Abstract
Enterohemorrhagic Escherichia coli (EHEC) 0157:H7 is a food-borne pathogen that can cause bloody diarrhea and, occasionally, acute renal failure as a consequence of Shiga toxin (Stx) production by the organism. Stxs are potent cytotoxins that are lethal to animals at low doses. Thus, Stxs not only harm the host but, as reported here, also significantly enhance the capacity of EHEC O157:H7 to adhere to epithelial cells and to colonize the intestines of mice. Tissue culture experiments showed that this toxin-mediated increase in bacterial adherence correlated with an Stx-evoked increase in a eukaryotic receptor for the EHEC O157:H7 attachment factor intimin.
Keywords: adherence, nucleolin
Enterohemorrhagic Escherichia coli (EHEC) are commensal bacteria of cattle that can cause serious disease in humans who come in direct contact with the animals or who ingest undercooked ground beef or cow manure-contaminated water, cider, vegetables, or other products (1). The disease presents as mild diarrhea, hemorrhagic colitis, or, in a subset of patients, a sequela called the hemolytic uremic syndrome (HUS) that is characterized by thrombocytopenia, hemolytic anemia, and kidney lesions (2). Indeed, EHEC serotype O157:H7, which is a member of a larger group of Shiga toxin (Stx)-producing E. coli (STEC), is estimated to cause ≈73,000 cases of disease per year in the United States (3) and is the most frequent cause of acute renal failure in children in the United States (2). Stxs are responsible for HUS (4) and are categorized into two antigenically distinct groups, Stx1 and Stx2 (5). Stxs are potent cytotoxins that are composed of a single A polypeptide and five B polypeptides. The toxins are internalized by clathrin-dependent endocytosis after binding of the B pentamer to the target cell glycolipid receptor globotriaosylceramide (Gb3) (5). The A polypeptide of the toxins is an N-glycosidase that removes a single adenine from the 28S eukaryotic rRNA and inhibits protein synthesis in susceptible cells (5). Although the production of Stxs by STEC is correlated with pathology in the host, identification of a mechanism by which Stx, or, for that matter, any bacterial toxin, is of direct benefit to the survival of the bacterium has eluded microbiologists.
One possible connection between toxin production and EHEC O157:H7 persistence might relate to how the organism is maintained in the gut. Colonization of the large intestine by EHEC O157:H7 results in an “attaching-and-effacing” lesion characterized by an actin-rich pedestal formed by the host cell around the bacteria, destruction of brush border microvilli, and intimate adhesion of the pathogen to the enterocyte surface (6, 7). Two bacterial genes expressed from a chromosomal pathogenicity island termed the locus for enterocyte effacement (LEE) contribute to this pattern of adherence. Intimin, the protein product of the eae gene, is a major outer membrane surface adhesin (8). The translocated intimin receptor (Tir) is a transmembrane protein secreted through the type III secretion system of the LEE locus that is inserted into the host cell membrane to serve as a receptor for intimin (6, 7).
We previously reported that intimin recognizes not only the bacterially encoded Tir as a receptor but also the eukaryotic protein nucleolin localized on the surface of tissue culture cells (9). Nucleolin is a multifunctional protein that is present in abundance in the nucleus and primarily functions in ribosome biogenesis. However, nucleolin can be expressed at the cell surface in many cell types (10) and serves as a receptor for some viruses (11, 12). In our earlier study (9), we demonstrated that (i) intimin interacts with nucleolin that is present in HEp-2 cell extracts, (ii) purified intimin and nucleolin colocalize on the surface of HEp-2 cells, (iii) the sites of EHEC O157:H7 microcolonies coincide with regions of surface-expressed nucleolin, and (iv) anti-nucleolin serum reduces EHEC O157:H7 adherence when added before or at the time of infection. We hypothesized that Stx produced during infection may promote increased adherence of EHEC O157:H7 to enterocytes and, consequently, colonization of the bowel by enhancing cell surface expression of nucleolin.
Results
Comparison of Growth, Protein Expression, and Cytotoxicity.
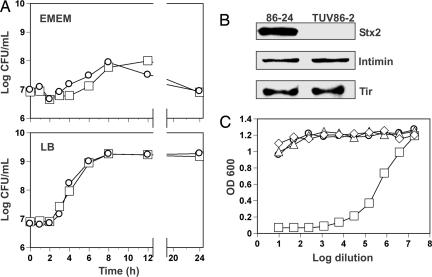
Strain 86-24 is a clinical isolate of EHEC O157:H7 that produces Stx2. EHEC O157:H7 strain TUV86-2, an isogenic stx2 mutant, was constructed (13) and kindly provided by A. Donohue-Rolfe (Tufts University, North Grafton, MA). These strains were used to compare levels of adherence to HEp-2 epithelial cells in culture [an established model for evaluating adherence of EHEC O157:H7 (14)] and colonization of mice. Before these assays, we conducted comparative analyses of the relevant properties of the two strains. We found that the growth rates of 86-24 and TUV86-2 in minimal or rich culture media were indistinguishable (Fig. 1A) and that Stx2 was detectable by immunoblot in lysates of 86-24 but not TUV86-2 (Fig. 1B). We also noted Vero cell cytotoxic activity in association with the 86-24 lysates (50% cytotoxic dose = 4 × 106 per ml) but not the TUV86-2 lysates (Fig. 1C). The finding that lysates of strain TUV86-2 were not toxic for Vero cells is solely attributable to the deleted sequence in the stx2 operon and not the absence of the bacteriophage (see Fig. 5 and Supporting Text, which are published as supporting information on the PNAS web site). Additionally, we compared Tir and intimin expression levels in 86-24 and TUV86-2 by immunoblot and found them to be comparable for both proteins (Fig. 1B).
