Abstract
Somatic mutations in exons encoding the tyrosine kinase domain of the epidermal growth factor receptor (EGFR) gene are found in human lung adenocarcinomas and are associated with sensitivity to the tyrosine kinase inhibitors gefitinib and erlotinib. Nearly 90% of the EGFR mutations are either short, in-frame deletions in exon 19 or point mutations that result in substitution of arginine for leucine at amino acid 858 (L858R). To study further the role of these mutations in the initiation and maintenance of lung cancer, we have developed transgenic mice that express an exon 19 deletion mutant (EGFRΔL747–S752) or the L858R mutant (EGFRL858R) in type II pneumocytes under the control of doxycycline. Expression of either EGFR mutant leads to the development of lung adenocarcinomas. Two weeks after induction with doxycycline, mice that express the EGFRL858R allele show diffuse lung cancer highly reminiscent of human bronchioloalveolar carcinoma and later develop interspersed multifocal adenocarcinomas. In contrast, mice expressing EGFRΔL747–S752 develop multifocal tumors embedded in normal lung parenchyma with a longer latency. With mice carrying either EGFR allele, withdrawal of doxycycline (to reduce expression of the transgene) or treatment with erlotinib (to inhibit kinase activity) causes rapid tumor regression, as assessed by magnetic resonance imaging and histopathology, demonstrating that mutant EGFR is required for tumor maintenance. These models may be useful for developing improved therapies for patients with lung cancers bearing EGFR mutations.
Keywords: EGFR, lung adenocarcinoma, mice, tyrosine kinase inhibitor
The epidermal growth factor receptor (EGFR) is a membrane-bound receptor tyrosine kinase that belongs to a subfamily of four closely related receptors: HER1/EGFR/ERBB1, HER2/NEU/ERBB2, HER3/ERBB3, and HER4/ERBB4. Upon ligand binding, these receptors form homo- or heterodimers leading to autophosphorylation of key tyrosine residues in the cytosolic domains of the proteins. The phosphorylated tyrosine residues become docking sites for signaling molecules, thereby activating cellular signaling pathways that regulate cell proliferation and survival.
EGFR is either mutated or shows altered expression in a variety of human cancers. Deletions of exons 2–7 of EGFR, which encode part of the extracellular domain of the protein, have been observed in gliomas (Ekstrand et al. 1992), and mutations in exons encoding the tyrosine kinase domains of EGFR are found in 10% of lung adenocarcinomas (Lynch et al. 2004; Paez et al. 2004; Pao et al. 2004). High levels of EGFR have been described in many human tumors including gliomas and carcinomas of the head and neck, lung, breast, ovary, and bladder. Moreover, gliomas and lung cancers frequently exhibit increased copies of EGFR (Wong et al. 1987; Hirsch et al. 2003).
Nearly 90% of the lung adenocarcinoma-associated somatic mutations in the kinase-encoding portion of the EGFR gene fall into one of two classes: in-frame deletions in exon 19 that eliminate the conserved LREA motif and a T-to-G base substitution in exon 21 that substitutes arginine for leucine at position 858 (L858R). Patients whose tumors contain either of these two classes of mutations have similar clinical characteristics; they frequently are female, Asian, and never-smokers, and their adenocarcinomas often show bronchioloalveolar features. Furthermore, these and other less common mutations in exons encoding the kinase domain of EGFR are associated with sensitivity to the tyrosine kinase inhibitors (TKIs) gefitinib and erlotinib. Mutations have been detected in ∼85% of patients who have had clinical or radiographic responses to these agents, but in only 5% of patients refractory to treatment (Huang et al. 2004; Lynch et al. 2004; Paez et al. 2004; Mitsudomi et al. 2005; Pao et al. 2005a; Tokumo et al. 2005).
It is still unclear whether a response to TKI treatment translates into increased survival for these patients. Patients with lung tumors bearing EGFR mutations and treated with TKIs show an improved overall survival when compared with patients with tumors without detectable EGFR mutations (Cappuzzo et al. 2005; Chou et al. 2005; Han et al. 2005; Mitsudomi et al. 2005; Tokumo et al. 2005), but EGFR-mutant lung cancer may be a less aggressive disease, on average, than lung cancer with wild-type EGFR. In support of this, adding erlotinib to chemotherapy does not appear to improve overall survival in patients with EGFR-mutant lung tumors (Eberhard et al. 2005).
In most patients with tumors bearing EGFR mutations who initially respond to erlotinib and gefitinib with symptomatic improvement and reduction in tumor size, the cancer resumes detectable growth within 6 mo to 2 yr. In ∼50% of these resistant tumors, the mutant EGFR allele has acquired a secondary mutation in exon 20, which leads to substitution of methionine for threonine at position 790 (T790M) in the kinase domain (Kobayashi et al. 2005; Pao et al. 2005b). The secondary change is predicted to block binding of drug to the ATP-binding pocket, strengthening the hypothesis that EGFR is the main target of gefitinib and erlotinib when these drugs induce tumor regression (Kobayashi et al. 2005; Kwak et al. 2005; Pao et al. 2005b).
The rapid response to TKIs observed in non-small-cell lung cancer (NSCLC) patients with tumors bearing EGFR mutations suggests that the viability of the cancer cells depends on the continued activity of mutant EGFR. These observations are supported by experiments in vitro; TKIs and mutant allele-specific siRNAs induce apoptosis in human lung adenocarcinoma cell lines carrying mutant EGFR (Sordella et al. 2004; Tracy et al. 2004).
The oncogene dependence of tumors has been studied most extensively in the mouse using tetracycline-regulated transgenic oncogenes in mouse models of cancer (Felsher 2004; Varmus et al. 2005). Tumors induced by such regulated oncogenes generally regress rapidly—by apoptosis or differentiation—when expression of the oncogene is reduced. For example, we have previously described a tetracycline-inducible model of lung adenocarcinoma in which lung tumors that arise as a result of oncogenic Kras are dependent on sustained expression of the mutant transgene for tumor maintenance (Fisher et al. 2001).
To understand further the role of mutant EGFRs in initiation, progression, and maintenance of lung adenocarcinomas in vivo, we have generated tetracycline-inducible transgenic mice that express either an exon 19 deletion or the L858R mutant in lung epithelial cells. We regulated the expression of these EGFR mutants by the administration and withdrawal of doxycycline and tested the ability of erlotinib to treat lung tumors that developed in these mice.
Results
Generation of mice carrying inducible lung-specific mutant EGFR transgenes
To generate mice in which expression of mutant EGFR can be regulated in type II alveolar epithelial cells, we used a tetracycline-inducible system known to regulate expression of oncogenic Kras in such cells under the control of the reverse tetracycline transactivator (rtTA), which itself is driven by the Clara cell secretory protein (CCSP) promoter (Tichelaar et al. 2000; Fisher et al. 2001). EGFR transgenic constructs were generated by cloning human cDNAs encoding either EGFRL858R or EGFRΔL747–S752 (Pao et al. 2004) downstream of a tetracycline-responsive element (TetO), followed at the 3′ end by an intron and polyadenylation sequence derived from the mouse protamine-1 gene to ensure stability of the mRNA (Fig. 1A).
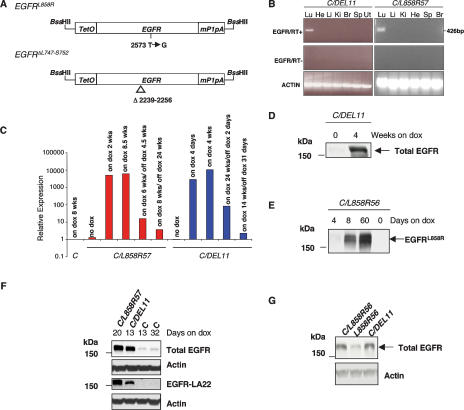
Figure 1.
