Abstract
Yeast Abp1p is a cortical actin cytoskeleton protein implicated in cytoskeletal regulation, endocytosis, and cAMP-signaling. We have identified a gene encoding a mouse homologue of Abp1p, and it is identical to SH3P7, a protein shown recently to be a target of Src tyrosine kinases. Yeast and mouse Abp1p display the same domain structure including an N-terminal actin-depolymerizing factor homology domain and a C-terminal Src homology 3 domain. Using two independent actin-binding domains, mAbp1 binds to actin filaments with a 1:5 saturation stoichiometry. In stationary cells, mAbp1 colocalizes with cortical F-actin in fibroblast protrusions that represent sites of cellular growth. mAbp1 appears at the actin-rich leading edge of migrating cells. Growth factors cause mAbp1 to rapidly accumulate in lamellipodia. This response can be mimicked by expression of dominant-positive Rac1. mAbp1 recruitment appears to be dependent on de novo actin polymerization and occurs specifically at sites enriched for the Arp2/3 complex. mAbp1 is a newly identified cytoskeletal protein in mice and may serve as a signal-responsive link between the dynamic cortical actin cytoskeleton and regions of membrane dynamics.
INTRODUCTION
The dynamic regulation of the actin cytoskeleton is controlled by the Rho family of small GTPases, including Cdc42, Rac1, and RhoA (reviewed in Hall, 1998; Aspenström, 1999). Rac1 controls formation of lamellipodia (Ridley et al., 1992), sheet-like membrane protrusions containing highly dynamic networks of short, orthogonally arranged actin filaments representing the leading edge of motile cells (reviewed in Mitchison and Cramer, 1996). Actin dynamics in lamellipodia are thought to be facilitated by proteins that mediate the nucleation, polymerization, capping, severing, depolymerization, and cross-linking of actin filaments (reviewed in Hartwig and Kwiatkoswki, 1991; Mitchison and Cramer, 1996; Machesky and Gould, 1999). Despite this structural and functional complexity, nature uses a surprisingly small variety of actin-interacting domains (reviewed in Puius et al., 1998). Recently, we suggested a novel module, the actin-depolymerizing factor homology (ADF-H) domain (Lappalainen et al., 1998), found in a family of functionally diverse proteins. By sequence analysis, we subdivided this family into three classes. The first class of ADF-H proteins consists of cofilin, ADF, and other closely related proteins that are ∼15 kDa in size and promote actin filament disassembly (for review, see Moon and Drubin, 1995; Carlier, 1998; Mciver, 1998). The second class, the twinfilins, contain a simple repeat of the ADF-H domain and bind to and sequester actin monomers (Goode et al., 1998). The third class consists of Abp1s (Drubin et al., 1988; Drubin et al. 1990; Lange et al. 1994) and drebrins (Shirao and Obata, 1985). Abp1s and drebrins contain an N-terminal ADF-H domain, and Abp1s also contain a C-terminal Src homology 3 (SH3) domain. Drebrins bind to F-actin but are unable to bind to monomeric actin (Ishikawa et al., 1994). Saccharomyces cerevisiae Abp1p was identified by F-actin affinity chromatography and localizes to cortical actin patches, structures that are concentrated at growing surfaces of yeast cells (Drubin et al., 1988). Overexpression of Abp1p results in a depolarized distribution of cortical actin patches and loss of spatial regulation of cell growth. In contrast, deletion of the ABP1 gene does not cause any obvious defects (Drubin et al., 1988). This lack of a null phenotype reflects functional redundancy within the cytoskeleton, because Abp1p becomes essential when any one of the cytoskeletal genes SAC6 (yeast fimbrin), SLA1, SLA2, and PRK1 is deleted (Holtzman et al., 1993; Cope et al., 1999). Mutants of these genes show defects in actin cytoskeleton organization and in the uptake step of receptor-mediated endocytosis (reviewed in Geli and Riezman, 1998).
A variety of findings suggest an intimate functional connection between the actin cytoskeleton and membrane dynamics: the involvement of the actin cytoskeleton in exocytosis in yeast and mammals is established, although not fully understood (Muallem et al., 1995; Vitale et al., 1995; Bi et al., 1997; Finger and Novick, 1998). Additionally, the actin cytoskeleton has been implicated in different steps of both receptor-mediated and fluid phase endocytosis by the use of actin-disrupting drugs, ultrastructural and biochemical analysis, and the analysis of yeast mutants (reviewed in Geli and Riezman, 1998). Nevertheless, a connection between the actin cytoskeleton and membrane dynamics remains elusive, because the proteins underlying such connections remain to be identified. Yeast Abp1p is a molecular candidate for such a link, because this cytoskeletal component is genetically redundant with proteins important for endocytosis and also interacts physically with Rvs167p, a yeast protein implicated in endocytosis (Lila and Drubin, 1997; Wesp et al. 1997). Rvs167 is related to amphiphysin, a vertebrate protein involved in receptor-mediated endocytosis (Lichte et al., 1992). Because yeast and mammalian cells have complementary advantages for studies of cell biology, we searched for mammalian homologues of yeast Abp1p. Our hope was that studies of a mammalian Abp1p homologue might yield novel insights into the functions of yeast Abp1 and an opportunity to test whether principles from studies of Abp1p in yeast would apply in mammalian cells.
Here we report the identification of Abp1s in different species and show that Abp1s, together with drebrins, form a functionally distinct class of ADF-H domain proteins. We demonstrate that mAbp1 is expressed in most if not all tissues of the mouse and functions as an F-actin-binding protein in vitro and in vivo. mAbp1 uses two different, independent actin-binding modules, the ADF-H domain and a novel actin-binding motif, a highly charged helical domain. Our results suggest that through its participation in dynamic cortical actin rearrangements triggered by Rac-activated pathways, mAbp1 is involved in polarized cell growth and cell motility.
