Abstract
This study reports magnitudes and the orientation of the 13Cα chemical shift anisotropy (CSA) tensors of peptides obtained using quantum chemical calculations. The dependency of the CSA tensor parameters on the energy optimization of hydrogen atom positions and hydrogen bonding effects and the use of zwitterionic peptides in the calculations are examined. Our results indicate that the energy optimization of the hydrogen atom positions in crystal structures is necessary to obtain accurate CSA tensors. The inclusion of intermolecular effects such as hydrogen bonding in the calculations provided better agreement between the calculated and experimental values; however, the use of zwitterionic peptides in calculations, with or without the inclusion of hydrogen bonding, did not improve the results. In addition, our calculated values are in good agreement with tensor values obtained from solid-state NMR experiments on glycine-containing tripeptides. In the case of peptides containing an aromatic residue, calculations on an isolated peptide yielded more accurate isotropic shift values than the calculations on extended structures of the peptide. The calculations also suggested that the presence of an aromatic ring in the extended crystal peptide structure influences the magnitude of the δ22 which the present level of ab initio calculations are unable to reproduce.
Introduction
Determination of high-resolution three-dimensional structure and dynamics of peptides and proteins is one of the major goals of structural proteomics. The knowledge of protein structure is essential in order to effectively manipulate and regulate its function, making the determination of protein structure indispensable to virtually every field of structural biology and proteomics. High-resolution NMR spectroscopy has been extensively used to solve the structures of globular proteins. Similarly, solid-state NMR techniques have been used to study membrane-associated proteins, fibrils, and microcrystalline proteins. These applications of NMR spectroscopy utilize CSA tensors and also indicate that it is essential to understand the variation of the CSA tensor.1-7 Particularly, interpretations of results from applications of TROSY8 and PISEMA9 and experiments that measure relaxation parameters require well-characterized CSA tensors.10 In addition, CSA tensors are essential for NMR experimental studies on fully or partially aligned samples.11 Therefore, accurately determined CSA tensors are absolutely essential, and understanding the variation of CSA tensors would be of considerable importance in determining the structure, dynamics, and topology of proteins using NMR spectroscopy.
However, using NMR spectroscopy alone, it is often difficult to completely characterize and understand the variation of the CSA tensor. On the other hand, quantum chemical calculations can be utilized in the determination of the CSA tensor of an NMR active nucleus. Quantum chemical calculations can be used to efficiently observe the behavior of CSA tensors when spectroscopic methods are difficult to carry out. In addition, this method can be used as a predictive tool for protein structure, or as an aid to both solution and solid-state NMR studies. In this paper, we report 13Cα CSA tensors determined from quantum chemical calculations and the calculated results are compared with experimentally determined values.
Recently, NMR studies have reported the CSA tensors of 13Cα on a few short peptides.2,12-17 It has been shown that the isotropic chemical shift value of 13Cα depends on the backbone conformation of a peptide. This is due to changes in individual CSA tensor elements, as has been shown that the variation in the span of 13Cα CSA correlates with the protein secondary structure.1 Theoretical studies reported in the literature have shown that the 13Cα CSA tensor is influenced by a number of structural factors.18-20 In addition to the identity of the side chain, the backbone and side chain conformation (where applicable) have been shown to have the largest influence on the magnitudes of the principal components of the tensor,18,20 while bond lengths and bond angles between 13Cα and its neighbors have a lesser effect.18
All these previous theoretical studies on 13Cα tensors were based on single amino acids or N-formyl amino acid amide fragments.18,20 The effects of neighboring residues and intermolecular interactions, such as hydrogen bonding, have been largely ignored. Hydrogen bonding has been shown to have a significant effect on the amide-15N nucleus.21,22 It is likely that the 13Cα also experiences a similar effect as a result of hydrogen bonding. It is our intention to systematically investigate the 13Cα CSA tensor in peptides using quantum chemical calculations. We will specifically focus on the effects of intermolecular and intramolecular interactions on the 13Cα CSA tensor. In addition, we show that our calculated values are in good agreement with the experimentally determined CSA tensor data and are more accurate than the previously reported ab initio values.
Method
Calculation of 13Cα chemical shift tensors was carried out using the gauge-including atomic orbitals (GIAO) method23 and DFT level of theory in the Gaussian98 program.24 B3PW91 hybrid functionals were used for density functional calculations. Several different basis sets were used, ranging from 4-31G to 6-311++G(2d,p), to understand the effect of the basis set size on the accuracy of CSA tensors. These calculations were performed on several different peptides. Except for α-glycyl-glycine (*GG), which was obtained from a neutron diffraction study,25 structures for all other peptides were obtained from reported X-ray data: l-alanyl-l-serine (*AS),26 l-alanyl-glycine (*AG and A*G),27 l-alanyl-l-aspartic acid (A*D and *AD),28 glycyl-l,d-phenylalanine (*GF),29 d,l-alanyl-l,d-methionine (*AM),30 glycyl-l-asparagine (*GN),31 l-alanyl-l-alanyl-l-alanine (*AAA and A*AA),32 α-l-glutamyl-glycine (E*G),33 glycyl-l-alanyl-l-leucine trihydrate (G*AL•3H2O),34 glycyl-glycyl-glycine (G*GG),35 glycyl-glycyl-l-valine dihydrate (G*GV•2H2O and GG*V•2H2O),36 l-alanyl-glycyl-glycine monohydrate (A*GG•H2O),37 l-valyl-glycyl-glycine (V*GG),38 l-phenylalanyl-glycyl-glycine (F*GG),39 l-prolyl-glycyl-glycine (P*GG),40 tryptophanyl-glycyl-glycine dihydrate (W*GG•2H2O),41 l-tyrosyl-glycyl-glycine monohydrate (Y*GG•H2O),42 and N-acetyl valine (Ac-V)43 (where * indicates the amino acid residue of interest). Since the position of hydrogen atoms in X-ray crystal structures are inherently inaccurate, energy minimization was performed on the hydrogen atoms of the crystal structures using the PM344 semiempirical level of theory in the program CAChe.45 This step leaves the positions of the heavy atoms unaltered. Gaussian98 calculations provide absolute shielding values, which were arbitrarily assigned such that σ33 ≥ σ22 ≥ σ11. The absolute shielding values obtained from all calculations were converted to chemical shifts (δ11 ≥ δ22 ≥ δ33) relative to the absolute shielding of liquid TMS at room temperature of 184.1 ppm46 such that
The CSA tensor orientation is described relative to the peptide backbone. The tensor orientation can be described either as a set of Cartesian axes with respect to the Cα—H bond vector or as another set of axes with respect to the Cα—N bond vector; hence, data for two different sets of axes are reported.
