Abstract
An important consideration in transgenic research is the choice of promoter for regulating the expression of a foreign gene. In this study several tissue-specific and inducible promoters derived from Japanese flounder Paralichthys olivaceus were identified, and their promoter activity was examined in transgenic zebrafish. The 5′ flanking regions of the Japanese flounder complement component C3, gelatinase B, keratin, and tumor necrosis factor (TNF) genes were linked to green fluorescence protein (GFP) as a reporter gene. The promoter regulatory constructs were introduced into fertilized zebrafish eggs. As a result we obtained several stable transgenic zebrafish that displayed green fluorescence in different tissues. Complement component C3 promoter regulated GFP expression in liver, and gelatinase B promoter regulated it in the pectoral fin and gills. Keratin promoter regulated GFP expression in skin and liver. TNF gene promoter regulated GFP expression in the pharynx and heart. TNF promoter had lipoplysaccharide-inducible activity, such that when transgenic embryos were immersed lipopolysaccharide, GFP expression increased in the epithelial tissues. These 4 promoters regulated the expression of GFP in different patterns in transgenic zebrafish.
Keywords: Japanese flounder, tissue-specific promoter, inducible promoter, green fluorescence protein (GFP), transgenic zebrafish
Introduction
Transgenic technology is a powerful tool not only for analyzing molecular biological functions but also for obtaining commercially important traits for aquaculture. In the last decade fish transgenesis has become common and has progressed considerably (Chen et al., 1996; Hackett and Alvarez, 2000). Various transgenic fish that possess commercially important traits, such as growth enhancement (Zhang et al., 1990) or disease resistance (Sarmasik et al., 2002), have been generated. One of the most critical factors in transgenic research is the regulation of foreign gene expression. Early fish transgenic research was conducted using housekeeping gene promoters such as β-actin (Liu et al., 1990; Higashijima et al., 1997; Hwang et al., 2003; Noh et al., 2003) and elongation factor (EF) (Gao et al., 1997; Kinoshita et al., 2000). These promoters resulted in excess expression in various tissues. However, tissue-specific or inducible promoters are needed to regulate foreign genes in different situations. The activities of tissue-specific promoters were assessed in zebrafish using green fluorescence protein (GFP) as a reporter gene (Udvadia and Linney, 2003). This technique has provided a powerful tool for analyzing the regulation of gene expression in living fish. The pattern of expression of GFP in these transgenic zebrafish was the same as that of the gene from which the promoter was derived. However, promoters derived from most marine aquaculture species have not been described. To generate transgenic aquaculture fish, tissue-specific and stress-inducible promoters from these species need to be identified.
Japanese flounder Paralichthys olivaceus is widely cultured and one of the most important food fish in East Asia. Because this industry is seriously affected by disease, the development of disease-resistant strains is a high priority. Previous studies by our laboratory have used expressed sequence tags (ESTs) (Aoki et al., 1999, 2000; Nam et al., 2000, 2003) and molecular genetic analyses, especially of immune-related genes in Japanese flounder (Hirono et al., 2003). In these reports we focused on cDNA clones that displayed tissue-specific expression or inducible expression, and 4 Japanese flounder genes were chosen for this study: complement component C3, gelatinase B, keratin, and tumor necrosis factor (TNF) genes. These genes were chosen because they were revealed to be expressed in specific tissues of Japanese flounder by EST analysis. We assumed that the promoter region of these genes would show tissue-specific or inducible activities.
