Abstract
pex mutants are defective in peroxisome assembly. The mutant strain pex23-1 of the yeast Yarrowia lipolytica lacks morphologically recognizable peroxisomes and mislocalizes all peroxisomal matrix proteins investigated preferentially to the cytosol. pex23 strains accumulate vesicular structures containing both peroxisomal matrix and membrane proteins. The PEX23 gene was isolated by functional complementation of the pex23-1 strain and encodes a protein, Pex23p, of 418 amino acids (47,588 Da). Pex23p exhibits high sequence similarity to two hypothetical proteins of the yeast Saccharomyces cerevisiae. Pex23p is an integral membrane protein of peroxisomes that is completely, or nearly completely, sequestered from the cytosol. Pex23p is detected at low levels in cells grown in medium containing glucose, and its levels are significantly increased by growth in medium containing oleic acid, the metabolism of which requires intact peroxisomes.
INTRODUCTION
Peroxisomes, together with the glyoxysomes of plants and the glycosomes of trypanosomes, constitute the microbody family of organelles. Peroxisomes are the site of a diverse set of metabolic reactions, which vary depending on the organism and its physiological conditions. Functions that have been conserved in peroxisomes from yeasts to humans include the β-oxidation of fatty acids and the decomposition of hydrogen peroxide by catalase (reviewed by Lazarow and Fujiki, 1985; Subramani, 1993). The necessity of peroxisomes for normal human development and physiology is demonstrated by the lethality of a group of genetic disorders, including Zellweger syndrome, in which peroxisomes fail to assemble normally (Lazarow and Moser, 1994). Accordingly, a great deal of attention has been paid in recent years to the question of how peroxisomes assemble in an attempt to understand the molecular bases of these disorders. Much of the progress in this area has come from the use of yeasts as model systems.
Complementation of peroxisome assembly mutants, collectively known as pex mutants, in yeasts has contributed to the identification of 22 PEX genes coding for a group of proteins known as peroxins that are required for peroxisome assembly (Subramani, 1997, 1998; Götte et al., 1998; Purdue et al., 1998; Titorenko et al., 1998; Koller et al., 1999). Yeast PEX genes have been used to identify 13 human orthologues through screening of the Expressed Sequence Tags databases, and of these, 8 have been shown to complement the peroxisome deficiencies of cells of patients with peroxisome biogenesis disorders (Dodt et al., 1995; Subramani, 1997, 1998). Peroxisome biogenesis, therefore, is a highly conserved process in eukaryotic cells, and yeasts are admirably suited for the identification of the proteins involved in this process and for the elucidation of the overall pathway of assembly of peroxisomes in cells.
Protein targeting to peroxisomes is compromised in pex mutants. Peroxisomal proteins are encoded in the nucleus and synthesized on cytosolic polysomes (Lazarow and Fujiki, 1985; Subramani, 1993, 1998). Most soluble proteins of the matrix are targeted by one of two types of peroxisomal targeting signal (PTS). PTS1 is a carboxyl-terminal tripeptide (SKL and conserved variants) (Gould et al., 1987, 1989) found in a large number of matrix proteins (Gould et al., 1989; Motley et al., 1995; Elgersma et al., 1996b), whereas PTS2 is a sometimes cleaved amino-terminal nonapeptide found in a smaller subset of matrix proteins (Swinkels et al., 1991; Glover et al., 1994; Waterham et al., 1994). Pex5p and Pex7p are the receptors for PTS1 and PTS2, respectively, and various peroxins, including Pex13p and Pex14p, form a docking complex at the peroxisomal membrane for these receptors (reviewed by Erdmann et al., 1997; Subramani, 1998). Sequences involved in the sorting of peroxisomal membrane proteins have been identified for a few proteins and, in general, appear to be defined as a stretch of basic amino acid residues (McCammon et al., 1994; Dyer et al., 1996; Wiemer et al., 1996). The machinery for targeting proteins to the peroxisomal membrane is apparently different from that involved in the import of matrix proteins, because although most pex mutants are compromised in the import of matrix proteins, they do target peroxisomal membrane proteins and possess peroxisomal structures called “ghosts” that contain peroxisomal membrane proteins (Santos et al., 1988; Subramani, 1993, 1998). Recently, cells from a Zellweger syndrome patient with a mutation in the PEX16 gene coding for a peroxin integral to the peroxisomal membrane were shown to be unable to import peroxisomal membrane proteins, implicating Pex16p in this process (South and Gould, 1999).
Here we report the isolation and characterization of a novel PEX gene, PEX23, from the yeast Yarrowia lipolytica encoding the peroxin Pex23p. Mutants of PEX23 lack peroxisomes and mislocalize matrix proteins preferentially to the cytosol. pex23 strains accumulate vesicles that contain both peroxisomal matrix and membrane proteins. Pex23p is an integral membrane protein of peroxisomes that is sequestered from the cytosol and whose levels are increased by growth of cells in oleic acid.
MATERIALS AND METHODS
Strains and Culture Conditions
The Y. lipolytica strains used in this study are listed in Table 1. Growth was at 30°C. Strains containing plasmids were grown in minimal medium (YND or YNO). Strains not containing plasmids were grown in rich medium (YEPD or YPBO). Media components were as follows: YND, 1.34% yeast nitrogen base without amino acids, Complete Supplement Mixture (Bio 101, Vista, CA) minus the appropriate amino acids at twice the manufacturer's recommended concentration (2× CSM), 2% glucose; YNO, 1.34% yeast nitrogen base without amino acids, 2× CSM, 0.05% (wt/vol) Tween 40, 0.1% (wt/vol) oleic acid; YEPD, 1% yeast extract, 2% peptone, 2% glucose; YPBO, 0.3% yeast extract, 0.5% peptone, 0.5% K2HPO4, 0.5% KH2PO4, 1% Brij-35, 1% (wt/vol) oleic acid. Escherichia coli was grown as described previously (Ausubel et al., 1994).
Table 1.
Y. lipolytica strains used in this study
| Straina | Genotype |
|---|---|
| E122 | MATA, ura3-302, leu2-270, lys 8-11 |
| 22301-3 | MATB, ura3-302, leu2-270, his1 |
| pex23-1 | MATA, ura3-302, leu2-270, lys 8-11, pex23-1 |
| P23TR | MATA, ura3-302, leu2-270, lys8-11, p23E4(LEU2) |
| pex23KOA | MATA, ura3-302, leu2-270, lys8-11, pex23∷URA3 |
| pex23KOB | MATB, ura3-302, leu2-270, his1, pex23∷URA3 |
| D1-23 | MATA/MATB, ura3-302/ura3-302, leu2-270/leu2-270, lys8-11/+, +/his1, pex23-1/+ |
| D2-23 | MATA/MATB, ura3-302/ura3-302, leu2-270/leu2-270, lys8-11/+, +/his1, pex23∷URA3/+ |
| D3-23 | MATA/MATB, ura3-302/ura3-302, leu2-270/leu2-270, lys8-11/+, +/his1, pex23-1/pex23∷URA3 |
| P23-Myc | MATA, ura3-302, leu2-270, lys8-11, pPEX23-Myc(LEU2) |
Strains E122 and 22301-3 were from C. Gaillardin (Thiverval-Grignon, France). All other strains were from this study.