To efficiently calculate CSA values on the peptides of interest, four parameters were considered in our calculations: (1) basis set size, (2) energy optimization of hydrogen atom position, (3) the effect of hydrogen bonding on peptides, and (4) the use of zwitterionic structures in calculations. Our results on the basis set size dependence of the CSA tensor is given in the Supporting Information. These results are in good agreement with previous studies.47-49 The locally dense basis set method,50 which involved using a larger basis set, 6-311++G(2d,2p), on the neighboring atoms of Cα and 4-31G on all other atoms, was used in all other calculations presented in this paper.
Results and Discussion
Effect of Energy Optimization of H Atom Positions in Crystal Structures. Previous ab initio studies on small N-formyl amino acid amide fragments of the peptide backbone suggested that the use of ab initio geometry optimized structures has a very small effect on the observed 13C shieldings.6,18,20,51 However, a recent study on melanostatin that compared the results of quantum chemical calculations with accurately determined 13Cα and 15N CSA tensors from solid-state NMR experiments suggested that energy optimization significantly improved the calculated values.52 In this study, peptide structures were derived from X-ray diffraction data. Since X-ray crystal structures do not accurately define the location of hydrogen atoms, energy minimization using PM344 parameters was used to optimize hydrogen atom positions without altering the coordinates of other atoms in the peptide. Calculations were performed on nonoptimized structures (hydrogen positions were obtained directly from X-ray crystal data) and compared to the data obtained from the optimized structures of the same peptides (see Supporting Information). Calculated chemical shift values for nonoptimized structures tend to be significantly lower than those for optimized structures. The difference is as large as 35 ppm. Comparison of calculated and experimental data for A*AA, A*G, E*G, and Ac-*V (see the last table in the Supporting Information) suggests that calculations on optimized structures considerably improved the accuracy of CSA values. Our calculations also suggested that, like the magnitudes of the principal components of the tensor, the angles defining the orientation of the 13Cα CSA tensor in the molecular frame are also affected by the optimization of hydrogen atom positions in the crystal structures (see Supporting Information). The tensor orientation relative to the Cα—H bond, in particular, differs greatly between optimized and nonoptimized structures because the positions of the hydrogen atom differ between optimized and nonoptimized structures. These results suggest that energy optimization of hydrogen atom positions is necessary in the calculation of CSA tensors when using structures derived from X-ray crystal data.
Effects of Intermolecular Interactions. Protein structure is greatly influenced by intermolecular effects such as hydrogen bonding, van der Waals forces, and hydrophobic interactions. Previous studies on the quantum chemical calculations of amide-15N CSA tensors of peptides suggested that hydrogen bonding interactions significantly influence the magnitudes but not the orientation of the principal elements of the tensor.21,53,54 In this study, calculations were performed on extended and isolated crystal structures of the peptides in order to understand the effect of intermolecular forces on the 13Cα CSA tensor. To keep the calculation time within reasonable limits, only the functional groups of nearby molecules having atoms within 3 Å of the 13Cα center of interest were considered. The resulting simplified version of the crystal structure for GAL•3H2O along with the isolated peptide structure is shown in Figure 1.
Figure 1.
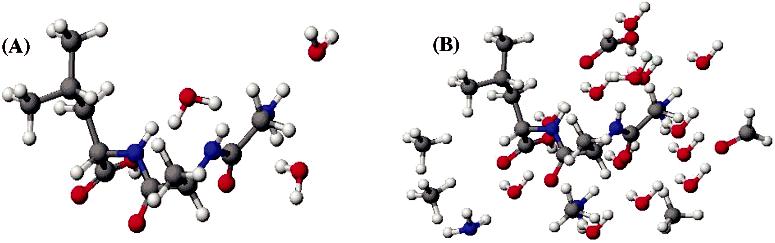
X-ray crystal structure34 of single-unit (A) and extended (B) forms of GAL•3H2O.
13Cα CSA tensor values calculated from isolated peptides and extended structures are compared in Tables 1 and 2. Both the magnitude and orientation of the principal elements of the tensor significantly differ between the two structures. There is, however, no uniformity in the change between CSA tensors of single peptide structures or extended crystal structures for the peptides studied, indicating that the effects of intermolecular interactions vary from peptide to peptide. These results suggest that inclusion of intermolecular interactions is important in determining accurate 13Cα CSA tensors of peptides. This prediction is in good agreement with a recent study that compared 13C and 15N CSA data from quantum chemical calculations and solid-state NMR experiments on melanostatin.52
Table 1.