Fig. 1.
Growth comparison, expression profiles, and characterization of cytotoxicity associated with WT EHEC and its stx2 isogenic mutant. (A) Growth curves of strains 86-24 (□) and TUV86-2 (○) in Eagle’s minimal essential medium (EMEM) and LB. (B) Immunoblot evaluation of lysates of cultures of 86-24 or TUV86-2 for Stx2, intimin, and Tir. (C) Cytotoxic effects of serially diluted lysates of DH5α (▵), 86-24 (□), TUV86-2 (○), or medium (◇) alone on Vero cells after 48 h of incubation.
Stx2 Contributes to Increased Adherence to Epithelial Cells in Culture.
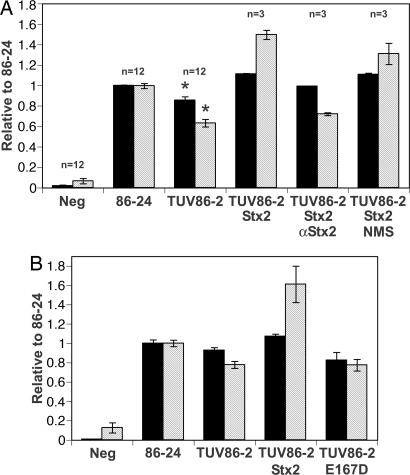
Next, we compared the pattern of adherence to HEp-2 cells by 86-24 and TUV86-2; each strain expressed GFP equivalently from a plasmid (data not shown). As expected, both strains adhered to HEp-2 cells under standard conditions and exhibited a typical localized pattern of adherence that is characteristic of attaching-and-effacing lesions, including microcolony formation and condensed actin staining (see Fig. 6, which is published as supporting information on the PNAS web site). However, microcolony formation appeared to be less pronounced for TUV86-2 than for 86-24 (Fig. 6). These observations prompted us to analyze adherence of the GFP-expressing strains to HEp-2 cells by a quantitative flow cytometric assay. Those epithelial cells to which bacteria adhered exhibited green fluorescence. The intensity of fluorescence was considered reflective of the number of adhered bacteria per epithelial cell and thus the magnitude of microcolony formation. The percentage of HEp-2 cells infected with TUV86-2 was modestly (≈15%) but significantly (P = 0.003) reduced compared with cells infected with the WT 86-24 (Fig. 2A). Additionally, there was a nearly 40% decrease (P < 0.001; Fig. 2A) in microcolony formation of the population infected with TUV86-2. These quantitative adherence results suggest that Stx2 production provides an advantage to 86-24 by increasing the organism’s capacity to adhere to cells.
Fig. 2.
Quantitation of in vitro adherence to epithelial cells of WT EHEC and its stx2 isogenic mutant. Adherence to HEp-2 cells infected with GFP-expressing bacteria was assessed by flow cytometry. Black bars represent the proportion of HEp-2 cells to which any 86-24 or TUV86-2 adhered (the infected cell populations). Gray hatched bars indicate the relative number of bacteria adhered to an individual HEp-2 cell in the respective infected populations when compared with 86-24-infected samples (set at 1). Asterisks indicate statistical significance in the 95% confidence interval analyzed by ANOVA. (A) Purified Stx2 (100 ng/ml) was added to HEp-2 cells in the presence or absence of anti-Stx2 neutralizing mAb or normal mouse serum (NMS) where indicated. Findings for the addition of Stx2 or antibodies show a single representative experiment performed in triplicate (n = 3) ± SE. Data for the negative control, 86-24, and TUV86-2 alone are the geometric means of four individual experiments performed in triplicate (n = 12) ± SE. (B) Purified Stx2-6H or the E167D mutant derivative (100 ng/ml) was added to HEp-2 cells where indicated as in A. Results shown correspond to a single representative experiment performed in triplicate (n = 3) ± SE.
Because Stx2 was produced by 86-24 under conditions of the adherence assay (see Fig. 7, which is published as supporting information on the PNAS web site) and could account for the difference exhibited between 86-24 and TUV86-2, we investigated whether the presence of Stx2 during TUV86-2 adherence could restore adherence to WT levels. Governmental restrictions prevent complementation of stx2 genes by recombinant techniques, so purified Stx2 was added to HEp-2 cells 2 h before infection. The presence of Stx2 before and during 6 h of infection fully restored adherence of infected epithelial cells to levels equivalent to or greater than those obtained with 86-24 (Fig. 2A). The addition of anti-Stx2 neutralizing mAb BC5 to the Stx2-containing media returned levels of TUV86-2 adherence to those observed in the absence of toxin, but the presence of normal mouse serum did not abrogate the effects of Stx2 (Fig. 2A). In a separate experiment, we investigated the effect on adherence of an E167D mutation in the Stx2 active site (15) that reduced cytotoxic activity 1,000-fold (see Fig. 8, which is published as supporting information on the PNAS web site). This mutant toxin did not increase adherence of TUV86-2 (Fig. 2B). These results indicate that the presence of Stx2 can specifically complement the toxin-negative TUV86-2 strain for the capacity to adhere to epithelial cells at a level comparable to the WT strain and that the enzymatic activity of the toxin is required for this effect.