Cloning, Sequencing, and Integrative Disruption of the PEX23 Gene
The pex23-1 mutant strain was isolated from randomly mutagenized Y. lipolytica strain E122 as described previously (Nuttley et al., 1993). The PEX23 gene was isolated by functional complementation of the pex23-1 strain with a Y. lipolytica genomic DNA library in the autonomously replicating E. coli shuttle vector pINA445 (Nuttley et al., 1993). Leu+ transformants were replica plated onto selective YNO agar plates and screened for their ability to use oleic acid as a sole carbon source. Total DNA was isolated from colonies that recovered growth on YNO and used to transform E. coli for plasmid recovery. Restriction fragments prepared from the genomic insert were subcloned and tested for their ability to functionally complement the pex23-1 strain. The smallest genomic DNA fragment capable of complementation was sequenced in both directions.
Targeted integrative deletion of the PEX23 gene was performed with the URA3 gene of Y. lipolytica. A 1.7-kilobase pair (kbp) SalI fragment containing the URA3 gene was inserted into a plasmid containing the PEX23 gene locus cut with EcoRV and StuI, thereby replacing a 2.2-kbp fragment containing the entire PEX23 ORF with the URA3 gene. This construct was then cleaved with BamHI and XbaI to liberate the URA3 gene flanked by 1068 and 1407 base pairs of the 5′ and 3′ regions, respectively, of the PEX23 gene. The resultant linear construct was used to transform Y. lipolytica strains E122 and 22301-3 to uracil prototrophy. Ura+ transformants were selected and screened for their inability to grow on YNO agar. Correct integration of the URA3 gene at the PEX23 gene locus was confirmed by Southern blot analysis. Deletion strains were crossed with wild-type strains and the pex23-1 mutant strain, and the resultant diploids were checked for growth on YNO agar.
Microscopic Analysis
Electron microscopy (Goodman et al., 1990) and indirect immunofluorescence microscopy (Szilard et al., 1995) were performed as described.
Epitope Tagging of Pex23p
Pex23p was tagged at its carboxyl terminus with three tandem copies of the human c-Myc epitope consisting of the amino acid sequence EQKLISEEDL (Kolodziej and Young, 1991). The ORF and termination codon of the PEX23 gene, along with ∼1.6 kbp of genomic DNA 5′ to the ORF, were amplified by PCR with the use of primers 706 (5′) and 805 (3′) (Table 2). The amplified product was digested with BamHI and SalI and inserted into the plasmid pSP73 (Promega, Madison, WI) cut with the same enzymes to yield plasmid pPEX23-5. Approximately 1.8 kbp of the 3′ flanking region of the PEX23 gene was also amplified by PCR with the use of primers 806 (5′) and 807 (3′) (Table 2). This PCR product was digested with XhoI and XbaI and inserted into the same sites of pGEM 7Zf (+) (Promega) to yield the plasmid pPEX23-3. The insert of pPEX23-5 was liberated by cleavage with BamHI and XhoI and ligated into the same sites of pPEX23-3 to make pPEX23-53. Next, a DNA fragment coding for three tandem copies of the antigenic region of the c-Myc protein was excised from the plasmid pCR2.1 (a kind gift of D. Stuart, University of Alberta) with SalI and XhoI and inserted in frame and downstream of the PEX23 gene ORF in pPEX23-53 to make the plasmid pPEX23-Myc expressing the chimeric protein Pex23p-Myc. pPEX23-Myc was tested for its ability to restore growth on oleic acid and peroxisome assembly to the pex23-1 and pex23KOA mutant strains. Pex23p-Myc was detected immunologically with mouse mAb 9E10 (Santa Cruz Biotechnology, Santa Cruz, CA).
Table 2.
PCR primers used in this study
| Number | Sequence |
|---|---|
| 706 | 5′-TTCACACAGGAAACAGCTATGACCATG |
| 805 | 5′-CTCGAGGACTCCGTCGACTCTCTTAGAGTCCTCCTCGAATCTAATGC |
| 806 | 5′-GTCGACGGAGTCCTCGAGTAATTAAATATATGAATGTATCAT |
| 807 | 5′-ATTCTAGATCGACTGGTCCAAAGTTGTGG |
Cell Fractionation, Peroxisome Subfractionation, and Flotation Gradient Analysis
Fractionation of oleic acid–grown cells was performed as described previously (Szilard et al., 1995) and included the differential centrifugation of lysed and homogenized spheroplasts at 1,000 × g for 8 min at 4°C in a model JS13.1 rotor (Beckman, Fullerton, CA) to yield a postnuclear supernatant fraction. The postnuclear supernatant fraction was further subjected to differential centrifugation at 20,000 × g for 30 min at 4°C in a JS13.1 rotor to yield a pellet (20KgP) fraction enriched for peroxisomes and mitochondria and a supernatant (20KgS) fraction enriched for cytosol. Peroxisomes were purified from the 20KgP fraction by isopycnic centrifugation on a discontinuous sucrose gradient (Titorenko et al., 1996).
Peroxisome subfractions were prepared from purified peroxisomes essentially as described (Eitzen et al., 1997). Briefly, 150 μg of purified peroxisomes was lysed by the addition of 10 volumes of ice-cold Ti8 buffer (10 mM Tris-HCl, pH 8.0, 5 mM EDTA, 1 mM PMSF, 5 mM NaF, and pepstatin, leupeptin, and aprotinin each at 1 μg/ml) and subjected to centrifugation at 100,000 × g for 30 min at 4°C. Half of the resultant pellet was then treated with 0.1 M Na2CO3 (pH 11.5), followed by centrifugation as described above. Proteins were precipitated by the addition of trichloroacetic acid to 10% and washed with 80% (vol/vol) acetone.
The 20KgP fraction from the pex23KOA mutant strain was subjected to a two-step flotation gradient analysis to detect the presence of vesicular structures containing peroxisomal proteins. The 20KgP fraction was resuspended in 100 μl of 30% (wt/wt) sucrose and 0.5 M sorbitol in buffer M [5 mM 2-(N-morpholino)ethanesulfonic acid, pH 5.5, 1 mM KCl, 0.5 mM EDTA, 0.1% (vol/vol) ethanol, 1 mM PMSF, and leupeptin, pepstatin, and aprotinin each at 1 μg/ml] and mixed with 300 μl of 65% (wt/wt) sucrose in buffer M. The sample was transferred to a 5-ml centrifuge tube and overlaid with 2.3 ml of 50% (wt/wt) sucrose and 2.3 ml of 20% (wt/wt) sucrose (both in buffer M). Gradients were subjected to centrifugation in a SW50.1 rotor (Beckman) at 200,000 × g for 18 h at 4°C. Gradients were fractionated from the top, and 18 fractions of ∼275 μl each were collected.
The 20KgS fraction from the pex23KOA mutant strain was subjected to centrifugation at 200,000 × g for 30 min at 4°C to yield a pellet (200KgP) fraction and a supernatant (200KgS) fraction consisting essentially of cytosol. The 200KgS fraction was divided into two equal aliquots. The first aliquot was incubated for 2 h at 75°C. Under these conditions, all cytosolic proteins formed insoluble aggregates, as judged by light scattering at 320 nm and as confirmed by SDS-PAGE followed by Coomassie staining. Aggregates of cytosolic proteins were pelleted by centrifugation at 20,000 × g for 30 min at 4°C, resuspended in 100 μl of 30% (wt/wt) sucrose and 0.5 M sorbitol in buffer M, and mixed with 300 μl of 65% (wt/wt) sucrose in buffer M. This material was subjected to flotation on a two-step sucrose gradient as described above. The second aliquot of the 200KgS fraction (in buffer M supplemented with 1 M sorbitol) was concentrated to a final volume of 50 μl by centrifugation through a Biomax-30 filter (Millipore, Bedford, MA) at 7,200 × g for 40 min at 4°C. The concentrated soluble proteins were mixed with 50 μl of 60% (wt/wt) sucrose and then with 300 μl of 65% (wt/wt) sucrose (both in buffer M) and subjected to flotation on a two-step sucrose gradient as described above. Gradient fractions were assayed for protein and sucrose density and for the presence of peroxisomal proteins.