MATERIALS AND METHODS
DNA Constructions
The following murine expressed sequence tag (EST) clones were obtained from the American Type Culture Collection (Manassas, VA): GenBank accession numbers AA000682 (ESTm1), W98470 (ESTm2), AA119182 (ESTm3), AA017916 (ESTm5), and W09622 (ESTm8). The plasmid DNA was isolated and sequenced. These EST clones and the full-length cDNA of SH3P7/mAbp1 in pEXlox (kindly provided by Brian Kay, University of Wisconsin-Madison, Madison, WI) were used to generate the different constructs used in this study. The serine 235 codon for which we found a polymorphism was included in all generated plasmids containing this region. To construct glutathione S-transferase (GST)-mAbp1 fusion plasmids for expression in bacteria, DNA sequences encoding either the full-length protein (aa 1–433) or truncations were amplified by PCR using primers that generate BamHI and HindIII sites at the 5′ and 3′ ends, respectively. The PCR products were cut with BamHI and HindIII and ligated into the pGAT2 vector (a GST fusion derivative of pBAT; Peränen et al., 1996) cut with the same enzymes to generate pGAT2-mAbp1(1–146), pGAT2-mAbp1(1–163), pGAT2-mAbp1(1–281), pGAT2-mAbp1(165–433), pGAT2-mAbp1(282–370), pGAT2-mAbp1(282–433), and pGAT2-mAbp1(371–433). All generated constructs were sequenced to ensure that no mutations were introduced during the construction of the plasmids.
Production of Polyclonal Anti-mAbp1 Antibodies
Polyclonal anti-mAbp1 antibodies were raised against two different regions of mAbp1. First, a peptide comprising the N-terminal 26 amino acids of the protein plus a C-terminal glycine-cysteine linker was synthesized by Dr. David King (University of California, Berkeley, CA). The purity of the peptide was confirmed by HPLC. The peptide was dissolved in 0.2 mM Tris, pH 8.2, containing 20% dimethylformamide and triterated with a pipette. The C-terminal cysteines were oxidized by stirring the peptide solution in contact with air for 18 h. The resulting disulfide-bridged peptide dimers were mixed with Ribi adjuvant (R-700; Ribi ImmunoChem Research, Hamilton, MT) and subcutanously injected into three Hartley guinea pigs (100 μg of peptide per injection). After the initial injection, boosts were given every 3 wk. All animals showed an immune response. The crude sera were tested by immunoblotting against a dilution series of purified, Escherichia coli-expressed GST-mAbp1 ranging from 500 to 5 ng, against E. coli-expressed mAbp1 fragments in crude E. coli homogenates, and against mouse tissue homogenates. After six injections, antisera were collected, and the crude antiserum from guinea pig 1 (GP1) was affinity purified according to a blot purification protocol (Qualmann et al., 1999) using GST-mAbp1(1–146) fusion protein as a matrix. Second, polyclonal guinea pig antibodies were raised against the so-called flexible domain of mAbp1 (aa 282–370), which was expressed as a GST fusion protein in E. coli. The protein was purified on glutathione-agarose, cleaved with thrombin (Sigma, Saint Louis, MO), and purified away from the GST by gel filtration. Pure mAbp1(aa 282–370) protein was injected into three guinea pigs (110 μg per injection). All three animals showed an immune response. The sera were screened as described above. After five injections, antisera were collected, and the crude antiserum GP5 was purified by blot affinity purification using mAbp1(aa282–370) as a matrix.
Northern Blot Analysis of mAbp1 mRNA Levels
Northern blot analyses of mAbp1 mRNA levels were performed using blots containing either RNA from multiple mouse tissues or RNA from mouse embryos at multiple embryonic stages. The RNA blots (Clontech, Palo Alto, CA) contained 2 μg of poly(A)+ RNA per lane. The clone ESTm1, which we determined to contain a 456-bp oligonucleotide insert corresponding to aa 282–433 of mAbp1, was used as a PCR template to generate a DNA fragment corresponding to aa 282–382. This DNA fragment was subsequently used to generate a probe labeled with [α-32P]CTP (Amersham, Bukinghamshire, United Kingdom) using an oligonucleotide labeling kit, which uses Klenow polymerase (Pharmacia, Piscataway, NJ). Unincorporated nucleotides were removed using MicroSpin S-200 columns (Pharmacia); 37.5 ng of the probe with a specific activity of 4.7 × 105 cpm/ng were used for the incubations of each prehybridized RNA blot in ExpressHyb solution (Clontech) at 68°C for 75 min. The filters were then washed three times for 1 min each and three times for 10 min each with 300 mM NaCl, 30 mM sodium citrate, pH 7.0, containing 0.05% SDS at room temperature and were subsequently washed twice (20 min each) with prewarmed 15 mM NaCl, 1.5 mM sodium citrate, pH 7.0, containing 0.1% SDS at 50°C. mAbp1 RNAs were detected by autoradiography (exposure time, 48 h). To control for RNA loadings and integrity, a probe derived from a human β-actin cDNA (Clontech) was used. Blots were reused after stripping them in 0.5% SDS at 90–100°C for 10 min under frequent rocking and testing for the absence of any remaining radioactive probe by autoradiography.
Immunoblot Analysis of mAbp1 Expression and Subcellular Fractionation
Tissues for immunoblot analysis were harvested from adult female Swiss Webster mice and immediately frozen in liquid nitrogen. The excised organs and tissues were cut into small pieces, which were then homogenized at 4°C with Dounce glass homogenizers (size AA, ≈20 strokes) in 3 ml/g tissue homogenization buffer A (250 mM sucrose, 10 mM EGTA, 2 mM EDTA, 20 mM, Tris pH 7.5) containing 10 μg/ml leupeptin, 10 μg/ml aprotinin, 10 μg/ml soybean trypsin inhibitor, and 1 mM PMSF as protease inhibitors (Sigma). The samples were spun 20 min at 4000 rpm in a microcentrifuge, and the resulting postnuclear supernatants were used for the Western blot analyses. Equal amounts of protein per lane were loaded.
Cell line homogenates were prepared by growing cells to confluence, rinsing them with ice-cold PBS, scraping the cells off the plates, collecting them by centrifugation, and lysing the cell pellets in 350 μl per 5 × 106 cells ice-cold homogenization buffer B (150 mM NaCl, 2 mM EDTA, 1% NP-40, 50 mM HEPES, pH 7.5) supplemented with the same protease inhibitors as described above. The homogenization was subsequently completed using a Dounce homogenizer at 4°C. The resulting homogenates were spun for 5 min at 5000 rpm at 4°C in a microcentrifuge.
