Abstract
The DEAD-box RNA helicase Xp54 is an integral component of the messenger ribonucleoprotein (mRNP) particles of Xenopus oocytes. In oocytes, several abundant proteins bind pre-mRNA transcripts to modulate nuclear export, RNA stability and translational fate. Of these, Xp54, the mRNA-masking protein FRGY2 and its activating protein kinase CK2α, bind to nascent transcripts on chromosome loops, whereas an Xp54-associated factor, RapA/B, binds to the mRNP complex in the cytoplasm. Over-expression, mutation and knockdown experiments indicate that Xp54 functions to change the conformation of mRNP complexes, displacing one subset of proteins to accommodate another. The sequence of Xp54 is highly conserved in a wide spectrum of organisms. Like Xp54, Drosophila Me31B and Caenorhabditis CGH-1 are required for proper meiotic development, apparently by regulating the translational activation of stored mRNPs and also for sorting certain mRNPs into germplasm-containing structures. Studies on yeast Dhh1 and mammalian rck/p54 have revealed a key role for these helicases in mRNA degradation and in earlier remodelling of mRNP for entry into translation, storage or decay pathways. The versatility of Xp54 and related helicases in modulating the metabolism of mRNAs at all stages of their lifetimes marks them out as key regulators of post-transcriptional gene expression.
INTRODUCTION
The first members of the DEAD-box RNA helicase subfamily, referred to here as ‘DDX6-like’, were discovered in diverse situations: the gene encoding Drosophila Me31B was found to be expressed in oocytes and nurse cells (1); the gene encoding a human orthologue rck/p54 occurred at a chromosomal breakpoint in the human cell line RC-K8, derived from a diffuse large B-cell lymphoma (2), to be later described as a putative proto-oncogene in both human (3) and mouse (4) cells; the gene encoding Schizosaccharomyces pombe Ste13 was cloned by functional complementation of the sterility mutant ste13 (5); and the gene encoding Saccharomyces cerevisiae Dhh1 was found to be required for sporulation (6). Studies on an orthologue expressed in Xenopus oocytes, Xp54, clearly identified this protein as an integral component of messenger ribonucleoprotein (mRNP) particles and as a factor involved in translational control (7). This provided a focus for the interpretation of earlier observations and subsequent studies have placed this subfamily of helicases as a key component in the metabolism of mRNA. Recent additions to the list of orthologues include CGH1, which is expressed in the germ cells of Caenorhabditis and modulates physiological germline apoptosis (8), and p47, which is a component of non-translating mRNP particles in oocytes and early embryos of the clam Spisula (9). The occurrence and main functions of the DDX6 members are summarized (Table 1).
Table 1.
Occurrence and proposed functions of DDX6-like RNA helicases
| Organism | Protein | Expression | Location | Function |
|---|---|---|---|---|
| Xenopus laevis | Xp54 | Oocytes and early embryos | Maternal mRNP particles | mRNP assembly |
| Translation regulation | ||||
| Balbiani body | Required for progression of meiosis | |||
| Mouse | p54 | Gametes and early embryos | Maternal mRNP particles | Translation regulation |
| Drosophila melanogaster | Me31B | Oocytes and early embryos | Maternal mRNP particles | Translation repression |
| Sponge body | ||||
| Caenorhabditis elegans | CGH-1 | Germ cells and early embryos | mRNP particles P granules | Modulation of physiological germ cell apoptosis |
| Spisula solidissima | p47 | Oocytes and early embryos | Maternal mRNP particles | Translation repression |
| S.pombe | Ste13p | Essential for progression of meiosis | ||
| S.cerevisiae | Dhh1p | mRNP particles | Required for sporulation | |
| P bodies | Translation repression | |||
| mRNA degradation | ||||
| G1/S checkpoint control | ||||
| Mammals | Rck/p54 | Somatic cells | P bodies | mRNA degradation |
| SGs | mRNP remodelling | |||
| Mammals | Rck/p54 | Over-expression in tumour cells | Cytoplasmic particles | Candidate proto-oncogene |
| Translation regulation |
See text for references.
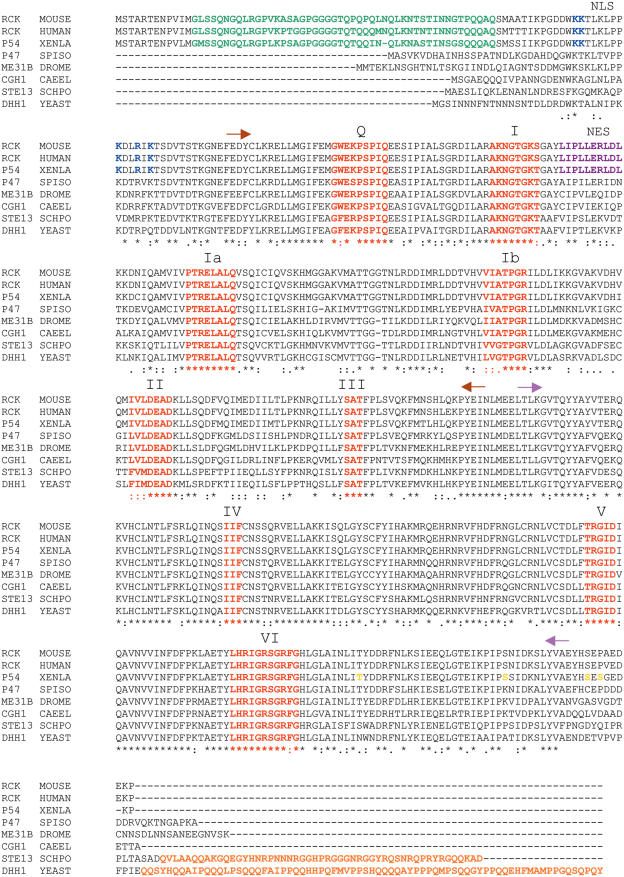
In addition to the high levels of protein sequence homology observed across a wide range of organisms, e.g. 72% identity between yeast Ste13 and Drosophila Me31B and 75% identity between Me31B and vertebrate p54 (Figure 1), equivalence of function has been demonstrated by complementation of mutant ste13 with ME31B cDNA (5) and mutant dhh1 with XP54 cDNA (10) and RCK/P54 cDNA (11).
Figure 1.
Alignment of the amino acid sequences of eight members of the DDX6 subfamily of DEAD-box RNA helicases. Identical residues (asterisk) and conserved substitutions (colon or stop) are indicated. Alignments were made using the Clustal W algorithm. Conserved motifs, shared by other DEAD-box helicases are highlighted (red). N-terminal extensions (green) or C-terminal extensions (orange) are present in only some members. In Xp54, single residues, S or T (yellow), are indicated as potential phosphorylation sites. Basic residues acting as an NLS are highlighted (blue) as are residues constituting a leucine-rich NES (purple). Extent of the two RecA-like domains are indicated by arrows: domain 1 (brown), domain 2 (pink).
In this review, we describe the structure of Xp54, its developmental expression and its presence and activity in mRNP particles and relate these results to recent studies on the other members of the DDX6-like subfamily.
STRUCTURE AND UNWINDING ACTIVITY
DDX6-like proteins may have unique structural features
In amino acid sequence and location of conserved DEAD-box motifs, DDX6 helicases are most closely related to the mRNA-binding translation factor eIF4A and the mRNA export factor Dbp5 [reviewed in Ref. (12)]. In this respect, they all contain sufficient sequence information for ATP binding and hydrolysis and RNA unwinding activity. The most significant differences within this whole group relate to extensions of the N- and C-termini of the DDX6-like members. Whereas the vertebrate proteins, Xenopus Xp54, human rck/p54 and mouse p54, have a long extension (∼70 residues) at the N-terminus and only a short extension at the C-terminus, the yeast proteins, Ste13p and Dhh1p, have a long extension (>70 residues) at the C-terminus and only a short extension at the N-terminus; the invertebrate proteins, Drosophila Me31B, Caenorhabditis CGH1 and Spisula p47, have only short extensions at each end (Figure 1). These asparagine/glutamine extensions may contribute to additional features of protein–protein interactions, although no specific interaction motifs have been identified.
Recent structural analysis has confirmed that, like other DEAD-box proteins, the core of yeast Dhh1p contains two RecA-like domains (13). However, unlike eIF4A, Dhh1p has a unique domain arrangement which permits novel interactions of motif V with motif I and the Q motif (14,15), thereby linking together the two domains and forming a prominent RNA-binding channel. Similar to eIF4A and other DEAD-box helicases, ATP binding enhances an RNA-induced conformational change, but, in addition, this change is disrupted by mutation of conserved motifs, especially those located at the interface between the two domains. The conformational switch, induced by ATP hydrolysis, is thought to be required for the function of Dhh1p and, presumably, of the other DDX6-like proteins.
