Abstract
The objective of this study was to investigate whether geographic distance was correlated with genetic homology among isolates of Porcine reproductive and respiratory syndrome virus (PRRSV) from a single pork-producing company. We analyzed geographic distance, temporal distance, and percentage similarity in the PRRSV nucleotide sequence among 62 farms, applying the Mantel test for correlation between distance matrices and PRRSV sequencing. Genomic similarity had a significant (P < 0.01) negative (rM = −0.217) correlation with geographic distance. These findings indicate that, under the conditions of this study, the greater the distance between farms, the less the genetic homology among PRRSV isolates.
Résumé
La présente étude visait à vérifier si la distance géographique était corrélée à l’homologie génétique parmi les isolats du virus du syndrome reproducteur et respiratoire porcin (PRRSV) provenant d’une seule entreprise de production porcine. Une analyse de la distance géographique, de la distance temporelle et du pourcentage de similarité des séquences nucléotidiques de PRRSV pour 62 fermes fut effectuée en appliquant le test de Mantel pour la corrélation entre les matrices de distance et le séquençage du PRRSV. La similarité génomique avait une corrélation négative (rM = −0.217) significative (P < 0,01) avec la distance géographique. Ces résultats indiquent que, dans les conditions expérimentales de la présente étude, plus la distance entre les fermes est grande, moins il y a d’homologie génétique entre les isolats de PRRSV.
(Traduit par Docteur Serge Messier)
Porcine reproductive and respiratory syndrome (PRRS) is the most important disease of swine worldwide. In the United States, losses are calculated to be $560 million yearly (1). Transmission of Porcine reproductive and respiratory syndrome virus (PRRSV) into a herd may occur by several routes, including pigs (2), semen (3), contaminated fomites, and infected vectors such as mosquitoes and flies (4). Transport can also play an important role in the transmission of virus between sites if trucks are not properly disinfected (5). Aerial spread as a mode of PRRSV transmission among farms is controversial. A way to measure the similarity between PRRSV isolates from different farms is sequencing the open reading frame 5 (ORF-5) region of the PRRSV genome. Goldberg and colleagues (6) did not detect a correlation between geographic proximity and genomic similarity on the basis of the ORF-5 region in 55 PRRSV isolates submitted by veterinarians from Illinois and eastern Iowa. This lack of correlation does not support the hypothesis of aerial spread, a distance-related phenomenon. However, Mortensen and associates (7), from a study of 1071 sow herds, reported that exposure from virus-infected neighboring herds and increased herd size significantly increased the risk of PRRSV infection. They suggested that virus spread via aerosols is a frequent method of PRRSV transmission among herds. Other authors have also suggested that aerial transmission among herds is probable, mainly in regions with a dense swine population (8,9). The purpose of our study was to investigate whether there was any correlation between geographic distance, temporal distance, and nucleotide similarity in the PRRSV genetic sequence among isolates within a single pork-producing company.
We obtained 62 PRRSV ORF-5 nucleotide sequences, 1 from each of 62 farms, processed in the Animal Disease Research and Diagnostic Laboratory of South Dakota State University. The isolates had been submitted from a pork-producing company for diagnostic testing from January 2002 to May 2003. Genomic sequences were aligned with Clustal X (available at http://bips.u-strasbg.fr/fr/Documentation/ClustalX/) (10), with the use of default settings, and the resultant alignment file was saved in multiple sequence format and loaded into the DNAStar program (Lasergne, Madison, Wisconsin, USA) to generate a matrix of genomic distances between isolates.
Geographic coordinates of the 62 farms in decimal degrees for longitude and latitude were obtained from the company’s database. Distance between sites was calculated in an Excel spreadsheet with application of a great circle formula (11), as follows:
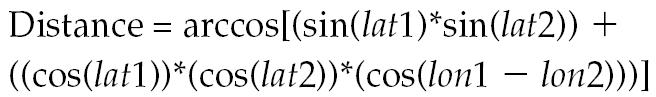 |
where lat1 = latitude point 1, lat2 = latitude point 2, lon1 = longitude point 1, and lon2 = longitude point 2.
With this formula we obtained distances in radians, which we multiplied by [(180*60)/π] to obtain distances in nautical miles, which we then multiplied by 1.562 to obtain distances in kilometers (11). A matrix of n*(n − 1)/2 pairs of distances was obtained applying these calculations.
The matrix of genomic distances generated by the DNAStar program was compared with the geographic distances from the Excel spreadsheet to analyze the relationship between geographic distance and genomic distance among the 62 farms.
The analysis for correlation between the variables in the study was based on the Mantel test, which applies the method of permutation (12). The Mantel statistic (rM) is derived as follows.