13Cα Chemical Shift Tensors Obtained from ab Initio Calculations of Glycine-Containing Tripeptidesa
| peptide | δ11 | δ22 | δ33 | Ω | δiso | C-H δ11b | C-H δ22 | C-H δ33 | C-N δ11c | C-N δ22 | C-N δ33 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GGG I | 61.7 | 49.5 | 20.3 | 41.4 | 43.8 | 141(77) | 52(113) | 99(153) | 39 | 58 | 71 |
| II | 60.9 | 50.5 | 21.5 | 39.4 | 44.3 | 16(121) | 105(141) | 94(110) | 103 | 82 | 16 |
| III | 62.2 | 50.2 | 19.5 | 42.7 | 44.0 | 143(125) | 54(119) | 98(131) | 40 | 56 | 72 |
| IV | 60.5 | 50.9 | 21.3 | 39.2 | 44.2 | 12(106) | 95(150) | 101(65) | 98 | 42 | 49 |
| experimentale | 58.1 | 48.9 | 22.9 | 35.2 | 43.3 | ||||||
| GGV·2H2O I | 71.5 | 43.1 | 18.8 | 52.7 | 44.5 | 126(54) | 88(36) | 36(88) | 55 | 145 | 87 |
| II | 70.8 | 45.5 | 18.8 | 52.0 | 45.0 | 125(42) | 109(56) | 42(65) | 65 | 141 | 118 |
| III | 75.0 | 41.5 | 20.8 | 54.2 | 45.8 | 126(47) | 83(43) | 37(88) | 62 | 152 | 86 |
| IV | 73.4 | 43.2 | 21.0 | 52.4 | 45.9 | 123(48) | 111(52) | 41(66) | 61 | 138 | 117 |
| experimentale | 70.6 | 42.0 | 21.7 | 48.9 | 44.8 | ||||||
| AGG·H2O I | 71.2 | 43.1 | 9.9 | 61.3 | 41.4 | 102(74) | 94(19) | 167(81) | 40 | 127 | 79 |
| II | 69.7 | 45.4 | 16.1 | 53.6 | 43.7 | 110(65) | 126(127) | 137(48) | 46 | 53 | 113 |
| III | 74.1 | 40.4 | 11.6 | 62.5 | 42.0 | 103(78) | 105(12) | 160(91) | 35 | 120 | 74 |
| IV | 73.7 | 40.7 | 16.3 | 57.4 | 43.6 | 108(66) | 143(111) | 121(32) | 45 | 63 | 122 |
| experimentale | 69.9 | 41.8 | 16.9 | 53.0 | 42.9 | ||||||
| PGGd I | 71.5 | 40.2 | 10.3 | 61.2 | 40.7 | 123(63) | 75(34) | 37(109) | 45 | 131 | 73 |
| II | 70.2 | 42.7 | 13.0 | 57.2 | 42.0 | 115(119) | 26(121) | 92(45) | 18 | 83 | 74 |
| experimentald | 69.4 | 40.8 | 18.8 | 50.6 | 43.0 | ||||||
| VGG I | 58.5 | 48.5 | 17.5 | 41.0 | 41.5 | 103(84) | 13(102) | 94(14) | 31 | 99 | 119 |
| II | 59.3 | 46.2 | 19.1 | 40.2 | 41.5 | 89(85) | 135(28) | 45(63) | 39 | 98 | 128 |
| III | 60.3 | 51.4 | 18.7 | 41.6 | 43.5 | 100(80) | 20(89) | 106(9.7) | 36 | 109 | 119 |
| IV | 60.9 | 48.3 | 19.6 | 41.3 | 42.9 | 83(86) | 140(33) | 51(57) | 43 | 92 | 133 |
| experimentald | 57.8 | 48.1 | 23.1 | 34.7 | 43.0 | ||||||
| FGG I | 71.9 | 43.4 | 19.6 | 52.3 | 45.0 | 107(99) | 28(87) | 112(9.8) | 14 | 100 | 100 |
| II | 71.4 | 50.0 | 26.7 | 44.7 | 49.4 | 125(76) | 38(85) | 103(15) | 33 | 90 | 123 |
| III | 75.7 | 42.1 | 19.5 | 56.2 | 45.8 | 102(97) | 23(87) | 110(7.7) | 20 | 105 | 102 |
| IV | 74.8 | 47.8 | 26.5 | 48.3 | 49.7 | 131(75) | 44(83) | 102(16) | 35 | 85 | 125 |
| experimentald | 69.6 | 43.3 | 20.5 | 49.1 | 44.5 | ||||||
| YGG·H2O I | 69.0 | 44.7 | 21.2 | 47.8 | 45.0 | 58(48) | 98(73) | 33(133) | 134 | 45 | 81 |
| II | 67.5 | 46.2 | 25.8 | 41.7 | 46.5 | 111(24) | 76(112) | 26(80) | 87 | 34 | 124 |
| III | 73.1 | 43.8 | 21.8 | 51.3 | 46.2 | 57(53) | 92(66) | 33(133) | 143 | 55 | 81 |
| IV | 71.2 | 43.8 | 27.0 | 44.2 | 47.3 | 113(19) | 78(105) | 27(80) | 92 | 35 | 125 |
| experimentald | 66.3 | 41.3 | 25.0 | 41.3 | 44.2 | ||||||
| WGG·2H2O I | 76.2 | 46.1 | 16.9 | 59.3 | 46.4 | 113(125) | 97(47) | 156(64) | 22 | 69 | 94 |
| II | 68.8 | 48.3 | 18.4 | 50.4 | 45.2 | 123(115) | 57(83) | 130(26) | 21 | 69 | 88 |
| III | 81.4 | 43.8 | 19.2 | 62.2 | 48.1 | 124(122) | 86(62) | 146(45) | 31 | 60 | 97 |
| IV | 72.6 | 45.8 | 19.6 | 53.0 | 46.0 | 116(127) | 92(120) | 26(128) | 28 | 118 | 87 |
| experimentald | 69.5 | 44.2 | 19.2 | 50.3 | 44.3 |
Results obtained from calculations on the central glycine residue from neutral isolated (I), neutral extended crystal structure (II), isolated zwitterionic (III), and zwitterionic extended crystal structure (IV) peptides. The magnitudes of the principal components are given with respect to TMS (at 184.1 ppm).46
C-H δ11, C-H δ22, and C-H δ33 are determined relative to the C-H bond vector and given in degrees. Angles with respect to the other hydrogen C-H bond for C-H δ11, C-H δ22, and C-H δ33 are given in parentheses.