Protease Protection
For protease protection experiments, peroxisomes purified by isopycnic centrifugation were diluted with 4 volumes of 0.5 M sucrose in buffer H [5 mM 2-(N-morpholino)ethanesulfonic acid, pH 5.5, 1 mM KCl, 0.5 mM EDTA, 0.1% (vol/vol) ethanol]. Peroxisomes were sedimented onto a 150-μl cushion of 2 M sucrose in buffer H by centrifugation at 200,000 × g for 20 min at 4°C in a model TLA120.2 rotor (Beckman). The sedimented peroxisomes were resuspended in 850 μl of buffer H containing 1 M sorbitol. Aliquots of 100 μg of protein were incubated with 0, 100, 200, and 500 μg of trypsin for 1 h on ice, either in the presence or the absence of Triton X-100 at 0.5% (vol/vol) final concentration. The reaction was terminated by the addition of trichloroacetic acid to a final concentration of 10%, and the protein precipitates were washed with acetone as described above. Equivalent fractions from each reaction were subjected to SDS-PAGE followed by immunoblotting.
Analytical Procedures
Whole cell lysates were prepared as described (Eitzen et al., 1997). Enzymatic activities of the peroxisomal marker catalase (Luck, 1963) and of the mitochondrial marker cytochrome c oxidase (Douma et al., 1985) were measured by established procedures. SDS-PAGE (Laemmli, 1970) and immunoblotting with the use of semidry electrophoretic transfer (Kyhse-Andersen, 1984) were performed as described. Antigen–antibody complexes in immunoblots were detected by ECL (Amersham Life Sciences, Arlington Heights, IL). Protein concentration was determined with a protein assay kit (Bio-Rad Laboratories, Richmond, CA) with BSA as the standard. Total nucleic acid was isolated by glass bead lysis and phenol extraction as described previously (Eitzen et al., 1995). Southern blot analysis was performed as described by Ausubel et al. (1994).
RESULTS
Isolation and Characterization of the PEX23 Gene
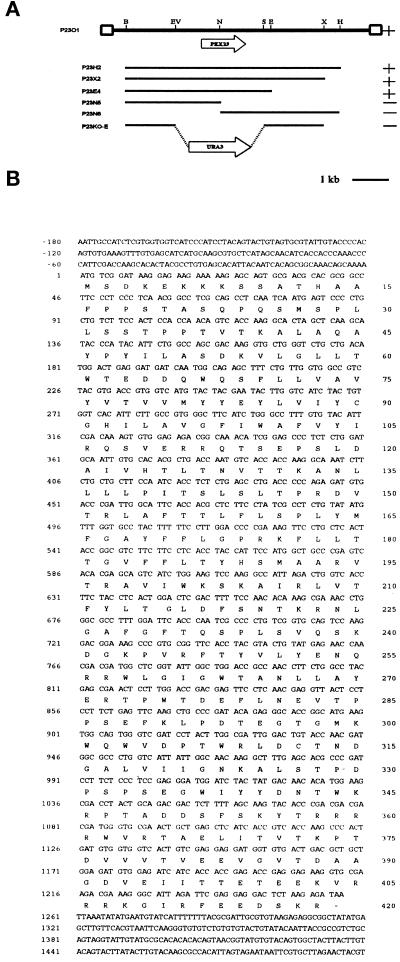
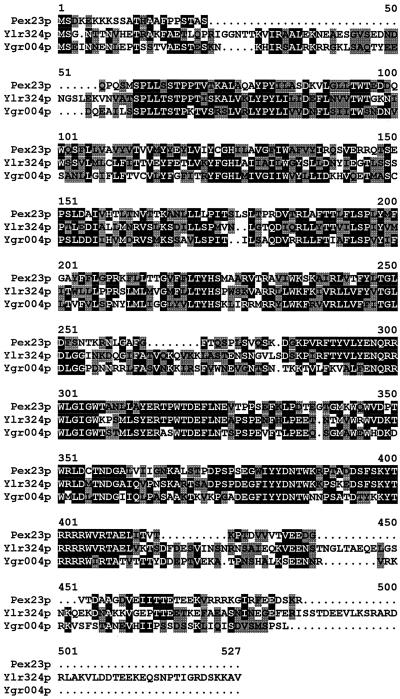
The pex23-1 mutant strain (Table 1) was isolated from randomly mutagenized Y. lipolytica cells by its inability to grow on agar plates containing oleic acid as the sole carbon source (ole− phenotype). Subsequent biochemical and morphological analyses (data presented below) demonstrated that this strain was affected in the peroxisome assembly pathway. The PEX23 gene was isolated from a library of Y. lipolytica genomic DNA by functional complementation of the pex23-1 strain. Screening of ∼2 × 105 leucine prototrophy (Leu+) transformants led to the identification of one strain that had recovered growth on oleic acid (ole+ phenotype). Total DNA was isolated from this strain, and the complementing plasmid was recovered by transformation of E. coli. The plasmid insert was mapped by restriction endonuclease digestion, and fragments of the insert resulting from the various digestions were cloned and introduced by transformation into the pex23-1 strain to delineate the region of complementation (Figure 1A). This region localized to a unique NdeI site within the initial complementing insert. DNA sequencing revealed an ORF of 1254 nucleotides coding for a protein of 418 amino acids, Pex23p, and having a predicted molecular weight of 47,558 (Figure 1B). Pex23p does not contain PTS1 or PTS2 motifs, and although a carboxyl-terminal SKR tripeptide is present, it is not necessary for function (see below). A search of protein databases with the use of the GENINFO(R) BLAST Network Service of the National Center for Biotechnology Information revealed two highly homologous proteins coded for by the ORFs YLR324W and YGR004W of the S. cerevisiae genome (Figure 2).
Figure 1.
Cloning and analysis of the PEX23 gene. (A) Complementing activity of inserts, restriction map analysis, and targeted gene deletion strategy for the PEX23 gene. The original complementing insert DNA in the plasmid p23O1 is denoted by the thick black line. (Solid lines) Y. lipolytica genomic DNA; (boxes) vector DNA. The ORFs of the PEX23 and URA3 genes and their directionality are indicated by the wide arrows. (+) Ability and (−) inability of an insert to confer growth on oleic acid to strain pex23-1. B, BamHI; E, EcoRI; EV, EcoRV; H, HindIII; N, NcoI; S, StuI; X, XbaI. (B) Nucleotide sequence of the PEX23 gene and deduced amino acid sequence of Pex23p. These sequence data have been deposited in the DDBJ/EMBL/GenBank databases under accession number AF160511.
Figure 2.
Sequence alignment of Pex23p with the hypothetical proteins Ylr324p and Ygr004p encoded by the ORFs YLR324W and YGR004W, respectively, of the S. cerevisiae genome. Amino acid sequences were aligned with the use of the ClustalW program (EMBL, Heidelberg, Germany). Identical residues (black) and similar residues (gray) in at least two of the proteins are shaded. Similarity rules: G = A = S; A = V; V = I = L = M; I = L = M = F = Y = W; K = R = H; D = E = Q = N; and S = T = Q = N. Dots represent gaps.