EGFR transgenes and their expression patterns. (A) Constructs used to generate tetracycline-inducible EGFR transgenic mice. Mutant human EGFR cDNAs (EGFRL858R and EGFRΔL747–S752) were cloned between a tetracycline operator sequence and the mouse protamine-1 polyadenylation sequence. (TetO) Tetracycline operator; (mP1pA) mouse protamine-1 gene polyadenylation sequence. (B) Expression of EGFRΔL747–S752 (DEL11, left panels) and EGFRL858R (L858R57, right panels) is restricted to lung tissue in bitransgenic mice maintained on doxycycline for 8 wk and 3 wk, respectively. PCR reactions were carried out in the presence (top) and absence (middle) of RT and products were visualized after electrophoresis in a 2% agarose gel. Amplification of actin mRNA by RT–PCR confirmed the presence of RNA in all samples. (Lu) Lung; (He) heart; (Li) liver; (Ki) kidney; (Br) brain; (Sp) spleen; (Ut) uterus. (C) Quantitative RT–PCR analysis of transgene expression in C/L858R57 and C/DEL11 bitransgenic mice and C transgenic control mice before, during, and after doxycycline treatment. (D–G) Mutant EGFR protein is expressed at high levels in lungs from induced mice. (D) Lung extracts from C/DEL11 mice maintained on doxycycline for the times indicated were immunoblotted with an antibody that recognizes human and mouse EGFR (Total EGFR). (E) Lung protein extracts from C/L858R bitransgenic mice maintained on doxycycline for the times indicated were immunoblotted with an EGFRL858R-specific antibody (see Materials and Methods for details). (F) Western analysis of lung protein extracts from mice maintained on doxycycline for 13–32 d using the total EGFR antibody and an antibody that recognizes human EGFR with a higher affinity than mouse EGFR (EGFR-LA22, third panel). (G) Western analysis of lung protein extracts from bitransgenic mice maintained on doxycycline for 4 d using the total EGFR antibody. Anti-actin blots are shown as loading controls.
Eighteen TetO-EGFRL858R (L858R) and 11 TetO-EGFRΔL747–S752 (DEL) founders were identified among the 83 and 43 pups obtained, respectively, from the injection of each transgene. We bred the founders to CCSP-rtTA (C) mice and placed bitransgenic offspring from 13 L858R and eight DEL founders on a diet containing doxycycline within 1 wk of weaning. After 2–16 wk, progeny were assayed for transgene expression using RT–PCR. We identified animals that expressed the transgene in the lung and further characterized two L858R lines (L858R56 and L858R57) and two DEL lines (DEL9 and DEL11).
Analysis of transgene expression
To test whether the transgene was expressed specifically in the lung, we performed RT–PCR on multiple tissues from C/DEL and C/L858R bitransgenic mice that received doxycycline. Expression of the EGFR transgenes was restricted to the lung in C/DEL11, C/L858R56, and C/L858R57 mice (Fig. 1B; data not shown). However, in C/DEL9 mice, transgenic EGFR RNA expression was also detected in the brain (data not shown).
Next, to determine whether expression of the transgenes could be regulated by doxycycline, we performed quantitative RT–PCR on RNA samples derived from lungs of C/L858R56, C/L858R57, and C/DEL11 mice. Transgene expression was induced 103- to 104-fold in lungs of C/L858R57 and C/DEL11 mice upon administration of doxycycline (Fig. 1C). We also observed deinduction of transgene expression upon doxycycline withdrawal, as described further below. Similar results were obtained with C/L858R56 mice (Supplementary Fig. 1). The levels of endogenous mouse Egfr RNA did not appear to change significantly when doxycycline was added or withdrawn (data not shown).
To measure induction of EGFR proteins encoded by the transgenes, we performed Western blots on extracts of lungs from bitransgenic mice maintained on a normal diet and from mice fed with doxycycline-impregnated chow for varying amounts of time (Fig. 1D,E). After 4 wk on doxycycline, the amount of total EGFR was dramatically increased in lungs from C/DEL11 animals (Fig. 1D). In addition, using an antibody specific to the L858R mutant (see Materials and Methods; Supplementary Fig. 2), we readily detected EGFR encoded by the transgene in the lungs of C/L858R56 mice that had received doxycycline for more than a week, and not in lungs of uninduced mice (Fig. 1E). After a 4-d induction, very low levels of EGFR were detected compared with the amounts observed at 8 d (Fig. 1E), suggesting that cells expressing high levels of the EGFR transgene may have a growth advantage even this early in the tumorigenic process. Similar results were obtained for C/L858R57 mice (data not shown). Overall, these results indicate that we have identified transgenic lines in which we can achieve lung-specific expression of the mutant EGFR transgenes after doxycycline induction.
In a preliminary effort to compare the amount of transgenic EGFR to endogenous mouse Egfr, we performed Western blots on lung extracts from bitransgenic C/L858R and C/DEL mice using an antibody that recognizes both human and mouse EGFR (Fig. 1F,G). After a 2- to 3-wk induction with doxycycline, the amount of total EGFR in C/L858R and C/DEL bitransgenic mice was significantly higher than in control mice (Fig. 1F). When incubated with an antibody that specifically recognizes human EGFR, a parallel blot showed transgenic EGFR in samples from the C/L858R and C/DEL bitransgenic mice and, as expected, did not reveal endogenous Egfr (Fig. 1F). Since the lungs of the bitransgenic animals used for these experiments showed significant proliferation (as described below), we reasoned that the high levels of EGFR in these samples are likely to reflect a high proportion of transgene-expressing tumor cells in the samples. To diminish this problem, we examined lung extracts after a short induction with doxycycline (4 d), prior to the appearance of any detectable lung pathology; under these conditions, extracts from lungs of C/L858R56 and C/DEL11 bitransgenic mice contained a three- to fourfold increase in total EGFR compared with extracts from control animals expressing only endogenous Egfr (Fig. 1G). However, when we stained lung tissue from C/L858R mice with a pro-surfactant C antibody (SP-C) that allows the identification of type II pneumocytes, we observed a 1.7-fold increase in the number of type II pneumocytes in samples from doxycycline-induced C/L858R bitransgenic mice versus control samples (data not shown). These findings indicate that even by 4 d of doxycycline induction, the number of type II pneumocytes had nearly doubled, which may account in part for the increase in the levels of EGFR observed in the Western blots. Additional experiments at early time points, with additional antisera and possibly with sorted cells, will be required to determine conclusively the comparative levels of transgenic EGFR and endogenous Egfr in cells that express the transgene in the presence of doxycycline. The findings in Figure 1, F and G, also indicate that similar amounts of EGFR were observed in C/DEL and C/L858R bitransgenic mice 4 d or 2–3 wk after induction, supporting the results obtained using quantitative RT–PCR (Fig. 1C).
C/L858R and C/DEL mice develop lung tumorson doxycycline
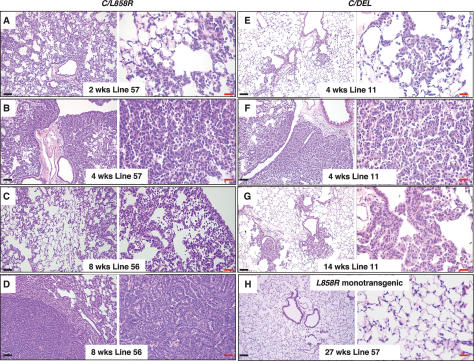
To determine the pathological effects of expressing the two mutant forms of EGFR in the lung, we fed doxycycline to C/L858R and C/DEL bitransgenic mice from the time of weaning. Twenty C/L858R bitransgenic mice from Lines 56 and 57 were followed for development of lung tumors (Supplementary Table 1). Histological changes in the lungs of these mice were observed as early as 2 wk after induction with doxycycline (Fig. 2A) and consist mainly of diffuse thickening of the alveolar walls due to proliferation of atypical pneumocytes (Fig. 2A–C). This lepidic type of cell growth resembles bronchioloalveolar carcinoma (BAC), a subtype of human lung adenocarcinoma characterized by a growth pattern that does not disrupt normal tissue architecture and by a lack of stromal, vascular, or pleural invasion (Travis et al. 1999). Infiltration of the lungs of these mice with macrophages was commonly observed, as has been seen in other models (Ji et al. 2006a), with macrophages generally occupying alveolar spaces in areas of BAC. After receiving doxycycline for >4 wk, mice developed multifocal invasive adenocarcinomas of varying sizes, embedded in the abnormal lung parenchyma (Fig. 2D), sometimes accompanied by progressive thickening of the alveolar walls with compression of air spaces (Fig. 2B). The invasive adenocarcinoma cells contain moderately pleomorphic nuclei with prominent nucleoli and exhibit a papillary growth pattern without lepidic spread. As controls, nine C/ L858R bitransgenic animals (two from Line 57, seven from Line 56) were maintained without doxycycline and sacrificed between 3 wk and 6 mo of age; all exhibited normal lung morphology (data not shown). In addition, monotransgenic L858R mice did not develop tumors, even when fed doxycycline for long periods of time (Fig. 2H).
Figure 2.