C-N δ11, C-N δ22, and C-N δ33 are determined relative to the C-N bond vector and given in degrees.
Due to the unique nature of the proline residue, a zwitterionic structure is not possible.
Experimental data obtained from ref 17.
Table 2.
13Cα Chemical Shift Tensors Obtained from ab Initio Calculations on Peptidesa
| peptide | δ11 | δ22 | δ33 | Ω | δiso | C-H δ11b | C-H δ22 | C-H δ33 | C-N δ11c | C-N δ22 | C-N δ33 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| *AAA I | 78.6 | 59.4 | 39.0 | 39.6 | 59.0 | 148 | 87 | 58 | 77 | 18 | 102 |
| II | 66.2 | 59.2 | 35.8 | 30.4 | 53.7 | 130 | 47 | 70 | 68 | 117 | 36 |
| A*AA I | 68.6 | 56.6 | 30.1 | 38.5 | 51.8 | 19 | 104 | 102 | 108 | 126 | 42 |
| II | 67.4 | 55.7 | 28.1 | 39.3 | 50.4 | 39 | 118 | 116 | 111 | 49 | 132 |
| experimental16 | 70.2 | 54.9 | 23.6 | 46.6 | 49.6 | ||||||
| *AG I | 67.8 | 60 | 30.6 | 37.2 | 52.8 | 102 | 151 | 65 | 144 | 54 | 90 |
| II | 65.8 | 61.8 | 26.6 | 39.2 | 51.4 | 167 | 88 | 103 | 59 | 112 | 141 |
| Zw | 70.9 | 61.4 | 23.1 | 47.8 | 51.8 | 166 | 103 | 84 | 56 | 139 | 70 |
| experimental16 | 58.7 | 56.7 | 34.0 | 24.7 | 49.8 | ||||||
| A*G I | 69.5 | 47.7 | 15.5 | 54.0 | 44.2 | 117(88) | 69(41) | 35(130) | 124 | 137 | 113 |
| II | 71.1 | 47.9 | 16.4 | 54.7 | 45.1 | 106(21) | 78(111) | 20(87) | 127 | 135 | 112 |
| Zw | 78.0 | 40.3 | 18.4 | 59.7 | 45.5 | 104(30) | 77(120) | 20(87) | 135 | 126 | 113 |
| experimental16 | 66.6 | 43.7 | 24.7 | 41.9 | 45.0 | ||||||
| *AD I | 67.7 | 62.2 | 27.7 | 40.0 | 52.5 | 124 | 41 | 110 | 85 | 87 | 6 |
| II | 65.2 | 63.2 | 25.6 | 39.6 | 51.3 | 113 | 50 | 131 | 84 | 64 | 26 |
| experimental16 | 57.4 | 55.9 | 34.5 | 22.9 | 49.3 | ||||||
| A*D I | 70.4 | 56.2 | 28.0 | 42.4 | 51.5 | 75 | 133 | 133 | 35 | 62 | 72 |
| II | 70.8 | 58.7 | 26.8 | 44.0 | 52.1 | 66 | 80 | 154 | 56 | 64 | 46 |
| experimental16 | 71.8 | 57.9 | 28.4 | 43.4 | 52.7 | ||||||
| *AM I | 65.3 | 61.4 | 34.9 | 30.4 | 53.9 | 95 | 107 | 163 | 141 | 52 | 81 |
| II | 77.4 | 54.7 | 35.5 | 41.9 | 55.9 | 104 | 136 | 131 | 147 | 76 | 61 |
| *AS I | 60.7 | 58.8 | 46.7 | 14.0 | 55.4 | 74 | 151 | 114 | 78 | 45 | 133 |
| II | 61.4 | 55.2 | 39.0 | 22.3 | 51.9 | 109 | 66 | 148 | 99 | 47 | 44 |
| *GG I | 51.3 | 27.1 | -19.8 | 71.1 | 19.5 | 122(34) | 100(61) | 146(106) | 74 | 151 | 67 |
| II | 56.7 | 40.9 | 22.4 | 34.3 | 40.0 | 104(88) | 16(104) | 83(166) | 137 | 121 | 64 |
| experimentald | 55.8 | 39.5 | 23.5 | 32.3 | 39.6 | ||||||
| *GN I | 67.2 | 54.5 | 15.1 | 52.1 | 45.6 | 81(30) | 11(120) | 95(87) | 122 | 100 | 34 |
| II | 65.2 | 53.7 | 13.4 | 51.7 | 44.1 | 100(33) | 13(103) | 99(120) | 82 | 98 | 11 |
| *GF I | 58.2 | 40.8 | -6.8 | 64.9 | 30.7 | 21(102) | 108(54) | 81(39) | 129 | 141 | 94 |
| II | 73.8 | 52.6 | 25.6 | 48.2 | 50.7 | 105(103) | 146(65) | 60(152) | 123 | 88 | 147 |
| E*G I | 68.0 | 38.1 | 18.6 | 49.4 | 41.6 | 138(94) | 132(56) | 92(146) | 30 | 120 | 91 |
| II | 79.1 | 36.0 | 10.7 | 68.4 | 41.9 | 36(90) | 80(53) | 56(37) | 49 | 125 | 120 |
| experimentald | 78.0 | 37.5 | 10.5 | 67.5 | 42.0 | ||||||
| Ac-V I | 85.4 | 50.6 | 36.3 | 49.1 | 57.4 | 105 | 59 | 144 | 16 | 101 | 102 |
| II | 86.6 | 52.5 | 35.8 | 50.9 | 58.3 | 29 | 80 | 117 | 123 | 33 | 92 |
| experimental4 | 82.6 | 53.2 | 36.1 | 46.5 | 57.3 | 22 | 36 | ||||
| G*AL·2H2O I | 74.2 | 47.8 | 36.2 | 38.0 | 52.7 | 82.1 | 157 | 112 | 101 | 94 | 12 |
| II | 74.1 | 48.3 | 38.0 | 36.1 | 53.5 | 68.1 | 135 | 127 | 147 | 69 | 114 |
| Zw | 79.1 | 48.2 | 41.6 | 37.5 | 56.3 | 64 | 130 | 129 | 154 | 77 | 112 |
| experimental15 | 70.0 | 51.0 | 35.0 | 35.0 | 52.0 | 116 | 46 | 70 | 108 | 40 | 56 |
| GG*V·2H2O I | 79.7 | 61.3 | 48.1 | 31.6 | 63.0 | 59.8 | 144 | 73 | 60 | 89 | 150 |
| II | 82.2 | 61.3 | 50.7 | 31.5 | 64.7 | 42.8 | 70 | 126 | 65 | 110 | 32 |
| Zw | 81.1 | 69.1 | 54.3 | 26.8 | 68.2 | 54 | 117 | 48 | 58 | 42 | 114 |
| experimental15 | 75.0 | 70.0 | 51.0 | 24.0 | 65.3 | 155 | 89 | 115 | 63 | 28 | 98 |
Results obtained from isolated peptide with geometry optimization (I), extended crystal structure of the peptide (II), and extended crystal structures of zwitterionic peptides (Zw) are compared. The magnitudes of the principal tensor components are given with respect to TMS (at 184.1 ppm).46