The putative PEX23 gene was deleted by targeted integration of the Y. lipolytica URA3 gene to make the strains pex23KOA and pex23KOB in the A (E122) and B (22301-3) mating types, respectively (Table 1). The PEX23 deletion strains were unable to grow on oleic acid and possessed the same morphological and protein-targeting defects as the original pex23-1 strain (see below). The diploid strains D1-23 and D2-23 from the mating of strains pex23-1 and pex23KOA with the wild-type strain 22301-3 could grow on oleic acid–containing medium, demonstrating the recessive nature of the original pex23-1 mutation and the PEX23 gene deletion. The diploid strain D3-23 made by mating the original pex23-1 strain to strain pex23KOB (Table 1) was unable to grow on oleic acid–containing medium, demonstrating that the authentic PEX23 gene had been cloned and that the ability to use oleic acid as the sole carbon source required at least one intact copy of the PEX23 gene.
pex23 Cells Lack Normal Peroxisomes but Do Have Vesicular Structures Containing Peroxisomal Matrix and Membrane Proteins
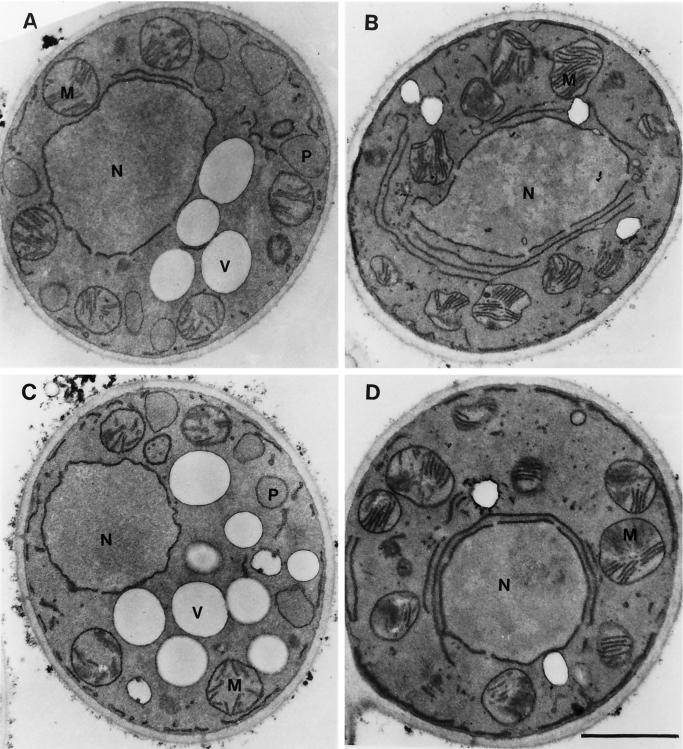
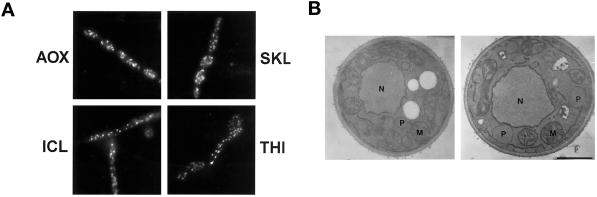
In electron micrographs, normal peroxisomes of Y. lipolytica appear as round vesicular structures, 0.2–0.5 μm in diameter, with a granular electron-dense core and a single unit membrane (Figure 3A). The original mutant strain pex23-1 (Figure 3B) and the deletion strain pex23KOA (Figure 3D) grown in oleic acid–containing medium lacked normal peroxisomes. Both mutant strains accumulated small vesicular structures that were rarely seen in wild-type cells and showed evidence of large membrane sheets surrounding the nucleus. The strain P23TR transformed with the PEX23 gene had the appearance of the wild-type strain and showed normal peroxisome morphology (Figure 3C).
Figure 3.
Ultrastructure of wild-type, pex23 mutant, and PEX23-transformed strains. The E122 (A), pex23-1 (B), P23TR (C), and pex23KOA (D) strains were grown in glucose-containing YEPD medium (YND medium for strain P23TR) for 16 h, transferred to oleic acid–containing YPBO medium (YNO medium for strain P23TR), and grown for an additional 8 h in oleic acid–containing medium. Cells were fixed in 1.5% KMnO4 and processed for electron microscopy. M, mitochondrion; N, nucleus; P, peroxisome; V, vacuole. Bar, 1 μm.
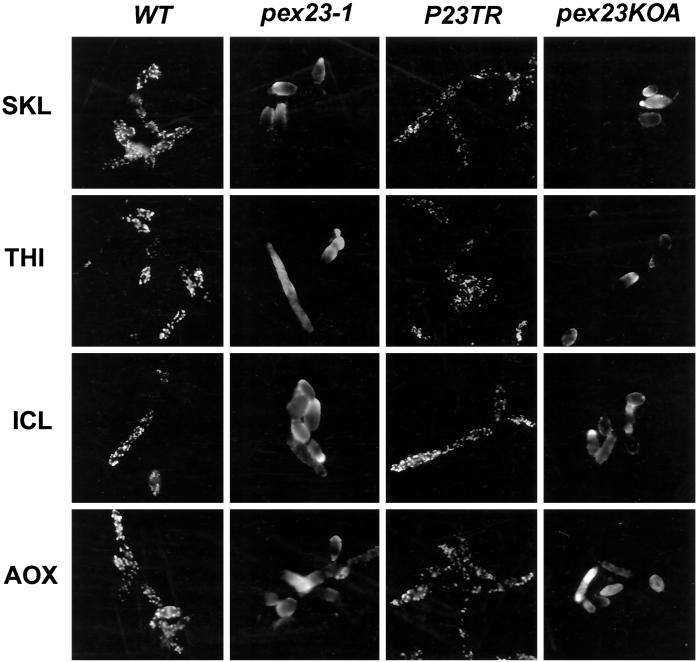
Immunofluorescence analysis of oleic acid–grown wild-type cells with anti-SKL antibodies and antibodies to the matrix proteins thiolase (THI), isocitrate lyase (ICL), and acyl-CoA oxidase (AOX) showed a punctate pattern of staining characteristic of peroxisomes (Figure 4). In contrast, pex23-1 cells stained with the same antibodies showed a more generalized pattern of fluorescence throughout the cell characteristic of cytosolic localization (Figure 4). The strain P23TR transformed with the PEX23 gene showed characteristic peroxisomal punctate staining with the four different antibodies, whereas the gene deletion strain pex23KOA displayed general cytosolic fluorescence like that of the original pex23-1 strain (Figure 4).
Figure 4.
Indirect immunofluorescence analysis of wild-type, pex23 mutant, and PEX23-transformed strains. Wild-type (WT) strain E122, mutant strains pex23-1 and pex23KOA, and transformed strain P23TR were grown in YPBO medium (YNO medium for strain P23TR). Cells were processed for immunofluorescence microscopy with antibodies to the PTS1 tripeptide SKL (SKL), thiolase (THI), isocitrate lyase (ICL), and acyl-CoA oxidase (AOX). Rabbit primary antibodies (SKL, ICL, AOX) were detected with fluorescein-conjugated goat anti-rabbit immunoglobulin G secondary antibodies, and guinea pig primary antibodies (THI) were detected with rhodamine-conjugated donkey anti-guinea pig immunoglobulin G secondary antibodies.