For subcellular fractionations, 0.7–0.8 g of brain or spleen tissue was homogenized in homogenization buffer (1:1 wt/vol) and spun at 3000 × g for 20 min. P1 contains unbroken cells, nuclei, mitochondria, and large plasma membrane pieces. The supernatants (S1) were collected and spun 30 min at 25,000 × g to generate P2 (small plasma membrane pieces, endoplasmic reticulum, and endosomes) and S2. Supernatant 2 was spun at 176,000 × g for 1 h to generate P3 containing light microsomes and small vesicles and S3 containing soluble proteins. Fractions containing equal amounts of protein were examined by SDS-PAGE and immunoblotting.
SDS-PAGE was conducted using a buffer system based on the method of Laemmli (1970). Proteins were transferred to nitrocellulose membranes. After blocking with 5% nonfat milk in PBS containing 0.05% Tween 20, mAbp1 was detected with the affinity-purified anti-mAbp1 antibodies GP1 and GP5. The primary antibodies were visualized by horseradish peroxidase-conjugated anti-guinea pig secondary antibodies (ICN Pharmaceuticals, Aurora, OH) and ECL development (Amersham).
In Vitro Actin Interaction Assays
Actin Filament Cosedimentation Assays.
Actin filaments were assembled as described by Goode et al. (1999) with minor alterations. Briefly, 0.1 final volume of 10× initiation mix (20 mM MgCl2, 0.5 M KCl, 5 mM ATP) was added to human, nonmuscle monomeric actin (Cytoskeleton, Denver, CO) diluted in G buffer (5 mM Tris, pH 7.5, 0.2 mM ATP, 0.2 mM DTT, 0.2 mM CaCl2) and incubated for 30 min at 20°C. The cosedimentation assays were performed using 50-μl reactions containing variable concentrations of actin (see Figure 2). In other experiments, the actin concentration was held constant at 10 μM, and the concentrations of actin-binding mAbp1 fragments were varied from 0.25 to 8 μM. These experiments permit the determination of the kD and the saturation stoichiometry. All reaction mixtures were centrifuged for 25 min at 90,000 rpm, 20°C in a TLA100 rotor (Beckman Instruments, Palo Alto, CA). Equivalent portions of pellets and supernatants were analyzed by SDS-PAGE, and proteins were stained with Coomassie blue and quantified using an IS-100 densitometer (Alpha Innotech, San Leandro, CA). The different mAbp1 fragments used for the analysis were expressed as GST fusion proteins in E. coli BL21(DE3) cells using the pGAT2 constructs described under DNA Constructs. The expression of the fragments was induced by addition of 400 mM isopropyl-thio-β-d-galactoside (Sigma). Cells were lysed by freeze–thawing followed by sonication and addition of 0.2 mg of lysozyme/ml of cell suspension in PBS containing 0.5 mM PMSF and 10 mM EDTA. GST fusion proteins were subsequently purified using glutathione-agarose beads as described by Ausubel et al. (1990).
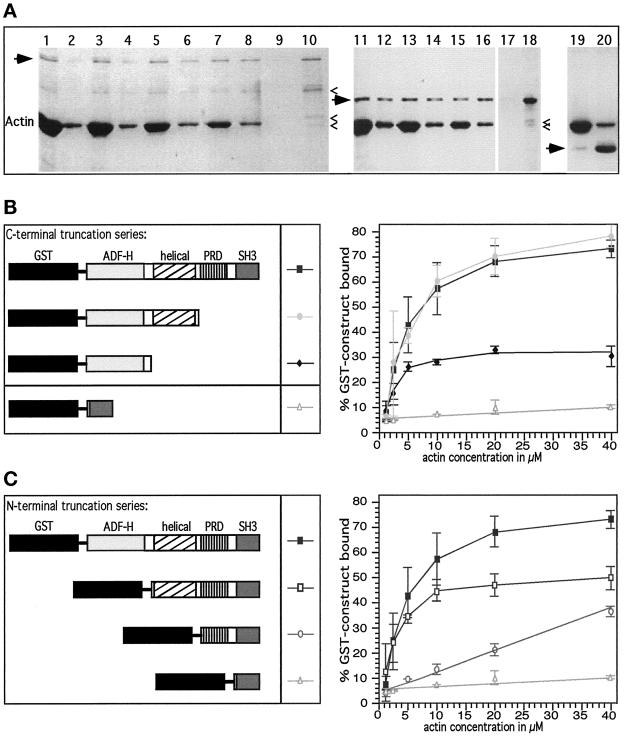
Figure 2.
Evidence that mAbp1 is an F-actin-binding protein and contains two different N-terminal F-actin-binding modules. (A) Examples of cosedimentation assays. Odd lanes are pellets; even lanes are supernatants. Coomasie blue-stained gel. Arrows mark mAbp1 fusion proteins, and carets indicate degradation products. Note that the largest degradation product of full-length mAbp1 (lanes 1–10) is the result of degradation of the flexible domain as judged by immunoblottings and that it still binds to F-actin. Lanes 1–8, 1 μM GST-mAbp1 pelleting with different amounts of F-actin (5, 10, 20, and 40 μM). Lanes 9 and 10, GST-mAbp1 alone. Lanes 11–18, pelleting of the N-terminal half of mAbp1 (ADF-H and helical domain) with 5, 10, and 20 μM F-actin. Lanes 19 and 20, 3 μM GST-SH3 domain fusion protein with 20 μM F-actin. (B and C) C-terminal truncation series and N-terminal truncation series of mApb1 fusion proteins, respectively (left panels), which were used for actin cosedimentation experiments with different actin concentrations (right panels). Cosedimentation assays were quantified densitometically. Percentages of mAbp1 bound to actin were determined. Values are given as mean and SD of two to five independent experiments.
Nucleotide Exchange Assays.
Effects of mAbp1 and fragments of mAbp1 on actin nucleotide exchange were tested fluorometrically. Forty microliters of monomeric actin (2.5 μM) in nucleotide exchange buffer (10 mM Tris, pH 7.5, 0.2 mM CaCl2, 0.5 mM DTT, 25 μM ATP) containing 0, 1.25, 2.5, 5, or 7.2 μM of the proteins to be tested were mixed with 10 μl of 1 mM etheno-ATP (Molecular Probes, Eugene, OR). The increase of the fluorescence during the nucleotide exchange reaction was followed for 30 min at 25°C using 360 nm for excitation and 410 nm for emission in an F-4010 fluorescence spectrophotometer (Hitachi Instruments, Tokyo, Japan). The t1/2 values were determined and compared.