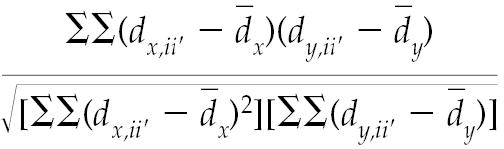 |
This statistic is equivalent to a Pearson correlation coefficient. However, since the values in a distance matrix are not independent, the Mantel statistic is tested by the method of permutation: the value of the obtained test statistic is compared not with any standard statistical distribution but with a reference distribution randomly generated from the same data (13). In this case the null hypothesis states that there is no correlation between the variables of genetic similarity and geographic distance. If the null hypothesis is true, all possible pairings of the variables (including the already calculated value for the test statistic) will not differ. The number of random permutations determines the precision of the test, with 1000 randomizations for α = 0.05 and a minimum of 5000 randomizations for α = 0.01 (14). Our analysis was performed with 10 000 permutations to estimate a significance level of 0.01, with the use of XLStat software (Kovach, Pentraeth, Wales).
The Spearman nonparametric correlation coefficient was applied in the Mantel test, given that the original technique is a linear model, and some nonlinear relationships could be underestimated (15). A variation of the Mantel test, the partial Mantel test, was used to compare 3 variables (16).
Among the 62 PRRSV isolates, 40 (64%) conformed to 4 major groups, with no more than 5% divergence between isolates within groups. The percentage of nucleotide similarity between isolates ranged from 88% to 100%, geographic distance from 638 to 854 km, and temporal distance from 1 to 499 d. Three distance matrices, each with 1861 measurements, were generated for pairwise comparison of the 3 variables in the study.
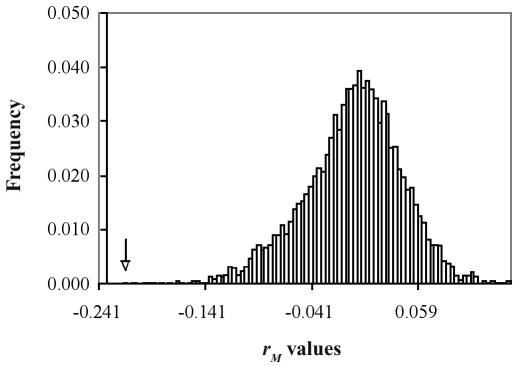
The statistic from the partial Mantel test for the correlation between geographic distance and nucleotide similarity was rM = −0.217. Statistical analysis of 10 000 permutations indicated a significant relationship (P < 0.01); Figure 1 shows the frequency distribution of the rM values. Mantel statistics do not need to be as large as Pearson correlation coefficients to reach significance (16). There was a nonsignificant correlation between temporal distance and genomic distance (rM = −0.090, P > 0.01). These findings suggest that PRRSV genomic variation among sites in this study was more related to increments in spatial distance than to increments in temporal distance.
Figure 1.
Mantel statistic values for the null hypothesis of no correlation between percentage of genetic similarity among isolates of Porcine reproductive and respiratory syndrome virus and geographic distance between the source of the isolates. The arrow indicates the position of the statistic value (rM = −0.217, in the zone of null hypothesis rejection; P = 0.0002) calculated from the original, nonpermuted data in the frequency distribution curve generated from 10 000 random permutations of the same data.
In recent years, there has been a substantial increase in knowledge of the main characteristics of PRRSV, its interaction with its host, the pig, and the response to infection. Such knowledge has been critical to the development of methods for eliminating PRRSV from individual farms (17). However, control of the disease in geographically wider areas is still a challenge for the pork industry.
A main obstacle to controlling PRRSV within regions is the lack of a complete understanding of the mechanisms of transmission between farms. Although the role of airborne transmission is not completely understood, there is consensus that farms in pig-dense areas are at increased risk of having PRRSV (2). The term “area spread” is used to describe the situation in which virus appears to move among farms within an area (18). Some factors frequently associated with presumed area spread or lateral transmission of PRRSV between farms, such as geographic distance and regional animal density, may influence the genomic variability among the farms in a region or a company. Such variability may be due to the introduction of external virus into the farms, with consequent replacement of the original virus population or exchange of genetic information between the 2 populations.
The Mantel test and its derived forms have been applied in the biologic and environmental sciences for investigating relationships among variables (19). Our application revealed that genetic variation was correlated only with geographic distance, indicating that the further apart farms were from each other, the lower was the genetic similarity of their PRRSV isolates.
