Abstract
We report here the design, synthesis and application of pyrene binary oligonucleotide probes for selective detection of cellular mRNA. The detection strategy is based on the formation of a fluorescent excimer when two pyrene groups are brought into close proximity upon hybridization of the probes with the target mRNA. The pyrene excimer has a long fluorescence lifetime (>40 ns) compared with that of cellular extracts (∼7 ns), allowing selective detection of the excimer using time-resolved emission spectra (TRES). Optimized probes were used to target a specific region of sensorin mRNA yielding a strong excimer emission peak at 485 nm in the presence of the target and no excimer emission in the absence of the target in buffer solution. While direct fluorescence measurement of neuronal extracts showed a strong fluorescent background, obscuring the detection of the excimer signal, time-resolved emission measurements indicated that the emission decay of the cellular extracts is ∼8 times faster than that of the pyrene excimer probes. Thus, using TRES of the pyrene probes, we are able to selectively detect mRNA in the presence of cellular extracts, demonstrating the potential for application of pyrene excimer probes for imaging mRNAs in cellular environments that have background fluorescence.
INTRODUCTION
The detection of specific mRNAs in vivo with high sensitivity and selectivity is of great importance for the study of the molecular interactions triggered by specific stimuli (1–3). We have been interested in studying the role of localized neuronal mRNA in the formation of new synapses and memory consolidation. A model organism that has been widely used for such studies is the mollusk Aplysia californica (4,5) due to its simple nervous system, the relatively large size of its neurons, and the easy identification of individual neuronal types (6). In particular, we are interested in detecting and imaging sensorin mRNA in sensory neurons because of its important role in synaptic plasticity (2).
The detection and study of a specific mRNA both in vitro and in vivo require the use of state-of-the-art molecular probes that can detect target sequences selectively and sensitively. A common choice is the adaptation of molecular beacons (MBs), which are oligonucleotide probes that present a strong fluorescence response upon hybridization with target (7). Typical and characteristic MBs possess a stem–loop structure in which a fluorescent dye and quencher are in close proximity in the absence of target but separate when hybridized with the target sequence (7–9). Although this kind of probe has been widely used to detect assorted oligonucleotide sequences in different environments, recent studies suggest that non-specific opening of MB can occur in cells, causing false possitive signals (10,11). In addition to non-specific detection, another factor that is important for in vivo studies is the autofluorescent cellular background (12). Typically, analyses of Aplysia neurons using confocal microscopy, and neuronal extracts using a spectrofluorometer show background fluorescence arising from the sample, which can obscure the specific fluorescent signal due to hybridization with the target. As an alternative method, to avoid false positive signals and to overcome the cellular background fluorescence, we report here the construction and application of pyrene binary probes (Py-BPs) for selective mRNA detection using time-resolved fluorescence spectroscopy.
Binary probes (BPs) consist of two single-stranded (ss) fluorescent-labeled oligonucleotides that can selectively bind to adjacent regions on a target sequence (13,14). When the BP hybridizes as a pair to the corresponding sequences of the same target, a distinctive signal different from that of the non-hybridized probes is produced (15). Each unit of a BP contains two basic components: an oligonucleotide chain, which is complementary to the target sequence of interest and the chromophore that serves as a fluorescent reporter. Unique fluorescence responses of BPs upon hybridization have been attained by mean of two well-known photophysical processes: Fluorescence Resonance Energy Transfer (FRET) (13–15) and excited dimer (excimer) formation (16–18). FRET-BPs possess a donor and an acceptor fluorophore that come into close proximity only upon hybridization to the target sequence, promoting FRET. Ideally, exciting the probes at the donor dye absorption band will produce only donor fluorescence for the free probes (unhybridized), whereas the probes hybridized with target will produce significant acceptor fluorescence. In the case of excimer probes, both strands are labeled with a pyrene molecule (Py-BP) (Figure 1). The excitation of the free probes in solution (350 nm) is expected to yield a well defined fluorescence spectrum with maxima at 390 and 410 nm (monomer emission); addition of target, which brings the two pyrenes into close contact due to hybridization is expected to yield excimer emission. The excimer emission with a maximum at ∼480 nm is a broad band that results from the emission of a transient excited state dimer which is produced when an excited pyrene forms a complex with a ground state pyrene. Advantages of Py-BPs include a large Stokes shift between monomer and excimer emission, and their relatively long lifetime (30–60 ns) in comparison with the lifetime of the neuronal autofluorescence background (∼8 ns). The latter feature implies that the fluorescence from pyrene can be selectively detected if it is taken after the fluorescence from the cellular background has decayed.