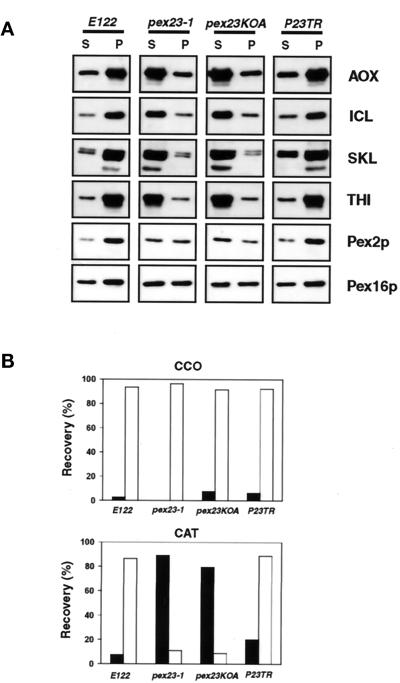
The different strains grown in oleic acid–containing medium were subjected to subcellular fractionation to give a 20,000 × g pellet (20KgP) enriched for peroxisomes and mitochondria and a 20,000 × g supernatant (20KgS) enriched for cytosol. As expected, peroxisomal matrix proteins recognized by anti-SKL antibodies and the matrix proteins THI, ICL, and AOX (Figure 5A), as well as the classic peroxisomal matrix enzymatic marker catalase (CAT) (Figure 5B), were preferentially localized to the 20KgP of wild-type E122 cells grown in oleic acid–containing medium. The peroxisomal integral membrane peroxin Pex2p and the peripheral membrane peroxin Pex16p were also both preferentially localized to the 20KgP of wild-type cells (Figure 5A). In contrast, in the original mutant strain pex23-1 and in the gene disruption strain pex23KOA, all matrix proteins were preferentially mislocalized to the 20KgS (Figure 5, A and B), although they could also be detected to a much lesser extent in the 20KgP. In contrast, Pex2p and Pex16p were distributed approximately equally between the 20KgP and 20KgS in pex23 mutant strains (Figure 5A). In the wild-type and pex23 mutant strains, the mitochondrial marker cytochrome c oxidase (CCO) was preferentially localized to the 20KgP (Figure 5B). Because in pex23 mutant strains all matrix proteins investigated mislocalized preferentially to the 20KgS enriched for cytosol and gave a general fluorescence characteristic of the cytosol, pex23 mutants are compromised in the import of PTS1 (ICL and anti-SKL proteins), PTS2 (THI), and non-PTS1, non-PTS2 proteins (AOX) (Wang et al., 1999).
Figure 5.
Peroxisomal matrix proteins are mislocalized in pex23 mutant strains. The wild-type strain E122, the original mutant strain pex23-1, and the gene deletion strain pex23KOA were grown in oleic acid–containing YPBO medium and subjected to subcellular fractionation to yield a 20KgP fraction enriched for peroxisomes and mitochondria and a 20KgS fraction enriched for cytosol. (A) Equal portions of the 20KgP (P) and 20KgS (S) were analyzed by immunoblotting to the indicated proteins. (B) The activities of catalase (CAT) and cytochrome c oxidase (CCO) were assayed enzymatically, and the percentages of enzymatic activity recovered in the 20KgS (▪) and the 20KgP (□) relative to the total enzymatic activity in the postnuclear supernatant fraction are reported.
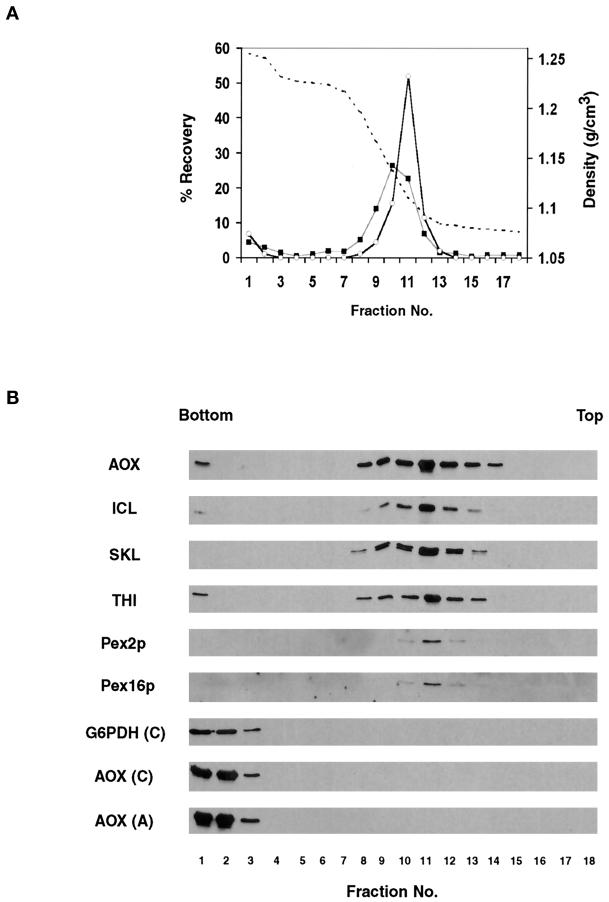
We performed a two-step flotation gradient analysis of the 20KgP fraction from the pex23KOA strain to determine whether the peroxisomal matrix and membrane proteins recovered in this fraction were membrane associated or simply represented large protein aggregates and/or cytosolic contamination of the 20KgP. Flotation of the 20KgP revealed that all peroxisomal proteins floated out of the most dense sucrose and concentrated at the interface between 50 and 20% sucrose (Figure 6, A and B). In contrast, both soluble cytosolic proteins and temperature-induced protein aggregates of cytosolic proteins remained at the bottom of the gradient (Figure 6B). Therefore, the peroxisomal matrix and membrane proteins recovered in the 20KgP fraction from the pex23KOA mutant are present in membrane-associated form, i.e., they localize to vesicular structures.
Figure 6.
Peroxisomal matrix and membrane proteins recovered in the 20KgP fraction of the pex23KOA mutant are associated with vesicular structures. The 20KgP fraction and cytosolic (C) and heat-aggregated (A) proteins from the 200KgS fraction of the pex23KOA strain grown in oleic acid–containing YPBO medium were subjected to flotation on a two-step sucrose density gradient as described in MATERIALS AND METHODS. (A) Sucrose density (g/cm3) (dashed line), and percentage recovery of loaded protein (▪) and catalase activity (○) in gradient fractions. (B) Equal volumes of gradient fractions were analyzed by immunoblotting with antibodies to peroxisomal matrix (AOX, ICL, SKL, THI) and membrane (Pex16p, Pex2p) proteins and with antibodies to the cytosolic protein glucose-6-phosphate dehydrogenase (G6PDH).
Pex23p Is an Integral Membrane Protein Sequestered from the Cytosolic Face of the Peroxisome
Pex23p was tagged at its carboxyl terminus with the c-Myc epitope (Pex23p-Myc) to allow its detection in cells. Expression of Pex23p-Myc complemented the pex23 mutant phenotype and reestablished peroxisome formation and the import of peroxisomal matrix proteins, as judged by immunofluorescence (Figure 7A), electron microscopy (Figure 7B), and subcellular fractionation (Figure 8A). Therefore, Pex23p-Myc mimics faithfully the biological activity of wild-type Pex23p.
Figure 7.
Expression of Pex23p-Myc restores peroxisome formation. Immunofluorescence (A) and electron microscopic (B) analysis of strain P23-Myc expressing the protein Pex23p-Myc. (A) Cells were grown in oleic acid–containing YNO medium and processed for immunofluorescence microscopy with antibodies to acyl-CoA oxidase (AOX), isocitrate lyase (ICL), the PTS1 tripeptide SKL (SKL), and thiolase (THI) as described in the legend to Figure 4. The characteris-tic punctate pattern of staining of peroxisomes is seen. (B) Cells were grown in YNO medium and processed for electron microscopy as described in the legend to Figure 3. Typical peroxisomes are seen. Abbreviations are as in Figure 3. Bar, 1 μm.