DDX6-like proteins have in vitro RNA-unwinding activity
Both Xp54 and rck/p54 have been shown to have RNA-unwinding activity in vitro. Native Xp54, biochemically isolated from oocyte mRNP particles, has ATP-dependent RNA helicase activity (7). Duplexed sequences of both plasmid and maternal mRNA origin were unwound from 5′ single-strand ends, unwinding being specifically blocked by the addition of anti-Xp54 IgG (7). Electron-microscopical and surface plasmon resonance studies have shown that recombinant rck/p54 binds c-myc RNA transcripts in the presence of ATP with a Kd of 18 nM and unwinds intra-strand duplex structures (16). Deletion of the C-terminal 184 amino acid residues, essentially domain 2, from rck/p54 results in loss of unwinding activity. However, neither of these results excludes a requirement for partner proteins in regulating helicase activity in vivo. Furthermore, recent developments favour the view that RNA unwinding, per se, is not essential for modulation of RNA–protein interactions.
NUCLEAR ACTIVITIES: BINDING TO NASCENT TRANSCRIPTS AND MRNA EXPORT
Function of N-terminal and C-terminal ancillary motifs
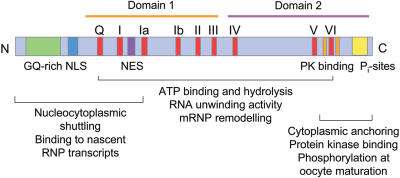
In addition to the conserved helicase motifs, Xp54 contains motifs at the N- and C-termini that are involved in additional functions (Figure 2). Both a lysine-rich nuclear localization signal (NLS) and a leucine-rich nuclear export signal (NES) are present in the N-terminal region, suggesting that Xp54 might be able to shuttle between nucleus and cytoplasm. Although a nuclear presence of native Xp54 is noticeable in oocyte nuclei only at early stages, when transcriptional activity is maximal, an N-terminal truncation of recombinant Xp54 fused to green fluorescent protein (NT-GFP) was shown to be capable of shuttling when expressed in later stage oocytes and in Xenopus or mammalian culture cells (17). The shuttling activity was shown to be sensitive to leptomycin B (LMB), a potent inhibitor of the CRM1 nuclear export pathway and was compromised by mutation of key leucine residues. In addition to its shuttling activity, this fusion protein had the ability to bind to the nascent RNP transcripts present on the loops of lampbrush chromosomes. However, the N-terminal region used contained only conserved motifs I and Q: since the proposed RNA-binding sequence of motif V was absent, it is possible that the observed binding is due to protein–protein interactions. Whatever the mode of interaction, once bound to transcripts, Xp54 is exported to the cytoplasm with mRNA by a CRM1-independent pathway (17) and remains associated with the RNA until translation activation. These observations on the behaviour of Xp54 are similar to those described for other DEAD–box helicases involved in nuclear export of mRNA, such as An3 (18) and Dbp5 (19).
Figure 2.
Diagram showing the location of sequence motifs and proposed functional regions within Xp54. Blocks indicate the conserved DEAD-box helicase motifs (red), the N-terminal region with a high content of glycine and glutamine residues (GQ-rich; green), the putative NLS (blue), the identified NES (purple) and putative acidophilic protein kinase (PK) sites (yellow). Functional analysis was carried out using vectors expressing truncations of recombinant Xp54 (17).
The N-terminal nucleocytoplasmic shuttling motifs characterized in Xp54 (17) are conserved only in vertebrates (Figure 1). This does not exclude the possibility that other members of the DDX6 subfamily also have a nuclear presence and have the potential to bind nascent RNA polymerase II transcripts. For instance, Spisula p47 is detected in the nuclei of early embryos (9). The helicase may be transported into the nucleus as part of a protein complex with appropriate import/export signals being present on another component. Xp54, itself, appears to be a component of a preformed protein complex which is active in binding nascent transcripts in Xenopus oocytes. Such RNA-free particles have been isolated from oocytes by rate-zonal and density centrifugation (20,21) and the kinetics and sites of Xp54 binding to nascent transcripts are coincident with those of the other protein components of the preformed particle (22,23). In S.cerevisiae, Dhh1p was detected as part of the Ccr4/Pop2/Not multiprotein complex that is implicated in transcription regulation (24,25). However, Ccr4p is a catalytic subunit of the yeast deadenylation system, which also contains Pop2p and Not proteins (26,27); therefore the possibility exists that the Ccr4/Pop2/Not complex, including Dhh1p, accompanies mRNA sequences from their initial transcription to their eventual degradation. A role for Dhh1p at the end of the lifetime of mRNA is confirmed by the findings that it interacts with the decapping factor Dcp1p to stimulate removal of the 5′ cap structure (28,29).
That other DDX6-like proteins are not normally detected in the nucleus is not unexpected because the C-terminal region of Xp54 over-expressed in mid-sized (stage IV) oocytes appears to act as a cytoplasmic anchor: replacement of the C-terminal 82 amino acid residues of Xp54 with an unrelated plasmid-encoded sequence permits nuclear uptake and rapid nucleocytoplasmic shuttling in both oocytes and transfected somatic cells (17). The deleted region contains potential CK2 phosphorylation sites; however, we have not detected any turnover of phosphates at these sites at times of mRNP assembly, although they do appear to be phosphorylated at times coincident with mRNA translation activation (J. Sommerville and D. A. Smillie, unpublished data). The molecular basis of the cytoplasmic retention of over-expressed Xp54 is not known.
Potential role of Xp54 in nuclear assembly of pre-mRNP particles
The most obvious nuclear presence of Xp54 is seen in early (stage I–III) oocytes when transcriptional activity is at its most intense and in early embryos when zygotic transcription is activated at mid-blastula: between these stages, there is an apparent nucleoplasmic absence of Xp54 (17). Over-expressed Xp54 is restricted to the cytoplasm of oocytes and culture cells and only enters the mRNP assembly pathway on hyperactivation of transcription, for instance by treatment of oocytes with trichostatin A (17), which has been shown to stimulate histone acetylation and global chromatin remodelling in oocytes (30). However, endogenous Xp54, together with the other major protein constitutents of mRNP storage particles, are always detected on the nascent transcripts of most chromosomal loops (22). Indeed, it is rare to detect any difference in loop proteins by immunofluorescence (31). This uniformity has prompted the suggestion that these nascent RNP transcripts carry all of the components required for their processing and determination of their translational fate, omnia mecum porto (32). Perhaps all mRNPs start, by default, in a pre-masked state; their translation, immediate or later, being determined by mRNA-specific reorganization. The association of Xp54 with RNA from the level of transcription, raises the question of an early requirement for RNA helicase activity: it is not known whether RNA unwinding is required to accommodate RNP proteins on nascent transcripts or whether it is incorporated as a passenger for later remodelling events. An early and extended requirement for DDX6-like helicases in mRNA metabolism is suggested by demonstration that deletion of yeast DHH1 is synthetically lethal with mutations in DBP5 and DED1, RNA helicases with roles in mRNA export and translation initiation (10).
Effects of over-expression, mutation and knockdown of Xp54 on mRNA export
The involvement of Xp54 in the nuclear export of mRNA can be assessed by interfering with Xp54 expression. By comparing the steady-state nuclear:cytoplasmic ratios of a range of mRNA species, an impression can be gained on the involvement of Xp54 in the export of individual mRNAs. Examples shown here (Table 2) indicate that the effects of perturbation of Xp54 levels are not simple. (i) Over-expression of Xp54 had little effect, except on those mRNAs in demand for immediate translation, such as mRNAs encoding ribosomal proteins (L1 and S1). (ii) Mutation of the DEAD motif to DQAD reversed this effect and inhibited export, not only of L1 and S1 mRNAs, but also of those stored mRNAs that are only translated at oocyte maturation (cyclin B1, cB1) or in embryos (FRGY1/YB1). (iii) Knockdown of Xp54 translation by prior injection of an antisense morpholino (MO) had a similar negative effect on some mRNAs (4). Although both translating and stored mRNAs can be affected, some mRNAs, namely those encoding translating histone H4 and stored linker histone B4, remain unaffected. The differences that can be detected between translating and stored mRNAs emphasize that, although Xp54 may bind to all classes of pre-mRNA and function in mRNA export, its presence results in mRNA-specific effects, perhaps by inducing mRNA sequence-dependent changes in the selection of bound proteins.