Stx2 Production Enhances Colonization of Mice.
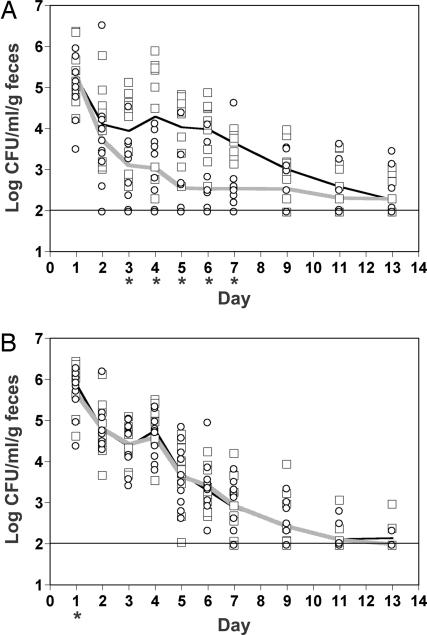
We then investigated whether production of Stx2 by strain 86-24 affected the colonization of BALB/c mice. A significant difference in colonization [geometric mean colony-forming units (cfu)/ml per g of feces] between strains 86-24- and TUV86-2-infected animals was evident by day 3 after infection (Fig. 3A). This difference in colonization was maintained through day 7, and an overall significant difference between strains for all days studied was exhibited (P = 0.004).
Fig. 3.
Comparison of the capacity of WT EHEC and its stx2 isogenic mutant to colonize mice. (A) Single-strain challenges: BALB/c mice were orally infected twice with an inoculum that contained ≈2.5 × 109 cfu of 86-24 (□) or TUV86-2 (○). On subsequent days after infection, fecal pellets were collected, homogenized, and serially diluted for plating on MacConkey agar that contained sorbitol. Results from each mouse and the geometric mean of 10 mice infected with 86-24 (black line) or TUV86-2 (gray line) are presented. The horizontal line represents the limit of detection. Asterisks below the x axis indicate days in which a statistical difference (P ≤ 0.05) between strains was established. (B) Mixed-strain challenge: BALB/c mice were orally infected as in A with an inoculum that contained ≈2.5 × 109 cfu of 86-24 and TUV86-2 in equal ratios. Individual colonies were differentiated by screening for an stx2 sequence or Stx2. The level of colonization reflected by each strain in an individual mouse and the geometric mean of 10 mice are presented as in A.
Next, we considered whether 86-24 could out-compete TUV86-2 in a mixed infection. To address this possibility, mice were orally infected with an equal ratio of 86-24 and TUV86-2. There was no significant difference in colonization between strains (P = 0.69). These results suggest that Stx2 production by 86-24 enhances TUV86-2 colonization in a mixed-strain infection. This contention is further supported by the observation that, on some days, the level of colonization of TUV86-2 was increased by more than one order of magnitude compared with the single-strain infections (Fig. 3B). Furthermore, these results are consistent with in vitro adherence results (Fig. 2).
Stx2 Increases Nucleolin Surface Expression on Epithelial Cells in Culture.
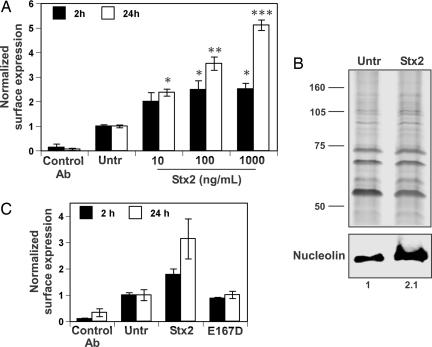
Previous unpublished work from our laboratory suggested that exposure of epithelial cells to Stx1 and Stx2 resulted in an increase in surface-expressed nucleolin. This increase should, in theory, provide a greater number of receptors available for EHEC attachment before the translocation of Tir. Therefore, we quantified the amount of nucleolin on the surface of HEp-2 cells over time in response to treatment with increasing concentrations of purified Stx2. Our immunodetection method was limited to the cell surface, as indicated by the lack of significant surface labeling of the isotype-matched control antibody specific for a cytosolic antigen (Fig. 4A). By contrast, nucleolin-specific mAb bound to the surface of HEp-2 cells treated with Stx2 for 2 h at a 2-fold or greater level compared with untreated HEp-2 cells (Fig. 4A). This increase in nucleolin surface expression was further enhanced by 24 h of treatment with Stx2 that also corresponded with a dose-dependent increase in cell death (Fig. 4A). This latter observation was not unexpected; induction of apoptosis by Stx is a feature of intoxication (16). To further substantiate this apparent Stx2-evoked increase in surface-localized nucleolin, membrane-associated proteins were prepared by high-salt extraction from epithelial cells treated with or without Stx2 (100 ng/ml) for 2 h, and nucleolin was detected by immunoblot. The immunodetection protocol revealed that epithelial cells treated with Stx2 displayed an increase in nucleolin at the cell surface (Fig. 4B) that was consistent with an augmentation of TUV86-2 adherence in the presence of purified Stx2 (Fig. 2). In a separate experiment, we demonstrated that HEp-2 cells treated with the purified Stx2 toxoid that contained the E167D mutation did not exhibit increased surface localization of nucleolin (Fig. 4C), a finding that is consistent with the results depicted in Fig. 2B.
Fig. 4.