C/L858R and C/DEL mice fed doxycycline develop lung tumors. Hematoxylin and eosin (H&E)-stained sections of lung samples derived from C/L858R (A–D) and C/DEL (E–G) bitransgenic mice after treatment with doxycycline for the indicated times. Labels indicate the transgenic EGFR line and time on doxycycline. C and D contain images derived from the same lung, illustrating different pathology in different regions. (H) Lung tissue from a doxycycline-treated L858R57 monotransgenic mouse after prolonged treatment with doxycyline. Black bars, 100 μm. Red bars, 20 μm.
Nine of 10 C/DEL mice from lines DEL9 and DEL11 fed doxycycline also developed pathological changes in the lungs (Supplementary Table 2). Lungs from two mice were examined within 1–2 wk on doxycycline: One showed early proliferative lesions resembling atypical adenomatous hyperplasia (AAH) at the lung periphery, and the other showed tumor cells diffusely throughout the lung field, resembling BAC as seen in mice with the L858R transgene. Five out of eight animals examined after longer treatment with doxycycline (>28 d) showed multifocal tumors arising from the bronchioloalveolar duct junctions (BADJs), surrounded by normal lung parenchyma without areas of BAC (Fig. 2E,G); two animals showed AAH and one animal did not show abnormalities in the lungs (this animal received doxycycline for only 6 wk) (see Supplementary Table 2). The lesions arising from the BADJs consist of fronds of tumor cells radiating into the adjacent lung tissue. In the more advanced lesions, the tumor cells form a compact mass with a papillary growth pattern. In addition to the focal tumors associated with central BADJs, BAC was found (Fig. 2F), mainly at the periphery of the lungs. Three C/DEL11 bitransgenic mice were maintained on a normal diet, and lungs from these animals appeared histologically normal at 4.5 mo of age (data not shown).
In summary, both EGFRL858R and EGFRΔL747–S752 cause the formation of lung adenocarcinomas with BAC features in transgenic mice. Despite similar levels of transgene induction, lung tumors from EGFRL858R-expressing mice develop faster and exhibit a preponderance of BAC and a more aggressive nature, compared with lung tumors in EGFRΔL747–S752-expressing mice. These differences are unlikely to be due to transgene position effects, since two transgenic lines for each EGFR mutant were studied.
Lung tumors in bitransgenic mice are composedof EGFR- and SP-C-positive cells
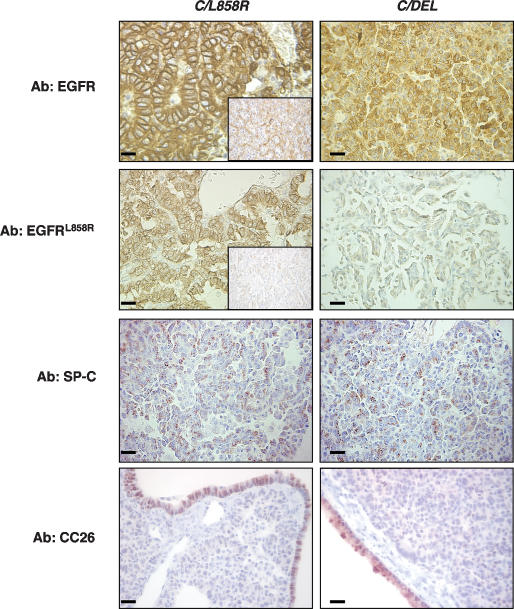
We stained lung tumor tissue from C/L858R and C/DEL bitransgenic mice with antibodies to total EGFR or EGFRL858R (Fig. 3). Both EGFRL858R- and EGFRΔL747–S752induced tumors stained strongly with the EGFR antibody (Fig. 3). As expected, tumors induced by EGFRL858R, but not those induced by EGFRΔL747–S752, stained with the L858R-specific antibody. Tumors induced by both EGFRL858R and EGFRΔL747–S752 were positive for SP-C and negative for the Clara cell protein CC26, consistent with a type II-like phenotype of the tumor cells (Fig. 3).
Figure 3.
Lung tumors induced in C/L858R and C/DEL mice contain abundant EGFR protein and are composed mainly of type II pneumocytes. Sections were stained with antibodies to total EGFR, EGFRL858R, SP-C, and CC26 as indicated. The insets in samples from EGFRL858R-induced tumors show the tissue stained without the primary antibody. Bars, 20 μm.
Magnetic resonance imaging (MRI) of murinelung tumors
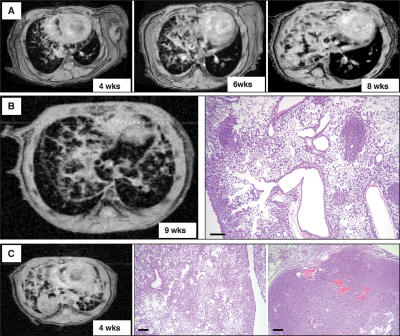
We tested whether MRI of mouse lungs, previously used in our laboratory to follow KrasG12D-dependent lung tumors (Fisher et al. 2001), could also be used to detect and monitor tumors in animals carrying regulated EGFR transgenes (Fig. 4). Within 4 wk of exposure to doxycycline, when C/L858R mice have developed diffuse tumors, consolidations suggestive of a reticulonodular growth pattern were visible in scans of one or both lungs of these mice (Fig. 4A). With longer exposure to doxycycline, we observed increasingly dense reticulonodular patterns and consolidations filling the whole lung or a lung lobe (Fig. 4A), indicating that tumor progression can be followed in individual mice using MRI. To confirm that the consolidations observed in MR images indicate the presence of histologically visible lung tumors, we sacrificed additional mice within 2 d after the MRI and processed lung tissue for histology (Fig. 4B,C): The pattern suggestive of a reticulonodular infiltrate observed on the scans reflects diffuse BAC, and dense white masses in the MR scans correspond to solid invasive adenocarcinomas (Fig. 4B,C). In summary, MRI can be used to detect tumors, and the patterns observed by MRI correlate with tumor histopathology.
Figure 4.
Lung tumor growth can be monitored by MRI. (A) Serial MRI of a C/L858R57 mouse after 4, 6, and 8 wk on doxycycline shows a pattern consistent with a diffuse reticulonodular infiltrate in the right lung that progressively increased in size and became more consolidated. (B, left) MR image of the lungs from a C/L858R57 mouse fed doxycycline for 9 wk. (Right) The reticulonodular pattern observed by MRI reflects diffuse BAC and tumor nodules throughout the affected lung field, as shown in the H&E-stained section. (C, left) MR image of the lungs from a C/L858R56 bitransgenic mouse fed doxycycline for 4 wk showing reticulonodular consolidation and a large solid nodule. These features on MRI correspond to diffuse BAC (middle) and an invasive adenocarcinoma (right) observed histologically. Bars, 200 μm.
Mutant EGFR is required for tumor maintenance
In most cases, human lung cancers with somatic mutations in EGFR regress by radiological criteria after treatment with the TKIs gefitinib and erlotinib, implying that the kinase activity of the mutant receptor is required to maintain viability of the tumor cells. We tested whether the mutant EGFR-induced lung tumors in the models described here also depend on mutant EGFR for cell viability and proliferation, using two strategies: (1) withdrawing doxycycline from mice to down-regulate transgene expression or (2) treating the mice with erlotinib to inhibit the kinase activity of mutant EGFR.
Doxycycline withdrawal leads to tumor regression
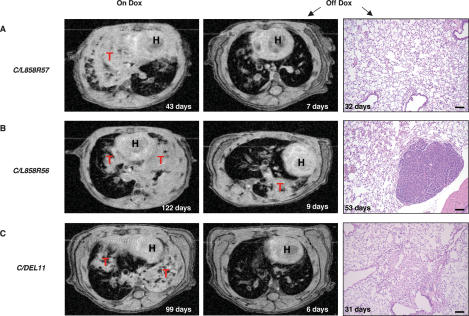
To examine the dependence of mutant EGFR-induced lung tumors on continued expression of the mutant EGFR transgenes, we identified six C/L858R and two C/DEL mice with significant consolidations evident by MRI after 43–180 d on doxycycline (Supplementary Table 3). After removal of doxycycline from the diet, mice were imaged and sacrificed between 2 d and 6 mo after deinduction to look for residual neoplastic disease. After a 2- to 4-d deinduction, a rapid change in the MR image was observed, with the disappearance (complete response) or shrinkage (partial response) of most of the consolidations in the lungs (Supplementary Table 3). However, tumors were still visible histologically when mice were sacrificed at this stage (data not shown). In contrast, lung tissue from all four mice sacrificed more than a month after removal of doxycycline was mostly normal histologically (Fig. 5), suggesting that elimination of lung cancer cells occurs progressively over time. In two mice, focal areas of scarring were also observed (e.g., see Fig. 5C). In one C/L858R56 mouse a single adenocarcinoma nodule was still embedded in normal lung epithelium 53 d after doxycycline was withdrawn (Fig. 5B). Whether this tumor nodule was present originally and never regressed or whether it re-emerged in the absence of doxycycline cannot presently be determined.