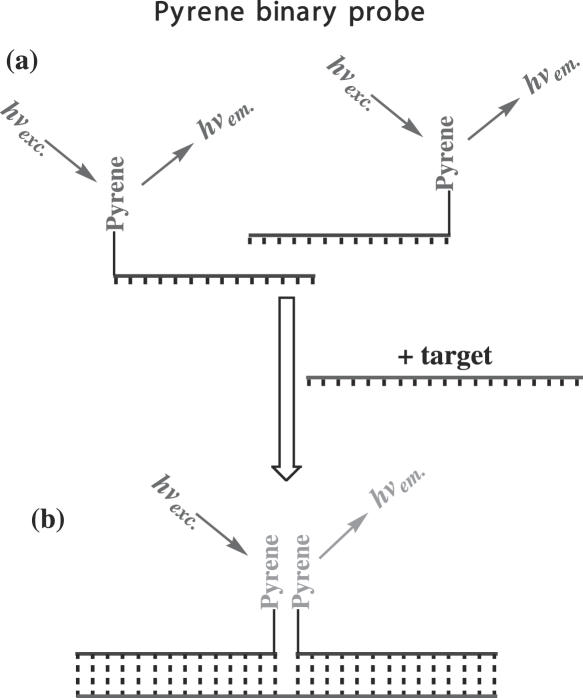
Figure 1.
Binary pyrene probes in the absence of target (a) and after hybridizing with target (b).
The use of Py-BPs for nucleic acid detection has been reported by two different groups. Masuko et al. (17) synthesized Py-BP with pyrenyl groups attached to the 3′ and 5′ ends of 16mer oligonucleotide strands. These studies showed strong excimer formation after target addition and high specificity for the detection of Vibrio mimicus 16S rRNA in vitro. However, this system requires 20% (v/v) dimethylformamide (DMF) in the hybridization buffer (16,17), which is incompatible with in vivo applications. Paris et al. (18) reported the synthesis of Py-BP in which pyrene chromophores replaced the standard base at the 1′ position of the nucleotide monomer. The probes constructed using this novel pyrene-ribose nucleotide yield a high excimer emission. However, this approach requires preparation of the phosphoramidite pyrene-ribose nucleoside (19). Other non-BP systems based on pyrene monomer-excimer signal ratio have been described in the literature; they include detection of single base mutations (20–22), mRNA secondary structure (23–25) and ion sensing (26). The use of the pyrene monomer–excimer emission in a variety of different applications demonstrates its robustness in selective target detection. Yet, although much research has been devoted to pyrene-based oligonucleotide systems, exploitation of the pyrene excimer's long-lived emission lifetime for target detection has only recently been explored for the selective detection of proteins (27).
In this report, we describe the synthesis and characterization of Py-BPs for the detection of endogenous mRNA. Specifically, we targeted sensorin mRNA, which has an important role in synaptic plasticity. The synthesis of the Py-BPs is straightforward and involves only standard, widely used techniques and reagents. To demonstrate the utility of Py-BP's long fluorescence lifetime in the detection of mRNA in a cell mimicking environment (neuronal protein extracts) with a high fluorescent background, we used time-resolved fluorescence spectroscopy for the detection of the pyrene excimer emission after the decay of the background fluorescence, allowing selective detection of the target mRNA molecule.
MATERIALS AND METHODS
Synthesis of pyrene labeled 2′-methoxy RNA probes
3′-Amino group modified 2′-methoxy RNA was synthesized on a DNA synthesizer (Expedite 8909, Applied Biosystems, CA) by phosphoramidite chemistry with a set of 2′-OMe Abz, Gdmf, Cbz and U-CE phosphoramidite nucleosides, and 3′-PT-amino-modifier C3 CPG or 3′-PT-amino-modifier C6 CPG (Glen Research, Sterling, VA). 5′-Amino group modified 2′-methoxy RNA was synthesized using 5′-amino-modifier C3 TFA or 5′-amino-modifier C6 TFA (Glen Research, Sterling, VA) as a final coupling monomer. Then deprotection was carried out as recommended by the manufacturer. The deprotected RNAs were purified by OPC cartridge (ABI, Foster City, CA) or high-performance liquid chromatography (HPLC) (Waters system, Milford, MA) followed by labeling with the appropriate pyrene derivative. The purified 2′-OMe RNAs were conjugated with the pyrene at either the 3′-amino or 5′-amino end by 5 h of incubation in 0.1 M sodium bicarbonate/sodium carbonate buffer (pH 8.5) mixed with a 15-fold excess of dimethyl sulfoxide (DMSO)-dissolved succinimidyl esters of 1-pyrene butanoic acid or 1-pyrene acetic acid (Invitrogen, Carlsbad, CA). The unreacted pyrene derivative was removed by size exclusion chromatography on a PD-10 column (Amersham Life Sciences, Piscataway, NJ). After desalting with an OPC, the products were lyophilized and purified by reverse-phase HPLC using a C-18 reverse column (Xterra MS C18, 4.6 × 50 mm) at a flow rate of 0.5 ml/min and with a linear gradient of 12–34.5% methanol in buffer A [8.6 mM triethylammonium and 100 mM hexafluoroisopropyl alcohol aqueous solution (pH 8.1)] over 40 min. The eluate was detected at 260 nm and fractions containing the desired product were collected, evaporated to dryness under vacuum and characterized by MALDI-TOF mass spectrometry. The sequences of the pyrene labeled oligonucleotide probes are shown in Figures 2a and 3a.