Quantification of nucleolin surface expression on epithelial cells after exposure to Stx2 or Stx2 toxoid. (A) HEp-2 cells were treated with or without Stx2 at indicated concentrations for 2 (■) or 24 (□) h. Surface-expressed proteins were labeled with monoclonal isotype-matched control or a nucleolin-specific antibody. The results were normalized by expressing values corresponding to all sample groups relative to untreated cells. Data presented are arithmetic means ± SE from a representative experiment with samples performed in triplicate. Asterisks above Stx2-treated sample groups indicate statistical difference in the 95% confidence interval from the untreated control within a time series established by independent sample t tests; sample groups with an equivalent number of asterisks are not significantly different from each other. (B) HEp-2 cells were untreated (Untr) or treated with 100 ng/ml Stx2 for 2 h. Membrane-associated proteins were extracted in detachment buffer that contained 1 M NaCl for 2 min on ice. Membrane-associated proteins were separated by SDS/PAGE and stained with silver (Upper) or subjected to immunoblotting with nucleolin-specific mAb (Lower). Immunolabeled band densities expressed relative to the untreated control were quantified with image j software. (C) HEp-2 cells were treated with or without Stx2-6H or the E167D mutated derivative (100 ng/ml) for 2 or 24 h, and surface-expressed proteins were labeled with monoclonal isotype-matched control or a nucleolin-specific antibody. Data are presented as in A.
Discussion
Stx production by EHEC O157:H7 was linked to the development of the life-threatening hemolytic uremic syndrome more than 2 decades ago (Stx was called Vero toxin in ref. 17). However, the advantage, if any, that synthesis of Stx provides to this microbe that normally resides in the bovine host without causing apparent illness (2) remained a subject of speculation. Here, we sought to address this issue by asking whether Stx synthesis might aid EHEC O157:H7 in establishing infection. We previously reported that nucleolin interacts with intimin and is associated with sites of EHEC attachment (9). Therefore, we reasoned that an Stx-mediated enhancement of cell surface-localized nucleolin might provide an adherence advantage to toxin-producing EHEC O157:H7 strains. In this report, we provide several lines of evidence to suggest that this hypothesis is correct. Specifically, we show that epithelial cells exposed to enzymatically active purified Stx2 exhibited increased surface expression of nucleolin, that the WT EHEC O157:H7 strain 86-24 did in fact adhere better to epithelial cells in tissue culture than did its stx2 isogenic mutant, and that 86-24 colonized mice at a higher level than did the isogenic mutant.
That Stx2 plays an important role in augmenting the adherence of EHEC O157:H7 strain 86-24 was further demonstrated by the finding that exposure of epithelial cells to Stx2 for 2 h before infection and maintenance of toxin in tissue culture medium throughout the infection enhanced the adherence levels of the stx2 isogenic mutant strain TUV86-2 to an even greater extent than that of WT 86-24. The observation that the extent of TUV86-2 adherence when Stx2 was added was greater than that of 86-24 might reflect differences in toxin concentrations between the two types of experiments (100 ng/ml in the complementation study and ≈25–75 ng/ml in supernatants that contained WT bacteria during the adherence assay) and/or the pretreatment of HEp-2 cells with toxin that was necessary to fully restore adherence of the toxin-negative mutant strain to at least WT levels. The finding that Stx-neutralizing antibody and an active site mutation abrogated this increased adherence supported the specificity of the toxin effect on bacterial adherence. This enhancement of adherence by the addition of enzymatically active Stx2 to the culture system paralleled the Stx2-mediated increases in surface-expressed nucleolin (shown in Fig. 4). From these data, we inferred that when epithelial cells were exposed to Stx2, more nucleolin became localized at the cell surface, and this increase allowed for greater intimin-driven EHEC O157:H7 adherence.
The in vitro adherence assay is a useful system for evaluating the specific steps in bacterial:host cell interactions but does not fully reflect the conditions that must be overcome by the bacterium to successfully attach to enterocytes in the intestinal milieu. Therefore, we compared the colonizing capacities of strain 86-24 with those of TUV86-2 in conventionally bred mice that harbor normal intestinal flora. When these animals were infected individually with these bacteria, the Stx2-expressing WT strain colonized the animals to a greater extent than did the stx2 isogenic mutant. When such mice were infected with an inoculum that contained equivalent concentrations of 86-24 and TUV86-2, toxin produced from 86-24 appeared to complement the colonization defect of TUV86-2. This result was consistent with the capacity of purified Stx2 to augment TUV86-2 adherence in vitro (Fig. 2).
Two reports in the literature contain data, albeit limited, that support our contention that Stx expression positively impacts EHEC O157:H7 colonization. First, in a signature-tagged mutagenesis study aimed at identifying factors that influence colonization of the bovine gut, Dziva et al. (18) failed to recover a mutant of the EHEC 0157:H7 strain EDL933 that contained a transposon inserted into a gene located between the stx1 genes and bacteriophage genes involved in bacterial cell lysis. This observation was not discussed in detail, nor was the toxin phenotype of that strain described, but we would predict that Stx1 production or release could likely be abrogated in such a mutant. Second, Sjogren et al. (19) showed that the rabbit diarrheal pathogen E. coli strain RDEC-1 transduced with an Stx1-converting bacteriophage caused a more severe infection than did the nontransduced strain and also noted that the transduced strain colonized rabbits 1–2 days earlier.