Actin Monomer Binding Assays.
Interactions of mAbp1 and its fragments (3.3 μM final concentration) with monomeric actin (6.6 μM final) were monitored by native gel electrophoresis as described by Safer (1989). Various acrylamide concentrations and running times were used to achieve optimal resolutions of G-actin from the different mAbp1 protein fusions. Recombinant yeast cofilin was used as a positive control. The proteins were detected by Coomassie blue staining.
Actin Assembly and Disassembly Assays.
Actin assembly was followed by light scattering as described previously (Goode et al., 1999), except that reactions were carried out at 10°C and were followed for 30 min. The actin disassembly assays were performed as described by Lappalainen et al. (1997), except that human nonmuscle actin was diluted to 0.25 μM and that the reactions were followed for 10 min.
Cell Culture
The human lung fetal fibroblast cell line IMR90, NIH 3T3 fibroblasts, Cos-7 cells, and Raw264.7 macrophages were maintained in Dulbecco's modified Eagle's medium (DMEM; Life Technologies, Gaithersburg, MD) containing 10% fetal bovine serum.
Labeling of Endosomal Compartments in Cos-7 Cells
The endosomes of the clathrin-dependent, receptor-mediated endocytic pathway were labeled by either continuous uptake or by pulse–chase of transferrin. For these assays, Cos-7 cells of ∼30% confluence were serum starved for 45 min at 37°C and subsequently were incubated with 25 μg/ml human transferrin labeled with Texas Red (Molecular Probes) for either 1 h at 37°C or 2 h at 15°C, washed three times for 5 min with ice-cold PBS containing 1 mg/ml BSA and fixed (continuous uptakes). For the pulse–chase experiments, the cells were incubated with the transferrin for 1 h at 4°C, washed as above, and then fixed with an ice-cold solution of 4% formaldehyde and 0.1% glutaraldehyde (0 min) or warmed to 37°C for 2, 5, 15, 30, or 60 min before they were washed with ice-cold PBS containing BSA and fixed. The fixed cells were then double stained for mAbp1 and examined by confocal microscopy as described under Indirect Immunofluorescence Microscopy.
Fibroblast Wound-healing Assay
IMR90 cells were grown to confluence on glass coverslips and maintained in DMEM containing serum for 1 more day. The cell layers were then wounded using a needle. Wounds of ∼500 μm in width were created. Upfolding of cell sheets was avoided. When single cells remained in the wound, these cells were removed under microscopic observation. The cells were incubated in DMEM containing serum at 37°C for 2, 4, or 6 h, processed for immunofluorescence, and stained for mAbp1, actin, paxillin (a marker for focal adhesions), and/or dynamin II to determine the position of the Golgi apparatus.
Treatment of NIH3T3 Cells with Different Growth Factors and Latrunculin A
NIH 3T3 cells were grown to confluence. The medium was replaced, and the cells were maintained for 3 more days. The quiescent fibroblasts were then serum starved in DMEM supplemented with 20 mM HEPES, pH 7.55, for 16–20 h. The cells were washed, trypsinized, and replated onto glass coverslips (either uncoated or fibronectin-coated) to achieve an optimal cell density for immunofluorescence. The fibroblasts were allowed to attach for 1–2 h in DMEM containing HEPES at 37°C and then treated with 5 ng/ml human recombinant platelet-derived growth factor (PDGF; Sigma) for 10 min, with 300 ng/ml phorbol 12-myristate 13-acetate (PMA; Sigma) for 10 min, with 200 ng/ml bradykinin (Sigma) for 10 min, or with 100 ng/ml lysophosphatidic acid (Sigma) for 15 min. Activation of the cells by PDGF and PMA was also studied at different time points (1–10 min). For some experiments, PDGF and PMA treatments were combined with a disruption of the actin cytoskeleton by addition of Latrunculin A. For this purpose, cells were either pretreated with 2 μM Latrunculin A (a gift from P. Crews, University of California, Santa Cruz, CA) for 20 min, or the drug was added together with PDGF or PMA for varying times. A subset of cells was left untreated or was allowed to recover by adding serum-containing DMEM. All cells were rapidly washed with PBS after the times indicated, fixed, and processed for fluorescence microscopy.
Transfection of NIH 3T3 Cells with Dominant-Positive Rac1
Cells were grown as for growth factor treatment and were then replated onto glass coverslips to achieve a lower cell density for immunofluorescence microscopy. These cells were transfected with 0.5 μg/well (24-well cell culture plate) mycRac1L61-carrying pRK5 plasmid generously provided by Dr. Alan Hall (University College London, London, United Kingdom) using the LipofectAMINE method according to the supplier (Life Technologies) for 4 h in serum-free Optimem cell culture medium. Control cells were incubated with the transfection solution lacking DNA. The transfection solution was removed, and the cells were washed and maintained for 3 additional hours in serum-free DMEM before they were washed with 50 mM 2-[N-morpholino]ethanesulfonic acid (MES) pH 6.1, 5 mM MgCl2, 3 mM EGTA, 5 mM glucose and fixed. mycRac1L61 expression was detected using an anti-myc antibody (monoclonal 9E10) followed by Alexa 350-labeled anti-mouse. FITC- and rhodamine-labeled secondary antibodies were used to detect mAbp1 and F-actin, as described under Indirect Immunofluorescence Microscopy.