Figure 8.
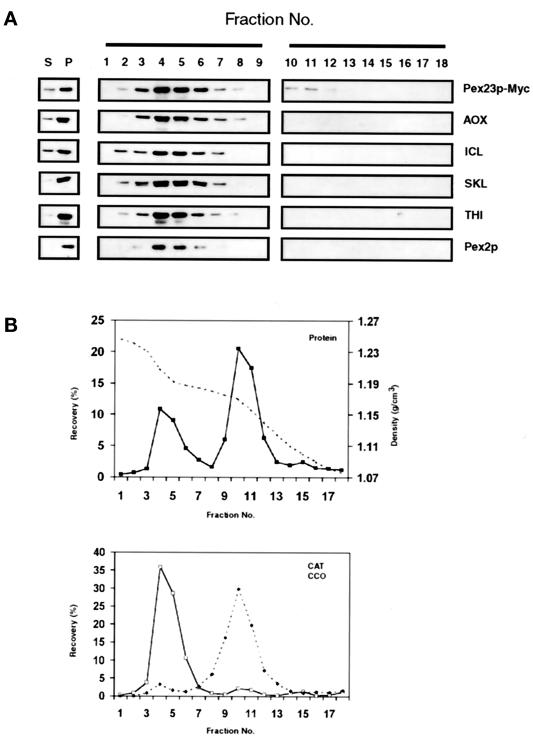
Pex23p-Myc is a peroxisomal protein. (A) Immunoblot analysis of the 20KgS (S) and 20KgP (P) fractions and of the fractions of a sucrose density gradient (numbered 1–18) of the 20KgP fraction from the strain P23-Myc expressing Pex23p-Myc. Equal proportions of the 20KgS and 20KgP fractions, and of each of the fractions of the gradient, were analyzed by immunoblotting with antibodies to the indicated proteins. Pex23p-Myc was detected with mouse mAb 9E10 to the c-Myc epitope. (B) Distribution of protein (▪), catalase (□), and cytochrome c oxidase (⋄) across the density gradient. The dashed line in the top panel shows the density profile (g/cm3) of the gradient.
The pex23KOA strain expressing Pex23p-Myc (Table 1, strain P23-Myc) was grown in oleic acid–containing medium and subjected to subcellular fractionation. Pex23p-Myc preferentially fractionated to the 20KgP (Figure 8A), as did peroxisomal matrix and membrane proteins (Figure 8, A and B). Peroxisomes were isolated from the 20KgP fraction by isopycnic centrifugation on a discontinuous sucrose gradient. Immunoblot analysis demonstrated that Pex23p-Myc localized to fractions enriched for peroxisomes and showed essentially the same distribution across the gradient as peroxisomal matrix (AOX, ICL, THI, CAT, anti-SKL) and peroxisomal integral membrane protein (Pex2p) markers, peaking in fraction 4 of the gradient at a sucrose density of 1.21 g/cm3 (Figure 8A) and being well separated from fractions enriched for mitochondria (Figure 8B), which peaked in fraction 10 at a density of 1.17 g/cm3.
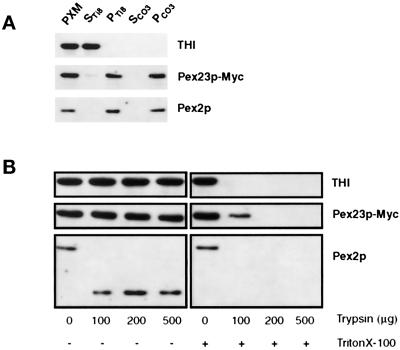
Lysis of peroxisomes with Ti8 buffer followed by high-speed centrifugation showed Pex23p-Myc to be localized exclusively to the pellet fraction enriched for membranes, as was the peroxisomal membrane protein Pex2p (Figure 9A, lane PTi8). This treatment liberated the matrix protein THI to the supernatant (Figure 9A, lane STi8). Treatment of the PTi8 with 0.1 M Na2CO3, pH 11.5, followed by high-speed centrifugation, showed that Pex23p-Myc colocalized with Pex2p to the pellet fraction (Figure 9A, lane PCO3), consistent with Pex23p-Myc being an integral protein of peroxisome membranes.
Figure 9.
Pex23p-Myc is an integral membrane protein sequestered from the cytosolic face of the peroxisome. (A) Immunoblot analysis of whole peroxisomes (PXM) separated into pellet (P) and supernatant (S) fractions by treatment with Ti8 buffer or sodium carbonate buffer (CO3). The top blot was probed with antibodies to thiolase (THI) to detect peroxisomal matrix proteins. The middle blot was probed with mouse mAbs to the c-Myc epitope to detect Pex23p-Myc. The bottom blot was probed with antibodies to the peroxisomal integral membrane protein Pex2p. (B) Protease protection analysis. Purified peroxisomes from the P23-Myc strain were incubated with increasing amounts of the protease trypsin in the absence (−) or presence (+) of the detergent Triton X-100.
A protease protection assay was performed on isolated peroxisomes to obtain some idea of the orientation of Pex23p-Myc in the peroxisome membrane. Aliquots of peroxisomes were treated with increasing amounts of trypsin in the absence or presence of the nonionic detergent Triton X-100. Immunoblot analysis showed no detectable degradation of Pex23p-Myc by trypsin in the absence of detergent, similar to the matrix protein THI (Figure 9B). In contrast, Pex2p showed cleavage by trypsin in the absence of detergent, as has been demonstrated previously (Titorenko and Rachubinski, 1998). Therefore, trypsin was indeed active in the absence of detergent. Addition of increasing amounts of trypsin in the presence of Triton X-100 led to the complete degradation of Pex23p-Myc, THI, and Pex2p. These results are consistent with Pex23p-Myc being localized preferentially away from the cytosolic surface of the peroxisome. Because Pex23p was tagged at its carboxyl terminus with the c-Myc epitope, our data are also consistent with the carboxyl terminus of Pex23p being localized away from the cytosolic face of peroxisomes. It should be noted that although Pex23p ends in the tripeptide SKR, which shows some similarities to a PTS1 sequence, this sequence is apparently not required for targeting to the peroxisome, because Pex23p-Myc, which ends in the tripeptide EDL (which does not resemble a PTS1 motif), is still targeted to peroxisomes.
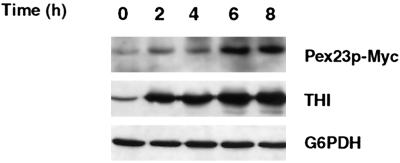
Synthesis of Pex23p-Myc Is Induced by Growth of Yeast in Oleic Acid–containing Medium
Immunoblotting showed that Pex23p-Myc was barely detectable in cells grown in glucose-containing medium, but its levels were increased significantly after cells were shifted to oleic acid–containing medium (Figure 10). Under these conditions, THI also showed increased levels of synthesis when cells were grown in oleic acid–containing medium, as has been reported previously (Titorenko et al., 1998). In contrast, there was no change in the levels of the cytosolic enzyme glucose-6-phosphate dehydrogenase under the same conditions.
Figure 10.
Synthesis of Pex23p-Myc is induced by growth of Y. lipolytica in oleic acid–containing medium. Strain P23-Myc was grown to an OD600 of 2.0 in glucose-containing YND medium (0 h) and then transferred at a dilution of 1:4 to oleic acid–containing YNO medium. Samples were taken from YNO medium at the times indicated. At each time point, equal amounts of protein of total cell lysates were analyzed by SDS-PAGE, followed by transfer to nitrocellulose and immunoblotting with mouse mAbs to the c-Myc epitope to detect Pex23p-Myc (top blot) and with antibodies to THI (middle blot) and the cytosolic enzyme glucose-6-phosphate dehydrogenase (G6PDH) (bottom blot).