Because as few as 100 EHEC O157:H7 organisms are sufficient to cause human disease (2), it seems unlikely that the concentration of toxin produced from such a small number of bacteria would be sufficient to alter the surface expression of nucleolin in the large bowel during the initial attachment phase of infection. Thus, Stx2 may not provide a significant advantage for bacterial attachment in the earliest stages of a low-dose infection of humans. Although much higher doses of EHEC O157:H7 are required to establish infection in mice, our general supposition that the impact of toxin from the inoculum is not critical to initial colonization is in keeping with our finding in single-strain mouse challenge experiments. In those studies, 86-24 did not colonize demonstrably better than did TUV86-2 during the first 2 days of infection. Thus, we propose that only when EHEC is intimately attached to the intestinal epithelium do subsequent progeny benefit from an increase in surface receptors available for attachment. Although toxin produced during infection is likely diffusible throughout the intestinal mucosa, we suspect that, at specific regions of bacterial attachment and attaching-and-effacing lesion formation, concentrations of toxin are transiently much higher than the net concentration throughout the large bowel. Therefore, we hypothesize that, at local sites of adherence where toxin concentration is expected to be higher, an environment suited for enhanced adherence is created that stimulates the enlargement of microcolonies. This prediction is supported by adherence micrographs and quantitative results that typically show microcolony formation to be more developed when epithelial cells are infected with the toxin-producing strain.
Whether the effect of Stx2 on nucleolin surface localization is the sole explanation for the clear benefit that toxin production plays in the capacity of EHEC O157:H7 to adhere to epithelial cells and colonize mice remains to be determined. We recognize that, in addition to nucleolin, other eukaryotic proteins may also bind intimin and promote bacterial adhesion. However, we have no evidence as yet that the distribution of such theoretical intimin receptors is affected by Stx. Moreover, a close association between localization of nucleolin and adherent EHEC O157:H7 has been demonstrated in the mouse intestine (20).
The details of the cellular mechanism involved in Stx-mediated enhancement in surface-expressed nucleolin are still unknown. Certainly, several explanations are possible. One particularly appealing idea is that the increase in nucleolin at the cell surface that is reported here might be part of a stress response to intoxication and a pre-apoptotic mechanism. Indeed, increased surface expression of nucleolin has also been described in leukemia cells undergoing apoptosis (21). However, it is also noteworthy that nucleolin and Stx share an affinity for the same substrate, ribosomal RNA. Thus, it is conceivable that there is a direct mechanistic link between the Stx mode of action and an increase in surface expression of nucleolin.
Materials and Methods
Strains, Antibodies, and Primers.
EHEC O157:H7 strain 86-24 and EHEC O157:H7 strain TUV86-2, an isogenic stx2 mutant, are both nalidixic acid resistant and were kindly provided by A. Donahue-Rolfe (13). Antibodies used in this study were mouse monoclonal anti-nucleolin C23 (MS-3; Santa Cruz Biotechnology), mouse monoclonal anti-glyceraldehyde phosphate dehydrogenase (Santa Cruz Biotechnology), mouse monoclonal anti-intimin-γ EE4IIE2 (9), rabbit polyclonal anti-Tir (9), mouse monoclonal anti-Stx2 11E10 (22), mouse monoclonal anti-Stx2 BC5 (ref. 23; kindly provided by Nancy Strockbine, Centers for Disease Control and Prevention, Atlanta), horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Bio-Rad), and HRP-conjugated goat anti-rabbit IgG (Bio-Rad). The PCR primers used in this study are listed in Table 1, which is published as supporting information on the PNAS web site.
Colony PCR.
PCR performed directly from bacterial colonies lysed in sterile water at 95°C for 5 min was performed in a 50-μl reaction that contained 5 μl of lysed bacterial suspension, 50 mM Tris·HCl (pH 8.3), 10 mM KCl, 5 mM [NH4]2SO4, 2 mM MgCl2, 1 μM forward and reverse primers, 0.4 mM dNTPs, and FastStart TaqDNA polymerase (2 units; Roche Applied Science, Indianapolis). Reaction cycling was done in a PTC-150 Minicycler (MJ Research, Cambridge, MA).
Construction of Stx2-6-His Tag and Protein Purification.
The plasmid construct pMJS50 that permitted expression of Stx2 with a C-terminal 6-histidine tag on the B subunit (designated Stx2-6H), was generated by consecutive rounds of PCR by standard techniques. Initially, amplification from pMJS2 (24) was done with primers 2A51 and C12B. Subsequent PCRs were performed with primer pairs 2A52 and C22B, followed by 2A52 and BC3. The final amplicon included an optimized Shine–Dalgarno sequence upstream of the stxA2 gene and was ligated into the expression vector pTrcHis2C (Invitrogen). The E167D mutation was generated in pMJS50 (creating pCMR1) by internal ligation after restriction at a common NsiI site of PCR fragments amplified with the primer pairs TRC-F and 167R for the upstream fragment or 167-F and TRC-R for the downstream fragment. The purification of Stx2-6H and Stx2(E167D)-6H from E. coli L172 (25) was done by nickel-affinity column chromatography (Qiagen, Valencia, CA), essentially according to the manufacturer’s instructions. Stx2 was purified by affinity chromatography as described in ref. 26 by using 11E10 mAb.
Vero Cell Cytotoxicity Assay.
The cytotoxic activity for Vero cells of clarified bacterial lysates or adherence assay culture supernatants that contained Stx2, Stx2-6H, or Stx2(E167D)-6H or purified preparations of these proteins was determined as detailed in ref. 24.