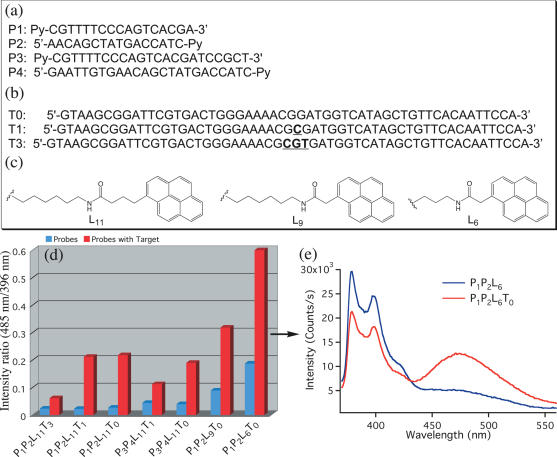
Figure 2.
Parameters for the optimization of excimer emission in Py-BP: probe sequences (a), target sequences (b) and pyrene linkers used to optimize Py-BP excimer emission (c). Monomer to excimer ratio for the different probes studied with (red) and without (blue) target (d); emission spectra of P1P2L6 with (red) and without (blue) target (T0) (e). [Probes] = 0.1 µM; Buffer: 20 mM Tris–HCl (pH 7.5), 500 mM NaCl, 60 mM MgCl2; hybridization time = 30 min.
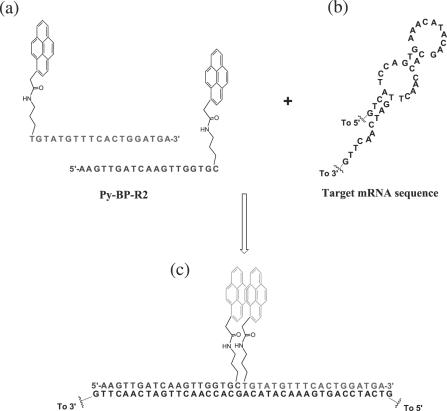
Figure 3.
Pyrene probe pairs Py-BP-R2 (a), region of sensorin mRNA target sequence complementary to Py-BP-R2 (b) and Py-BP-R2 hybridized with target mRNA (c).
Fluorescence emission spectra were obtained on a Fluorolog-3 spectrometer FL3–22 (J. Y. Horiba, Edison, NJ) using quartz cuvettes with a path length of 0.4 cm. The experiments were performed by mixing 1 equivalent of each probe ([Probe] = 0.1 µM) with 1 equivalent of target ([Probe] = 0.1 µM) in a buffer solution. The excitation wavelength for the steady state measurements was 350 nm. The spectra were corrected for monochromator and PMT efficiencies. The signal to background ratio was determined by using Equation 1:
| 1 |
where E and M are the excimer and monomer maxima, respectively. Time-resolved fluorescence measurements were performed on an OB920 single-photon counting fluorometer (Edinburgh Analytical Instruments) with a pulsed nanosecond nitrogen lamp as excitation source (λexc.= 337 nm). Sample concentrations were determined by ultraviolet (UV)–Vis spectrometry on an Agilent 8456 spectrophotometer using quartz cuvettes with an optical path of 1 cm.
Protein extracts were isolated from A.californica pleural ganglia. Extracts were prepared by homogenization in the extraction buffer [25 mM Tris (pH 7.4), 525 mM NaCl, 25 mM EDTA and protease inhibitor cocktail from Roche] at 4°C. The product was centrifuged at 10 000 r.p.m. for 10 min followed by collection of the supernatant. Full-length capped polyadenylated sensorin mRNA as well as its antisense copy were generated by in vitro transcription in the presence of cap analog m7G(5′)ppp(5′GTP) (Ambion, TX) using as a template a PCR-amplified fragment of sensorin cDNA flanked by the T7- and T3-promoters. Upon the completion of the in vitro transcription, 100–120 base long poly(A) tails were introduced at the 3′ ends of the RNA transcripts with Escherichia coli poly(A) RNA polymerase (Ambion, TX). The RNA fragments were purified on MEGAclear cartridges (Ambion, TX) and characterized by UV-spectroscopy and glyoxal agarose gel electrophoresis.
The secondary structure of target sensorin mRNA was modeled with the program mFold (28) using a published A.californica sensorin mRNA sequence (29). Up to 30 possible mRNA conformations were analyzed to find target regions with a high probability of being mostly ss, to assure that the Py-BP can hybridize with the native mRNA. Oligonucleotides for the selected regions were synthesized using the procedure described above.