Table 2.
Effects on nuclear export and stability of various mRNA species in stage IV oocytes assayed after over-expression or knockdown of Xp54 or over-expression of a DEAD-box mutant
| mRNA | Translated | EFFECT ON mRNA EXPORT | EFFECT ON mRNA STABILITY | ||||
|---|---|---|---|---|---|---|---|
| Over-expression of | Injection of | Over-expression of | Injection of | ||||
| Xp54-wt | Xp54-DQAD | Antisense MO | Xp54-wt | Xp54-DQAD | Antisense MO | ||
| HistoneB4 | Embryo | 0 | 0 | 0 | 0 | 0 | 0 |
| FRGY1 | Embryo | 0 | − | − | + | − | − |
| Cyclin B1 | Maturation | 0 | − | − | + | − | 0 |
| HistoneH4 | Oocyte | 0 | 0 | 0 | − | 0 | + |
| rp L1 | Oocyte | + | − | 0 | 0 | + | + |
| rp S1 | Oocyte | + | − | − | 0 | + | + |
Nuclei and cytoplasms were isolated at 0 and 24 h after injection of wild-type (wt) or mutant (DQAD) expression vectors or of antisense or control morpholinos (MO). Steady-state levels of specific mRNAs were measured in all samples by semi-quantitative RT–PCR. Effects on mRNA export were calculated from changes in nuclear:cytoplasmic ratios and effects on mRNA stability were calculated from changes in total oocyte amounts. Changes are indicated as significant increase (+), significant decrease (−), insignificant change (0), where significance is set a >2-fold difference. Data were collected from three separate experiments.
REGULATION OF TRANSLATION
DDX6-like proteins interact with non-translating mRNA
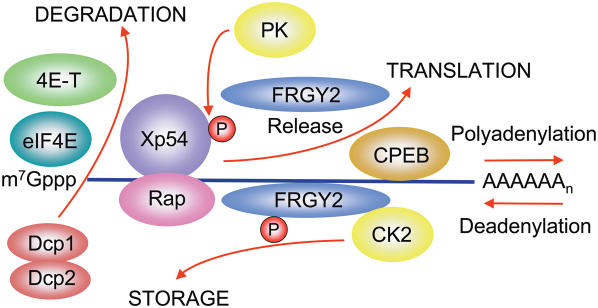
Oocytes from a wide variety of organisms are highly active in transcription and accumulate mRNA to be translated at various times during oogenesis, oocyte maturation and early embryogenesis. Similarly, spermatocytes accumulate mRNA species required for sperm maturation (33). At any one time in germ cell development only a small percentage of the available mRNA is present in polysomes. Thus most of the mRNA of germ cells exists in a translationally repressed state as ‘masked’ mRNP particles. Details of the masking mechanism may differ in different species: for instance, in Drosophila many mRNP proteins play roles in the transport and localization of different mRNA to specific regions of the oocyte [reviewed in Ref. (34)]. In Xenopus oocytes the DDX6 helicase Xp54 is an abundant protein found bound to non-translating mRNA together with other abundant proteins: the cold-shock domain (CSD) protein FRGY2, the α-subunit of the protein kinase CK2 and poly(A)-binding protein (Figure 3). A similar complement of proteins, including a p54 helicase, is reported for mRNP particles of mouse oocytes (35,36). CSD proteins bind single-stranded RNA by way of an OB-fold [a 5-β-barrel structure with an RNA-binding surface similar to that of the RNA-recognition motif (RRM) of other RNP proteins (23)], together with an ancillary C-terminal domain consisting of basic/aromatic ‘islands’ or RGG repeats (23). A common feature of non-translating mRNP particles in a wide range of organisms is the relative abundance of such CSD proteins in germ cells and early embryos: FRGY2 in Xenopus (23); MSY2 in mouse (33); Ypsilon Schachtel (yps) in Drosophila (37); and CEY-2,-3 and -4 in Caenorhabditis (38). In addition to their presence in germ cells and embryos, CSD proteins are also key constituents of mRNP particles of mammalian somatic cells [p50/YB-1; reviewed in Ref. (39)] and Chironomus salivary gland cells [p40/50; (40)]. In all situations there appears to be an intimate relationship between DEAD-box RNA helicases and CSD proteins, one possibility being that the helicase component is required to stabilize the interaction of the CSD protein with single-stranded RNA. Subsequent stabilization, throughout the period of mRNP storage in Xenopus oocytes, is maintained by continuous phosphorylation of FRGY2 by its associated CK2α subunit (20,23,41).
Figure 3.
Diagram indicating possible interactions of mRNA-bound and associated proteins involved with Xp54 helicase in the remodelling of mRNP for storage, translation and degradation. FRGY2 is the major mRNA masking protein that avidly binds single-stranded RNA. Stability of RNA binding is maintained by continuous phosphorylation (P red) of FRGY2 oligomers by CK2α, which is an integral component of stored mRNP particles. The RNA helicase Xp54 can be efficiently crosslinked to an abundant mRNP component Rap, which is an RNA-associated protein that may regulate helicase activity. Furthermore, translation repression may involve formation of a 3′–5′ molecular bridge between the protein that binds to the cytoplasmic polyadenylation element (CPEB), oligomerized Xp54 and the cap-binding protein eIF4E. The eIF4E regulatory protein 4E-T may be required to complete the bridge. In translation activation, the poly(A) tail is generally extended and most, but not all, of FRGY2 is released from the mRNA, apparently through interaction with the acidic chaperone, nucleoplasmin (51) and/or phosphorylation by the protein kinase Akt (52). In addition, the translation initiation factor eIF4G may displace the co-repressor 4E-T and form a bridge with poly(A)-binding protein bound to the extended tail (data not shown). Coincident with translation activation is phosphorylation of Xp54 (P red) by an unidentified protein kinase (PK). During mRNA degradation, studies on other systems (71) indicate that deadenylation is followed by decapping (Dcp1/2) and exonuclease digestion by Xrn1 (data not shown).
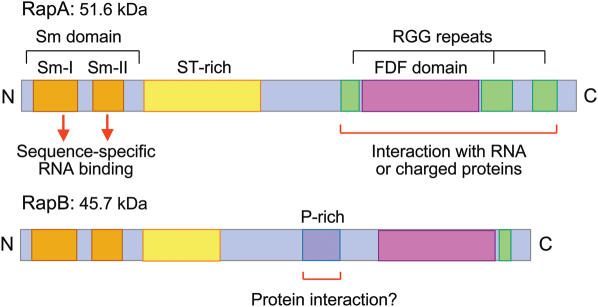
An additional component that may play a role in cytoplasmic stabilization of mRNP is RAP55, first described as an mRNA-associated protein found in oocytes of the amphibian Pleurodeles (42). Orthologues of RAP55 are also expressed in other developmental systems, RapA/RapB in Xenopus oocytes, Tral in Drosophila oocytes (38,43) and CAR-1 in Caenorhabditis (38,44,45). RAP55, RapA/B, Tral and CAR-1 form part of a highly conserved protein family, called Scd6p (46), which contains structural features probably involved in RNA- and protein-binding activities (Figure 4). At the N-terminus is an Sm-like domain forming a 5-β-fold that may form specific contacts with oligo(U) through conserved residues in the loops connecting beta strands (47). Towards the C-terminus of each protein is a novel FNF domain that is enriched in polar and charged residues forming a long alpha-helical structure with multiple exposed hydrophilic loops likely to interact with RNA or highly charged peptides (46). Also present is a variable number of RGG repeats that may contribute to further interactions. Like the Lsm1-7 proteins that are involved in mRNA degradation, Scd6 proteins can form multimers that may also adopt a toroidal structure (46). In each instance studied, the Scd6 protein has been shown to interact or co-localize with its corresponding DDX6-like protein: RapA/B with Xp54, Tral with Me31B (43) and CAR-1 with CGH-1 (38). Little is known about the functional consequences of these interactions although one possibility is that Scd6 proteins compete with Lsm1-7 proteins on the mRNA to prevent entry into the degradation pathway [processing bodies (P-bodies) and mRNA degradation]. However, recent studies indicate that Scd6 proteins are involved in processes beyond mRNA metabolism alone: in Caenorhabditis, CAR-1 is found to play an essential role in endoplasmic reticulum organization and late cytokinesis (44,45), while in Drosophila embryos, Tra1 is required for formation of normal endoplasmic reticulum exit sites and proper secretion of proteins involved in key developmental events (43). These connections, between mRNP complexes and membrane trafficking, may add further to the versatility in function of the conserved families of mRNP proteins. The different members of the conserved families of mRNA-associated proteins found in germ cells are listed in Table 3.