Indirect Immunofluorescence Microscopy
Cells grown on glass coverslips were washed with 50 mM MES, pH 6.1, 5 mM MgCl2, 3 mM EGTA, 5 mM glucose and were then incubated with 4% formaldehyde, 0.1% glutaraldehyde in the same buffer for 30 min. The samples were then quenched with 1 mg/ml NaBH4 (two times for 5 min) and blocked with Tris-buffered saline (TBS; 20 mM Tris pH 7.5, 154 mM NaCl, 2 mM EGTA, 2 mM MgCl2) containing 2% BSA and 0.02% saponin for 1.5 h. Subsequently, the cells were incubated on droplets of primary antibody dilutions in TBS, BSA and saponin for 1.5 h. In separate experiments, NIH 3T3 cells were extracted with 0.1% Triton X-100 in 50 mM MES, pH 6.1, 5 mM MgCl2, 3 mM EGTA, 5 mM glucose for 3–5 s before fixation. The affinity-purified anti-mAbp1 guinea pig antibodies were usually used at 1:50; the monoclonal anti-paxillin antibody (Transduction Laboratories, Lexington, KY) was used at 1:200; the monoclonal anti-dynamin II antibody (Transduction Laboratories) was used at 1:100; the monoclonal anti-tubulin antibody (ICN) was used at 1:200; the monoclonal anti-Rab8 antibody (Transduction Laboratories) was used at 1:50; the affinity-purified polyclonal anti-Arp3 rabbit antibody (kindly provided by Dr. Matthew D. Welch, University of California, Berkeley, CA) was used at a dilution of 1:100; and the monoclonal anti-myc antibody 9E10 (Santa Cruz Biotechnology, Santa Cruz, CA) was used at 1:200. After incubation with the antibodies, the coverslips were washed three times for 10 min with TBS containing BSA and saponin and incubated with secondary antibodies (FITC goat anti-guinea pig [ICN; 1:750], rhodamine goat anti-guinea pig [ICN; 1:600], FITC donkey anti-mouse [Jackson ImmunoResearch, West Grove, PA; 1:100], rhodamine donkey anti-mouse [Jackson; 1:500], FITC goat anti-rabbit [ICN; 1:750], and/or rhodamine-phalloidin [Molecular Probes; 1:1200], and Alexa 350 goat anti-mouse [Molecular probes; 1:100]) in TBS and BSA for 1 h. The samples were then washed with TBS containing BSA (10 min) and with TBS (two times for 10 min), liberated from excess of buffer, mounted onto glass slides using VectaShield mounting medium containing or lacking DAPI, respectively (Vector Laboratories, Burlingame, CA), sealed with nail polish, and viewed with an inverted Nikon (Tokyo, Japan) Eclipse TE300 fluorescence microscope or with a Leica (Wetzlar, Germany) TCS NT laser confocal microscope with a Leica TCS software package. Images were recorded digitally (ImageProPlus; Phase3 Imaging Systems, Glen Mills, PA) and processed using Adobe (Mountain View, CA) Photoshop software.
Miscellaneous
Protein concentrations were determined by using the following 280-nm extinction coefficients calculated from the amino acid composition: ε = 5.99 mM−1cm−1 for mAbp1(aa 282–370); ε = 90.00 mM−1cm−1 for GST-mAbp1(full-length, aa 1–433); ε = 42.67 mM−1 cm−1 for GST-mAbp1(aa 1–281); ε = 55.91 mM−1cm−1 for GST-mAbp1(aa 371–433); ε = 55.86 mM−1cm−1 for GST-mAbp1(aa 1–146); ε = 56.94 mM−1cm−1 for GST-mAbp1(aa 1–163); ε = 72.93 mM−1cm−1 for GST-mAbp1(aa 165–433); ε = 60.59 mM−1cm−1 for GST-mAbp1(aa 282–433); and ε = 15.9 mM−1cm−1 for yeast cofilin. The extinction coefficient for actin at 290 nm is ε = 28.8 mM−1cm−1. In addition, protein concentrations were determined by the method of Bradford (1976).
RESULTS
Identification and Sequence Analysis of a Mouse Homologue of Yeast Abp1p
We performed a database query and identified several EST clones from different species encoding proteins of similarity to S. cerevisiae Abp1p. These EST clones encoded drebrins, neuronal Abp1-like proteins, and a single novel protein in mouse, rat, and human. Different EST clones encoding the novel protein were obtained, and their full sequences were determined. All mouse clones corresponded to SH3P7, a gene found in a phage display screen for SH3-containing proteins (Sparks et al., 1996). We noticed a small polymorphism: the codon for serine 235 was absent in two of three clones containing this region. Neither splice variants nor isoforms of SH3P7 have been identified thus far. For all further experiments, we used either plasmids from suitable EST clones or the full-length SH3P7 cDNA (kindly provided by Dr. Brian Kay) containing serine 235. We will henceforth refer to SH3P7 as mAbp1 (for mouse or mammalian actin-binding protein 1, respectively) in deference to the previously identified yeast protein (Drubin et al. 1988). Furthermore, we identified an open reading frame in Caenorhabditis elegans (GenBank accession number Z81568), which encodes an uncharacterized protein closely resembling S. cerevisiae and mouse Abp1 (cAbp1; Figure 1).
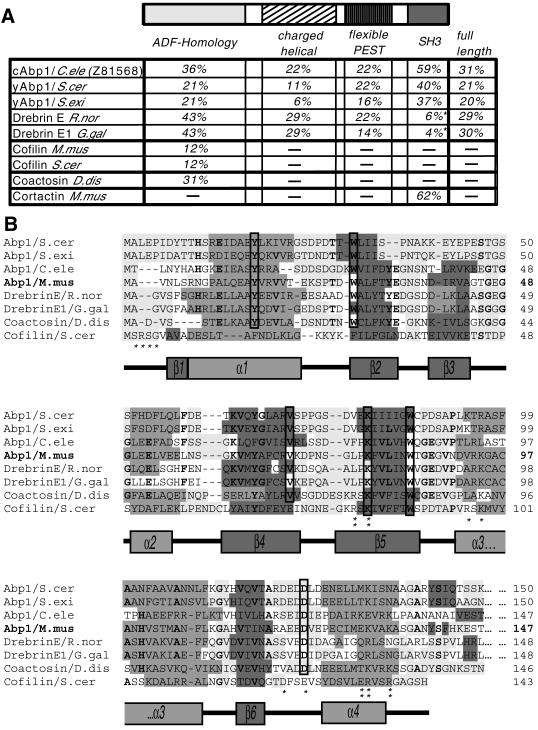
Figure 1.