It is possible to generate an effective transgenic fish using tissue-specific or inducible promoters. Indeed, such promoters can regulate foreign gene expression faithfully. In contrast, use of a housekeeping gene promoter would result in nonspecific expression (i.e., overexpression) of foreign genes that may be harmful to the host fish. In this study we developed several Japanese flounder promoters that showed tissue-specific or inducible gene expression with the aim of using these in Japanese flounder. Although we have succeeded in introducing a foreign gene into Japanese flounder fertilized eggs and shown transient expression in Japanese flounder embryos, it has been difficult to conduct promoter assays in vivo because the technology has not been sufficiently developed to produce transgenic Japanese flounder with the necessary efficiency. However, the transgenic zebrafish model system has the advantages of shorter generation time, better optical transparency, and easier treatment schemes when compared with Japanese flounder. In this study, promoter regions of the complement component C3, gelatinase B, keratin and TNF genes were isolated and their promoter activities were characterized in transgenic zebrafish.
Materials and Methods
mRNA Expression in Japanese Flounder Tissues
Total RNA was extracted from spleen, fin, liver, head kidney, post kidney, skin, blood, heart, intestine, ovary, gill, leukocyte, and peripheral blood leukocytes (PBLs) stimulated with lipopolysaccharide (LPS) using TRIzol (Invitrogen). The purified total RNA (10 μg) was reverse transcribed into cDNA using an avian myeloblastosis virus (AMV) transcriptase First-strand cDNA synthesis kit (Amersham Bioscience). The reverse-transcribed sample (1 μl) was used in 50 μl of PCR mixture. The PCR primers for Japanese flounder complement component C3 mRNA detection were (sense) 5′-GCTGGAGAAAG TCGTCTTGG-3′ and (anti) 5′-GGATACCTCTCAACTCTG CC-3′, gelatinase B mRNA detection, (sense) 5′-GCA GGAGCCACCAGTCAAAA-3′ and (anti) 5′-GGTCCAGTG TTCATCATCGT-3′, keratin mRNA detection, (sense) 5′-ACTCCGTCGCACAATGCAGA-3′ and (anti) 5′-CTGCA ATTTCCATCTCCAGC-3′, and TNF mRNA detection, (sense) 5′-CCCTATGAACTGTAACAGTTTG-3′ and (anti) 5′-GTCAGGTACTTAACCCTCAT-3′. The ß-actin primer set for an internal control was (sense) 5′-TTTCCCT CCATTGTTGGTCG-3′ and (anti) 5′-GCGACTCTCAGCTC GTTGTA-3′. The PCR was performed with an initial denaturation step of 2 minutes at 95°C and then 20 cycles were run as follows: 30 seconds of denaturation at 95°C, 30 seconds of annealing at 55°C, and 1 minute of extension at 72°C. The reaction products were electrophoresed in a 2.0% agarose gel.
Identification of 5′ Flanking Region Sequences
The 5′ flanking regions of 4 genes were isolated from a Japanese flounder genomic BAC library (Katagiri et al., 2000) using cDNA clones of complement component C3 (AB021653), gelatinase B (AU091035), keratin (AB079729), and TNF (AB040448), as probes for gene screening. To determine the transcription start site, the primer extention method was performed according to a modified version of that in Hikima et al. (2001). A 20mer synthetic oligonucleotide corresponding to the complementary sequence of the first exon was labeled with fluorochrome. The fluorochrome-labeled primer was hybridized to 10 μg of kidney total RNA (extracted using Trizol, Invitrogen) and then extended using an AMV reverse transcriptase first-strand cDNA synthesis kit (Amersham Bioscience). Subcloned plasmid DNA, containing the first exon of each gene, was adopted as a template for the sequencing using the same fluorochrome-labeled primer. The sequence products were analyzed with primer-extended cDNA fragment by the cycle sequencing method.
Construction of Promoter-EGFP Plasmid
The reporter gene vector pEGFP-1 (Clontech) was used for the construct. Four different Japanese flounder promoters including 5′ untranslated region (UTR) were amplified by specific PCR primers and inserted into the multiple cloning site of pEGFP-1. GFP reporter gene was the endogenous initiation codon directly replaced with that of GFP. To confirm the sequence and direction of insert, the recombinant plasmid was sequenced using a Thermo Sequenase II kit and automated DNA sequencer 373A with a primer designed for pEGFP-1.