Table 3.
Identity of proteins associated with DDX6-like helicases in mRNP particles from oocytes of different organisms
| DDX6-like DEAD box protein | Y-box (CSD) protein | Scd6 (Sm domain) protein | Cytoplamic poly-adenylation element binding protein | |
|---|---|---|---|---|
| X.laevis | Xp54 | FRGY2a/b | RapA/B | CPEB |
| D.melanogaster | Me31B | Yps | Tra1 | Orb |
| C.elegans | CGH-1 | CEY-2/3/4 | CAR-1 | CPB-1/2/3/4 |
For references see text.
Involvement of DDX6-like proteins in translation control
FRGY2 and Xp54 are both present in non-translating mRNP particles as protein multimers (48,49) possibly arranged along much of the length of the mRNA as indicated by RNase protection and cross-linking studies (48). In addition to mRNA masking by FRGY2, stored mRNA generally has a short poly(A) tail [reviewed in Ref. (50)]. The switch from stored mRNP to recruitment into polysomes has been studied primarily in full-grown (stage VI) oocytes and has been shown to involve cytoplasmic extension of the poly(A) tail and removal of most of the FRGY2 from translationally activated mRNA (50,51). Factors that have been implicated in the destabilization of FRGY2 binding to mRNA include the acidic chaperone protein, nucleoplasmin (51) and a protein kinase that phosphorylates the C-terminal region of Xp54 (Figure 3). It has been shown recently that association of mammalian YB-1 with the 5′ cap structure of mRNA is regulated by phosphorylation by the S/T protein kinase Akt, leading to relief of translational repression of YB1-bound mRNAs (52). These results have relevance to the resumption of meiosis in oocytes because it was shown previously that Akt is essential for insulin-stimulated cell-cycle progression of Xenopus oocytes (53).
In addition to the major masking proteins, other factors regulating translation are located at the 5′ and 3′ ends of mRNA. At the 3′ end the protein CPEB is bound to the cytoplasmic polyadenylation element (CPE) and appears to have dual functions: in directing polyadenylation to activate translation and in interacting with maskin (a translation inhibitor expressed at late oogenesis) to repress translation [reviewed in Ref. (50)]. Both Spisula p47 and Xp54 have been shown to associate with CPEB, but not through direct binding: whereas p47 associates with CPEB through RNA binding (9), Xp54 associates with CPEB in the absence of RNA, probably via another protein. It has been shown that Xp54, tethered to the 3′-untranslated region (3′-UTR) of a poly(A)−reporter, results in a 3- to 5-fold suppression of translation (9). However, tethered DQAD and HRIGQ mutants actually stimulate translation, whereas both wild-type and mutated Xp54 tethered to a poly(A)+ reporter had no effect on translational efficiency (9). These results indicate that Xp54 activity is required for repression of mRNA with short poly(A) tails. The mechanism of repression appears to rely on RNA-dependent Xp54 oligomerization initiated either by tethering Xp54 to the 3′-UTR or by interaction of Xp54 with a CPEB-associated factor (49). Oligo-Xp54 can then interact with the cap-binding protein eIF4E, possibly through an inhibitory complex containing CPEB and co-repressor maskin, to repress translation initiation [reviewed in Ref. (50)]. However, maskin is expressed only late in oogenesis (stage VI) and an alternative bridge component would be required to repress translation at earlier times.
These observations have been extended in studies on mammalian cells which indicate that rck/p54 interacts with eIF4E and an associated factor, eIF4E-transporter (4E-T) which may substitute for eIF4G to form a translationally repressed mRNP (Figure 3), or replace eIF4G to direct the mRNP towards degradation (54,55). However, such a mechanism of translation repression and activation appears not to be universal. In yeast cells, Dhh1p physically interacts with Pat1p (28). Although both proteins carry out similar functions, they now appear to operate through separate mechanisms, which are not known in detail. Nevertheless, over-expression of Dhh1p has been shown to be sufficient to drive translational repression of mRNAs in yeast cells and both Dhh1 and its human orthologue, rck/p54, can exert this effect on translation in vitro (56). Furthermore, Dhh1p can work to repress translation independently of the presence of a 5′ cap structure on the mRNAs, being capable of repressing those mRNAs whose translation initiation is dependent on the presence of an IRES (56). Translation activation might then be expected to result from a knockdown of the DDX6-like component alone.
Global activation of translation is observed by injecting oocytes with antisense morpholinos that are complementary to the translation start site of XP54 mRNA. Compared with injected control morpholino, translation levels become noticeably greater as knockdown of available Xp54 protein takes effect. This activation of translation apparently applies to different classes of mRNA, for instance, those encoding ribosomal proteins and proteins bound to poly(A)+ RNA (J. Sommerville and A. Weston, unpublished data). However no common regulatory element has been identified in these two classes of mRNA. The extent to which increased synthesis results from release from translation repression, rather than stabilization of polysomal mRNA, remains to be determined.
A similar situation to that in Xenopus oocytes occurs in Drosophila oocytes. Here Me31B, the RNA-binding protein Tral, the CSD protein yps, the CPEB orthologue Orb and RNA localization factor Exuperantia (Exu) assemble with mRNAs to form translationally repressed mRNPs (37,38,57,58). Consistent with its proposed role, Me31B knockout results in premature translation of oskar and bicoid mRNAs in nurse cells, instead of later translation at their positional destinations in oocytes (58) (Figure 5).
Effects of over-expression, mutation and knockdown of Xp54 on mRNA stability
It has been assumed that individual maternal mRNAs are stable over the time range (several months) of oogenesis, whereas what are stable are steady-state amounts due to continuous transcriptional activity. Rates of turnover can be measured by inhibiting transcription or by interfering with the expression of potential stabilizing factors. By manipulating the level of available Xp54, effects on the steady-state levels of a variety of mRNAs can be assayed. As with mRNA export, these effects are not simple (Table 2). (i) Whereas over-expression of Xp54 had little effect on the stability of translationally active mRNAs encoding ribosomal proteins L1 and S1, stored mRNAs encoding cyclin B1 and FRGY1 appeared to be further stabilized. (ii) Over-expression of the DQAD mutant reversed cyclin B1 and FRGY1 stabilization but had diverse effects on translating mRNAs (cf. L1, S1 and H4). (iii) Knockdown of Xp54, by injection of an antisense morpholino (MO), stabilized mRNAs encoding L1, S1 and H4 and tended to destabilize mRNAs encoding cyclin B1 and FRGY1. In all treatments, the level of stored mRNA encoding the embryonic linker histone, B4, remained unchanged and served as an internal control in all assays. As with the assay of mRNA export efficiency, these results point to the importance of available Xp54 and indicate its diverse effects on different mRNAs. The molecular mechanisms involved in significant levels of mRNA decay in oocytes are not known.
Recent studies, on mouse gametes and embryos, have shown that over-expression of rck/p54, by injection of an expression plasmid into fertilized eggs, delays the progression of early embryogenesis and that generation of RCK knockout mice results in significant lethality of offspring, indicating that perturbation of rck/p54 production can have major effects on the development of mammals (36).
REMODELLING AND DEGRADATION OF mRNP
Localization and function of DDX6-like proteins in cytoplasmic bodies
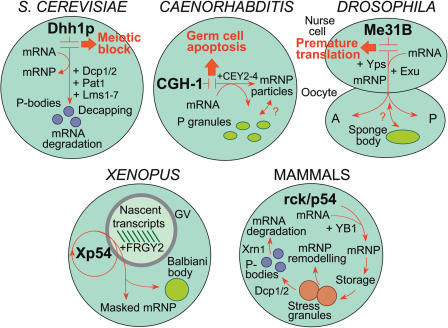
In addition to a diffuse cytoplasmic distribution reflecting free mRNP particles, DDX6-like proteins are found to be concentrated in distinct cytoplasmic foci. These foci take three forms: (i) structures containing germ plasm in oocytes of Caenorhabditis [P-granules; (8,38,59)], Drosophila [sponge bodies; (58)] and Xenopus [Balbiani body/germinal granules; (17)]; (ii) structures associated with mRNA degradation, the P-bodies of yeast (60–62) and mammalian cells (63–67); and (iii) the stress granules (SGs) of mammalian cells (66–68). The function of these structures in widely different organisms may be comparable (Figure 5).