DISCUSSION
Pex23p is a 418-amino acid protein with a predicted molecular mass of 47,588 Da. Sodium carbonate extraction and protease protection analyses of a c-Myc–tagged version of Pex23p that complements the pex23 mutation showed that Pex23p is an integral membrane protein that for the most part is localized away from the cytosolic surface of the peroxisome. How Pex23p is targeted to peroxisomes is not known, but a SKR tripeptide at the carboxyl terminus of Pex23p, which shows some resemblance to a PTS1 motif, is not required for targeting Pex23p to peroxisomes, because Pex23p with the c-Myc epitope at its carboxyl terminus is found in the peroxisomal membrane. Additionally, preliminary yeast two-hybrid analysis has failed to detect any interaction between Pex23p and the PTS1 receptor Pex5p. To date, we have not detected a two-hybrid interaction between Pex23p and Pex1p, Pex2p, Pex6p, Pex8p, Pex9p, Pex16p, or Pex20p (data not presented).
Pex23p shows high homology to two putative proteins encoded by the ORFs YLR324W and YGR004W of the S. cerevisiae genome. These proteins remain uncharacterized. Possible functional redundancy among these three proteins may have prevented their ready identification as PEX genes in S. cerevisiae by means of selection procedures involving random mutagenesis.
In contrast to wild-type Y. lipolytica cells, pex23 strains are unable to use oleic acid as their sole source of carbon. Growth of pex23 cells in oleic acid–containing medium leads to the appearance of a large number of small vesicular structures that are rarely seen in wild-type cells grown under the same conditions (see Figure 3). Independent biochemical analysis with flotation gradients confirmed the presence of vesicular structures containing both peroxisomal matrix and membrane proteins in the PEX23 gene disruption strain pex23KOA. Because these vesicles contain both peroxisomal matrix and membrane proteins, they are not classic “peroxisome ghosts,” which, as originally defined, are membranous structures containing peroxisomal membrane proteins but lacking peroxisomal matrix proteins (Santos et al., 1988). Whether the vesicular structures present in pex23 strains represent precursors to mature peroxisomes or are simply small peroxisomes lacking their full complement of peroxisomal proteins is unknown at present. We are currently conducting experiments in an attempt to answer this question.
In the cytosol, PTS1-targeted proteins are recognized by the PTS1 receptor Pex5p, whereas PTS2-targeted proteins are recognized by the PTS2 receptor Pex7p (reviewed by Subramani, 1993, 1998). Although separation of these two matrix protein pathways exists at this initial stage, convergence of the two pathways has been proposed to occur at the level of Pex14p, an integral peroxisomal membrane peroxin that has been demonstrated to bind both Pex5p and Pex7p (Albertini et al., 1997; Brocard et al., 1997; Huhse et al., 1998; Girzalsky et al., 1999; Shimizu et al., 1999). Pex13p, another peroxisomal integral membrane peroxin that was initially identified as the docking protein for the PTS1 receptor (Elgersma et al., 1996a; Erdmann and Blobel, 1996; Gould et al., 1996), has also been shown recently to bind the PTS2 receptor and to be required for the peroxisomal association of Pex14p (Girzalsky et al., 1999), suggesting that the point of convergence for the PTS1- and PTS2-dependent protein import pathways is at the level of the peroxisomal membrane and consists of a protein complex that contains both Pex13p and Pex14p. Because all peroxisomal matrix proteins investigated, including AOX, which has neither PTS1 nor PTS2 motifs (Wang et al., 1999), are mislocalized to the cytosol in pex23 mutant strains, Pex23p may act downstream of this point and thereby affect the import of all matrix proteins. Such a scenario would suggest that eventually all matrix proteins enter the peroxisome by a common import pathway, although the existence of such a common pathway remains to be demonstrated experimentally. Dysfunction and/or absence of Pex23p could also be proposed to lead to major structural alterations in the peroxisomal membrane that would prevent the correct assembly of the translocation machinery or machineries required for the import of matrix proteins, thereby leading to mislocalization of matrix proteins in general to the cytosol. Future analyses of the interacting partners of Pex23p should provide insight into which, if either, of these two scenarios is correct and whether Pex23p forms an integral part of the molecular machinery required for the translocation of peroxisomal matrix proteins.
ACKNOWLEDGMENTS
We thank Honey Chan for help with electron microscopy. R.A.R. is a Senior Scientist of the Medical Research Council of Canada and an International Research Scholar of the Howard Hughes Medical Institute. This work was supported by Medical Research Council of Canada grant MT-9208 (to R.A.R).
REFERENCES
- Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel JAKW, Veenhuis M, Kunau W-H. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89:83–92. doi: 10.1016/s0092-8674(00)80185-3. [DOI] [PubMed] [Google Scholar]
- Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1994. [Google Scholar]
- Brocard C, Lametschwandtner G, Koudelka R, Hartig A. Pex14p is a member of the protein linkage map of Pex5p. EMBO J. 1997;16:5491–5500. doi: 10.1093/emboj/16.18.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G, Braverman N, Wong C, Moser A, Moser HW, Watkins P, Valle D, Gould SJ. Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat Genet. 1995;9:115–125. doi: 10.1038/ng0295-115. [DOI] [PubMed] [Google Scholar]
- Douma AC, Veenhuis M, de Koning W, Evers M, Harder W. Dihydroxyacetone synthase is localized in the peroxisomal matrix of methanol-grown Hansenula polymorpha. Arch Microbiol. 1985;143:237–243. [Google Scholar]
- Dyer JM, McNew JA, Goodman JM. The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J Cell Biol. 1996;133:269–280. doi: 10.1083/jcb.133.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen GA, Aitchison JD, Szilard RK, Veenhuis M, Nuttley WM, Rachubinski RA. The Yarrowia lipolytica gene PAY2 encodes a 42-kDa peroxisomal integral membrane protein essential for matrix protein import and peroxisome enlargement but not for peroxisome membrane proliferation. J Biol Chem. 1995;270:1429–1436. doi: 10.1074/jbc.270.3.1429. [DOI] [PubMed] [Google Scholar]
- Eitzen GA, Szilard RK, Rachubinski RA. Enlarged peroxisomes are present in oleic acid-grown Yarrowia lipolytica overexpressing the PEX16 gene encoding an intraperoxisomal peripheral membrane peroxin. J Cell Biol. 1997;137:1265–1278. doi: 10.1083/jcb.137.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Kwast L, Klein A, Voorn-Brouwer T, van den Berg M, Metzig B, America T, Tabak HF, Distel B. The SH3 domain of the Saccharomyces cerevisiae peroxisomal membrane protein Pex13p functions as a docking site for Pex5p, a mobile receptor for the import of PTS1 containing proteins. J Cell Biol. 1996a;135:97–109. doi: 10.1083/jcb.135.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Vos A, van den Berg M, van Roermund CW, van der Sluijs P, Distel B, Tabak HF. Analysis of the carboxyl-terminal peroxisomal targeting signal 1 in a homologous context in Saccharomyces cerevisiae. J Biol Chem. 1996b;271:26375–26382. doi: 10.1074/jbc.271.42.26375. [DOI] [PubMed] [Google Scholar]