RESULTS AND DISCUSSION
Probe design and hybridization evaluation using oligonucleotide target sequences
The BP oligonucleotide probes shown in Figure 2a were synthesized using standard phosphoramidite solid phase chemistry. The sequence of the probes was selected to be complementary to a portion of M13 vector sequence. To optimize the excimer formation upon hybridization with target, the probes were varied with respect to the linker lengths between the oligonucleotide chain and the pyrene moiety, the oligonucleotide chain length, and/or different number of bases between the two pyrene moieties after hybridization of the two probes to the target. Figure 2a–c shows the three parameters that were varied to optimize the pyrene excimer fluorescence emission. Figure 2d shows the excimer to monomer ratio for the different probes. A clear increase in the excimer emission is found as shorter linkers between the pyrene and the oligonucleotide chain are used. As an example, the fluorescence spectra for P1P2L6T0, in the presence or absence of target are shown in Figure 2e (where P1 and P2 are the Py-BPs, L6 is the length of the pyrene linker, and T0 is the target sequence with no nucleotide separation between the pyrene groups when the probes are hybridized to the target). Kostenko et al. (23) suggested that short linkers between the pyrene and the oligonucleotide chains are effective in pyrene excimer formation, possibly because they avoid strong interactions of the chromophores with the DNA strand. Intercalation of pyrene within DNA has been extensively reported to produce quenching of the pyrene monomer excited singlet state (30–32). It has also been observed that two pyrene molecules linked together with a pentyl chain present strong excimer emission in solution until double-stranded (ds) DNA is added to the solution (33). Excimer emission depletion due to interaction with ss or ds oligonucleotides has been attributed to intercalation of the pyrene chromophores between the bases of the duplex, causing a spatial separation of the pyrenes and precluding excimer formation (21,22,24,33). The use of short linkers limits the translational freedom of the pyrene probes and can prevent or hinder their intercalation between the nucleic acid bases.
On the other hand, we observed that increasing the number of nucleotides in the BP did not increase the amount of excimer, though it did increase the hybridization time required to reach maximum excimer fluorescence emission. This is expected since longer chains should require longer times to diffuse and hybridize with their target sequence. Our evaluation also indicated that decreasing the number of bases between the probes increased the excimer emission. This is in agreement with studies from Paris et al. (18) and Ebata et al. (16,17) which showed that close distances between pyrenes are required for efficient excimer formation. Based on these results it was concluded that the best conditions for excimer fluorescence emission are probes of 17 nt in length (P1 and P2), short linkers between the oligonucleotide strand and the pyrene (L6), and no spacing bases between the hybridized probes (P1P2L6T0).
Hybridization studies of Py-BP with sensorin mRNA
The detection of mRNA in solution presents additional challenges compared with the detection of linear DNA strands in solution. Cellular mRNA possesses a secondary structure consisting of stem and loop regions that are important for protein translation and binding processes (34). Avoidance of regions with intrinsic strong secondary structure such as the stems is important to allow the probes to be accessible to the mRNA sequence. The target sequences were selected from the calculated 2D structure of sensorin mRNA using the program mFold. Three regions were selected which possess a high probability of being accessible to the probe to allow the hybridization of Py-BP with the target mRNA.Figure 3 shows the structure of the two pyrene probes Py-BP-R2 and one of the possible secondary structures of the target region in sensorin mRNA.
Pyrene binary probe Py-BP-R2 complementary to the selected region of sensorin mRNA was synthesized using 2′-methoxy modified nucleoside phosphoramidites. These modified 2′-methoxy nucleosides are more stable than standard DNA nucleosides being resistant to RNase and DNase, and have been shown to increase the stability of the duplex (35). We first tested the hybridization of Py-BP-R2 with full-length sensorin mRNA in a buffer solution; the fluorescence spectra are shown in Figure 4a. The spectra indicated that there was no excimer emission in the absence of mRNA, and a strong increase in the excimer emission peak at 485 nm after the addition of sensorin mRNA. The signal to background ratio for this probe is ∼8, 40 min after the addition of target, and increases up to 18.5 after a 2 h hybridization period (Supplementary Figure S1). It is also known that the ds formation rate of oligonucleotides is highly dependent on the concentration of MgCl2, which is absent in the hybridization buffer used in these experiments. Since the MgCl2 concentration within the sensory neurons of Aplysia is in the range of 40 mM, the hybridization rate may be even faster. The extremely low excimer fluorescence of Py-BP-R2 in the absence of the mRNA target and its dramatically increased excimer fluorescence in response to the addition of full-length sensorin mRNA makes this probe an excellent candidate for neuronal studies.