Figure 5.
Lung opacities disappear in MR images, and the tumors regress histologically in the lungs of bitransgenic mice after withdrawal of doxycycline. (A–C) Serial MR images from the lungs of C/L858R and C/DEL mice maintained on doxycycline for the times indicated (left panels) and after deinduction (middle panels). (Right panels) H&E-stained sections from the mice after deinduction for the times indicated on the panels. (H) Heart; (T) tumor. Bars, 100 μm (see Supplementary Table 3 for additional information on these mice, and the text for a discussion of the peristent nodule shown in B, right panel).
Tumor regression after treatment with erlotinib
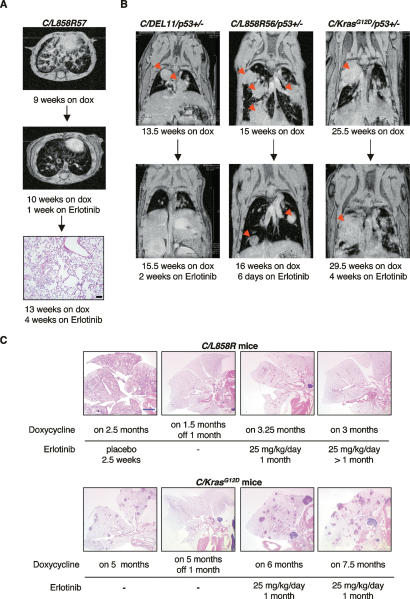
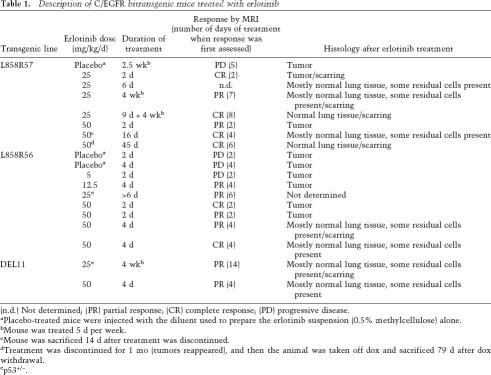
To ask whether TKIs can also produce tumor regression in our mouse models, we treated 14 EGFRL858R- and two EGFRΔL747–S752-induced tumors with erlotinib for varying amounts of time, at doses ranging from 5 to 50 mg/kg/d, while the mice were still being fed doxycycline (Fig. 6; Table 1). All of the animals that received erlotinib at doses ≥12.5 mg/kg/d showed partial to complete responses by MRI as early as 2 d after erlotinib treatment (Table 1). The amount of tumor regression as assayed histologically correlated with the length of treatment (Table 1). After animals were on drug for 2–4 d, tumor cells were still observed throughout the lung sections. Very few tumor cells were detected in lung tissue from animals treated with erlotinib for ≥2 wk (Fig. 6A, bottom panel). Emphysematous changes and scarring were observed in the lungs of treated mice, presumably in areas where adenocarcinomas were eliminated or were being eliminated. Tumors in two mice that were heterozygous for p53, generated in the context of ongoing experiments to determine the effect of tumor suppressor gene deficiencies on mutant EGFR-dependent tumorigenesis, also regressed upon erlotinib treatment (Fig. 6B; Table 1). An animal treated for 2 d at the lowest dose of drug (5 mg/kg/d) did not show any detectable response by MRI (Table 1). Three placebo-treated mice did not show a response by MRI or histologically (Table 1).
Figure 6.
Lung tumors in C/L858R or C/DEL, but not C/KrasG12D bitransgenic mice on doxycycline disappear during treatment with erlotinib. (A) Axial MR images of lungs from a C/L858R57 mouse after 9 wk on doxycycline (top) and after 1 wk of erlotinib treatment (25 mg/kg/d) plus continued doxycycline (middle). The mouse was sacrificed after 4 wk of erlotinib treatment, and an H&E-stained section of the lung is shown in the bottom panel. Black bar, 100 μm. (B) Coronal MR images of the lungs of C/DEL11/p53+/− (left), C/L858R56/p53+/− (center), and C/KrasG12D/p53+/− (right) mice before (top) and after (bottom) receiving erlotinib for the indicated times. Red arrows indicate tumors. (C) H&E-stained lung sections from C/L858R (top) and C/KrasG12D (bottom) mice, placebo-treated, erlotinib-treated, or deinduced as indicated. These images provide a qualitative, but not quantitative, assessment of the effect of erlotinib on mice that express oncogenic Kras. A quantitative assessment would require knowledge of the initial tumor burden in individual mice prior to erlotinib therapy. Blue bar, 2 mm.
Table 1.
Description of C/EGFR bitransgenic mice treated with erlotinib
(n.d.) Not determined; (PR) partial response; (CR) complete response; (PD) progressive disease.
aPlacebo-treated mice were injected with the diluent used to prepare the erlotinib suspension (0.5% methylcellulose) alone.
bMouse was treated 5 d per week.
cMouse was sacrificed 14 d after treatment was discontinued.
dTreatment was discontinued for 1 mo (tumors reappeared), and then the animal was taken off dox and sacrificed 79 d after dox withdrawal.
ep53+/−.
Prolonged erlotinib treatment caused slight weight loss and loss of some hair, but the mice were active and appeared to be eating normally. To exclude the possibility that tumor regression in erlotinib-treated mice resulted from ingestion of insufficient doxycycline-containing food to maintain transgene expression, we measured transgene expression by quantitative RT–PCR in lungs after long-term treatment and found no reduction in transgenic RNA (data not shown).
Collectively, these data demonstrate that EGFRL858R- and EGFRΔL747–S752-induced lung tumors are both sensitive to the TKI erlotinib. These results suggest that inhibition of the kinase activity of the mutant receptor is sufficient to elicit responses similar to those observed upon deinduction of the EGFR oncogene in mice and after TKI treatment of most human lung tumors with EGFR mutations. In addition, both histological subtypes, BAC and solid invasive adenocarcinomas, respond to treatment of mice with erlotinib.
In human lung tumors, mutations in KRAS are associated with primary resistance to tyrosine kinase inhibitors (Pao et al. 2005a). To verify this result in murine lung tumors carrying oncogenic mutations in Kras, we treated four bitransgenic C/KrasG12D mice and one C/KrasG12D/p53+/− mouse (Fisher et al. 2001) that had been on doxycycline for 5–6.5 mo with erlotinib (25 mg/kg/d). In comparison to tumors initiated by oncogenic EGFR, KrasG12D-induced tumors were not eliminated by erlotinib treatment as assessed by MRI (Fig. 6B, cf. the right panel and the left and middle panels) or histologically (Fig. 6C, cf. the lungs from C/L858R mice and those from C/KrasG12D). Others (Fujimoto et al. 2005) have shown that gefitinib leads to a decrease in the number (but not the disappearance) of lung surface nodules in murine lung adenocarcinomas with Kras mutations. Future experiments in which the pretreatment and post-treatment tumor burdens are carefully measured will help determine whether erlotinib has even a partial effect on oncogenic Kras-induced tumors in our mice.
Downstream targets of EGFR are phosphorylatedin both EGFR and KrasG12D-induced lung tumors
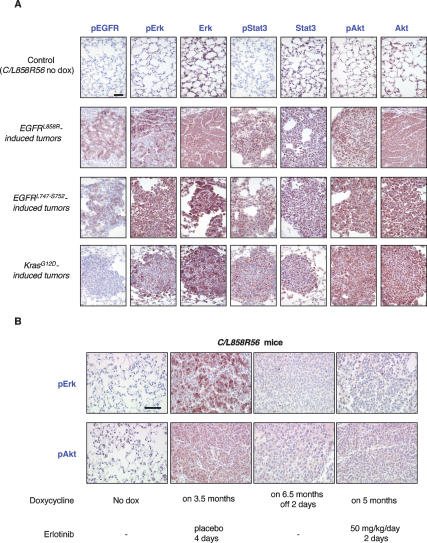
To begin to examine the consequences of mutant EGFR expression on signaling pathways implicated in signaling downstream of the mutant EGFR receptors (Sordella et al. 2004; Tracy et al. 2004; Engelman et al. 2005; Greulich et al. 2005; Jiang et al. 2005) and to test whether any of these pathways are differentially activated when mutant EGFR- and KrasG12D-induced lung tumors are compared, we immunostained lung sections from C/L858R, C/DEL, and C/KrasG12D mice with antibodies for phosphorylated EGFR (pEGFR), phosphorylated Erk (pErk), phosphorylated Akt (pAkt), and phosphorylated Stat3 (pStat3) (Fig. 7). All of the lung tumors were positive for pErk, pAkt, and pStat3, but strong pEGFR staining was detected only in mutant EGFR-induced tumors and was either absent or weak in tumors caused by mutant Kras (Fig. 7A). These results suggest that the MAPK, PI3K, and STAT3 pathways are all stimulated by mutant EGFR and oncogenic Kras during the tumorigenic process, although we have not determined which pathways are essential for oncogenesis.
