Skok J.A., Medina K.L., Riblet R., Le Beau M.M., Fisher A.G., Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Laroche T., Martin S.G., Gotta M., Gorham H.C., Pryde F.E., Louis E.J., Gasser S.M. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 1998;8:653–656. doi: 10.1016/s0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- Osborne C.S., Chakalova L., Brown K.E., Carter D., Horton A., Debrand E., Goyenechea B., Mitchell J.A., Lopes S., Reik W., et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- Parada L.A., McQueen P.G., Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004;5:R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy T., Telling A., Sawado T., Groudine M., Kosak S.T. A genetic analysis of chromosome territory looping: Diverse roles for distal regulatory elements. Chromosome Res. 2003;11:513–525. doi: 10.1023/a:1024939130361. [DOI] [PubMed] [Google Scholar]
- Sawado T., Halow J., Bender M.A., Groudine M. The β-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes & Dev. 2003;17:1009–1018. doi: 10.1101/gad.1072303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Arib G., Laemmli C., Nishikawa J., Durussel T., Laemmli U.K. Nup-PI: The nucleopore–promoter interaction of genes in yeast. Mol. Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Schubeler D., Groudine M., Bender M.A. The murine β-globin locus control region regulates the rate of transcription but not the hyperacetylation of histones at the active genes. Proc. Natl. Acad. Sci. 2001;98:11432–11437. doi: 10.1073/pnas.201394698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skok J.A., Brown K.E., Azuara V., Caparros M.L., Baxter J., Takacs K., Dillon N., Gray D., Perry R.P., Merkenschlager M., et al. Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat. Immunol. 2001;2:848–854. doi: 10.1038/ni0901-848. [DOI] [PubMed] [Google Scholar]
- Socolovsky M., Nam H., Fleming M.D., Haase V.H., Brugnara C., Lodish H.F. Ineffective erythropoiesis in Stat5a−/−5b−/− mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- Su R.C., Brown K.E., Saaber S., Fisher A.G., Merkenschlager M., Smale S.T. Dynamic assembly of silent chromatin during thymocyte maturation. Nat. Genet. 2004;36:502–506. doi: 10.1038/ng1351. [DOI] [PubMed] [Google Scholar]
- Trimborn T., Gribnau J., Grosveld F., Fraser P. Mechanisms of developmental control of transcription in the murine α- and β-globin loci. Genes & Dev. 1999;13:112–124. doi: 10.1101/gad.13.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T., Belmont A.S. Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat. Cell Biol. 2001;3:134–139. doi: 10.1038/35055033. [DOI] [PubMed] [Google Scholar]
- Volpi E.V., Chevret E., Jones T., Vatcheva R., Williamson J., Beck S., Campbell R.D., Goldsworthy M., Powis S.H., Ragoussis J., et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 2000;113:1565–1576. doi: 10.1242/jcs.113.9.1565. [DOI] [PubMed] [Google Scholar]
- Wijgerde M., Grosveld F., Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- Williams R.R., Broad S., Sheer D., Ragoussis J. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 2002;272:163–175. doi: 10.1006/excr.2001.5400. [DOI] [PubMed] [Google Scholar]
- Williams R.R., Azuara V., Perry P., Sauer S., Dvorkina M., Jorgensen H., Roix J., McQueen P., Misteli T., Merkenschlager M., et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- Wu R., Terry A.V., Singh P.B., Gilbert D.M. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol. Biol. Cell. 2005;16:2872–2881. doi: 10.1091/mbc.E04-11-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Ermakova O.V., Riblet R., Birshtein B.K., Schildkraut C.L. Replication and subnuclear location dynamics of the immunoglobulin heavy-chain locus in B-lineage cells. Mol. Cell. Biol. 2002;22:4876–4889. doi: 10.1128/MCB.22.13.4876-4889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D., Amaral M.D., Englmann A., Lang S., Clarke L.A., Rudolph C., Alt F., Luther K., Braz C., Sadoni N., et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J. Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]