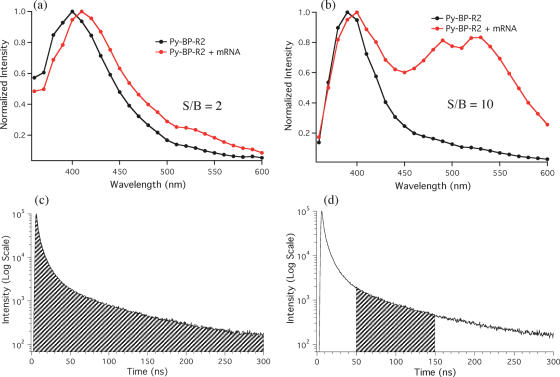
Figure 4.
Steady-state fluorescence spectra of Py-BP-R2 in buffer solution (a) and cellular protein extract (b). [Py-BP-R2] = 0.1 µM; Buffer: 10 mM Tris (pH 7.5), 400 mM NaCl; hybridization time = 2 h; normalized at 400 nm.
As a negative control to test for non-specific interactions, we added the complementary sequence of sensorin mRNA to Py-BP-R2; this yielded no excimer emission. To test this probe in a neuron-like environment, neuronal protein extracts were used to mimic in vivo neuronal conditions. The absorption spectra of the cell extract and of the cell extract plus Py-BP-R2 reveal the high abundance of proteins in the extract (∼0.41 mg/ml), which are in high excess compared to the amount of probe (Supplementary Figure S2). Addition of target mRNA to the neuronal extract with Py-BP-R2 produced only a small increase in the excimer, which was equivalent to a ratiometric enhancement of ∼2 (Figure 4b). This small increase can be attributed to the high fluorescence neuronal background that effectively masks the emission output of the probes. To overcome this limitation, time-resolved fluorescence measurements were performed.
Time-resolved fluorescence measurements to selectively detect sensorin mRNA in cellular extract
Studies in the neuronal protein extract revealed a strong background fluorescence that obscures the fluorescence signals of Py-BP-R2 and hence the detection of target sensorin mRNA. Time-resolved fluorescence measurements of Py-BP-R2 show biexponential decays both in the presence and in the absence of the mRNA target. This biexponential decay can be explained by the heterogeneous environment induced by the probe's oligonucleotide chain. The fluorescence lifetimes of Py-BP-R2 under different conditions are presented in Table 1. The fluorescence lifetime of the monomer and excimer components of Py-BP-R2 are ∼65 and 39 ns, respectively, while the lifetime for the cell extract alone is ∼7 ns. An interesting observation from Table 1 is that the fluorescence lifetime of the pyrene monomer increases in the neuronal protein extract when mRNA target is added, in contrast with its behavior in buffer solution. This suggests that the fluorescence of the Py-BP-R2 monomer is being quenched by interaction of the probe in its random coil conformation with the proteins in the extract; this quenching is apparently inhibited when Py-BP-R2 binds to its target.
Table 1.
Lifetime of Py-BP-R2 and buffer extract
| Target | aτ398 nm, ns (%) | bτ480 nm, ns (%) | |||
|---|---|---|---|---|---|
| Py-BP-R2 in Tris–HCl (pH 8.1) | No | 64.5 (81) | 10.9 (19) | 38.9 (57) | 7.7 (43) |
| Sensorin mRNA | 48.5 (73) | 8.0 (27) | 35.3 (73) | 9.9 (28) | |
| Py-BP-R2 in protein extract | No | 38.7 (36) | 7.3 (64) | 36.6 (57) | 8.0 (43) |
| Sensorin mRNA | 53.1 (60) | 7.8 (40) | 39.0 (50) | 7.9 (50) | |
| Protein extract | No | 41.2 (9) | 7.4 (91) | 20.5 (12) | 7.2 (88) |
aτ398 nm and bτ480 nm are the biexponential fluorescence lifetimes at 398 and 480 nm, respectively. The two columns represent the two lifetimes obtained from the iterative reconvolution of the experimental decays. The contribution of each lifetime compared to the fluorescence signal intensity is in parentheses.
The fast fluorescence decay (7 ns) of the protein extract together with the slower (>39 ns) fluorescence decay of Py-BP-R2 suggest that gating the detection after the decay of the neuron background fluorescence should allow the extraction of the pyrene emission with a high signal to background (S/B) ratio. Time-resolved emission spectra (TRES) of Py-BP-R2 integrating the whole decay area (steady-state like condition, Figure 5c) and integrating only from 30 to 150 ns (Figure 5d) are shown in Figure 5a and b. The spectrum in Figure 5a is very similar to that in Figure 4b (Py-BP-R2 + mRNA), in which no clear excimer signal can be detected due to the large background fluorescence present in the spectra. In contrast, the excimer signal can be easily distinguished in the gated time-resolved spectra giving a S/B ratio of ∼10 (Figure 5b). These results indicate that time-resolved fluorescence measurements provide the possibility of discriminating the long-lived signals generated by the pyrene probes from the more quickly decaying background fluorescence elicited by native molecules in the neuron.