Figure 7.
Multiple signaling pathways are activated in mutant EGFR and KrasG12D-induced lung tumors and deactivated during tumor regression. (A) Immunohistochemical staining was used to detect pEGFR, pErk, Erk, pStat3, Stat3, pAkt, and Akt in L858R-, DEL-, and KrasG12D-induced lung tumors and in uninduced lungs (normal lung tissue derived from a bitransgenic C/L858R56 mouse maintained on normal chow was stained as a control) as indicated. (B) Immunohistochemical staining was used to detect pErk and pAkt in lung tumors from C/L858R56 mice after 2 d of erlotinib treatment or doxycycline withdrawal and in uninduced and placebo-treated lungs as indicated. Bar, 50 μm.
We have also begun to compare the effects of tyrosine kinase inhibitor treatment and doxycycline withdrawal on the activity of these signaling pathways. Diminished phospho-Erk and phospho-Akt staining was observed in tumor samples from C/L858R56 (Fig. 7B) and C/L858R57 transgenic mice as early as 2 d after deinduction with doxycycline or after starting erlotinib, indicating that activation of these signaling pathways is directly related to activity of mutant EGFR.
Apoptosis and arrest of proliferation during tumor regression
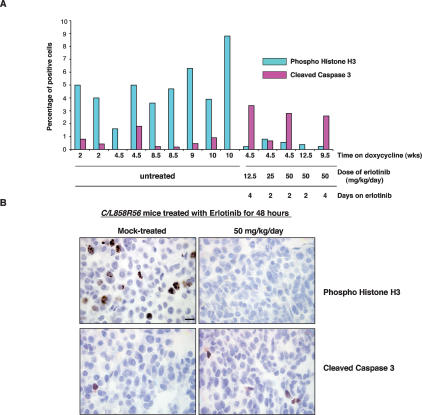
Tumor regression upon oncogene deinduction in other tetracycline-inducible transgenic models is due to apoptosis, differentiation, and/or proliferation arrest (Felsher 2004). In a preliminary effort to investigate the mechanism by which tumors regress during treatment with erlotinib, we stained tissue sections from drug-treated and untreated C/L858R mice with antibodies to phosphorylated histone H3 (to identify mitotic cells) and activated caspase 3 (to identify apoptotic cells). At least 400 cells were scored for each section analyzed. We consistently observed a drop in the number of mitoses upon drug treatment that was accompanied, in most cases, by an increase in the numbers of apoptotic cells (Fig. 8). These data suggest that cell division decreases acutely upon drug treatment and that cell death also increases. The apoptotic process appears to show more variability than mitotic arrest. Time-course studies may help to elucidate the sequence of events that occurs upon drug treatment.
Figure 8.
Decreased mitoses and increased apoptosis upon effective erlotinib treatment. (A) Quantification of phosphohistone H3 and cleaved caspase 3 in untreated and erlotinib-treated mice. (B) Phosphohistone H3 (top) and cleaved caspase 3 (bottom) staining of lung sections from untreated and erlotinib-treated C/L858R56 mice is shown. Bar, 10 μm.
Discussion
In this study, we describe the development of two new mouse models of lung adenocarcinoma. We generated tetracycline-inducible transgenic mice bearing EGFR mutants often found in human lung tumors: the L858R allele and an exon 19 deletion mutant. We show that both of these mutants give rise to lung adenocarcinomas upon induction with doxycycline. Importantly, deinduction of the transgene by doxycycline withdrawal or administration of the TKI erlotinib leads to tumor regression, demonstrating that continued function of the mutant receptor is required for tumor maintenance. Similar results have been obtained by another group that has also generated tetracycline-inducible transgenic mice encoding EGFR mutants (Ji et al. 2006b).
Lung tumor pathology
When expressed in type II pneumocytes (or possibly their precursors), EGFRL858R and EGFRΔL747–S752 both give rise to lung adenocarcinomas that show many common and some distinct features. C/L858R mice rapidly develop diffuse BAC, first seen at the lung periphery; at later stages, invasive adenocarcinoma nodules appear, embedded in BAC. In C/DEL mice, adenocarcinomas form more slowly, at or near BADJs, and BAC is found at the lung periphery.
Pure BAC is a rare subtype of lung adenocarcinoma that is characterized by a lepidic growth pattern of tumor cells along alveolar walls, the absence of invasion, and a better prognosis than other types of lung cancer (Barkley and Green 1996). In most cases, BAC is found in association with an invasive adenocarcinoma component (Ebright et al. 2002).
Adenocarcinomas with BAC features are more likely to respond to gefitinib and may have a higher frequency of EGFR mutations than adenocarcinomas alone (Miller et al. 2004; Marchetti et al. 2005). In view of these observations, the coexistence of BAC and invasive adenocarcinomas in our mouse models is especially provocative and can be hypothesized to arise in two ways. According to one hypothesis, two different cell types within the lung epithelium may be sensitive to and become transformed by mutant EGFR, giving rise to tumors with distinct histopathologies. For example, it is possible that in the presence of mutant EGFR, bronchioalveolar stem cells, described by Kim et al. at the BADJs (Kim et al. 2005), and type II pneumocytes in alveoli both give rise to tumors that, however, are phenotypically distinct. Alternatively, varying degrees of BAC and invasive adenocarcinoma may reflect sequential stages in the progression of lung cancer in which BAC represents an early stage in the tumorigenic process. Further studies with these mice may be useful to distinguish between these two possibilities.
The longer tumor latency and the histopathological differences observed in C/DEL mice compared with C/L858R mice, despite similar patterns of transgene expression, may be due to different biochemical properties of the two mutant receptor kinases. Indeed, autophosphorylation of the deletion mutant is lower in transient transfection assays (Pao et al. 2004; Amann et al. 2005). Patients with lung tumors harboring the different EGFR mutations also appear to have a different prognosis. Tumors with exon 19 deletion mutations have a higher response rate to TKIs, and the treated patients live longer on average than those with the L858R mutation (Mitsudomi et al. 2005; Riely et al. 2006). However, in surgically resected patients never treated with a TKI, those with an L858R mutation have a longer overall survival than those with an exon 19 deletion (Shigematsu et al. 2005). Our mouse models may be useful to investigate the observed clinical differences between the L858R and exon 19 deletion mutations.
The diffuse nature of the BAC observed in lungs from C/L858R mice contrasts very sharply with the focal nature of lung adenocarcinomas induced by oncogenic Kras (Fig. 6C, cf. images of doxycycline-induced animals) using the same methodology (Fisher et al. 2001), while EGFRΔL747–S752-induced tumors have an intermediate phenotype. Fisher et al. suggested that although oncogenic Kras is probably expressed in the majority of type II pneumocytes, only a small proportion of the cells respond and are sensitive to oncogene expression. If this is the case, it appears from our results that many more cells are sensitive to expression of mutant EGFR than oncogenic Kras. This result could in part be due to the type of stimulus provided by the different oncogenes: Mutant EGFR mimics a growth factor stimulus that drives cell proliferation and survival by engaging many signaling pathways, while mutant Kras can, in some settings, cause cell senescence (Collado et al. 2005) or apoptosis (Cox and Der 2003). Comparison of the proliferative, apoptotic, and senescent responses of type II pneumocytes or their precursors shortly after induction of oncogenic EGFR or Kras with doxycycline might help explain the histopathological differences we observe.