Production of Transgenic Zebrafish
Adult zebrafish (Danio rerio) and embryos were maintained at 28.5°C on a 14-hour light and 10-hour dark cycle. The recombinant plasmid was propagated in Escherichia coli JM109 and purified using a GFX Micro Plasmid prep kit (Amersham Biosciences). Purified plasmid DNA was adjusted to 50 ng/μl in distilled water and microinjected into one-celle-stage zebrafish embryos. Initially 400 to 500 embryos were injected with plasmid DNA in each promoter-GFP construct. Living embryos were examined under an SZX12 fluorescence microscope (Olympus), and GFP-positive zebrafish embryos were selected. After maturation the GFP-positive fish were mated with wild types to select fish for germline transmission. GFP expression in F1 embryos was also examined under the fluorescence microscope in each developmental stage.
LPS Treatment of TNF-GFP Transgenic Zebrafish Embryo
F2 generations of transgenic zebrafish injected with TNF-GFP at 30 hours postfertilization (hpf) were stimulated with LPS (Sigma) at 140 μg/ml for 30 minutes and then washed in sterilized water. GFP expression in F2 embryos was examined under the fluorescence microscope at 48 hpf.
Detection of GFP Gene Expression in Adult Transgenic Zebrafish by RT-PCR
Total RNA extraction and PCR was performed as described above. Total RNA was extracted from fin, skin, liver, kidney, muscle, and brain of adult transgenic zebrafish. The following PCR primers were designed for the detection of EGFP gene: GFP-F, 5′-GGTCG AGCTGGACGGCGACG-3′, and GFP-R, 5′-ACGAACTCC AGCAGGACCAT-3′. The zebrafish ß-actin primer set for internal control was zactin-F, 5′-TTTCCCTCCATT GTTGGTCG-3′, and zactin-R, 5′-GCGACTCTCAGCTCG TTGTA-3′.
Results and Discussion
The expression of complement component C3, gelatinase B, keratin, TNF and β-actin genes in Japanese flounder was detected by reverse transcriptase polymerase chain reaction (RT-PCR). The complement component C3 gene was expressed only in the liver. The gelatinase B gene was expressed in the kidney, blood, and leukocytes. The keratin gene was expressed in various tissues. The TNF gene was expressed only in PBLs when stimulated with LPS. The β-actin gene used as an internal control was expressed in all tissues (Figure 1). Thus the expression patterns of these genes were different in different Japanese flounder tissues. We assumed that the tissue-specific and inducible expression of these genes was regulated by their promoter region.
Fig. 1.
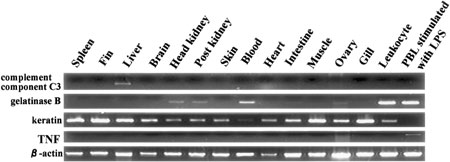
Expression of Japanese flounder tissue-specific genes in different tissues. Messenger RNA expression of complement component C3, gelatinase B, keratin, and TNF genes in spleen, fin, liver, head kidney, post kidney, skin, blood, heart, intestine, ovary, gills, leukocyte, and PBLs stimulated with LPS from Japanese flounder were detected by RT-PCR.
The 5′ flanking regions of the genes for complement component C3 (1381 bp), gelatinase B (1420 bp), keratin (1288 bp), and TNF (2351 bp) were isolated, and the respective transcriptional initiation sites were determined (Figure 2). There were some predicted transcriptional factor binding sites, such as TATA box, CAAT box or NF-κB binding site, in the 5′ flanking region. These sites were highly homologous with mammalian transcriptional factor binding sites. It is not certain that these predicted transcriptional factor binding sites can be applied to Japanese flounder promoters as there are few reports on fish transcriptional factor binding protein. It should be interesting to determine if these transcriptional factor binding sites are common to mammal and fish. These promoter regions were cloned into pEGFP-1 plasmid vector. These promoter regulatory constructs were designated C3-GFP, gelatinase-GFP, keratin-GFP, and TNF-GFP, respectively.