- Erdmann R, Blobel G. Identification of Pex13p, a peroxisomal membrane receptor for the PTS1 recognition factor. J Cell Biol. 1996;135:111–121. doi: 10.1083/jcb.135.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R, Veenhuis M, Kunau W-H. Peroxisomes: organelles at the cross-roads. Trends Cell Biol. 1997;7:400–407. doi: 10.1016/S0962-8924(97)01126-4. [DOI] [PubMed] [Google Scholar]
- Girzalsky W, Rehling P, Stein K, Kipper J, Blank L, Kunau W-H, Erdmann R. Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2-dependent protein import into peroxisomes. J Cell Biol. 1999;144:1151–1162. doi: 10.1083/jcb.144.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Andrews DA, Subramani S, Rachubinski RA. Mutagenesis of the amino targeting signal of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase reveals conserved amino acids required for import into peroxisomes in vivo. J Biol Chem. 1994;269:7558–7563. [PubMed] [Google Scholar]
- Goodman JM, Tramp SB, Hang H, Veenhuis M. Peroxisomes induced in Candida boidinii by methanol, oleic acid and d-alanine vary in metabolic function but share common integral membrane proteins. J Cell Sci. 1990;97:193–204. doi: 10.1242/jcs.97.1.193. [DOI] [PubMed] [Google Scholar]
- Götte K, Girzalsky W, Linkert M, Baumgart E, Kammerer S, Kunau W-H, Erdmann R. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol Cell Biol. 1998;18:616–628. doi: 10.1128/mcb.18.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Kalish JE, Morrell JE, Bjorkman J, Urquhart AJ, Crane DI. Pex13 is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytosolic PTS1 receptor. J Cell Biol. 1996;135:85–95. doi: 10.1083/jcb.135.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Keller G-A, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Keller G-A, Subramani S. Identification of a peroxisomal targeting signal at the carboxy terminus of firefly luciferase. J Cell Biol. 1987;105:2923–2931. doi: 10.1083/jcb.105.6.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhse B, Rehling P, Albertini M, Blank L, Meller K, Kunau W-H. Pex17p of Saccharomyces cerevisiae is a novel peroxin and component of the peroxisomal protein translocation machinery. J Cell Biol. 1998;140:49–60. doi: 10.1083/jcb.140.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller A, Snyder WB, Faber KN, Wenzel TJ, Rangell L, Keller GA, Subramani S. Pex22p of Pichia pastoris, essential for peroxisomal matrix protein import, anchors the ubiquitin-conjugating enzyme, Pex4p, on the peroxisomal membrane. J Cell Biol. 1999;146:99–112. doi: 10.1083/jcb.146.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziej PA, Young RA. Epitope tagging and protein surveillance. Methods Enzymol. 1991;194:508–519. doi: 10.1016/0076-6879(91)94038-e. [DOI] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Lazarow PB, Moser HW. Disorders of peroxisome biogenesis. In: Beaudet AL, Sly WS, Valle AD, editors. The Metabolic Basis of Inherited Disease. New York: McGraw-Hill; 1994. pp. 2287–2324. [Google Scholar]
- Luck H. Catalase. In: Bergmeyer H-U, editor. Methods of Enzymatic Analysis. New York: Academic Press; 1963. pp. 885–888. [Google Scholar]
- McCammon MT, McNew JA, Willy PJ, Goodman JM. An internal region of the peroxisomal membrane protein PMP47 is essential for sorting to peroxisomes. J Cell Biol. 1994;124:915–925. doi: 10.1083/jcb.124.6.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A, Lumb MJ, Oatey PB, Jennings PR, De Zoysa PA, Wanders RJ, Tabak HF, Danpure CJ. Mammalian alanine/glyoxylate aminotransferase 1 is imported into peroxisomes via the PTS1 translocation pathway: increased degeneracy and context specificity of the mammalian PTS1 motif and implications for the peroxisome-to-mitochondrion mistargeting of AGT in primary hyperoxaluria type 1. J Cell Biol. 1995;131:95–109. doi: 10.1083/jcb.131.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttley WM, Brade AM, Gaillardin C, Eitzen GA, Glover JR, Aitchison JD, Rachubinski RA. Rapid identification and characterization of peroxisomal assembly mutants in Yarrowia lipolytica. Yeast. 1993;9:507–517. [Google Scholar]
- Purdue PE, Yang X, Lazarow PB. Pex18p and Pex21p, a novel pair of peroxins essential for peroxisomal targeting by the PTS2 pathway. J Cell Biol. 1998;143:1859–1869. doi: 10.1083/jcb.143.7.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M, Imanaka T, Shio H, Small GM, Lazarow PB. Peroxisomal membrane ghosts in Zellweger syndrome–aberrant organelle assembly. Science. 1988;239:1536–1538. doi: 10.1126/science.3281254. [DOI] [PubMed] [Google Scholar]
- Shimizu N, et al. The peroxin Pex14p: cDNA cloning by functional complementation on a Chinese hamster ovary cell mutant, characterization, and functional analysis. J Biol Chem. 1999;274:12593–12604. doi: 10.1074/jbc.274.18.12593. [DOI] [PubMed] [Google Scholar]
- South ST, Gould SJ. Peroxisome synthesis in the absence of preexisting peroxisomes. J Cell Biol. 1999;144:255–266. doi: 10.1083/jcb.144.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. Protein import into peroxisomes and biogenesis of the organelle. Annu Rev Cell Biol. 1993;9:445–478. doi: 10.1146/annurev.cb.09.110193.002305. [DOI] [PubMed] [Google Scholar]
- Subramani S. PEX genes on the rise. Nat Genet. 1997;15:331–333. doi: 10.1038/ng0497-331. [DOI] [PubMed] [Google Scholar]
- Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol Rev. 1998;78:171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szilard RK, Titorenko VI, Veenhuis M, Rachubinski RA. Pay32p of the yeast Yarrowia lipolytica is an intraperoxisomal component of the matrix protein translocation machinery. J Cell Biol. 1995;131:1453–1469. doi: 10.1083/jcb.131.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Eitzen GA, Rachubinski RA. Mutations in the PAY5 gene of the yeast Yarrowia lipolytica cause the accumulation of multiple subpopulations of peroxisomes. J Biol Chem. 1996;271:20307–20314. doi: 10.1074/jbc.271.34.20307. [DOI] [PubMed] [Google Scholar]
- Titorenko VI, Rachubinski RA. Mutants of the yeast Yarrowia lipolytica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol Cell Biol. 1998;18:2789–2803. doi: 10.1128/mcb.18.5.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Smith JJ, Szilard RK, Rachubinski RA. Pex20p of the yeast Yarrowia lipolytica is required for the oligomerization of thiolase in the cytosol and for its targeting to peroxisomes. J Cell Biol. 1998;142:403–420. doi: 10.1083/jcb.142.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Le Dall M-T, Waché Y, Laroche C, Belin J-M, Gaillardin C, Nicaud J-M. Evaluation of acyl CoA oxidase (Aox) isozyme function in the n-alkane-assimilating yeast Yarrowia lipolytica. J Bacteriol. 1999;181:5140–5148. doi: 10.1128/jb.181.17.5140-5148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Titorenko VI, Haima P, Cregg JM, Harder W, Veenhuis M. The Hansenula polymorpha PER1 gene is essential for peroxisome biogenesis and encodes a peroxisomal matrix protein with both carboxy- and amino-terminal targeting signals. J Cell Biol. 1994;127:737–749. doi: 10.1083/jcb.127.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemer EAC, Lüers G, Faber KN, Wenzel T, Veenhuis M, Subramani S. Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J Biol Chem. 1996;271:18973–18980. doi: 10.1074/jbc.271.31.18973. [DOI] [PubMed] [Google Scholar]