Mutant EGFR is required for tumor maintenance
Studies using tetracycline-inducible transgenic models of cancer have demonstrated that deinduction of the initiating oncogene is sufficient to achieve sustained regression in a wide variety of tumor types (Felsher 2004; Varmus et al. 2006). In this study, we show that oncogenic EGFR is required for tumor maintenance with two different strategies: (1) when expression of the oncogene is switched off (by withdrawing doxycycline) or (2) the activity of the oncogene is inhibited (by drug treatment). It is especially significant that lung tumors induced in mice by mutant EGFR regress dramatically and promptly upon treatment with the FDA-approved drug erlotinib. Most patients upon treatment with mutation-positive lung tumors also respond to this drug, albeit without the nearly complete responses observed in our mice.
Are high levels of mutant EGFR required to elicitan oncogenic effect?
The relative contribution of qualitative (mutational) and quantitative (gene expression) changes to the process of oncogene activation remains an important and unresolved issue in many human and experimental cancers. Results with some genetically engineered mice indicate that, when a mutant oncogene is expressed from its endogenous locus, amplification of the mutant allele or loss of the wild-type allele can or must occur during tumor development (Andrechek et al. 2000; Johnson et al. 2001). Similarly, in human lung tumors, EGFR mutations are often accompanied by polysomy or gene amplification, suggesting that enhanced expression of the mutant gene may facilitate oncogenesis (Cappuzzo et al. 2005; V.A. Miller, pers. comm.).
We have attempted to explore this issue in the transgenic models reported here by comparing levels of RNA and protein produced from the mutant human EGFR transgenes and those from the endogenous mouse Egfr locus (see Fig. 1 and accompanying text). Our findings suggest that, while the induction ratios for the doxycycline-regulated transgenes are large (Fig. 1C–E), the transgenic products are similar to or only modestly higher than endogenous Egfr RNA and protein shortly after induction. Such measurements are complicated, however, by the semiquantitative methods, by uncertainty about the percentage of cells expressing the transgenes, and by the specificity and sensitivity of reagents for measuring human and mouse gene products. Furthermore, cells expressing higher levels of transgenic products may have a growth advantage during tumor progression, accounting for the apparently abundant EGFR protein in extracts from lungs with high tumor burden (Fig. 1D,E). Better definition of the contributions of quantitative and qualitative effects to oncogenic potential will require additional experiments with improved assays for EGFR and its effects, studies of mice in which mutant alleles have replaced endogenous Egfr exons (K. Lane and T. Jacks, pers. comm.) and transgenic mice expressing wild-type EGFR.
We have begun to generate tetracycline-inducible transgenic mice that express wild-type EGFR in lung epithelial cells. In one such line, we have found only minor pathological changes in two of 10 mice observed for 4–8 mo; however, the transgenic EGFR is constitutively expressed in many tissues (and is not inducible) in this line, presumably due to a transgene position effect. We are currently characterizing additional mice that carry the wild-type EGFR transgene to identify lines with doxycycline-responsive expression of EGFR. Such lines may be helpful for evaluating claims that amplification of wild-type EGFR in human lung cancer is associated with an increased overall survival as a result of tyrosine kinase inhibitor therapy (Cappuzzo et al. 2005; Tsao et al. 2005).
What is the mechanism of action of oncogenic EGFR?
Although it is clear that these NSCLC-associated EGFR mutations are gain-of-function oncogenic mutations (Greulich et al. 2005) that confer a selective growth advantage to lung cells, their effects on the functional properties of the receptor still remain to be determined. Likewise, although a crystal structure of the wild-type human EGFR kinase domain bound to erlotinib has been reported (Stamos et al. 2002), structures of the mutant EGFR kinases have not been described, with or without TKIs. Several studies in transfected cells and human lung cancer cell lines have shown that the mutant receptors lead to constitutive activation of the antiapoptotic AKT and STAT signaling pathways and mitogenic ERK signaling (Sordella et al. 2004; Tracy et al. 2004; Greulich et al. 2005; Jiang et al. 2005). Our lung tumor models permit the study of signaling downstream of mutant EGFRs in primary lung tumors. Thus far, we have been able to confirm activation of several signaling pathways in mutant EGFR-induced tumors (Fig. 7), consistent with the high mitotic and low apoptotic rates observed in the tumors (Fig. 8). The signaling requirements for oncogenic effects and the contribution of EGFR heterodimerization partners (Engelman et al. 2005) to the tumorigenic process can be assessed in these in vivo models. Moreover, easy access to tissues and blood from treated mice will allow us to identify molecular indicators of treatment response.
Mechanisms of resistance
We observed solitary residual tumor nodules in two mice in this study—one in which doxycycline was withdrawn and another in which the mouse was treated with erlotinib. We cannot say whether these represent lung tumors that failed to respond to EGFR down-regulation or kinase inhibitors (primary resistance) or whether most of the tumor cells initially disappeared, with a few residual cells acquiring changes that allowed them to escape a requirement for mutant EGFR (secondary resistance or relapse). This distinction between primary and secondary resistance is important because the molecular mechanisms may be different and relevant to treatment of patients with lung cancer.
Primary resistance
We know from human studies that primary resistance to TKIs in human lung tumors is associated with the presence of KRAS mutations (Pao et al. 2005a). However, KRAS mutations are rarely found in conjunction with EGFR mutations, and it still is unclear what contributes to primary resistance in tumors with EGFR mutations that do not respond to the drug. Our mouse models may help to determine why most tumors respond to treatment with erlotinib (or doxycycline withdrawal) and why other tumors are inherently resistant to the drug.
Secondary resistance
Since human cancers that initially respond to TKI treatment often relapse during therapy, it is critical to identify the mechanisms of drug resistance. Fifty percent of human lung tumors with EGFR mutations, examined thus far, that have progressed during drug treatment have a secondary mutation in EGFR (T790M) that is predicted to prevent drug binding (Kobayashi et al. 2005; Pao et al. 2005b). The mechanism of drug resistance in the remaining cases is unknown. Using the mouse models presented here, we can determine whether mice acquire resistance to erlotinib by treating them for extended periods of time. Moreover, the contribution of tumor suppressor deficiencies to TKI resistance, which is not yet well understood in humans, can easily be studied in mice. For example, in Wnt1-induced mammary tumorigenesis, p53 status affects the ability of tumors to recur after doxycycline withdrawal, and recurrence is independent of Wnt1 gene expression (Gunther et al. 2003). In preliminary experiments, we have observed tumor regression upon deinduction of mutant EGFR in the absence of Ink4A/Arf, indicating that this tumor suppressor does not confer primary resistance to the tumor cells. Whether the absence of Ink4A/Arf promotes tumor recurrence in this context is under investigation. Although very few studies have examined the mechanism of tumor relapse in tetracycline-inducible transgenic mice, it is likely that the molecular events that govern the process are highly dependent on the initiating oncogene (Gunther et al. 2003; Boxer et al. 2004; Moody et al. 2005). Our mouse models, in combination with mutations in tumor suppressor genes, may help uncover genetic events that contribute to the recurrence and progression of EGFR mutant tumors.
The recent clinical successes of the TKIs—imatinib for the treatment of chronic myelogenous leukemia and gastrointestinal stromal tumors, and gefitinib and erlotinib for lung adenocarcinoma—have helped to establish that targeted cancer therapy can be an effective method for controlling cancer. Genetically engineered mouse models like the lung cancer models presented here, in which an inhibitor of the product of a transgenic human oncogene is highly effective, are likely to be important research tools for optimizing targeted therapies in the future.
Materials and methods
Generation of transgenic mice
The Tet-op-mp1 vector (Fisher et al. 2001), containing the tetracycline operator and the mouse protamine intron and polyadenylation sequence, was linearized with EcoRV, which cuts between the promoter and the poly(A) sequence. The wild-type or mutant EGFR cDNA (Pao et al. 2004) was excised from pcDNA3.1 with PmeI and ligated into Tet-op-mp1 via the EcoRV site. Restriction digests and sequencing were used to identify clones in which the EGFR cDNA had inserted in the correct orientation. The specific mutations introduced into human EGFR to generate the mutants used in this study are as follows: a base substitution at position 2573 in the EGFR cDNA, which gives rise to an amino acid substitution of arginine for leucine at position 858 in the kinase domain of EGFR, and an in-frame deletion of nucleotides 2239–2256, which leads to the elimination of six amino acids, L747 to S752, in the kinase domain of EGFR. TetO-EGFR transgenic mice were genotyped using the following primers: EGFR3421F, 5′-ACT GTCCAGCCCACCTGTGT-3′; and mp-1R, 5′-GCCTGCGAC GGCGGCATCTGC-3′. Reactions were amplified with the following PCR protocol: denaturation for 5 min at 95°C, followed by 35 cycles of 30 sec at 95°C, 30 sec at 58°C, and 30 sec at 72°C, followed by a 5-min extension at 72°C. Additional details are supplied in the Supplemental Material.