Figure 5.
Time-resolved fluorescence spectra of Py-BP-R2 with and without target in neuronal protein extract, gating from 0–300 ns (a) or from 30–150 ns (b). Fluorescence decay of Py-BP-R2 (λem = 398 nm) showing the signal gating for the spectra in (a) (c) and in (b) (d). [Py-BP-R2] = 0.1 µM; hybridization time = 30 min.
Probes utilizing the pyrene monomer/excimer fluorescence have several advantages over most other commonly used fluorescence probes. The unusually long fluorescence lifetime of pyrene monomers and excimers allows for time gated background fluorescence rejection as described above. Fluorescence lifetime imaging microscopy, a technique that recently became available, (36,37) can take advantage of this long fluorescence lifetime. Another advantage of pyrene probes is the large spectral separation between the excitation wavelength (∼340 nm), pyrene monomer fluorescence (∼390 nm), and excimer fluorescence (∼470 nm). This large ‘Stoke's shift’ minimizes the spectral overlap of scattered excitation light, monomer fluorescence and excimer fluorescence. In microscopy, this large ‘Stoke's shift’ allows for the use of filters with a wider spectral bandpass for the detection channels, which enhances the signal intensity significantly.
The main disadvantage of the pyrene chromophore is its short excitation wavelength (∼340 nm), and its moderately low extinction coefficient (ɛ344nm ∼ 27 500 M−1 cm−1) (38). This near-UV excitation might be unfavorable for in vivo mRNA detection by microscopy, because of the reduced penetration depth of near-UV light and increased autofluorescence (background fluorescence) compared to the more commonly used visible excitation range. However, stronger light sources can compensate for the moderately low extinction coefficient and also increase the depth of penetration. Furthermore, Takahashi et al. have already demonstrated that pyrene probes can be used in conventional fluorescence microscopy for cell studies (39).
CONCLUSIONS
Pyrene binary probes have been designed to unambiguously detect sensorin mRNA under cellular conditions. Various Py-BPs were synthesized by adjusting various parameters. Probes of 17 nt chain length, containing short linkers between the pyrene and the oligonucleotide chain and no base spacing between the pyrene groups when hybridized with the target, yield the maximum excimer emission. The probes for sensorin mRNA were designed following these general criteria along with taking into account the sensorin mRNA secondary structure to minimize interaction of the probes with ds regions. Probes pair Py-BP-R2 designed for a specific region in sensorin mRNA showed strong excimer emission in the presence of sensorin mRNA. The fluorescence spectra of Py-BP-R2 showed an excellent S/B ratio in buffer solution (∼18.5) but a low S/B ratio (∼2) in the protein extract. Time-resolved fluorescence spectra obtained by gating the emission from 30 to 150 ns showed a S/B ratio of 10 for Py-BP-R2 in the protein extract. This demonstrates that it is possible to separate the long-lived fluorescence decay of pyrene from the short-lived background fluorescence decay of the protein extract. Also, Py-BP-R2 allows specific detection of sensorin mRNA even in the presence of cellular proteins where other probes (like MBs) can present non-specific binding signals. Thus, Py-BPs should have potential advantages in selectively imaging mRNA in cells when time-resolved fluorescence microscopy techniques are used to eliminate the cellular background fluorescence from the target detection signals.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
This work was supported by the Center of Excellence in Genomic Science Grant P50 HG002806 from the National Institutes of Health and NSF CHE-04-15516. Funding to pay the Open Access publication charges for this article was provided by the Center of Excellence in Genomic Science Grant P50 HG002806 from the National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hu J.-Y., Meng X., Schacher S. Target interaction regulates distribution and stability of specific mRNAs. J. Neurosci. 2002;22:2669–2678. doi: 10.1523/JNEUROSCI.22-07-02669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu J.-Y., Goldman J., Wu F., Schacher S. Target-dependent release of a presynaptic neuropeptide regulates the formation and maturation of specific synapses in Aplysia. J. Neurosci. 2004;24:9933–9943. doi: 10.1523/JNEUROSCI.3329-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knowles R.B., Sabry J.H., Martone M.E., Deerinck T.J., Ellisman M.H., Bassell G.J., Kosik K.S. Translocation of RNA granules in living neurons. J. Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin K.C., Casadio A., Zhu H., Yaping E., Rose J.C., Chen M., Bailey C.H., Kandel E.R. Synapse-specific, long-term facilitation of Aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91:927–938. doi: 10.1016/s0092-8674(00)80484-5. [DOI] [PubMed] [Google Scholar]