Fig. 2.

The 5′ flanking region sequence of the Japanese flounder gene: complement component C3 (A); gelatinase B (B); keratin (C); and TNF gene (D). We isolated 1381 bp of the 5′ flanking regions from transcription initiation codon for complement component C3; 1420 bp for gelatinase B; 1288 bp for keratin; and 2351 bp for TNF. The first exon is indicated in capital letters. The coding region and deduced amino acids are indicated in boldface capital letters. The transcription initiation site is shown by an asterisk. The predictive transcriptional factor binding site is underlined.
We obtained F1 transgenic individuals using each promoter regulatory constructs. An F2 strain was established with the C3-GFP, gelatinase-GFP, and keratin-GFP construcst, and two F2 strains from two different founders were established with the TNF-GFP construct. Similar GFP expression patterns were shown in F1 individuals and their F2 strains. GFP expression controlled by the complement component C3 promoter was observed in the yolk sac from the gastrula stage, in F2 transgenic zebrafish strains. In 10-day embryos strong GFP expression was observed only in the liver (Figure 3). GFP expression could not be observed in adults. The RT-PCR analysis showed that complement component C3 promoter also induced GFP expression in the adult liver (see Figure 7). Fish complement component C3 gene has the same function as the mammalian gene, and is expressed mainly in the liver (Abelseth et al., 2003). Japanese flounder complement component C3 was also expressed mainly in the liver (Figure 1). This was similar to the promoter activity observed in transgenic zebrafish. This suggests that Japanese flounder complement component C3 promoter could also induce expression of genes in the same tissues in zebrafish.
Fig. 3.
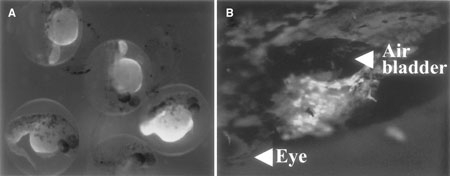
GFP expression in C3-GFP transgenic zebrafish embryos. The larval embryos at 24 hpf (A) and liver of the 10-day embryos (B) of F2 transgenic strain. Arrowheads indicate bladder and eye.
Fig. 7.
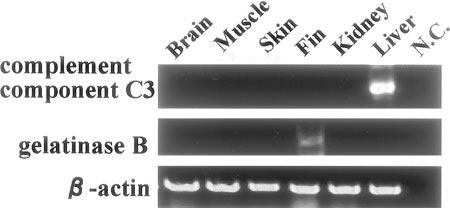
Detection of mRNA expression of GFP gene in fin, skin, liver, kidney, muscle, and brain from adult transgenic zebrafish by RT-PCR. GFP expression regulated by TNF promoter was not detected. N.C. indicates negative control.
GFP expression regulated by the gelatinase B promoter was observed after the one-cell stage, and was expressed strongly in the whole body until 48 hpf. However, GFP expression in the whole body decreased in the course of development. In 5-day embryos, strong GFP expression was detected only in the pectoral fins and gills (Figure 4). In adult transgenic fish gelatinase B promoter regulated GFP expression in the fin (see Figure 7). This result agrees with the GFP expression at embryonic stage. The GFP expression under the control of the Japanese flounder gelatinase B gene promoter in the transgenic zebrafish was not identical with the expression of the gelatinase B gene in Japanese flounder, in which it was expressed mainly in hematopoietic tissues (Figure 1). This promoter may lack a region essential for tissue-specific expression.
Fig. 4.