Animal husbandry and genotyping
All animals were kept in specific pathogen-free housing with abundant food and water under guidelines approved by the MSKCC Institutional Animal Care and Use Committee and Research Animal Resource Center. CCSP-rtTA tetracycline-dependent activator mice (Tichelaar et al. 2000) and TetO-KrasG12D (Fisher et al. 2001) and p53-null (Jacks et al. 1994) mice have been previously described. Doxycycline was administered by feeding mice with doxycycline-impregnated food pellets (625 ppm; Harlan-Teklad). Erlotinib (provided by Genentech) was suspended in 0.5% (w/v) methylcellulose (as per a protocol from the drug supplier) and injected intraperitoneally (Pollack et al. 1999) at the dose and times indicated in the experiments. Tail DNA was isolated using Qiaprep Tail DNeasy isolation kit (Qiagen) according to the manufacturer’s protocol. Detection of the rtTA activator transgene, the KrasG12D transgene, and p53 genes was performed as described previously (Jacks et al. 1994; Fisher et al. 2001).
Histology and immunohistochemistry
Animals were sacrificed with a lethal dose of CO2 per institutional guidelines. The lungs were excised; the left lung was flash-frozen in liquid nitrogen for molecular analyses, and the right lung was inflated with 4% paraformaldehyde in PBS. Lungs were fixed in 4% paraformaldehyde overnight at room temperature, placed in 70% ethanol, and sent for paraffin embedding and sectioning (Histoserv). All lungs were sectioned in the same fashion; 10 steps were taken, 100 μm apart. All 10 steps were evaluated to determine whether tumors were present. Slides were reviewed by a board-certified pathologist (M.F.Z.). The antibodies used for immunohistochemistry are listed in the Supplemental Material.
RT–PCR analysis
Tissue samples were crushed and RNA was extracted using Trizol (Invitrogen) reagent. RNA was treated with DNase I (Invitrogen) to eliminate any contaminating DNA. RT–PCR reactions were carried out using the SuperScript III One-Step RT–PCR with Platinum Taq system (Invitrogen). For quantitative RT–PCR, reactions were performed using the ABI7900 Sequence Detection System (Applied Biosystems). Primer sequences and amplification conditions are described in the Supplemental Material.
Antibodies and immunoblotting
The EGFRL858R antibody was generated by immunizing a rabbit against a 14-residue peptide [(C)TDFGRAKLLGAEE] containing the substituted amino acid (arginine instead of leucine) at position 858 in the region 854–866 (Zymed). Immunoblotting was carried out as described in the Supplemental Material.
MRI
Mice were anesthetized with 2% isofluorane oxygen gas. Respiratory-gated lung MR images were acquired on a Bruker 4.7T Biospec scanner (Bruker Biospin Inc.) using a custom-made 36-mm quadrature birdcage coil in the Small Animal Imaging MR Core Facility at MSKCC (further details are provided in the Supplemental Material). Human criteria always use measurable lesions to define response; the diffuse and reticular nature of the consolidations observed in images derived from most of the mice we studied make this difficult. Therefore, we established our own definition of response to treatment. Tumor-bearing mice were considered to have had a complete response if a scan was scored as negative for lung opacities after doxycycline withdrawal or drug treatment. A partial response call was made when lung opacities were decreased but still clearly present. We considered an animal to have progressive disease if the post-treatment MRI showed an increase in lung opacities compared with the pretreatment scan.
Acknowledgments
We thank Marissa Balak, Anthony Daniyan, Jennifer Doherty, Mary Ann Melnick, and Irina Linkov for expert technical assistance; Jason Koutcher, Carl Le, Mihaela Lupu, and Cornelia Matei for the MRI (support for the Magnetic Resonance Imaging Core was provided by a Small Animal Imaging Resource Program [SAIRP] grant ]NIH R24CA83084] to Jason Koutcher); Jeffrey Whitsett (University of Cincinnati) for the CCSP-rtTA mice; Luca Cartegni (MSKCC) for the ribosomal protein L37A primers; Genentech for providing erlotinib (Tarceva) and critical reading of the manuscript; Hongbin Ji (DFCI) for the human EGFR quantitative RT–PCR primer sequences and for sharing unpublished data; Tyler Jacks (MIT), Keara Lane (MIT), and Kwok-Kin Wong (DFCI) for sharing unpublished data; the MSKCC Lung Cancer Oncogenome Group for discussions; and members of the Varmus laboratory, especially Katrina Podsypanina, for discussions, support, and critical reading of the manuscript. K.P. is an American Cancer Society-Davidson Sinai Research Foundation Post-doctoral Fellow (PF-05-078-01-MGO) and received support from the Labrecque Foundation. W.P. received support from the NCI (K08-CA097980) and Joan’s Legacy Foundation. This work was supported in part by an anonymous donor and the Carmel Hill Fund.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1417406
References
- Amann J., Kalyankrishna S., Massion P.P., Ohm J.E., Girard L., Shigematsu H., Peyton M., Juroske D., Huang Y., Stuart Salmon J., et al. Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer. Cancer Res. 2005;65:226–235. [PubMed] [Google Scholar]
- Andrechek E.R., Hardy W.R., Siegel P.M., Rudnicki M.A., Cardiff R.D., Muller W.J. Amplification of the neu/erbB-2 oncogene in a mouse model of mammary tumorigenesis. Proc. Natl. Acad. Sci. 2000;97:3444–3449. doi: 10.1073/pnas.050408497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkley J.E., Green M.R. Bronchioloalveolar carcinoma. J. Clin. Oncol. 1996;14:2377–2386. doi: 10.1200/JCO.1996.14.8.2377. [DOI] [PubMed] [Google Scholar]
- Boxer R.B., Jang J.W., Sintasath L., Chodosh L.A. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell. 2004;6:577–586. doi: 10.1016/j.ccr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Cappuzzo F., Hirsch F.R., Rossi E., Bartolini S., Ceresoli G.L., Bemis L., Haney J., Witta S., Danenberg K., Domenichini I., et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J. Natl. Cancer Inst. 2005;97:643–655. doi: 10.1093/jnci/dji112. [DOI] [PubMed] [Google Scholar]
- Chou T.Y., Chiu C.H., Li L.H., Hsiao C.Y., Tzen C.Y., Chang K.T., Chen Y.M., Perng R.P., Tsai S.F., Tsai C.M. Mutation in the tyrosine kinase domain of epidermal growth factor receptor is a predictive and prognostic factor for gefitinib treatment in patients with non-small cell lung cancer. Clin. Cancer Res. 2005;11:3750–3757. doi: 10.1158/1078-0432.CCR-04-1981. [DOI] [PubMed] [Google Scholar]
- Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A.J., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., et al. Tumour biology: Senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- Cox A.D., Der C.J. The dark side of Ras: Regulation of apoptosis. Oncogene. 2003;22:8999–9006. doi: 10.1038/sj.onc.1207111. [DOI] [PubMed] [Google Scholar]
- Eberhard D.A., Johnson B.E., Amler L.C., Goddard A.D., Heldens S.L., Herbst R.S., Ince W.L., Janne P.A., Januario T., Johnson D.H., et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J. Clin. Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- Ebright M.I., Zakowski M.F., Martin J., Venkatraman E.S., Miller V.A., Bains M.S., Downey R.J., Korst R.J., Kris M.G., Rusch V.W. Clinical pattern and pathologic stage but not histologic features predict outcome for bronchioloalveolar carcinoma. Ann. Thorac. Surg. 2002;74:1640–1646. doi: 10.1016/s0003-4975(02)03897-3. [DOI] [PubMed] [Google Scholar]
- Ekstrand A.J., Sugawa N., James C.D., Collins V.P. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc. Natl. Acad. Sci. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman J.A., Janne P.A., Mermel C., Pearlberg J., Mokuhara T., Fleet C., Cichowski K., Johnson B.E., Cantley L.C. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc. Natl. Acad. Sci. 2005;102:3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsher D.W. Reversibility of oncogene-induced cancer. Curr. Opin. Genet. Dev. 2004;14:37–42. doi: 10.1016/j.gde.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Fisher G.H., Wellen S.L., Klimstra D., Lenczowski J.M., Tichelaar J.W., Lizak M.J., Whitsett J.A., Koretsky A., Varmus H.E. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes & Dev. 2001;15:3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto N., Wislez M., Zhang J., Iwanaga K., Dackor J., Hanna A.E., Kalyankrishna S., Cody D.D., Price R.E., Sato M., et al. High expression of ErbB family members and their ligands in lung adenocarcinomas that are sensitive to inhibition of epidermal growth factor receptor. Cancer Res. 2005;65:11478–11485. doi: 10.1158/0008-5472.CAN-05-1977. [DOI] [PubMed] [Google Scholar]