Bacterial Adherence Assay.
HEp-2 cells were maintained in complete Eagle’s minimal essential medium (EMEM) at 37°C in 5% CO2. Plasmid pEGFP (Clontech) was electroporated into EHEC O157:H7 strain 86-24 and EHEC O157:H7 strain TUV86-2 that had been made competent by standard techniques. EHEC strains were grown statically to stationary phase in LB supplemented with ampicillin (100 μg/ml), nalidixic acid (25 μg/ml), and isopropyl-β-d-thiogalactopyranoside (IPTG) (2 mM) at 37°C. Subconfluent monolayers of HEp-2 cells cultured in 12-well dishes (for flow cytometric analysis; Costar and Corning) or 8-well chamber slides (for microscopic analysis; Lab-Tek and Nalge Nunc) were infected at a multiplicity of infection of ≈50 (bacteria to epithelial cell) in adherence medium [EMEM supplemented with l-glutamine (2 mM)/sodium bicarbonate (0.4%)/d-mannose (1%)/ampicillin (100 μg/ml)/nalidixic acid (25 μg/ml)/IPTG (2 mM)] for 6 h. Supernatants from uninfected controls and infected cultures were saved for evaluation of toxicity toward Vero cells and for the presence of Stx2 protein by immunoblot.
For flow cytometric analysis, HEp-2 cells were washed extensively with PBS and harvested by treatment with 0.25% trypsin (Cambrex, East Rutherford, NJ). Intimate adherence of EHEC was not sensitive to the activity of trypsin. HEp-2 cells were suspended in PBS that contained 3% formalin, and fluorescent intensity for 10,000 gated events was quantified with an EPICS XL-MCL flow cytometer in the FL-1 channel. For experiments that included the addition of purified toxin, Stx2, Stx2-6H, or Stx2(E167D)-6H (100 ng/ml) was added to HEp-2 cells with or without equivalently diluted BC5 (mouse monoclonal Stx2 neutralizing antiserum) or normal mouse serum for 2 h before infection and maintained throughout the assay. For microscopic analysis, individual wells of chamber plates were washed extensively with PBS and fixed with PBS that contained 3.7% paraformaldehyde for 10 min. The epithelial cells were then permeabilized with 0.1% Triton X-100 for 5 min and blocked for 30 min with PBS containing 1% BSA. The epithelial cells were then stained with Alexa Fluor 594 phalloidin (Molecular Probes) diluted in PBS that contained 1% BSA for 20 min at room temperature. Slides were examined with a BX60 microscope (Olympus, Melville, NY) fitted with a BX-FLA reflected light fluorescence attachment. The images presented here were obtained with a SPOT RT charge-coupled device digital camera (Diagnostic Instruments, Sterling Heights, MI)
Mouse Infection.
Six-week-old BALB/c mice (Charles River Laboratories) were fasted for ≈18 h and deprived of water for 4 h before infection. Cultures (100 ml) of EHEC O157:H7 strain 86-24 and O157:H7 strain TUV86-2 were grown in LB at 37°C overnight with aeration and then concentrated 50-fold in PBS that contained 20% sucrose. For single-strain experiments, five mice were fed 50 μl of solution that contained ≈2.5 × 109 cfu of EHEC O157:H7 strain 86-24 or EHEC O157:H7 strain TUV86-2 followed by a second feeding 2 h later. The experiment was repeated, and the results from the two studies were combined for a total of 20 mice (10 animals per strain). For mixed-strain studies, five mice were fed 50 μl of solution that contained ≈2.5 × 109 cfu composed of equivalent amounts of EHEC O157:H7 strain 86-24 and EHEC O157:H7 strain TUV86-2. Two such mixed-strain experiments were done, and the data from the 5 animals per study were combined for a total of 10 doubly infected mice. Fecal pellets were collected on subsequent days, homogenized in sterile PBS to create an initial 10-fold dilution (wt/vol), and serially diluted for plating on sorbitol MacConkey agar supplemented with nalidixic acid (25 μg/ml). Individual sorbitol-fermenting colonies that grew after overnight incubation at 37°C were enumerated. For the mixed infection, colonies that grew on sorbitol MacConkey agar were patched onto LB agar that contained nalidixic acid (25 μg/ml) for differentiation according to stx2 sequence (by colony blot) or Stx2 production (by dot blot). When necessary, results were confirmed by colony PCR with the primer pair UB-3 and CR-1398 (Table 1). These primers generate amplification products from 86-24 and TUV86-2 that differ in size by ≈589 bp because of the deleted sequence in TUV86-2.
Colony Blots.
Colonies were lifted onto nylon membranes (Hybond-N+; Amersham Pharmacia Biosciences) that were subsequently placed upright on filter paper soaked in 0.5 M NaOH for 5 min. Nylon membranes were then washed twice in 5× SSC (75 mM Na3C6H5O7/0.75 M NaCl, pH 7) and air-dried, and the DNA was crosslinked to the membrane by exposure to UV light. The 517-bp DNA probe was generated by colony PCR with the primer pair TUVdel-F and TUVdel-R specific for the regions of the stx2AB genes deleted in TUV86-2. This probe was purified by agarose gel electrophoresis and then labeled with HRP (ECL direct nucleic acid labeling system; Amersham Pharmacia Biosciences) according to the manufacturer’s instructions. The labeled probe (200 ng per membrane) was then added directly to hybridization buffer (ECL Gold; Amersham Pharmacia Biosciences) that had been preincubated with the membrane for 1 h at 42°C. The probe in hybridization buffer and the membrane were then incubated at 42°C overnight with gentle shaking. Each membrane was then rinsed twice in primary wash buffer (6 M urea/0.4% SDS/0.5× SSC) for 20 min at 42°C and twice in secondary wash buffer (2× SSC) for 5 min at room temperature. Hybridized probe was detected by enhanced chemiluminescence (ECL direct nucleic acid labeling system).