Angles C-H δ11, C-H δ22, and C-H δ33 are determined relative to the C-H bond vector and given in degrees. Angles with respect to the other hydrogen C-H bond for C-H δ11, C-H δ22 and C-H δ33 are given in parentheses.
C-N δ11, C-N δ22, and C-N δ33 are determined relative to the C-N bond vector and given in degrees.
Unpublished data measured from 2D PASS solid-state NMR experiments on powder samples of peptides as explained in our previous publication.14
Comparison to Experimental Results. Experimentally obtained accurate magnitudes of the principal elements of the 13Cα CSA tensor of central glycine residues of several tripeptides enabled the examination of the accuracy of our calculated values.17 Single peptide structures and extended crystal structures of several glycine-containing tripeptides derived from X-ray crystal structures35-42 were used to calculate CSA tensors; the data are given in Table 1 and are compared in Figure 2. Calculated values agree well with experimental CSA tensor data of the peptides; calculated isotropic chemical shifts are within ±2.7 ppm for calculations on single peptide structures, and within ±1.5 ppm for calculations on extended crystal structures with the exception of F*GG. In addition, the magnitude of δ11 is overestimated for most of the centers examined, and the magnitude of δ33 is generally underestimated in these peptides, with the exception of calculations on extended crystal structures of the aromatic-residue-containing peptides. For F*GG, the single peptide structure yields a less accurate isotropic shift value. The reasoning can be traced back to the principal elements of the tensor, where the extended crystal structure overestimates δ22 and δ33 by over 6 ppm each. In fact, extended crystal structures of all aromatic-residue-containing peptides overestimate δ22 by 4-6.6 ppm, though the overestimation of δ33 in the F*GG extended crystal structure makes the isotropic shift less accurate than the other aromatic-residue-containing peptides. Single-peptide structures of the aromatic-residue-containing peptides yield more accurate isotropic shifts, within ±2.1 ppm. This implies that the presence of an aromatic ring in the extended crystal peptide structure influences the magnitude of the δ22 which the present level of ab initio calculations are unable to reproduce.
Figure 2.
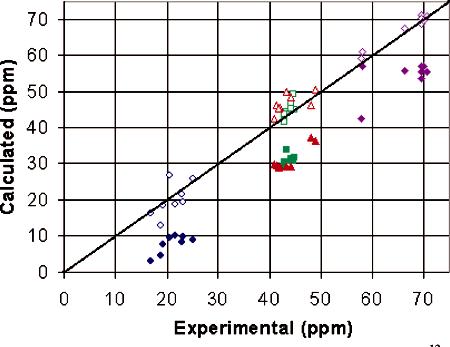
Comparison of the magnitudes of the principal components of the 13Cα CSA tensor obtained from previous20,55 (filled symbols) and present (open symbols) ab initio calculations (locally dense basis set) and solidstate NMR experiments.17 Values of δ11, δ22, δ33, and δiso are represented by diamonds, triangles, circles, and squares, respectively.
Since these are short peptides, charges on the terminal residues could affect the CSA tensor values. The peptide crystals from which structures were derived contained zwitterionic peptides.35-42 To determine if the zwitterionic nature of these peptides affects the 13Cα CSA tensor, calculations were carried out on zwitterionic structures of these tripeptides (both isolated peptide and extended crystal structures), and the results are compared with the values obtained from neutral peptides (Table 1). With the exception of G*GG, the calculations on zwitterionic peptides (both single and extended crystal peptides) yielded slightly more accurate results for the most shielded tensor element, δ33· However, the least shielded element, δ11, is overestimated to a greater extent in zwitterionic peptides than in neutral peptides. For peptides (F*GG, Y*GG•H2O, and W*GG•2H2O) containing an aromatic residue, calculations of extended crystal structures of zwitterionic structures do not grossly overestimate the δ22 component as neutral peptide extended crystal structure calculations did (although the δ22 component of F*GG is still 4.5 ppm greater than the experimentally observed value), indicating that the use of zwitterionic structures in calculations containing aromatic residues near the center of interest may yield more accurate results.