Germplasm-containing structures and the distribution of mRNP particles
In the early (stage I) oocytes of Xenopus, high concentrations of mRNP granules are located, along with proliferating mitochondria, in a large cytoplasmic structure located on the vegetal side of the nucleus known as the Balbiani body or ‘mitochondrial cloud’. The mRNP component constitutes the germplasm, which is later redistributed as germinal granules to the vegetal cortex and appears eventually to be involved in the determination of embryonic germ cells [reviewed in Ref. (69,70)]. The Balbiani body contains not only a distinct subset of mRNAs, but also a high concentration of Xp54 (17). A similar situation exists in sponge bodies and polar granules of Drosophila oocytes, which contain a subset of mRNAs together with Me31B helicase (58) and in P granules of Caenorhabditis germ cells (8,38,59), which contain mRNAs that specify germline fates together with CGH-1 helicase. It is tempting to speculate that Balbiani body/cortical granules, sponge bodies/polar granules and P granules are functionally equivalent. However, sponge bodies appear to be involved in the distribution of mRNP particles to different regions of the oocyte (58); and P granules are postulated to be involved in the stabilization of newly synthesized mRNAs by assembly with CGH-1 and associated proteins (8,38,59). In early (syncytial) stages of oogenesis, P granules have a perinuclear location in association with nuclear pores and, after entry into meiosis, newly synthesized mRNAs appears to pass through the P granules to accumulate as cytoplasmic mRNP particles containing CGH-1 and its associated RNA-binding protein CAR-1 (38). However, in the cgh-1 deletion mutant, CAR-1 is lost from P granules in meiotic cells, to accumulate as cytoplasmic aggregates (38). These results further support the importance of a functional association between CGH-1 and CAR-1 and indicate a role for CGH-1 in mRNP assembly. Therefore, in addition to sequestration of germline mRNPs, the Balbiani body may also have a more general early role in the assembly and distribution of mRNP particles. The Xp54/Me31B/CGH-1 proteins appear not to be essential for the formation or maintenance of any of these structures; rather they act as markers for mRNP complexes that reside within them or pass through them to be located elsewhere (Figure 5).
Processing bodies and mRNA degradation
The yeast helicase Dhh1p and the mammalian helicase p54/rck have been identified recently as key components in mRNA degradation. The major pathway of mRNA turnover is initiated with shortening of the 3′ poly(A) tail (deadenylation), followed by removal of the 5′-cap structure by the Dcp1p/Dcp2p decapping complex and 5′–3′ exonucleolytic decay (71). It has been shown that Dhh1p interacts physically with several proteins involved in this pathway: with Pop2p, a subunit of the mRNA deadenylation complex; with the decapping enzyme Dcp1p; and with Lsm1p and Pat1p, which are activators of decapping [(28,29); Figure 5]. Since Dhh1 increases the efficiency of mRNA decapping, one possible mechanism is that it remodels mRNP particles by destabilizing the translation initiation complex following deadenylation and facilitates access to the cap structure for Dcp1p/Dcp2p (28,29). An intriguing possibility is that some mRNP particles, instead of being fed into the decay pathway, are remodelled for re-entry to translation following storage, thus unifying the decay/storage functions of Dhh1p and Xp54 (29). It has been shown that mRNA decapping and 5′–3′ degradation, together with the protein components involved, locate to distinct cytoplasmic foci called P-bodies (60,61). Recent studies have shown that Dhh1 works redundantly with Pat1p to facilitate the formation of P-bodies, deletion of DHH1 in combination with PAT1 resulting in a significant decrease in the number of P-bodies (56). Furthermore, it has been confirmed that P-bodies are dynamic structures requiring not only mRNA for their formation (61), but also permitting re-entry of stored mRNA into the translation pathway (62). Since mRNA can be temporarily held in P-bodies in association with proteins related to those found in maternal mRNPs, mechanisms for developmental storage of non-translating mRNA may have been adapted from P-body function (62).
The P-bodies of mammalian cells appear to be similar to their yeast counterparts: in human cells, there is co-localization of degradation factors Dcp1/2, Ccr4, Lsm1-7 and Xrn1, together with rck/p54, the mammalian orthologue of RAP55 and accumulated poly(A)+ RNA (54,63–68). Recently it has been shown that rck/p54 is required for the accumulation of the mRNA degradation factors in P-bodies, indicating an additional, early role for the helicase in the decay pathway, perhaps in accommodating the degradation factors as part of larger mRNP complexes essential for P-body formation (54,55). Identification and analysis of additional components of P-bodies, namely eIF4E and its interaction partner 4E-T (but not translation initiation factors such as eIF4G), indicate key roles for these proteins in the remodelling of mRNPs for their targeting to P-bodies (54,55). An early stage in the transition of actively translating mRNAs to mRNAs destined for degradation in mammalian cells might involve the interaction of rck/p54 with eIF4E, perhaps through a 4E-T bridge, thus replacing the PABP/eIF4G/eIF4E bridge observed in translation complexes (54,55).
Stress granules and mRNP remodelling
Mammalian p54/rck and RAP55 are also present in SGs, which are temporary structures that accumulate in response to environmental stress. Basically, SGs are aggregates of stalled translational preinitiation complexes, assembled in response to eIF2α phosphorylation. (Phosphorylated eIF2α reduces the availability of the eIF–GTP–tRNA ternary complex that is required to initiate translation.) SGs therefore contain a complement of proteins different from P bodies: although both contain, for instance, p54/rck, RAP55, Xrn1, eIF4E and CPEB, SGs lack Dcp1/2, but contain eIF4G and PABP, which are absent in P bodies (63–65). As a consequence, SGs are not just involved in mRNA decay—they appear to be capable of remodelling mRNP for entry into alternative pathways: for reinitiation of translation; for translation repression and storage; or for transit to P-bodies (67) (Figure 5). Therefore, SGs may represent mRNP remodelling centres par excellence, bringing together all of the components required to determine the full range of mRNP transitions. It will be interesting to know to what extent rck/p54 is required in determining each of the transitions and to what extent these mechanism pertain to other systems. A recent study of mammalian RAP55 has shown that the C-terminal half, consisting of the two RGG domains and the FDF domain (Figure 4), is necessary and sufficient to target the protein to P-bodies, whereas only the second RGG domain is necessary and sufficient to target the protein to SGs (68). It is suggested that RAP55 may act as a shuttling protein, to target damaged mRNA present in SGs to P-bodies for degradation (68).
Figure 4.
Diagram showing the location of sequence motifs and proposed functional regions within Xenopus RapA and RapB which are members of the Scd6p family. Blocks indicate the conserved Sm-I and Sm-II motifs (orange) that constitute the Sm-like domain, the conserved FDF domain (pink) and the more variable blocks of RGG repeats (green), serine/threonine-rich regions (yellow) and proline-rich regions (violet). RAPA and RAPB genes are differentially expressed during oogenesis: RAPA during early oogenesis when mRNP synthesis is maximum, RAPB throughout oogenesis and into early embryogenesis when maternal mRNAs are being translated and degraded.
The mechanism of RNP remodelling by RNA helicases has received re-evaluation after the discovery that some DexH/D proteins do not need to unwind duplexed RNA in order to carry out their function of protein displacement. For instance, two quite distinct ‘RNA helicases’, NPH-II and DED1 have been shown to be capable of rearranging RNA–protein complexes independent of duplex unwinding (72,73). In essence, energy derived from RNA-stimulated ATP hydrolysis can be used to remodel RNP complexes by a mechanism that does not depend on changing the secondary structure of the RNA. In vivo specificity of a remodelling event could be derived from the immediate protein composition of the mRNP assembly. This ‘RNPase’ concept (72) has received widespread acceptance (72–76) and is in accord with most of the results discussed here on the DDX6-like subfamily.
EXPRESSION OF RCK/P54 IN TUMOUR CELLS
The human DDX6 gene, RCK, is the target of a chromosomal breakpoint translocation occurring in patients with B-cell lymphomas. The gene product, rck/p54, was found to be over-expressed in several malignant cell types, indicating that RCK is a candidate proto-oncogene (2,3,77–79). Over-expression of rck/p54 in colon tumour cells is frequently accompanied by over-expression of c-Myc, an observation that has led to the suggestion that rck/p54 contributes to the stabilization and increased translational efficiency of c-myc mRNA (77,78). That rck/p54 can directly affect c-myc mRNA has been indicated by in vitro-binding studies, which show that not only does rck/p54 protein bind to in vitro-transcribed c-myc RNA but it also converts a folded RNA structure to a linear one, but only in the presence of ATP (16). These results are suggestive of an RNA-unwinding activity that converts intrastrand duplexes to single-stranded RNA. However, this activity is not substrate-specific: in vitro-transcribed RCK RNA itself, is transformed in the same manner (16). Therefore, the proposed mode of action of rck/p54 complies with the consensus that all DDX6-like proteins act by regulating the metabolism of mRNA molecules. Where analogy with Xp54 and Me31B apparently breaks down, is that in oocytes these proteins are characterized as agents of translation repression (9,49,58), whereas in tumour cells rck/p54 apparently activates translation of factors associated with carcinogenesis. This difference can be resolved by postulating that a remodelling activity of DDX6-like helicases merely presents an opportunity to change the translational status of any particular mRNA.