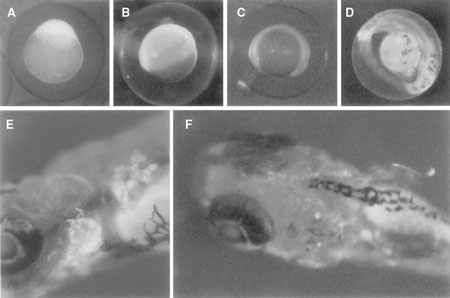
GFP expression of gelatinase B-GFP transgenic zebrafish. GFP expression at one-cell stage (A), 7 hpf (B), 12 hpf (C), 48 hpf (D), and 5-day embryo, lateral view (E) and dorsal view (F).
The keratin promoter regulated strong GFP expression from the embryo stage to the adult stage. In the embryo strong GFP expression was observed in the epithelial tissues and liver. In adult fish GFP expression was detected in skin, gills, fins, and liver (Figure 5). This expression pattern was similar to that of the Japanese flounder keratin gene (Figure 1). The zebrafish keratin 8 promoter was found to induce expression only in the skin (Gong et al., 2002). These results indicate that the promoter of the Japanese flounder keratin gene induces gene expression in the same tissues in zebrafish as in Japanese flounder. However, the sequences of the keratin promoter region of zebrafish and Japanese flounder are not the same (data not shown). This may be because some transcription factors that enhance skin-specific expression in zebrafish can bind and enhance the promoter activity of keratin promoter derived from Japanese flounder in transgenic zebrafish embryo.
Fig. 5.
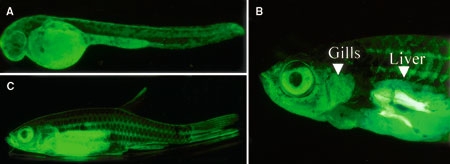
GFP expression in keratin-GFP F2 transgenic zebrafish under the fluorescence microscope. A: The 4-day embryo. B: Adult fish. C: Adult fish with operculum removed and abdominal muscle surgically opened. Arrowheads indicate gill and liver.
The TNF promoter regulated GFP expression only in the heart and pharynx in 5-day embryos. GFP expression was strongly induced in the epithelial tissues in 2-day transgenic embryos after LPS treatment (Figure 6). GFP expression was not detected in adult transgnic zebrafish by RT-PCR. This result suggested that TNF promoter controlled GFP expression in transgenic zebrafish under normal conditions (Figure 7). The Japanese flounder TNF promoter has inducible activity in zebrafish similar to that of Japanese flounder TNF gene expression in LPS-stimulated PBLs (Hirono et al., 2000). These results suggest that the Japanese flounder TNF promoter used in this study is sufficient to regulate the expression of a downstream gene.
Fig. 6.
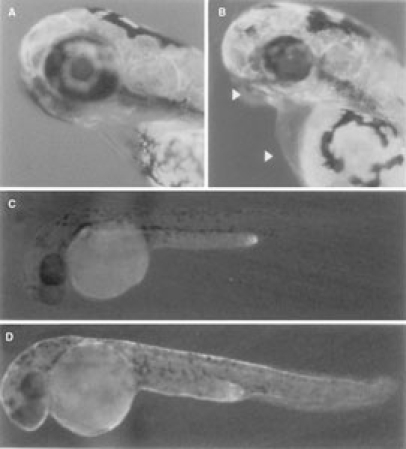
GFP expression in TNF-GFP transgenic zebrafish under the fluorescence microscope. A: The 5-day embryo of wild type. B: F2 transgenic strain, 5-day embryo. Arrowheads indicate heart and pharynx. C: Nontreated 3-day transgenic zebrafish embryo. D: LPS-treated 3-day transgenic zebrafish embryo.
In this study we showed that heterologous promoters derived from Japanese flounder worked in transgenic zebrafish. However, the expression patterns different from endogenous Japanese flounder expression patterns. Notably, the keratin and TNF promoters seemed to work faithfully in zebrafish embryos, but the gelatinase B promoter result differed from the results predicted from Japanese flounder. This may have been due to the short 5′ flanking region used in this study. Furthermore, we only observed the GFP expression pattern in a single transgenic line. Whether this GFP expression pattern is representative of gelatinase B promoter activity needs to confirmed in future studies by establishing multiple transgenic lines. Similarly, the fate of these promoters in Japanese flounder needs to be determined by production of transgenic lines of Japanese flounder.