Dot Blots.
Each colony was inoculated into a microfuge tube that contained LB with nalidixic acid (25 μg/ml) and grown overnight at 37°C. The tubes were then centrifuged at 4,000 × g for 5 min, and 200 μl of supernatant was vacuum-filtered onto nitrocellulose membranes (Schleicher & Schuell) by using a Minifold I microsample filtration manifold (Schleicher & Schuell). Membranes were blocked in Tris-buffered saline that contained 5% nonfat dry milk and 3% BSA overnight at 4°C and then probed with the 11E10 mAb specific for the Stx2 A polypeptide as described below.
ELISA-Based Quantification of Nucleolin Surface Expression on Epithelial Cells.
HEp-2 cells (105 per well) cultured in 24-well dishes were washed with Hanks’ balanced salt solution (BioWhittaker) and replaced with adherence medium with or without purified Stx2 at indicated concentrations. Cultures were incubated at 37°C in 5% CO2 for a total of 2 or 24 h. For the last hour of incubation, anti-nucleolin mAb C23 or anti-glyceraldehyde phosphate dehydrogenase mAb as an isotype-matched irrelevant control was added directly to appropriate wells. Surface-bound primary antibody was detected with HRP-conjugated goat anti-mouse IgG diluted in PBS with 3% BSA and visualized with TMB Peroxidase E1A substrate (Bio-Rad). Optical density (at 600 nm) for each well was measured and recorded with an ELx800 microplate reader. The colorimetric solution was then aspirated, and the formalin-fixed epithelial cells were stained with a solution that contained 0.13% (wt/vol) crystal violet and 5% ethanol to account for potential differences in cell density after 24 h of exposure to toxin.
Detachment of Membrane-Associated Proteins from Intact Cells.
Membrane-associated proteins were prepared as described in ref. 27 with minor modifications. Briefly, HEp-2 cells (2 × 107) seeded to subconfluency in T75 flasks were washed with Hanks’ balanced salt solution that was replaced with adherence medium with or without purified Stx2 (100 ng/ml) for 2 h. Media were removed, and the cells were washed with PBS. Detachment buffer (1 ml) (20 mM phosphate buffer, pH 7.5/500 mM l-proline/1 M NaCl/0.5 mM EDTA/10 μg/ml each aprotinin, leupeptin, and pepstatin A) was added for 2 min to each flask kept on ice. Flasks were tilted to collect the contents, and the solutions were clarified by centrifugation for 5 min at 200 × g at 4°C. Supernatants that contained membrane-associated proteins detached from the cell surface were collected and dialyzed in PBS overnight at 4°C. Dialysates were concentrated with Centricon concentrators (10-kDa molecular-mass cutoff; Amicon Bioseparations). Cell integrity was evaluated by monitoring for the presence of lactate dehydrogenase activity according to the manufacturer’s instructions (LDH cytoxicity kit; Roche Diagnostics) in both the extraction supernatant and concentrated dialysates.
Immunoblot Analysis.
Proteins were separated by molecular weight according to standard techniques by SDS/PAGE. Proteins were transferred to nitrocellulose membranes with a TransBlot SD SemiDry Electrophoretic Transfer Cell (Bio-Rad). Membranes were blocked in PBS that contained 5% powdered milk, 1% BSA, and 0.05% Tween 20. Primary and secondary antibodies were diluted in the same solution. Immunolabeled proteins were detected by enhanced chemiluminescence (ECL Plus; Amersham Pharmacia Biosciences).
Statistical Analysis.
For single-strain challenge mouse studies, repeated measures ANOVA with bacterial strain as a between-subjects factor and day as a within-subjects factor was used to assess any statistically significant (at P ≤ 0.05) differences in colonization levels between 86-24 and TUV86-2. Independent sample t tests were used to establish statistical significance at P ≤ 0.05 in colonization levels between bacterial strains for each day represented in the single-strain challenge mouse infection. For the dual strain challenge mouse infections, a mixed model for repeated measures ANOVA with strain and day as within-subject factors was used to define any statistically significant (at P ≤ 0.05) differences in colonization between bacterial strains. Paired sample t tests were used to analyze statistical significance at P ≤ 0.05 in colonization levels between bacterial strains for each day represented in these mixed-strain studies. For quantitative bacterial adherence to HEp-2 cell assays, ANOVA was used to analyze the statistical significance of differences among geometric means of triplicate samples from four individual experiments. For the quantification of nucleolin surface expression, independent sample t tests were used to evaluate statistical significance (P ≤ 0.05) of the fold change.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant AI20148-23.