The results of our study support the role of lateral, indirect transmission between neighboring sites for regional spread of PRRSV, in contrast to the results of Goldberg and colleagues (6). The opposing conclusions may reflect an effect of local conditions on area spread, given the differences in geographic location and extension between the areas. Future studies need to incorporate all the possible information about neighboring sites to bring more reliability to the results. As other producers and practitioners show interest in sharing their data, spatial analyses of regional PRRSV transmission will gain in accuracy.
It is important to understand the behavior of the variables affecting the movement of PRRSV among herds to develop effective strategies to control the disease. When the data available suggest regional spread of the disease, control strategies should be coordinated in time by common areas instead of having several isolated efforts at different times.
Acknowledgments
The authors wish to thank Dr. Jane Christopher-Hennings and her team from the Animal Disease Research and Diagnostic Laboratory of South Dakota State University for their valuable collaboration.
References
- 1.Neumann E, Kliebestein JB, Johnson CD, et al. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc. 2005;227:385–392. doi: 10.2460/javma.2005.227.385. [DOI] [PubMed] [Google Scholar]
- 2.Le Potier M-F, Blanquefort P, Morvan E, Albina E. Results of a control programme for the porcine reproductive and respiratory syndrome in the French “Pays de la Loire” region. Vet Microbiol. 1997;55:355–360. doi: 10.1016/s0378-1135(96)01318-1. [DOI] [PubMed] [Google Scholar]
- 3.Christopher-Hennings J, Nelson EA, Nelson JK, et al. Detection of porcine reproductive and respiratory syndrome virus in boar semen by PCR. J Clin Microbiol. 1995;33:1730–1734. doi: 10.1128/jcm.33.7.1730-1734.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otake S, Dee SA, Moon RD, Rossow KD, Trincado C, Pijoan C. Evaluation of mosquitoes, Aedes vexans, as biological vectors of porcine reproductive and respiratory syndrome virus. Can J Vet Res. 2003;67:265–270. [PMC free article] [PubMed] [Google Scholar]
- 5.Dee SA, Deen J, Otake S, Pijoan C. An experimental model to evaluate the role of transport vehicles as a source of porcine reproductive and respiratory syndrome virus to susceptible pigs. Can J Vet Res. 2004;68:128–133. [PMC free article] [PubMed] [Google Scholar]
- 6.Goldberg TL, Hahn EC, Weigel RM, Scherba G. Genetic, geographical and temporal variation of porcine reproductive and respiratory virus in Illinois. J Gen Virol. 2000;81:171–179. doi: 10.1099/0022-1317-81-1-171. [DOI] [PubMed] [Google Scholar]
- 7.Mortensen S, Stryhn H, Søgaard R, et al. Risk factors for infection of sow herds with porcine reproductive and respiratory syndrome (PRRS) virus. Prev Vet Med. 2002;53:83–101. doi: 10.1016/s0167-5877(01)00260-4. [DOI] [PubMed] [Google Scholar]
- 8.Kristensen CS, Bøtner A, Takai H, Nielsen JP, Jorsal SE. Experimental airborne transmission of PRRS virus. Vet Microbiol. 2004;99:197–202. doi: 10.1016/j.vetmic.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Lager KM, Mengeling WL, Wesley RD. Evidence for local spread of porcine reproductive and respiratory syndrome virus. J Swine Health Prod. 2002;10:167–170. [Google Scholar]
- 10.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams E. Aviation formulary V1.42. Available at http://williams.best.vwh.net/avform.htm#GCF (accessed 2006 May 6).
- 12.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 13.Legendre P. Spatial autocorrelation: Trouble or new paradigm? Ecology. 1993;74:1659–1673. [Google Scholar]
- 14.Manly BJF. Randomization, Bootstrap and Monte Carlo Methods in Biology. London: Chapman and Hall, 1997:172–205.
- 15.Dietz EJ. Permutation tests for association between two distance matrices. Syst Zool. 1983;32:21–26. [Google Scholar]
- 16.Legendre P. Comparison of permutation methods for the partial correlation and partial Mantel tests. J Stat Comput Simul. 2000;67:37–73. [Google Scholar]
- 17.Dee SA. Elimination of porcine reproductive and respiratory syndrome from 30 farms by test and removal. J Swine Health Prod. 2004;12:129–133. [Google Scholar]
- 18.Larochelle R, D’Allaire S, Magar R. Molecular epidemiology of porcine reproductive and respiratory syndrome virus (PRRSV) in Québec. Virus Res. 2003;96:3–14. doi: 10.1016/s0168-1702(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 19.Dutilleul P, Stockwell JD, Frigon D, Legendre P. The Mantel test versus Pearson’s correlation analysis: assessment of the differences for biological and environmental studies. J Agr Biol Environ Stat. 2000;5:131–150. [Google Scholar]