The Japanese flounder promoters developed in this study can be adapted for transgenic fish in a variety of situations. Keratin promoter induced expression in epithelial tissues, where it acts as a first line of defense against bacterial infection. It is possible to generate disease-resistant transgenic fish expressing an antibacterial or antiviral peptide only in epithelial tissues. Moreover, using the Japanese flounder TNF promoter, it is possible to generate transgenic fish expressing an antibacterial or antiviral peptide only in cases of infection with pathogens. Thus these promoters should be useful for production of disease-resistant transgenic fish. The overexpression of a foreign gene, especially the gene for an antimicrobial peptide, may suppress growth or maturity of the host fish. In addition, these transgenic fish are safe for human consumption, as the foreign gene is not expressed in the muscle. The Japanese flounder TNF gene promoter can also be used to monitor bacterial infection in live fish using GFP as an indicator. Furthermore, these transgenic zebrafish were suitable for developmental analysis of specific tissues or organs. It is possible to observe development of liver, gills, and skin in living embryos using GFP expression.
We also developed a method for introducing a gene into Japanese flounder fertilized eggs by particle gun bombardment (Yazawa et al., 2005). The development of tissue-specific, inducible promoters in this study will support the establishment of transgenic technology for Japanese flounder and should contribute to a better understanding of the biology and culture of this fish species.
Acknowledgments
This research was supported in part by a Grant-in Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Sciences and Technology of Japan.
References
- Abelseth TK, Stensvag K, Espelid S, Nygaard R, Ellingsen T, Bogwald J, Dalmo RA (2003). The spotted wolffish (Anarhichas minor Olafsen) complement component C3: isolation, characterisation and tissue distribution Fish Shellfish Immunol 15: 13–27 [DOI] [PubMed]
- Aoki T, Nam BH, Hirono I, Yamamoto E (1999) Sequences of 596 cDNA clones (565,977 bp) of Japanese flounder (Paralichthys olivaceus) leukocytes infected with Hirame rhabdovirus Mar Biotechnol 1: 477–488 [DOI] [PubMed]
- Aoki T, Hirono I, Kim MG, Katagiri T, Tokuda Y, Toyohara H, Yamamoto E (2000) Identification of viral induced genes in Ig+ leucocytes of Japanese flounder Paralichthys olivaceus, by differential hybridisation with subtracted and un-subtracted cDNA probes Fish Shellfish Immunol 10: 623–630 [DOI] [PubMed]
- Chen TT, Vrolijk NH, Lu JK, Lin CM, Reimschuessel R, Dunham RA. (1996) Transgenic fish and its application in basic and applied research Biotechnol Annu Rev 2: 205–236 [DOI] [PubMed]
- Gao D, Li Z, Murphy T, Sauerbier W. (1997) Structure and transcription of the gene for translation elongation factor 1 subunit alpha of zebrafish (Danio rerio) Biochim Biophys Acta. 1350: 1–5 [DOI] [PubMed]
- Gong Z, Ju B, Wang X, He J, Wan H, Sudha PM, Yan T (2002) Green fluorescent protein expression in germ-line transmitted transgenic zebrafish under a stratified epithelial promoter from keratin 8 Dev Dy. 223: 204–215 [DOI] [PubMed]