Analysis of the mAbp1 sequence. (A) Domain structure of mAbp1/SH3P7 (GenBank accession number U58884) and comparison of the sequence identities of individual domains and the full-length protein of mAbp1 to corresponding areas of related proteins of the Abp1/drebrin class of ADF-H domain proteins and cortactin. Note that drebrins, in contrast to what we reported earlier (Lappalainen et al., 1998), seem not to contain a C-terminal SH3 domain. Accession numbers: S. cerevisiae Abp1 (European Molecular Biology Laboratory, X51780; Swiss-Prot, P15891), Saccharomyces exiguus Abp1 (Swiss-Prot, P38479), Rattus norvegicus drebrin (Swiss-Prot, Q07266), Gallus gallus drebrinA, E1 and E2 (Swiss-Prot, P18302), Mus musculus cofilin1 (nonmuscle isoform) (Swiss-Prot, P18760), S. cerevisiae cofilin (Swiss-Prot, Q03048), C. elegans Abp1 (ORF) (GenBank, Z81568), Dictyostelium discoideum coactosin (Swiss-Prot, P34121), M. musculus cortactin (Swiss-Prot, Q60598). The three most similar proteins in each column are highlighted in bold. (B) Identification of a common fold among Abp1s, drebrins, coactosin, and cofilin by aligning their N termini and predicting their secondary structures (light gray, flexible regions; medium gray, α-helices; dark gray, β-sheets; no underlying shading, no prediction). Predictions were performed using the PHDsec program (Rost and Sander, 1994). Residues shown by mutational analysis of yeast cofilin to be important for G and F-actin-binding (Lappalainen et al., 1997) are marked by stars; those important for F-actin binding only are marked by double stars. Residues that are >70% conserved within the ADF-H domains of Abp1s, drebrins, and coactosin are in bold, whereas residues fully conserved are boxed.
Computer analyses revealed high sequence identities between mAbp1 and drebrins because of strong conservation of their N-terminal halves. Yeast Abp1p shows a lower sequence identity (Figure 1A), as expected from the phylogenetic distance. Importantly, yeast Abp1p is more similar to mAbp1 than to drebrins. Among all Abp1s and drebrins identified, the putative protein in C. elegans (cAbp1; ORF Z81568) shows the highest sequence homology to mAbp1 (31% identity over the whole amino acid sequence). In contrast to drebrins, similarity was found in all domains of the protein. Thus, Z81568 should be considered a C. elegans Abp1 and not a drebrin.
In addition, we discovered that secondary structure predictions for ∼150 amino acids at the N terminus of yeast Abp1s, drebrins, C. elegans Abp1, and mAbp1 all resemble that of the small actin-depolymerizing and -severing protein cofilin/ADF (Figure 1B). Accordingly, mAbp1 is predicted to display a fold consisting of a central six-stranded mixed β-sheet, sandwiched between two pairs of α-helices, one on each face. Six residues of mAbp1 (Tyr-16, Trp-29, Val-69, Lys-78, Trp-84, and Asp-122) are absolutely conserved among the entire Abp1/drebrin class and the related F-actin-binding protein coactosin (DeHostos et al., 1993). The charged residues Lys-78 and Asp-122 correspond to Lys-82 and Glu-126 shown to be crucial for binding of yeast cofilin to actin (Lappalainen et al., 1997). Other positions of charged side chains clustered in α-helices 3 and 4 that have been implicated in actin binding by mutational analysis are also maintained (Figure 1B). Using the ADF-H domain in database searches, we also identified a third class of proteins displaying a cofilin/ADF-like fold, the G-actin-binding twinfilins (Goode et al., 1998). Although the N terminus of mAbp1 shows homology to the ADF-H domains of ADF/cofilins and the twinfilins, phylogenetic analysis of the ADF-H domains of these proteins (our unpublished results) and analysis of domain organization (Figure 1A) clearly indicate that mAbp1 belongs in the drebrin/Abp1 class of ADF-H domain proteins.
The domain organization of mAbp1 is shown in Figure 1A: the N-terminal ADF-H domain is followed by a domain that is characterized by alternating positive and negative charges and that is predicted to be of α-helical conformation. This domain of mAbp1 (approximately aa 162–275) contains only two obvious regions (around aa 237 and 261) within which this helical conformation is predicted to be disrupted by turns. The charged helical domain of mAbp1 is followed by a flexible, hydrophilic region containing a proline-, glutamate-, serine-, and threonine-rich region, which is highly scored as a PEST protein degradation sequence (Rodgers et al., 1986). This region is indeed easily accessible for proteases and rapidly degraded (our unpublished results; Figure 2A). In this flexible region, the drebrin/Abp1 proteins show high diversity, but all contain putative PEST sequences. Mouse and C. elegans Abp1s also contain Src-tyrosine phosphorylation consensus sites, consistent with the observation that mAbp1 (SH3P7) was identified as a Src substrate (Lock et al. 1998, Larbolette et al. 1999). All Abp1s also contain a potential SH3 domain-binding sequence in this region. Finally, an SH3 domain is found at the C-terminus of every Abp1 protein. The similarity of the SH3 domains of mAbp1 and corresponding areas of drebrins is extremely low (4–6% identity depending on species), most likely because the C terminus of drebrins does not represent an SH3 domain. In contrast, the SH3 domain of mAbp1 shows significant similarity to that of the yeast Abp1s (40% identity for S. cerevisiae Abp1) and the putative C. elegans protein (59% identity) despite the phylogenetic distances separating these proteins. Additionally, with a 62% amino acid identity, the mAbp1 SH3 domain displays a very high similarity to the SH3 domain of the actin-binding protein cortactin (Wu and Parsons, 1993), a protein that does not belong to the ADF-H domain protein family (Figure 1A).
Biochemical Characterization of Interactions between mAbp1 and Actin
To investigate the interactions of mAbp1 with F- and G-actin and to determine which domain(s) mediate the interaction(s), we constructed series of N- and C-terminal domain truncation mutants of mAbp1 as GST fusion proteins (Figure 2, B and C, left panels). Proteolysis and expression problems precluded analysis of these expressed proteins in the absence of GST. The ability of these GST fusion proteins to bind to actin filaments was tested in F-actin cosedimentation assays (Figure 2).