- Greulich H., Chen T.H., Feng W., Janne P.A., Alvarez J.V., Zappaterra M., Bulmer S.E., Frank D.A., Hahn W.C., Sellers W.R., et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med. 2005;2:e313. doi: 10.1371/journal.pmed.0020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther E.J., Moody S.E., Belka G.K., Hahn K.T., Innocent N., Dugan K.D., Cardiff R.D., Chodosh L.A. Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes & Dev. 2003;17:488–501. doi: 10.1101/gad.1051603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.W., Kim T.Y., Hwang P.G., Jeong S., Kim J., Choi I.S., Oh D.Y., Kim J.H., Kim D.W., Chung D.H., et al. Predictive and prognostic impact of epidermal growth factor receptor mutation in non-small-cell lung cancer patients treated with gefitinib. J. Clin. Oncol. 2005;23:2493–2501. doi: 10.1200/JCO.2005.01.388. [DOI] [PubMed] [Google Scholar]
- Hirsch F.R., Varella-Garcia M., Bunn P.A., Jr., Di Maria M.V., Veve R., Bremmes R.M., Baron A.E., Zeng C., Franklin W.A. Epidermal growth factor receptor in non-small-cell lung carcinomas: Correlation between gene copy number and protein expression and impact on prognosis. J. Clin. Oncol. 2003;21:3798–3807. doi: 10.1200/JCO.2003.11.069. [DOI] [PubMed] [Google Scholar]
- Huang S.-F., Liu H.-P., Li L.-H., Ku Y.-C., Fu Y.-N., Tsai H.-Y., Chen Y.-T., Lin Y.-F., Chang W.-C., Kuo H.-P., et al. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin. Cancer Res. 2004;10:8195–8203. doi: 10.1158/1078-0432.CCR-04-1245. [DOI] [PubMed] [Google Scholar]
- Jacks T., Remington L., Williams B., Schmitt E., Halachmi S., Bronson R., Weinberg R. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Ji H., Houghton A.M., Mariani T.J., Perera S., Kim C.B., Padera R., Tonon G., McNamara K., Marconcini L.A., Hezel A., et al. K-ras activation generates an inflammatory response in lung tumors. Oncogene. 2006a;25:2105–2112. doi: 10.1038/sj.onc.1209237. [DOI] [PubMed] [Google Scholar]
- Ji H., Li D., Chen L., Shimamura T., Kobayashi S., McNamara K., Mahmood U., Mitchell A., Sun Y., Al-Hashem R., et al. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR targeted therapies. Cancer Cell. 2006b doi: 10.1016/j.ccr.2006.04.022. (in press) [DOI] [PubMed] [Google Scholar]
- Jiang J., Greulich H., Janne P.A., Sellers W.R., Meyerson M., Griffin J.D. Epidermal growth factor-independent transformation of Ba/F3 cells with cancer-derived epidermal growth factor receptor mutants induces gefitinib-sensitive cell cycle progression. Cancer Res. 2005;65:8968–8974. doi: 10.1158/0008-5472.CAN-05-1829. [DOI] [PubMed] [Google Scholar]
- Johnson L., Mercer K., Greenbaum D., Bronson R., Crowley D., Tuveson D., Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Kim C.F., Jackson E.L., Woolfenden A.E., Lawrence S., Babar I., Vogel S., Crowley D., Bronson R.T., Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Kobayashi S., Boggon T.J., Dayaram T., Janne P.A., Kocher O., Meyerson M., Johnson B.E., Eck M.J., Tenen D.G., Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Kwak E.L., Sordella R., Bell D.W., Godin-Heymann N., Okimoto R.A., Brannigan B.W., Harris P.L., Driscoll D.R., Fidias P., Lynch T.J., et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc. Natl. Acad. Sci. 2005;102:7665–7670. doi: 10.1073/pnas.0502860102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G., et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Marchetti A., Martella C., Felicioni L., Barassi F., Salvatore S., Chella A., Camplese P.P., Iarussi T., Mucilli F., Mezzetti A., et al. EGFR mutations in non-small-cell lung cancer: Analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J. Clin. Oncol. 2005;23:857–865. doi: 10.1200/JCO.2005.08.043. [DOI] [PubMed] [Google Scholar]
- Miller V.A., Kris M.G., Shah N., Patel J., Azzoli C., Gomez J., Krug L.M., Pao W., Rizvi N., Pizzo B., et al. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J. Clin. Oncol. 2004;22:1103–1109. doi: 10.1200/JCO.2004.08.158. [DOI] [PubMed] [Google Scholar]
- Mitsudomi T., Kosaka T., Endoh H., Horio Y., Hida T., Mori S., Hatooka S., Shinoda M., Takahashi T., Yatabe Y. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J. Clin. Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- Moody S.E., Perez D., Pan T.C., Sarkisian C.J., Portocarrero C.P., Sterner C.J., Notorfrancesco K.L., Cardiff R.D., Chodosh L.A. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Paez J.G., Janne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., et al. EGF receptor gene mutations are common in lung cancers from ‘never smokers’ and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc. Natl. Acad. Sci. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W., Wang T.Y., Riely G.J., Miller V.A., Pan Q., Ladanyi M., Zakowski M.F., Heelan R.T., Kris M.G., Varmus H.E. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005a;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W., Miller V.A., Politi K.A., Riely G.J., Somwar R., Zakowski M.F., Kris M.G., Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005b;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack V.A., Savage D.M., Baker D.A., Tsaparikos K.E., Sloan D.E., Moyer J.D., Barbacci E.G., Pustilnik L.R., Smolarek T.A., Davis J.A., et al. Inhibition of epidermal growth factor receptor-associated tyrosine phosphorylation in human carcinomas with CP-358,774: Dynamics of receptor inhibition in situ and antitumor effects in athymic mice. J. Pharmacol. Exp. Ther. 1999;291:739–748. [PubMed] [Google Scholar]
- Riely G.J., Pao W., Pham D., Li A.R., Rizvi N., Venkatraman E.S., Zakowski M.F., Kris M.G., Ladanyi M., Miller V.A. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin. Cancer Res. 2006;12:839–844. doi: 10.1158/1078-0432.CCR-05-1846. [DOI] [PubMed] [Google Scholar]
- Shigematsu H., Lin L., Takahashi T., Nomura M., Suzuki M., Wistuba I.I., Fong K.M., Lee H., Toyooka S., Shimizu N., et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J. Natl. Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- Sordella R., Bell D.W., Haber D.A., Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- Stamos J., Sliwkowski M.X., Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- Tichelaar J., Lu W., Whitsett J. Conditional expression of fibroblast growth factor-7 in the developing and mature lung. J. Biol. Chem. 2000;275:11858–11864. doi: 10.1074/jbc.275.16.11858. [DOI] [PubMed] [Google Scholar]
- Tokumo M., Toyooka S., Kiura K., Shigematsu H., Tomii K., Aoe M., Ichimura K., Tsuda T., Yano M., Tsukada K., et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin. Cancer Res. 2005;11:1167–1173. [PubMed] [Google Scholar]
- Tracy S., Mukohara T., Hansen M., Meyerson M., Johnson B.E., Janne P.A. Gefitinib induces apoptosis in the EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res. 2004;64:7241–7244. doi: 10.1158/0008-5472.CAN-04-1905. [DOI] [PubMed] [Google Scholar]
- Travis W.D., Colby T.V., Corrin B., Shimosato Y., Brambilla E., Sobin L.H. Springer Verlag; Berlin: 1999. Histological typing of lung and pleural tumors. [Google Scholar]
- Tsao M.S., Sakurada A., Cutz J.C., Zhu C.Q., Kamel-Reid S., Squire J., Lorimer I., Zhang T., Liu N., Daneshmand M., et al. Erlotinib in lung cancer—Molecular and clinical predictors of outcome. N. Engl. J. Med. 2005;353:133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- Varmus H., Pao W., Politi K., Podsypanina K., Du Y.-C.N. Oncogenes come of age. Cold Spring Harb. Symp. Quant. Biol. 2005;70:1–9. doi: 10.1101/sqb.2005.70.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.J., Bigner S.H., Bigner D.D., Kinzler K.W., Hamilton S.R., Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc. Natl. Acad. Sci. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]