A ROLE FOR DDX6-LIKE PROTEINS IN CELL-CYCLE REGULATION
In conditions unfavourable for cell division, eukaryotic cells undergo transient arrest at the G1-to-S phase transition. Initiation of arrest is known as the checkpoint response. Studies on G1/S-arrested yeast cells, following induced DNA damage or ectopic expression of the human tumour suppressor gene BRCA1, have shown that deleterious effects are suppressed by deletion of DHH1, implicating Dhh1p as a regulator at the G1/S transition, at which BRCA1 may have a checkpoint role in human cells (11). It will be interesting to know if the human orthologue, DDX6, interacts with BRCA1 in human cells and whether it is a potential breast cancer gene target. Further studies on DNA damage-induced arrest have identified Dhh1p as an essential component in release from checkpoint arrest (80). With localization of Dhh1 in mRNA-containing complexes (28,29) and biochemical isolation in mRNP particles (10), G1/S checkpoint recovery is likely to result from regulatory mechanisms acting at the level of mRNA stability and translatability. It is suggested that efficient recovery requires Dhh1p either to promote decay or to change the translational status of a subset of mRNAs (80). However, as with the discussion on rck/p54, Dhh1p need not only be associated with a specific subset of mRNA targets, the mRNPs involved in cell-cycle progression may simply be those that are remodelled in response to a particular stimulus.
PERSPECTIVE
Specificity versus ubiquity
The question remains as to the restriction of binding of DDX6-like proteins to a particular subset of mRNAs. If this were so, different subsets would be bound in different cell types, e.g. mRNAs encoding specialized meiotic functions in germ cells and mRNAs encoding proteins required for cell proliferation in tumour cells. So far there is little evidence of selective binding to particular mRNAs. It now seems unlikely that the DDX6-like proteins have a narrow range of specific mRNA targets and more likely that they bind any available mRNA molecules in a sequence-independent fashion. This does not mean that every mRNP would, at any one time, have to contain DDX6-like protein: the dynamic nature of mRNP composition and function would allow for temporary association of the helicase, perhaps by interacting with a sequence-specific-binding protein.
Coordination of mRNP metabolism
A case against temporary association of DDX6-like proteins with mRNAs can be argued from the detection of these proteins at practically every stage of life of the mRNAs. Other DEAD-box proteins associate with mRNA at specific stages—at splicing, or nuclear export or translation and tend not to persist beyond the site of their specific function: yet DXX6-like proteins are detected on mRNA throughout its lifetime, from transcription to degradation. Whether this represents a stable association or periodic rebinding is not known (given the dynamic nature of protein interaction with mRNA this distinction may be difficult to define). Nevertheless persistence of association with mRNA would give the helicase the opportunity of co-ordinating the activities of a particular mRNA through its metabolic transformations.
Molecular interactions
DDX6-like proteins can apparently interact with a range of different protein partners in a variety of cellular situations. Confirmation of the molecular nature of these interactions, particularly by identification of protein-binding sites, is required. For instance, several types of genetic interaction have been identified between DHH1 and other yeast genes or ectopically expressed genes: establishing corresponding molecular connections would greatly advance our understanding of the possible multiple outcomes of Dhh1p activity. Elucidating the molecular structure of multiprotein complexes involving DDX6-like proteins will be a major task, especially if their protein composition is labile.
Mode of action
The apparent dynamic transformation of structure and composition of mRNA particles calls into question the exact role of DDX6-like proteins in such transformations. Although double-strand unwinding activity can be demonstrated in vitro, it is not known how this activity relates to situations in vivo. The currently accepted view is that RNA helicases may operate to alter mRNP assemblies by their ‘remodelling’ (72–76). Whether remodelling takes the form of local RNA unwinding, or winding to create double-stranded RNA structures, or simply by stimulating exchange of protein-binding partners, remains largely unknown. Apart from using RNA-stimulated ATP hydrolysis as a motor for protein displacement, other influences, such as protein phosphorylation and competition for mRNP-binding sites, would determine the outcome of remodelling. Certainly, the varied changes that occur in mRNP complexes throughout their lifetimes mark out the DDX6-like proteins as classic remodelling factors.
Consequences of activity
An extension of the remodelling concept is that the outcome of DDX6-like helicase activity need not be uniform. For instance, association of the helicase with a group of mRNAs need not mark them all out for translation repression: some may be repressed, others may be activated. Nevertheless, the evidence so far available from studies on translation control during oogenesis and early development indicates that depletion of the DDX6-like protein results in widespread derepression of protein synthesis in oocytes.
Formation of mRNP assemblies
Noticeable cellular structures are generated by the amounts of mRNP being processed. Just as lampbrush chromosome loops impress by the density of nascent RNP transcripts present and masses of maternal mRNP can form large storage complexes in germ cells, P-bodies reflect a relatively high-throughput of mRNP for degradation and SGs form from aggregates of mRNPs and associated translation components in conditions that require a large-scale reassignment of mRNA metabolism. It is impressive that the various members of the DDX6 helicase subfamily appear to be major components in all of these structures and may even be key regulators of their formation. Perhaps equivalent structures, on vastly different scale, exist in all cell types to cover the diversity of mRNP transitions.
Figure 5.
Major pathways involving DDX6-like helicases in the metabolism of mRNA of different organisms. Different aspects of helicase activity are emphasized in different organisms. In S.cerevisiae, best studied is the decay pathway in which Dhh1p has been shown to associate both with the mRNA deadenylation complex containing Ccr4p and Pop2p and the mRNA decapping complex containing Dcp1/2p, Pat1p and Lsm1-7p. Decapping activity is located in P-bodies. Deletion of DHH1 results in a block in meiosis. In Caenorhabditis, CGH-1 locates to cytoplasmic mRNP particles and to P granules. Whereas CGH-1 particles are dynamic structures responsive to meiotic development, P granules may represent sites of mRNP assembly or remodelling. CEY2-4 are CSD proteins associated with mRNA. Deletion of CGH-1 results in physiological germ cell apoptosis. In Drosophila, Me31B binds to maternal mRNAs synthesized in nurse cells. Including the CSD protein Yps and the mRNA-localization factor Exu, mRNP is exported to the oocyte where localization between anterior (A) and posterior (P) regions may occur. In addition, Me31B locates to sponge bodies, which may represent the site of germplasm. Deletion of ME31B results in premature translation of mRNAs, normally located to and stored in the oocyte, in nurse cells. In Xenopus, Xp54 can shuttle between cytoplasm and nucleus (GV) but, after binding to nascent pre-mRNA transcripts together with the CSD protein FRGY2, exits the nucleus in mRNP particles, most of these being stored in a non-translating (maternal) form. In early oocytes, Xp54 also locates to the Balbiani body that contains the germplasm. In mammalian cells, rck/p54 is bound to mRNA together with the CSD protein YB1/p50. Some mRNP may be held in a non-translating form. rck/p54 is also a component of SGs and is implicated in the remodelling of mRNP for translation, storage or degradation in P-bodies. mRNP marked for degradation is associated with the decapping enzymes Dcp1/2 and the mRNA may be eventually degraded by the 5′ exonuclease Xrn1. For references see text.