Full-length mAbp1 bound to F-actin in a saturable, concentration-dependent manner (Figure 2, B and C, right panels). Tight binding (kD ≤ 2.25 ± 0.25 μM) was seen in experiments in which mAbp1 concentration was changed and the actin concentration was held constant. A more detailed analysis showed that even the smallest fragment tested in the C-terminal truncation series, the ADF-H domain alone, still bound to F-actin in a specific, saturable manner, although the affinity of binding was reduced compared with the larger fragments (Figure 2B, right panel). The reduction in affinity that resulted from removal of the helical domain (Figure 2B) suggests a contribution of this domain to the overall binding affinity of mAbp1, a conclusion supported by further analysis (below). Whereas neither of the two domains at the C terminus of mAbp1 (the flexible region and the SH3 domain) showed saturable F-actin binding (Figure 2C, right panel), an mAbp1 fragment comprising these domains plus the helical domain bound to F-actin in a saturable manner (Figure 2C, right panel). Thus, this charged helical domain alone appears to serve as an F-actin-binding module, although the percentage of binding at saturation was reduced compared with the full-length mAbp1 or the construct containing both N-terminal domains (ADF-H and helical; Figure 2C, right panel). Because the helical domain is characterized by a high content of alternating positive and negative charges, we doubled the salt concentration in our cosedimentation assays and reexamined the F-actin binding of this domain: We still observed a high level of specific and saturable binding (our unpublished results). Thus, mAbp1 contains two actin-binding motifs, both located in the N-terminal half of the protein: the ADF-H domain and the charged helical domain. The latter represents a novel actin-binding motif.
Because the members of both other classes of ADF-H domain-containing proteins, the cofilin/ADFs and the twinfilins, bind to and reduce the nucleotide exchange rate of G-actin, we next asked whether mAbp1 shares this property. Our nucleotide exchange assays, however, displayed no such activity for any construct tested, whereas a cofilin control showed a dramatic, concentration-dependent reduction in actin nucleotide exchange rate (our unpublished results; Table 1). Furthermore, we performed G-actin binding assays by native gel electrophoresis and detected no binding of GST-mAbp1 or different individual mAbp1 domains to G-actin, whereas cofilin used as a control comigrated with G-actin (our unpublished results). Because ADF/cofilins bind to F-actin and display F-actin-severing and depolymerizing activities, we next asked whether full-length mAbp1 or fragments of mAbp1 show similar activities. F-actin depolymerization assays did not reveal any such activities (our unpublished results). Similarly, we detected no effects of mAbp1 or any of its fragments tested on the polymerization kinetics of actin filament assembly (our unpublished results; Table 1).
Table 1.
Comparison of the activities of the ADF-H domain-containing proteins cofilin/ADFs, yeast twinfilin, and rat drebrin with those examined for GST-mAbp1 expressed in E. coli
| Mouse Abp1, E. coli expressed | Rat drebrin, Purified material | Yeast twinfilin, E. coli expressed | Cofilins/ADFs, E. coli expressed, Purified material | |
|---|---|---|---|---|
| F-actin binding | Yes | Yes | Yes | Yes |
| kD of F-actin binding (μM) | 2.2 ± 0.5 | 0.12 | No binding | ≈1 |
| Stoichiometry of actin binding | 1:5 | 1:5 | 1:1 | 1:1 |
| Number of actin-binding modules | Two | ? | ? | One |
| F-actin depolymerization and severing | No | No | No | Yes |
| F-actin bundling | No | No | No | No |
| F-actin capping | No | No | ? | ? |
| Actin nucleation | No | No | No | No |
| Regulation of actin binding activity | ? | ? | ? | pH; Ser3-P and PIPs inhibit |
| G-actin binding | No | No | Yes | Yes |
| Effect on actin nucleotide exchange | None | ? | Rate decreased | Rate decreased |
| Crystal structure | ADF-H domain solved recently (see below): central 6-stranded β-sheet with 2-α-helices on each side | ? | Work in progress | Solved for three different members of the family: central 6-stranded β-sheet with 2 α-helices on each side |
Yeast Abp1 (not included in table) is biochemically uncharacterized; however, it was shown to bind directly or indirectly to F-actin (Drubin et al., 1988), and the crystal structure of its ADF-H domain has recently been solved together with that of mAbp1 (Strokopytov and Almo, personal communication). Note that because of GST dimerization the actual kD for mAbp1 may be as low as half of the value determined. The saturation stoichiometry for GST-mAbp1 was 2.4 ± 0.3; thus the resulting value for mAbp1 (4.8 ± 0.6) was rounded up to 5. The data for drebrins were taken from Ishikawa et al. (1994), for twinfilin see Goode et al. (1998), and data for cofilins/ADFs were taken from a variety of studies as summarized by Moon and Drubin (1995), Carlier (1998), Mciver (1998), and Lappalainen et al. (1998).
Thus, as summarized in Table 1, recombinant mAbp1 does not depolymerize, cap, or bundle F-actin, nor does it bind to G-actin and/or inhibit the actin nucleotide exchange. The data from our study clearly indicate that mAbp1 displays activities typical for neither the ADF/cofilin class nor the twinfilin class of ADF-H domain proteins but functionally belongs to the drebrin/Abp1 class (see Table 1), as it does by sequence homology. This conclusion is further corroborated by the observation that both drebrins and mAbp1 bind to F-actin with a 1:5 M saturation stoichiometry (Table 1).
Expression Analysis of mAbp1 mRNA and Protein
Northern blot analysis of mouse embryos showed that mAbp1 expression is high during early development and drops during later developmental stages (Figure 3A). In contrast to drebrins (for review, see Shirao, 1995), mAbp1 mRNA is readily detectable in adult tissues and rather than being restricted to the brain is expressed ubiquitously (Figure 3). It should be noted, however, that levels of mAbp1 mRNA in testis, heart, and particularly skeletal muscle are low (Figure 3B). Two mAbp1 mRNAs of 1.8 and 2.9 kb were detected at approximately a 1:1 ratio in all tissues tested (Figure 3, A and B).
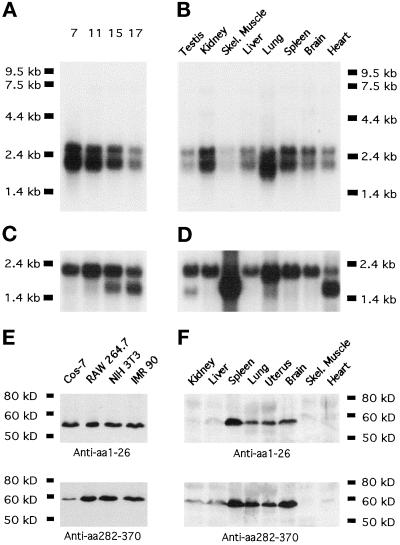
Figure 3.