When taken together, these data indicate that extended peptide structures of neutral peptides give the most accurate CSA tensor results as compared to experimental data with the exception of aromatic-residue-containing peptides. Single-peptide structures give slightly less accurate results than extended crystal structures, though they appear to be more accurate in the calculation 13Cα CSA tensors when the neighboring amino acid contains an aromatic ring. Calculated isotropic shifts are in excellent agreement with the experimental data, indicating that the summation of the errors in the calculated magnitudes of the principal elements cancel out.
Comparison with Previous ab Initio Data. The availability of solid-state NMR experimental17 and structural data on a large number of glycine-containing tripeptides35-42 allows for the comparison of theoretical data obtained in this study with the data obtained in previous ab initio studies of 13Cα CSA tensors on the N-formyl glycyl amide fragment.20 Data obtained from calculations on extended crystal structures as well as from the glycine shielding surfaces from the previous study20 are correlated with the experimental data17 in Figure 2. A recent solid-state NMR experimental study17 showed that the ab initio calculations of 13Cα CSA span from the previous report correlate well with the experimental data. However, as seen in Figure 2, the ab initio calculations of Sun et al. consistently underestimated the magnitudes of the principal components of the CSA tensor and the isotropic shift55 (absolute shielding values were converted to chemical shifts). Our calculations on the same peptides showed much better correlation between theoretical and experimental values of both the principal tensor elements and (especially) the isotropic shift. This implies that the energy optimization of the H atom positions in crystal structures and the inclusion of hydrogen bonding interactions are important in determining the CSA tensors accurately. Also, the identities of the neighboring residues of the 13Cα of interest are important to consider in the calculation of the magnitude of the CSA tensor: the presence of a neighboring residue cannot be substituted by an N-formyl or amide-protecting group on either side of the residue of interest.
Comparison to Additional Experimental Data. The availability of solid-state NMR experimental data on A*AA, *AG, A*G, *AD, A*D, *GG, E*G, GG*V•2H2O, G*AL•3H2O, and Ac-V allows for additional comparison of our calculation methods.12-14 For the 13Cα centers examined in this section, isotropic shift values show a high degree of accuracy with respect to experimentally determined values (Table 2). However, the magnitudes of the principal elements of the tensor differ greatly for some of the peptides examined. Most notable are the tensor elements of *AG, *AD, and GG*V extended crystal structures. The magnitude of δ11 is overestimated for most of the centers examined, and the magnitude of δ33 is generally underestimated in the glycine-containing peptides examined earlier; however, this trend is less clear among these peptides, especially in the results obtained for δ11 from calculations on single-peptide structures. Similarly, δ33 is not uniformly underestimated in these calculations. However, with some peptides, the individual tensor components are all close to experimental values, including the tensor span, Ω. A*D, *GG, E*G, and G*AL•3H2O all have tensor spans within 2.0 ppm (for extended crystal structures).
We were also interested in observing the effects that charge on terminal residues would have on CSA tensors in these peptides. Zwitterionic forms of the peptides were used in the calculations on the extended crystal structures of GG*V•2H2O, *AG, A*G, and G*AL. The results obtained show that there was no significant improvement in the isotropic shift as compared to calculations on neutral extended peptide structures. In addition, there is no improvement in the tensor span, and individual tensor elements are less accurate than calculations on neutral peptides, except for δ22 of GG*V. These results indicate that the calculation of zwitterionic peptides does not improve the calculated values of the principal tensor elements relative to experimentally obtained results, which is consistent with the results obtained from the glycine-containing tripeptides examined earlier.
The accuracy of the angles defining the tensor obtained by our calculation methods can be estimated by comparing calculated results to experimental results available for Ac-V, GG*V•2H2O, and G*AL•3H2O12,13 (Table 2). For Ac-V, tensor orientation angles between C—H and δ11 and C—N and δ22 measured using solid-state NMR experiments on a powder sample were reported to be 22°(158°) and 36°(144°), respectively.12 Angles calculated using an isolated Ac-V peptide are 75°(105°) and 80°(100°), respectively, differing greatly from the experimental values. However, calculations on the extended crystal structure provided tensor orientation angles that are much closer to the experimental values: 29°(151°) and 33°(147°). This indicates that it is likely that intermolecular effects, such as hydrogen bonding, influence tensor orientation angles. However, the tensor orientation angles calculated for GG*Vδ2H2O and G*ALδ3H2O extended crystal structures do not match experimentally obtained values. It should be noted that the values obtained from previous ab initio studies20 also do not match well with the experimental results as shown in ref 13. Since values obtained for all other peptides match well with the experimental data, the inability to reproduce the experimental results for GG*Vδ2H2O and G*ALδ3H2O may be because of the difference in the X-ray crystal structures used to calculate the CSA tensors and that of the powder samples used in the experimental studies. On the other hand, inclusion of long-range electrostatic interactions52 in the calculations may provide results that will better agree with the experimentally determined results. Also, the availability of more experimentally determined CSA tensor orientations would be useful to confirm the role of longrange electrostatic interactions. Nevertheless, results in this study suggest that the basis set size and selection have minimal influence on the angles that define the orientation of the tensor.
Conclusions
The goal of this study was to investigate the variation of 13Cα CSA tensors in small peptides and establish an accurate method to calculate them. A number of small peptides were studied with respect to several different variables using the quantum calculation method. The use of X-ray crystal structures as a basis for constructing the peptides for calculation, as well as the use of locally dense basis sets and geometry optimization of hydrogen atoms, proved to be a reliable technique. Calculations on extended crystal structures showed that the magnitudes of the principal tensor elements are sensitive to intermolecular effects, while the orientation of the tensor is very sensitive to such effects. The values obtained from different peptides suggest that the CSA dependency on intermolecular interactions is not uniform and is likely related to the location of hydrogen bonding partners. Calculated values from the extended crystal structures are in good agreement with experimental results. Use of zwitterionic structures, both single-peptide and extended crystal structures, in calculations do not improve the accuracy of the least and most shielded components, while they do improve the accuracy of the δ22 component of the CSA tensor. Based on the accuracy of our calculations of 13Cα CSA tensors, the method outlined in this paper establishes a reliable means to predict the magnitude of the principal elements of 13Cα CSA tensors. While the calculated angles defining the orientation of the tensor for N-acetyl-valine matches well with the experimental data, more experimental data are needed to evaluate the accuracy of the calculated values. We believe that the results reported in this paper will be useful in the structural studies of peptides and proteins using both solution and solid-state NMR techniques.