Acknowledgments
We thank Keith Blackwell, Jeff Coller, Mike Ladomery, Nancy Standart and Karsten Weis for helpful comments; The Wellcome Trust for financial support and the Biology and Biotechnology Science Research Council of Great Britain for a studentship award to A.W. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.de Valoir T., Tucker M.A., Belikoff E.J., Camp L.A., Bolduc C., Beckingham K. A second maternally-expressed Drosophila gene encodes a putative RNA helicase of the DEAD-box family. Proc. Natl Acad. Sci. USA. 1991;88:2113–2117. doi: 10.1073/pnas.88.6.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu D., Yunis J.J. Cloning, expression and localization of an RNA helicase gene from a human lymphoid cell line with chromosomal breakpoint 11q23.3. Nucleic Acids Res. 1992;20:1967–1972. doi: 10.1093/nar/20.8.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akao Y., Marukawa O., Morikawa H., Nakao K., Kamei M., Hachiya T., Tsujimoto Y. The rck/p54 candidate proto-oncogene product is a 54-kD DEAD box protein differentially expressed in human and mouse tissues. Cancer Res. 1995;55:3444–3449. [PubMed] [Google Scholar]
- 4.Seto M., Yamamoto K., Takahashi T., Ueda R. Cloning and expression of a murine cDNA homologous to the human RCK/P54, a lymphoma linked chromosomal translocation junction gene on 11q23. Gene. 1995;166:293–296. doi: 10.1016/0378-1119(95)00559-5. [DOI] [PubMed] [Google Scholar]
- 5.Maekawa H., Nakagawa T., Uno Y., Kitamura K., Shimoda C. The ste13+ gene encoding a putative RNA helicase is essential for nitrogen starvation-induced G1 arrest and initiation of sexual development in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 1994;244:456–464. doi: 10.1007/BF00583896. [DOI] [PubMed] [Google Scholar]
- 6.Strahl-Bolsinger S., Tanner W. A yeast gene encoding a putative RNA helicase of the ‘DEAD’-box family. Yeast. 1993;9:429–432. doi: 10.1002/yea.320090414. [DOI] [PubMed] [Google Scholar]
- 7.Ladomery M., Wade E., Sommerville J. Xp54, the Xenopus homologue of human RNA helicase p54, is an integral component of stored mRNP particles in oocytes. Nucleic Acids Res. 1997;25:965–973. doi: 10.1093/nar/25.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Navarro R.E., Shim E.Y., Kohara Y., Singson A., Blackwell T.K. Cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C.elegans. Development. 2001;128:3221–3232. doi: 10.1242/dev.128.17.3221. [DOI] [PubMed] [Google Scholar]
- 9.Minshall N., Thom G., Standart N. A conserved role of a DEAD-box helicase in mRNA masking. RNA. 2001;7:1728–1742. doi: 10.1017/s135583820101158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tseng-Rogenski S., Chong J.L., Thomas C.B., Enomoto S., Berman J., Chang T.H. Functional conservation of Dhh1p, a cytoplasmic DExD/H-box protein present in large complexes. Nucleic Acids Res. 2003;31:4995–5002. doi: 10.1093/nar/gkg712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westmoreland T.J., Olson J.A., Saito W.Y., Huper G., Marks J.R., Bennet C.B. DHH1 regulates the G1/S-checkpoint following DNA damage or BRCA1 expression in yeast. J. Surg. Res. 2003;113:62–73. doi: 10.1016/s0022-4804(03)00155-0. [DOI] [PubMed] [Google Scholar]
- 12.de la Cruz J., Kressler D., Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- 13.Cheng Z., Coller J., Parker R., Haiwei S. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA. 2005;11:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanner N.K., Cordin O., Banroques J., Doere M., Linder P. The Q motif: a newly identified motif in DEAD-box helicases may regulate ATP binding and hydrolysis. Mol. Cell. 2003;8:251–262. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 15.Cordin O., Tanner N.K., Doere M., Linder P., Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akao Y., Yoshida H., Matsumoto K., Matsui T., Hogetu K., Tanaka N., Usukura J. The tumour-associated DEAD-box protein, rck/p54 exhibits RNA unwinding activity toward c-myc RNAs in vitro. Genes Cells. 2003;8:671–6. doi: 10.1046/j.1365-2443.2003.00665.x. [DOI] [PubMed] [Google Scholar]
- 17.Smillie D.A., Sommerville J. RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles. J. Cell Sci. 2002;115:395–407. doi: 10.1242/jcs.115.2.395. [DOI] [PubMed] [Google Scholar]
- 18.Askjaer P., Rosendahl R., Kjems J. Nuclear export of the DEAD-box An3 protein by CRM1 is coupled to An3 helicase activity. J. Biol. Chem. 2000;275:11561–11568. doi: 10.1074/jbc.275.16.11561. [DOI] [PubMed] [Google Scholar]
- 19.Hodge C.A., Colot H.V., Stafford P., Cole C.N. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 2005;18:5778–5788. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cummings A., Sommerville J. Protein kinase activity associated with stored messenger ribonucleoprotein particles of Xenopus oocytes. J. Cell Biol. 1988;107:45–56. doi: 10.1083/jcb.107.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yurkova M.S., Murray M.T. A translation regulatory particle containing the Xenopus oocyte Y-box protein mRNP3+4. J. Biol. Chem. 1997;272:10870–10876. doi: 10.1074/jbc.272.16.10870. [DOI] [PubMed] [Google Scholar]
- 22.Sommerville J., Ladomery M. Transcription and masking of mRNA in germ cells: involvement of Y-box proteins. Chromosoma. 1996;104:469–478. doi: 10.1007/BF00352111. [DOI] [PubMed] [Google Scholar]
- 23.Sommerville J. Activities of cold-shock domain proteins in translation control. Bioessays. 1999;21:281–297. doi: 10.1002/(SICI)1521-1878(199904)21:4<319::AID-BIES8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 24.Hata H., Mitsui H., Liu H., Bai Y., Denis C.L., Shimizu Y., Sakai A. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics. 1998;148:571–579. doi: 10.1093/genetics/148.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maillet L., Collart M.A. Interaction between Not1p, a component of the Ccr4-Not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J. Biol. Chem. 2002;277:2835–2842. doi: 10.1074/jbc.M107979200. [DOI] [PubMed] [Google Scholar]
- 26.Tucker M., Valencia-Sanchez M.A., Staples R.R., Chen J., Denis C.L., Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 27.Tucker M., Staples R.R., Valencia-Sanchez M.A., Muhlrad D., Parker R. Ccr4 is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;20:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coller J.M., Tucker M., Sheth U., Valencia-Sanchez M.A., Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer N., Weis K. The DEAD-box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smillie D.A., Llinas A.J., Ryan J.T.P., Kemp G.D., Sommerville J. Nuclear import of histone deacetylase in Xenopus oocytes is regulated by phosphorylation. J. Cell Sci. 2004;117:1857–1866. doi: 10.1242/jcs.01008. [DOI] [PubMed] [Google Scholar]
- 31.Sommerville J. Immunolocalization and structural organization of nascent RNP. In: Busch H., editor. The Cell Nucleus. , Vol. 8. NY: Academic Press; 1981. pp. 1–57. [Google Scholar]
- 32.Spirin A.S. Storage of messenger RNA in eukaryotes: envelopment with proteins, translational barrier at 5′ side, or conformational masking by 3′side? Mol. Reprod. Dev. 1994;38:107–117. doi: 10.1002/mrd.1080380117. [DOI] [PubMed] [Google Scholar]
- 33.Yang J., Medvedev S., Reddi P.P., Schultz R.M., Hecht N.B. The DNA/RNA-binding protein MSY2 marks specific transcripts for cytoplasmic storage in mouse male germ cells. Proc. Natl Acad. Sci. USA. 2005;102:1513–8. doi: 10.1073/pnas.0404685102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.St.Johnston D. Moving messages: the intracellular localization of mRNAs. Nature Rev. Mol. Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- 35.Paynton B.V. RNA-binding proteins in mouse oocytes and embryos: expression of genes encoding Y-box, DEAD-box RNA helicases and poly(A)-binding proteins. Dev.Genet. 1998;23:285–298. doi: 10.1002/(SICI)1520-6408(1998)23:4<285::AID-DVG4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto K., Kwon O.Y., Kim H., Akao Y. Expression of rck/p54, a DEAD-box RNA helicase, in gametogenesis and early embryogenesis of mice. Dev Dyn. 2005;233:1149–1156. doi: 10.1002/dvdy.20429. [DOI] [PubMed] [Google Scholar]
- 37.Mansfield J.H., Wilhelm J.E., Hazelrigg T. Ypsilon schachtel, a Drosophila Y-box protein, acts antagonistically to Orb in the oskar mRNA localization and translation pathway. Development. 2002;128:197–209. doi: 10.1242/dev.129.1.197. [DOI] [PubMed] [Google Scholar]
- 38.Boag P.R., Nakamura A., Blackwell T.K. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C.elegans. Development. 2005;132:4975–4986. doi: 10.1242/dev.02060. [DOI] [PubMed] [Google Scholar]
- 39.Evdokimova V.M., Ovchinnikov L. Translational regulation by Y-box transcription factor: involvement of the major mRNA-associated protein p50. Int. J. Biochem. Cell Biol. 1999;31:139–149. doi: 10.1016/s1357-2725(98)00137-x. [DOI] [PubMed] [Google Scholar]