- 5.Mayford M., Barzilai A., Keller F., Schacher S., Kandel E.R. Modulation of an NCAM-related adhesion molecule with long-term synaptic plasticity in Aplysia. Science. 1992;256:638–644. doi: 10.1126/science.1585176. [DOI] [PubMed] [Google Scholar]
- 6.Kandel E.R. The molecular biology of memory storage: a dialog between genes and synapses. In: Jörnvall H., editor. Nobel Lectures, Physiology or Medicine 1996–2000. Singapore: World Scientific Publishing Co.; 2003. pp. 395–439. [Google Scholar]
- 7.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 8.Tan W., Fang X., Li J., Liu X. Molecular beacons: a novel DNA probe for nucleic acid and protein studies. Chem. Eur. J. 2000;6:1107–1111. doi: 10.1002/(sici)1521-3765(20000403)6:7<1107::aid-chem1107>3.3.co;2-0. [DOI] [PubMed] [Google Scholar]
- 9.Zhang P., Beck T., Tan W. Design of a molecular beacon DNA probe with two fluorophores. Angew. Chem. Int. Ed. Engl. 2001;40:402–405. doi: 10.1002/1521-3773(20010119)40:2<402::AID-ANIE402>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 10.Santangelo P.J., Nix B., Tsourkas A., Bao G. Dual FRET molecular beacons for mRNA detection in living cells. Nucleic Acids Res. 2004;32:e57. doi: 10.1093/nar/gnh062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanke H.J., Dirks R.W., Raap T. FISH and immunocytochemistry: towards visualising single target molecules in living cells. Curr. Opin. Biotechnol. 2005;16:49–54. doi: 10.1016/j.copbio.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 12.Mosiman V.L., Patterson B.K., Canterero L., Goolsby C.L. Reducing cellular autofluorescence in flow cytometry: an in situ method. Cytometry. 1997;30:151–156. [PubMed] [Google Scholar]
- 13.Cardullo R.A., Agrawal S., Flores C., Zamecnik P.C., Wolf D.E. Detection of nucleic acid hybridization by nonradiative fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 1988;85:8790–8794. doi: 10.1073/pnas.85.23.8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sei-Iida Y., Koshimoto H., Kondo S., Tsuji A. Real-time monitoring of in vitro transcriptional RNA synthesis using fluorescence resonance energy transfer. Nucleic Acids Res. 2000;28:e59. doi: 10.1093/nar/28.12.e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsourkas A., Behlke M.A., Xu Y., Bao G. Spectroscopic features of dual fluorescence/luminescence resonance energy-transfer molecular beacons. Anal. Chem. 2003;75:3697–3703. doi: 10.1021/ac034295l. [DOI] [PubMed] [Google Scholar]
- 16.Ebata K., Masuko M., Ohtani H., Kashiwasake-Jibu M. Nucleic acid hybridization accompanied with excimer formation from two pyrene-labeled probes. Photochem. Photobiol. 1995;62:836–839. doi: 10.1111/j.1751-1097.1995.tb09144.x. [DOI] [PubMed] [Google Scholar]
- 17.Masuko M., Ohtani H., Ebata K., Shimadzu A. Optimization of excimer-forming two-probe nucleic acid hybridization method with pyrene as a fluorophore. Nucleic Acids Res. 1998;26:5409–5416. doi: 10.1093/nar/26.23.5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paris P.L., Langenhan J.M., Kool E.T. Probing DNA sequences in solution with a monomer-excimer fluorescence color change. Nucleic Acids Res. 1998;26:3789–3793. doi: 10.1093/nar/26.16.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren R.X.-F., Chaudhuri N.C., Paris P.L., Rumney S.IV., Kool E.T. Naphthalene, phenanthrene, and pyrene as DNA base analogues: synthesis, structure, and fluorescence in DNA. J. Am. Chem. Soc. 1996;118:7671–7678. doi: 10.1021/ja9612763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamana K., Fukunaga Y., Ohtani Y., Sato S., Nakamura M., Kim W.J., Akaike T., Maruyama A. DNA mismatch detection using a pyrene-excimer-forming probe. Chem. Commun. 2005:2509–2511. doi: 10.1039/b502033f. [DOI] [PubMed] [Google Scholar]