Supplementary Material
Acknowledgment
This research was supported by the funds from the National Science Foundation (CAREER development award to A.R.) and partly by the NIH (AI054515-01A1).
Footnotes
Supporting Information Available: Calculated 13Cα CSA tensors obtained using various basis sets and values calculated from peptides with and without geometry optimization. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Tjandra N, Bax A. J. Am. Chem. Soc. 1997;119:9576–9577. [Google Scholar]
- (2).Hong M. J. Am. Chem. Soc. 2000;122:3762–3770. [Google Scholar]
- (3).Lee DK, Santos JS, Ramamoorthy A. J. Phys. Chem. B. 1999;103:8383–8390. [Google Scholar]
- (4).Fu RQ, Cross TA. Annu. ReV. Biophy. Biomol. Struct. 1999;28:235–268. doi: 10.1146/annurev.biophys.28.1.235. [DOI] [PubMed] [Google Scholar]
- (5).Asakawa N, Kurosu H, Ando I, Shoji A, Ozaki T. J. Mol. Struct. 1994;317:119–129. [Google Scholar]
- (6).Havlin RH, Le H, Laws DD, de Dios AC, Oldfield E. J. Am. Chem. Soc. 1997;119:11951–11958. [Google Scholar]
- (7).Case DA. Curr. Opin. Struct. Biol. 1998;8:624–630. doi: 10.1016/s0959-440x(98)80155-3. [DOI] [PubMed] [Google Scholar]
- (8).Wuthrich K. Nat. Struct. Biol. 2001;8:923–925. doi: 10.1038/nsb1101-923. [DOI] [PubMed] [Google Scholar]; Biosci. Rep. 2003;23:119–153. [Google Scholar]
- (9).Ramamoorthy A, Wu CH, Opella SJ. J. Magn. Reson. 1999;140:131–140. doi: 10.1006/jmre.1999.1827. [DOI] [PubMed] [Google Scholar]; Ramamoorthy A, Opella SJ. Solid-State NMR Spectrosc. 1995;4:387–392. doi: 10.1016/0926-2040(95)00054-t. [DOI] [PubMed] [Google Scholar]; Ramamoorthy A, Wei Y, Lee DK. Ann. Rep. NMR Spectrosc. 2004;52:1–52. [Google Scholar]
- (10).Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE. J. Am. Chem. Soc. 2003;125:10420–10428. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]; Kay LE. Nat. Struct. Biol. 1998;5:513–517. doi: 10.1038/755. [DOI] [PubMed] [Google Scholar]; Palmer AG. Curr. Opin. Struct. Biol. 1997;7:732–737. doi: 10.1016/s0959-440x(97)80085-1. [DOI] [PubMed] [Google Scholar]
- (11).Bax A. Protein Sci. 2003;12:1–16. doi: 10.1110/ps.0233303. [DOI] [PMC free article] [PubMed] [Google Scholar]; Sanders CR, Hare BJ, Howard KH, Prestegard JH. Prog. Nucl. Magn. Reson. Spectrosc. 1994;26:421–444. [Google Scholar]
- (12).Yao X, Hong M. J. Am. Chem. Soc. 2001;124:2730. doi: 10.1021/ja017137p. [DOI] [PubMed] [Google Scholar]
- (13).Yao X, Yamaguchi S, Hong M. J. Biomol. NMR. 2002;24:51–62. doi: 10.1023/a:1020626802472. [DOI] [PubMed] [Google Scholar]
- (14).Wei Y, Lee D-K, Ramamoorthy A. J. Am. Chem. Soc. 2001;123:6118–6126. doi: 10.1021/ja010145l. [DOI] [PubMed] [Google Scholar]
- (15).Heller J, Laws DD, Tomaselli M, King DS, Wemmer DE, Pines A, Havlin RH, Oldfield E. J. Am. Chem. Soc. 1997;119:7827–7831. [Google Scholar]
- (16).Takeda N, Kuroki S, Kurosu H, Ando I. Biopolymers. 1999;50:61–69. [Google Scholar]
- (17).Chekmenev EY, Xu RZ, Mashuta MS, Wittebort RJ. J. Am. Chem. Soc. 2002;124:11894–11899. doi: 10.1021/ja026700g. [DOI] [PubMed] [Google Scholar]
- (18).de Dios AC, Pearson JG, Oldfield E. J. Am. Chem. Soc. 1993;115:9768–9773. [Google Scholar]
- (19).Havlin RH, Laws DD, Bitter H-ML, Sanders LK, Sun H, Grimley JS, Wemmer DE, Pines A, Oldfield E. J. Am. Chem. Soc. 2001;123:10362–10369. doi: 10.1021/ja0115060. [DOI] [PubMed] [Google Scholar]
- (20).Sun H, Sanders LK, Oldfield E. J. Am. Chem. Soc. 2002;124:5486–5495. doi: 10.1021/ja011863a. [DOI] [PubMed] [Google Scholar]
- (21).Brender JR, Taylor DM, Ramamoorthy A. J. Am. Chem. Soc. 2001;123:914–922. doi: 10.1021/ja001980q. [DOI] [PubMed] [Google Scholar]
- (22).Le H, Oldfield E. J. Phys. Chem. 1996;100:16423–16428. [Google Scholar]
- (23).Ditchfield R. J. Chem. Phys. 1972;56:5688. [Google Scholar]