- 40.Soop T., Naschechekin D., Zhao J., Sun X., Alzhanova-Ericsson A.T., Bjorkoth B., Ovchinnikov L., Daneholt B. A p50-like Y-box protein with a putative translational role becomes associated with pre-mRNA concomitant with transcription. J. Cell Sci. 2003;116:1493–1503. doi: 10.1242/jcs.00353. [DOI] [PubMed] [Google Scholar]
- 41.Braddock M., Muckenthaler M., White M.R.H., Thorburn A.M., Sommerville J., Kingsman A.J., Kingsman S.M. Intron-less RNA injected into the nucleus of Xenopus oocytes accesses a regulated translation control pathway. Nucleic Acids Res. 1994;22:5255–5264. doi: 10.1093/nar/22.24.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieb B., Carl M., Hock R., Gebauer D., Scheer U. Identification of a novel mRNA-associated protein in oocytes of Pleurodeles waltl and Xenopus laevis. Exp. Cell Res. 1998;245:272–281. doi: 10.1006/excr.1998.4249. [DOI] [PubMed] [Google Scholar]
- 43.Wilhelm J.E., Buszcak M., Sayles S. Efficient protein trafficking requires Trailer Hitch, a component of a ribonucleoprotein complex located to the ER in Drosophila. Dev. Cell. 2005;5:675–685. doi: 10.1016/j.devcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 44.Audhya A., Hyndman F., McLeod I.X., Maddox A.S., Yates J.R., III, Desai A., Oegema K. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J. Cell Biol. 2005;171:267–79. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Squirrell J.M., Eggers Z.T., Luedke N., Saari B., Grimson A., Lyons G.E., Anderson P., White J.G. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER Organization in C.elegans embryos. Mol. Biol. Cell. 2005;17:336–344. doi: 10.1091/mbc.E05-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anantharaman V., Aravind L. Novel conserved domains in proteins with predicted roles in eukaryotic cell-cycle regulation, decapping and RNA stability. BMC Genomics. 2004;5:45. doi: 10.1186/1471-2164-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Achsel T., Stark H., Luhrmann R. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl Acad. Sci. USA. 2001;98:3685–3689. doi: 10.1073/pnas.071033998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marello K., LaRovere J., Sommerville J. Binding of Xenopus oocyte masking proteins to mRNA sequences. Nucleic Acids Res. 1992;20:5593–5600. doi: 10.1093/nar/20.21.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minshall N., Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 2004;32:1325–1334. doi: 10.1093/nar/gkh303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkie G., Dickson K.S., Gray N.K. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 51.Meric F., Matsumoto K., Wolffe A.P. Regulated unmasking of in vivo synthesized maternal mRNA at oocyte maturation: a role for the chaperone nucleoplasmin. J. Biol. Chem. 1997;272:12840–12846. doi: 10.1074/jbc.272.19.12840. [DOI] [PubMed] [Google Scholar]
- 52.Evdokimova V., Ruzanov P., Anglesio M.S., Sorokin A.V., Ovchinnikov L.P., Buckley J., Triche T.J., Sonenberg N., Sorenson P.H.B. Akt-mediated YB-1 phosphorylation activates translation of silent mRNA species. Mol. Cell. Biol. 2005;26:277–292. doi: 10.1128/MCB.26.1.277-292.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andersen C.B., Sakaue H., Nedachi T., Kovacina K.S., Clayberger C., Conti M., Roth R.A. Protien kinase B/Akt is essential for the insulin—but not progesterone—stimulated resumption of meiosis in Xenopus oocytes. Biochem. J. 2003;369:227–238. doi: 10.1042/BJ20021243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrei M.A., Ingelfinger D., Heintzmann R., Achsel T., Rivera-Pomar R., Luhrmann R. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–27. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferraiuolo M.A., Dostie J., Murray E.L., Schoenberg D.R., Sonenberg N. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 2005;170:913–24. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coller J., Parker R. General translation repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilhelm J.E., Mansfield J., Hom-Booher N., Wang S., Turck C.W., Hazelrigg T., Vale R.D. Isolation of a ribonucleoprotein complex involved in mRNA localization in Drosophila oocytes. J. Cell Biol. 2000;148:427–440. doi: 10.1083/jcb.148.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakamura A., Amikura R., Hanyu K., Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complexes during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- 59.Navarro R.E., Blackwell T.K. Requirement for P granules and meiosis for accumulation of the germline RNA helicase CGH-1. Genesis. 2005;42:172–180. doi: 10.1002/gene.20136. [DOI] [PubMed] [Google Scholar]
- 60.Sheth U., Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teixeira D., Sheth U., Valencia-Sanchez M.A., Brengues M., Parker R. Processing bodies require RNA for assembly and contain non-translating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brengues M., Teixeira D., Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ingelfinger D., Arndt-Jovin D.J., Luhrmann R., Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dep1/2 and Xrn1 in distinct cytoplasmic foci. RNA. 2001;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 64.Eystathioy T., Jakymiw A., Chan E.K.L., Seraphin B., Couget N., Fritzler M.J. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLms4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cougot N., Babajko S., Seraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilczynska A., Aigueperse C., Kress M., Dautry F., Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J. Cell Sci. 2004;118:981–992. doi: 10.1242/jcs.01692. [DOI] [PubMed] [Google Scholar]
- 67.Kedersha N., Stoeklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fitzler M., Scheuner D., Kaufman R.J., Golan D.E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodelling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang W.H., Yu J.H., Gulick T., Bloch K.D., Bloch D.B. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA. 2006:1–8. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Houston D.W., King M.L. Germ plasm and molecular determinants of germ cell fate. Curr. Top. Dev. Biol. 2000;50:155–181. doi: 10.1016/s0070-2153(00)50008-8. [DOI] [PubMed] [Google Scholar]
- 70.Kloc M., Etkin L.D. RNA localization mechanisms in oocytes. J. Cell Sci. 2005;118:269–82. doi: 10.1242/jcs.01637. [DOI] [PubMed] [Google Scholar]
- 71.Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nature Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 72.Jankowsky E., Gros C.H., Shuman S., Pyle A.M. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 73.Fairman M.E., Maroney P.A., Wang W., Bowers H.A., Gollnick P., Nilsen T.W., Jankowsky E. Protein displacement by DExH/D ‘RNA helicases’ without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 74.Tanner N.K., Linder P. DexD/H box RNA helicases: from generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 75.Schwer B. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nature Struct. Biol. 2001;8:113–116. doi: 10.1038/84091. [DOI] [PubMed] [Google Scholar]
- 76.Rocak S., Linder P. DEAD-box proteins: The driving forces behind RNA metabolism. Nature Rev. Mol. Cell Biol. 2004;11:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 77.Nakagawa Y., Morikawa H., Hirata I., Shiozaki M., Matsumoto A., Maemura K., Nishikawa T., Niki M., Tanigawa N., Ikegami M., et al. Overexpression of rck/p54, a DEAD box protein, in human colorectal tumours. Br. J. Cancer. 1999;80:914–917. doi: 10.1038/sj.bjc.6690441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hashimoto K., Nakagawa Y., Morikawa H., Niki M., Egashira Y., Hirata I., Katsu K., Akao Y. Co-overexpression of DEAD box protein rck/p54 and c-myc protein in human colorectal adenomas and the relevance of their expression in cultured cell lines. Carcinogenesis. 2001;22:1965–1970. doi: 10.1093/carcin/22.12.1965. [DOI] [PubMed] [Google Scholar]
- 79.Miyaji K., Nakagawa Y., Matsumoto K., Yoshida H., Morikawa H., Hongou Y., Arisaka Y., Kojima H., Inoue T., Hirata I., et al. Overexpression of a DEAD box/RNA helicase protein, rck/p54, in human hepatocytes from patients with hepatitis C virus-related chronic hepatitis and its implication in hepatocellular carcinogenesis. J. Viral Hepat. 2003;10:241–248. doi: 10.1046/j.1365-2893.2003.00447.x. [DOI] [PubMed] [Google Scholar]
- 80.Bergkessel M., Reese J.C. An essential role for the Saccharomyces cerevisiae DEAD-box helicase DHH1 in G1/S DNA-damage checkpoint recovery. Genetics. 2004;167:21–33. doi: 10.1534/genetics.167.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]