- 21.Yamana K., Iwai T., Ohtani Y., Sato S., Nakamura M., Nakano H. Bis-pyrene-labeled oligonucleotides: sequence specificity of excimer and monomer fluorescence changes upon hybridization with DNA. Bioconjugate Chem. 2002;13:1266–1273. doi: 10.1021/bc025530u. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto A., Ichiba T., Saito I. Pyrene-labeled oligodeoxynucleotide probe for detecting base insertion by excimer fluorescence emission. J. Am. Chem. Soc. 2004;126:8364–8365. doi: 10.1021/ja049061d. [DOI] [PubMed] [Google Scholar]
- 23.Kostenko E., Dobrikov M., Pyshnyi D., Petyuk V., Komarova N., Vlassov V., Zenkova M. 5′-bis-pyrenylated oligonucleotides displaying excimer fluorescence provide sensitive probes of RNA sequence and structure. Nucleic Acids Res. 2001;29:3611–3620. doi: 10.1093/nar/29.17.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahara A., Iwase R., Sakamoto T., Yamana K., Yamaoka T., Murakami A. Bispyrene-conjugated 2′-O-methyloligonucleotide as a highly specific RNA-recognition probe. Angew. Chem. Int. Ed. Engl. 2002;41:3648–3650. doi: 10.1002/1521-3773(20021004)41:19<3648::AID-ANIE3648>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 25.Lewis F.D., Zhang Y., Letsinger R.L. Bispyrenyl excimer fluorescence: a sensitive oligonucleotide probe. J. Am. Chem. Soc. 1997;119:5451–5452. [Google Scholar]
- 26.Nagatoishi S., Nojima T., Juskowiak B., Takenaka S. A pyrene-labeled G-quadruplex oligonucleotide as a fluorescent probe for potassium ion detection in biological applications. Angew. Chem. Int. Ed. Engl. 2005;44:5067–5070. doi: 10.1002/anie.200501506. [DOI] [PubMed] [Google Scholar]
- 27.Yang C.J., Jockusch S., Vincens M., Turro N.J., Tan W. Light switching excimer probes for rapid protein monitoring in complex biological fluids. Proc. Natl Acad. Sci. USA. 2005;102:17278–17283. doi: 10.1073/pnas.0508821102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 29.Brunet J.-F., Shapiro E., Foster S.A., Kandel E.R., Iino Y. Identification of a peptide specific for Aplysia sensory neurons by PCR-based differential screening. Science. 1991;252:856–859. doi: 10.1126/science.1840700. [DOI] [PubMed] [Google Scholar]
- 30.Telser J., Cruickshank K.A., Morrison L.E., Netzel T.L., Chan C. DNA duplex covalently labeled at two sites: synthesis and characterization by steady-state and time-resolved optical spectroscopies. J. Am. Chem. Soc. 1989;111:7226–7232. [Google Scholar]
- 31.Mann J.S., Shibata Y., Meehan T. Synthesis and properties of an oligonucleotide modified with a pyrene derivative at the 5′-phosphate. Bioconjugate Chem. 1992;3:554–558. doi: 10.1021/bc00018a015. [DOI] [PubMed] [Google Scholar]
- 32.Telser J., Cruickshank K.A., Morrison L.E., Netzel T.L. Synthesis and characterization of DNA oligomers and duplexes containing covalently attached molecular labels: comarison of biotin, fluorescein, and pyrene labels by thermodynamic and optical spectroscopic measurements. J. Am. Chem. Soc. 1989;111:6966–6976. [Google Scholar]
- 33.Kitamura M., Nimura A., Yamana K., Shimidzu T. Oligonucleotides with bis-pyrene adduct in the backbone: synthesis and properties of intramolecular excimer forming probe. Nucleic Acids Symp. Ser. 1991;25:67–68. [PubMed] [Google Scholar]
- 34.Voet D., Voet J.G. Biochemistry, 2nd edn. New York: Wiley and sons; 1995. pp. 915–958. [Google Scholar]
- 35.Lamond A.I., Sproat B.S. Antisense oligonucleotides made of 2′-O-alkylRNA: their properties and applications in RNA biochemistry. FEBS Lett. 1993;325:123–127. doi: 10.1016/0014-5793(93)81427-2. [DOI] [PubMed] [Google Scholar]
- 36.Carlsson K., Liljeborg A. Confocal fluorescence microscopy using spectral and lifetime information to simultaneously record four fluorophores with high channel separation. J. Microsc. 1996;185:37–46. [Google Scholar]
- 37.Squire A., Bastiaens P.I.H. Three dimensional image restoration in fluorescence lifetime imaging microscopy. J. Microsc. 1998;193:36–49. doi: 10.1046/j.1365-2818.1999.00427.x. [DOI] [PubMed] [Google Scholar]
- 38.Kierzek R., Li Y., Turner D.H., Bevilacqua P.C. 5′-Amino pyrene provides a sensitive, nonperturbing fluorescent probe of RNA secondary and tertiary structure formation. J. Am. Chem. Soc. 1993;115:4985–4992. [Google Scholar]
- 39.Takahashi S., Odani N., Tomokiyo K., Furuta K., Suzuki M., Ichikawa A., Negishi M. Localization of a cyclopentenone prostaglandin to the endoplasmic reticulum and induction of BiP mRNA. Biochem. J. 1998;335:35–42. doi: 10.1042/bj3350035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bratu D.P., Cha B.-J., Mhlanga M.M., Kramer F.R., Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl Acad. Sci. USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]