- (24).Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery JA, Jr., Stratmann RE, Burant JC, Dapprich S, Millam JM, Daniels AD, Kudin KN, Strain MC, Farkas O, Tomasi J, Barone V, Cossi M, Cammi R, Mennucci B, Pomelli C, Adamo C, Clifford S, Ochterski J, Petersson GA, Ayala PY, Cui Q, Morokuma K, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Cioslowski J, Ortiz JV, Baboul AG, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Gomperts R, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Gonzalez C, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Andres JL, Gonzalez C, Head-Gordon M, Replogle ES, Pople JA. Gaussian98. Gaussian, Inc.; Pittsburgh, PA: 1998. [Google Scholar]
- (25).Kvick A, Al-Karaghouli AR, Koetzle TF. Acta Crystallogr. 1977;B33:3796–3801. [Google Scholar]
- (26).Jones PG, Falvello L, Kennard O. Acta Crystallogr. 1978;B34:1939–1942. [Google Scholar]
- (27).Koch MHJ, Germain G. Acta Crystallogr. 1970;B26:410–417. [Google Scholar]
- (28).Eggleston DS, Hodgson DJ. Int. J. Peptide Protein Res. 1983;21:288–295. [PubMed] [Google Scholar]
- (29).Marsh RE, Ramakumar S, Venkatesan K. Acta Crystallogr. 1976;B32:66. [Google Scholar]
- (30).Stenkamp RE, Jensen LH. Acta Crystallogr. 1974;B30:1541. [Google Scholar]
- (31).Pasternak RA, Katz L, Corey RB. Acta Crystallogr. 1954;7:225. [Google Scholar]
- (32).Fawcett JK, Camerman N, Camerman A. Acta Crystallogr. 1975;B31:658. [Google Scholar]
- (33).Eggleston DS, Valente EJ, Hodgson DJ. Acta Crystallogr. 1981;B37:1430–1433. [Google Scholar]
- (34).Chaturvedi S, Go K, Parthasarathy R. Biopolymers. 1991;31:397–407. doi: 10.1002/bip.360310405. [DOI] [PubMed] [Google Scholar]
- (35).Srikrishnan N, Winiewicz N, Parthasarathy R. Int. J. Pept. Protein Res. 1982;19:103–113. doi: 10.1111/j.1399-3011.1982.tb02597.x. [DOI] [PubMed] [Google Scholar]
- (36).Lalitha V, Subramanian E, Bordner J. Int. J. Pept. Protein Res. 1984;24:437–441. doi: 10.1111/j.1399-3011.1984.tb03142.x. [DOI] [PubMed] [Google Scholar]
- (37).Lalitha V, Subramanian E, Bordner J. Indian J. Pure Appl. Phys. 1985;23:506–508. [Google Scholar]
- (38).Lalitha V, Murali R, Subramnian E. Int. J. Pept. Protein Res. 1986;27:472–477. doi: 10.1111/j.1399-3011.1986.tb01806.x. [DOI] [PubMed] [Google Scholar]
- (39).Subramnian E, Sahayamary JJ. Int. J. Pept. Protein Res. 1989;34:211–214. doi: 10.1111/j.1399-3011.1989.tb00232.x. [DOI] [PubMed] [Google Scholar]
- (40).Lalitha V, Subramanian E, Parthasarathy R. Int. J. Pept. Protein Res. 1986;27:223–228. doi: 10.1111/j.1399-3011.1986.tb01814.x. [DOI] [PubMed] [Google Scholar]
- (41).Subramnian E, Sahayamary JJ. Int. J. Pept. Protein Res. 1989;34:134–138. doi: 10.1111/j.1399-3011.1989.tb01502.x. [DOI] [PubMed] [Google Scholar]
- (42).Carson WM, Hackert ML. Acta Crystallogr. 1978;B34:1275–1280. [Google Scholar]
- (43).Carroll PJ, Stewart PL, Opella SJ. Acta Crystallogr. 1990;C46:243–246. [Google Scholar]
- (44).Stewart JJP. J. Comput. Chem. 1989;10:209. [Google Scholar]
- (45).CAChe Worksystem Pro, version 5.0; Fujitsu Ltd., 2001, Oxford Molecular Ltd., 1989-2000
- (46).Jameson AK, Jameson CJ. Chem. Phys. Lett. 1987;134:461–466. [Google Scholar]
- (47).Chesnut DB, Foley CK. J. Chem. Phys. 1986;84:852–861. [Google Scholar]
- (48).Schindler M. J. Am. Chem. Soc. 1987;109:5950–5955. [Google Scholar]
- (49).Schindler M, Kutzelnigg W. J. Am. Chem. Soc. 1983;105:1360–1370. [Google Scholar]
- (50).Chesnut DB, Moore KD. J. Comput. Chem. 1989;10:648–659. [Google Scholar]
- (51).Pearson JG, Le H, Sanders LK, Godbout N, Havlin RH, Oldfield E. J. Am. Chem. Soc. 1997;119:11941–11950. [Google Scholar]
- (52).Strohmeier M, Grant DM. J. Am. Chem. Soc. 2003;126:966–977. doi: 10.1021/ja037330e. [DOI] [PubMed] [Google Scholar]
- (53).Walling AE, Pargas RE, de Dios AC. J. Phys. Chem. A. 1997;101:7299–7303. [Google Scholar]
- (54).Scheurer C, Skrynnikov NR, Lienin SF, Straus SK, Bruüschweiler R, Ernst RR. J. Am. Chem. Soc. 1999;121:4242–4251. [Google Scholar]
- (55).http://feh.scs.uiuc.eduShift calculator can be obtained at http://feh.scs.uiuc.edu
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