Abbreviations
- EHEC
enterohemorrhagic Escherichia coli
- cfu
colony-forming units
- Stx
Shiga toxin
- Tir
translocated intimin receptor.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Griffin P. M. In: Infections of the Gastrointestinal Tract. Blaser M. J., Smith P. D., Ravdin J. I., Greenberg H. B., Guerrant R. L., editors. New York: Raven; 1995. pp. 739–761. [Google Scholar]
- 2.Paton J. C., Paton A. W. Clin. Microbiol. Rev. 1998;11:450–479. doi: 10.1128/cmr.11.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mead P. S., Slutsker L., Dietz V., McCaig L. F., Bresee J. S., Shapiro C., Griffin P. M., Tauxe R. V. Emerg. Infect. Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffin P. M., Tauxe R. V. Epidemiol. Rev. 1991;13:60–98. doi: 10.1093/oxfordjournals.epirev.a036079. [DOI] [PubMed] [Google Scholar]
- 5.Sandvig K. Toxicon. 2001;39:1629–1635. doi: 10.1016/s0041-0101(01)00150-7. [DOI] [PubMed] [Google Scholar]
- 6.LeBlanc J. J. Clin. Microbiol. Rev. 2003;29:277–296. doi: 10.1080/713608014. [DOI] [PubMed] [Google Scholar]
- 7.Nataro J. P., Kaper J. B. Clin. Microbiol. Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jerse A. E., Kaper J. B. Infect. Immun. 1991;59:4302–4309. doi: 10.1128/iai.59.12.4302-4309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinclair J. F., O’Brien A. D. J. Biol. Chem. 2002;277:2876–2885. doi: 10.1074/jbc.M110230200. [DOI] [PubMed] [Google Scholar]
- 10.Ginisty H., Sicard H., Roger B., Bouvet P. J. Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 11.Callebaut C., Blanco J., Benkirane N., Krust B., Jacotat E., Guichard G., Seddiki N., Svab J., Dam E., Muller S., et al. J. Biol. Chem. 1998;273:21988–21997. doi: 10.1074/jbc.273.34.21988. [DOI] [PubMed] [Google Scholar]
- 12.de Verdego U. R., Selinka H. C., Huber M., Kramer B., Kellermann J., Hofschneider P. H., Kandolf R. J. Virol. 1995;69:6751–6757. doi: 10.1128/jvi.69.11.6751-6757.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gunzer F., Bohn U., Sibylle F., Mühldorfer I., Hacker J., Tzipori S., Donohue-Rolfe A. Infect. Immun. 1998;66:2337–2341. doi: 10.1128/iai.66.5.2337-2341.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKee M. L., O’Brien A. D. Infect. Immun. 1995;63:2070–2074. doi: 10.1128/iai.63.5.2070-2074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson M. P., Deresiewicz R. L., Calderwood S. B. J. Bacteriol. 1990;172:3346–3350. doi: 10.1128/jb.172.6.3346-3350.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ching J. C., Jones N. L., Ceponis P. J., Karmali M. A., Sharman P. M. Infect. Immun. 2002;70:4669–4677. doi: 10.1128/IAI.70.8.4669-4677.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konowalchuk J., Speirs J. J., Stavric S. Infect. Immun. 1977;18:775–779. doi: 10.1128/iai.18.3.775-779.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dziva F., van Diemen P. M., Stevens M. P., Smith A. J., Wallis T. S. Microbiology. 2004;150:3631–3645. doi: 10.1099/mic.0.27448-0. [DOI] [PubMed] [Google Scholar]
- 19.Sjogren R., Neill R., Rachmilewitz D., Fritz D., Newland J., Sharpnack D., Colleton C., Fondacaro J., Gemski P., Boedeker E. Gastroenterology. 1994;106:306–317. doi: 10.1016/0016-5085(94)90587-8. [DOI] [PubMed] [Google Scholar]
- 20.Sinclair J. F., Dean-Nystrom E. A., O’Brien A. D. Infect. Immun. 2006;74:1255–1265. doi: 10.1128/IAI.74.2.1255-1265.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mi Y., Thomas S. D., Xu X., Casson L. K., Miller D. M., Bates P. J. J. Biol. Chem. 2003;278:8572–8579. doi: 10.1074/jbc.M207637200. [DOI] [PubMed] [Google Scholar]
- 22.Perera L. P., Marques L. R. M., O’Brien A. D. J. Clin. Microbiol. 1988;26:2127–2131. doi: 10.1128/jcm.26.10.2127-2131.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Downes F. P., Barrett T. J., Green J. H., Aloisio C. H., Spika J. S., Strockbine N. A., Wachsmuth I. K. Infect. Immun. 1988;56:1926–1933. doi: 10.1128/iai.56.8.1926-1933.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith M. J., Teel L. D., Carvalho H. M., Melton-Celsa A. R., O’Brien A. D. Vaccine. 2006;24:4122–4129. doi: 10.1016/j.vaccine.2006.02.035. [DOI] [PubMed] [Google Scholar]
- 25.Ganserhoff L. J., Wachtel M. R., O’Brien A. D. Infect. Immun. 1999;67:6409–6417. doi: 10.1128/iai.67.12.6409-6417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindgren S. W., Samuel J. E., Schmitt C. K., O’Brien A. D. Infect. Immun. 1994;62:623–631. doi: 10.1128/iai.62.2.623-631.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harms G., Kraft R., Grelle G., Volz B., Dernedde J., Tauber R. Biochem. J. 2001;360:531–538. doi: 10.1042/0264-6021:3600531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson M. P., Neill R. J., O’Brien A. D., Holmes R. K., Newland J. W. FEMS Microbiol. Lett. 1987;44:109–114. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.