- Hackett PB, Alvarez MC (2000) The molecular genetics of transgenic fish. In: Fingerman M, Nagabhushanam R, eds. Recent Advances in Marine Biotechnology: vol. 4 Aquaculture: Part B Fishes, New York, N.Y.: Science Publishers, 77–145
- Higashijima S, Okamoto H, Ueno N, Hotta Y, Eguchi G (1997) High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin Dev Biol 192: 289–299 [DOI] [PubMed]
- Hikima J, Minagawa S, Hirono I, Aoki T (2001) Molecular cloning, expression and evolution of the Japanese flounder goose-type lysozyme gene, and the lytic activity of its recombinant protein Biochim Biophys Acta 1520: 35–44 [DOI] [PubMed]
- Hirono I, Aoki T (2003) Immuno-related genes of Japanese flounder, Paralichthys olivaceus. In: Shimizu N, Aoki T, Hirono I, Takashima F, eds. Aquatic Genomics, Tokyo, Japan.: Springer-Verlag, 286–300
- Hirono I, Nam BH, Kurobe T, Aoki T (2000) Molecular cloning, characterization, and expression of TNF cDNA and gene from Japanese flounder Paralichthys olivaceus J Immunol 165: 4423–4227 [DOI] [PubMed]
- Hwang GL, Azizur RM, Abdul RS, Sohm F, Farahmand H, Smith A, Brooks C, Maclean N (2003) Isolation and characterisation of tilapia beta-actin promoter and comparison of its activity with carp beta-actin promoter Biochim Biophys Acta 1625: 11–18 [DOI] [PubMed]
- Katagiri T, Asakawa S, Hirono I, Aoki T, Shimizu N (2000) Genomic bacterial artificial chromosome library of the Japanese flounder Paralichthys olivaceus Mar Biotechnol 2: 571–576 [DOI] [PubMed]
- Kinoshita M, Kani S, Ozato K, Wakamatsu Y (2000) Activity of the medaka translation elongation factor 1alpha-A promoter examined using the GFP gene as a reporter Dev Growth Differ 42: 469–478 [DOI] [PubMed]
- Liu ZJ, Moav B, Faras AJ, Guise KS, Kapuscinski AR, Hackett PB (1990) Functional analysis of elements affecting expression of the beta-actin gene of carp Mol Cell Biol 10: 3432–3440 [DOI] [PMC free article] [PubMed]
- Nam BH, Yamamoto E, Hirono I, Aoki T (2000) A survey of expressed genes in the leukocytes of Japanese flounder, Paralichthys olivaceus, infected with Hirame rhabdovirus Dev Comp Immunol 24: 13–24 [DOI] [PubMed]
- Nam BH, Hirono I, Aoki T (2003) Bulk isolation of immune response-related genes by expressed sequenced tags of Japanese flounder Paralichthys olivaceus leucocytes stimulated with Con A/PMA Fish Shellfish Immunol 14: 467–476 [DOI] [PubMed]
- Noh JK, Cho KN, Han EH, Kim A, Lee JS, Kim DS, Kim CG (2003) Genomic cloning of mud loach Misgurnus mizolepis (Cypriniformes, Cobitidae) beta-actin gene and usefulness of its promoter region for fish transgenesis Mar Biotechnol 5: 244–252 [DOI] [PubMed]
- Sramasik A, Warr G, Chen TT (2002) Production of transgenic medaka with increased resistance to bacterial pathogens Mar Biotechnol 4: 310–322 [DOI] [PubMed]
- Udvadia AJ, Linney E (2003) Windows into development: historic, current, and future perspectives on transgenic zebrafish Dev Biol 256: 1–17 [DOI] [PubMed]
- Yazawa R, Hirono I, Yamamoto E, and Aoki T (2005) Development of a gene transfer technique for Japanese flounder fertilized eggs by particle gun bombardment, Fish Sci (in press)
- Zhang PJ, Hayat M, Joyce C, Gonzalez-Villasenor LI, Lin CM, Dunham RA, Chen TT, Powers DA (1990) Gene transfer, expression and inheritance of pRSV-rainbow trout-GH cDNA in the common carp, Cyprinus carpio (Linnaeus) Mol Reprod Dev 25: 3–13 [DOI] [PubMed]