Mammalian Abp1 mRNA and protein expression levels. (A) mAbp1 mRNA expression examined by Northern blotting during the course of mouse embryo development; the numbers on top of each lane represent days after fertilization. (B) Mouse Abp1 mRNA levels in different adult tissues. The Northern probe comprised the proline-rich domain (303 nucleotides corresponding to aa 282–382 of mAbp1). (C and D) Actin expression is shown as a control for RNA integrity and transfer efficiency. (E and F) Mammalian Abp1 protein on immunoblots is detected by both antibody GP1 (anti-N terminus) and antibody GP5 (anti-proline-rich domain) as a single band of 56 kDa. (E) Four representative cell lines are shown; 50 μg of protein from postnuclear cell homogenates were loaded per lane. (F) mAbp1 levels in different tissues from adult mice; 250 μg of protein were loaded per lane; both blots were incubated and developed together.
Affinity-purified polyclonal antibodies were raised against the extreme N terminus (aa 1–26) and against the proline-rich domain of mAbp1 (aa 282–370), regions of low sequence homology to related proteins. Both antibodies detected a single band of 56 kDa on immunoblots of extracts from cell lines and mouse tissues (Figure 3, E and F). This band was not detected by preimmune serum or by anti-mAbp1 antibodies that had been preincubated with their immunogen (our unpublished results). The fact that the 56-kDa band displays a higher apparent molecular mass in SDS-PAGE analysis than the calculated mass of mAbp1 (48.4 kDa) is likely a result of the high proline and acidic amino acid content. The recombinant protein expressed in E. coli migrated with the same apparent molecular mass by SDS-PAGE. Furthermore, analysis of the various mAbp1 deletion mutants showed that it was the proline- and glutamate-rich domain of mAbp1 that caused the increased apparent molecular mass (our unpublished results). Because antibody GP5 was generated against the same region of mAbp1 used for the Northern probe, and because antibody GP1 only detected the 56-kDa band, it is highly unlikely that the double band detected in the Northern blots represents two different slice variants of mAbp1; it is more likely that both mRNA species encode the same 56-kDa protein.
Our observation, made with two different antibodies, that mAbp1 is expressed ubiquitously and that levels in muscle tissues are especially low (Figure 3F), somewhat contradicts data of Larbolette et al. (1999), whose data suggested that levels in heart are approximately half of those in spleen and lung. Our mAbp1 protein levels (Figure 3F), however, correlated well with the mRNA levels we detected except in kidney (Figure 3B). The blots of Larbolette et al. (1999) showed a second band at 32 kDa in ovaries, testes, and muscles that we did not detect in muscle (Figure 3F; ovaries and testis were not tested), even though our antibody GP5 was raised against a region including an epitope of a Larbolette et al. (1999) antibody. Also, we did not detect any additional RNA species in whole-body RNA (Figure 3A), testis, or skeletal muscle (Figure 3B) using a Northern probe including the region corresponding to the epitope of one of their antibodies. It thus seems likely that the additional band detected by Larbolette et al. (1999) represents a cross-reactivity of their antipeptide antibodies with a protein not related to mAbp1.
In mammalian cell lines, a significantly higher signal for mAbp1 was detected compared with tissues (Figure 3, compare E and F), despite the fact that approximately five times less total protein from the cell lines was loaded in each gel lane. Curiously, mAbp1 expression levels did not vary significantly in 12 different cell lines screened, regardless of the tissue from which they were derived or whether the tested cell lines were fetal, cancer derived, or virally transformed (Figure 3E; our unpublished results). This observation may reflect high mAbp1 expression in all actively growing and dividing cells.
mAbp1 Shows a Punctate Intracellular Distribution in Fibroblasts and Associates with Cortical F-actin Structures at Sites of Cellular Growth
The subcellular distribution of mAbp1 was studied by immunofluorescence. Small mAbp1-positive dots are scattered throughout the cytoplasm, whereas nuclei were excluded (Figure 4A). This punctate subcellular distribution of mAbp1 was observed in all cell lines examined (Raw 264.7, PC12, Madin–Darby canine kidney, NIH 3T3, IMR90, and Cos-7) and under a variety of fixation conditions. This subcellular distribution is different from that recently presented by Larbolette et al. (1999), who reported that GFP-mAbp1 colocalized with actin stress fibers. We did not observe such a distribution in any of the cell lines we examined. Because we used two different antibodies to determine the localization of the endogenous protein, whereas Larbolette et al. (1999) used a GFP-mAbp1 construct to visualize a heterologous protein, overexpression and/or association of GFP with actin may account for the mAbp1 distribution reported by Larbolette et al. (1999).
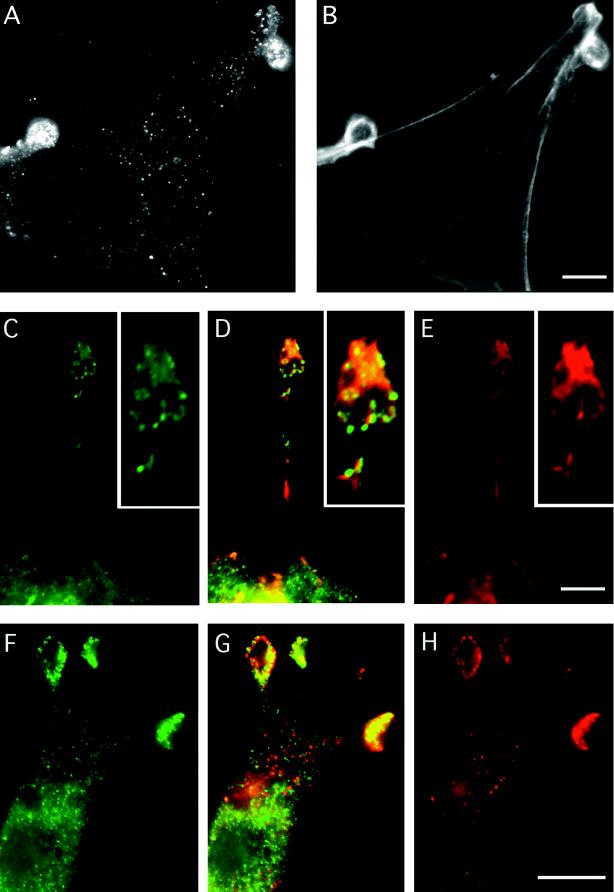
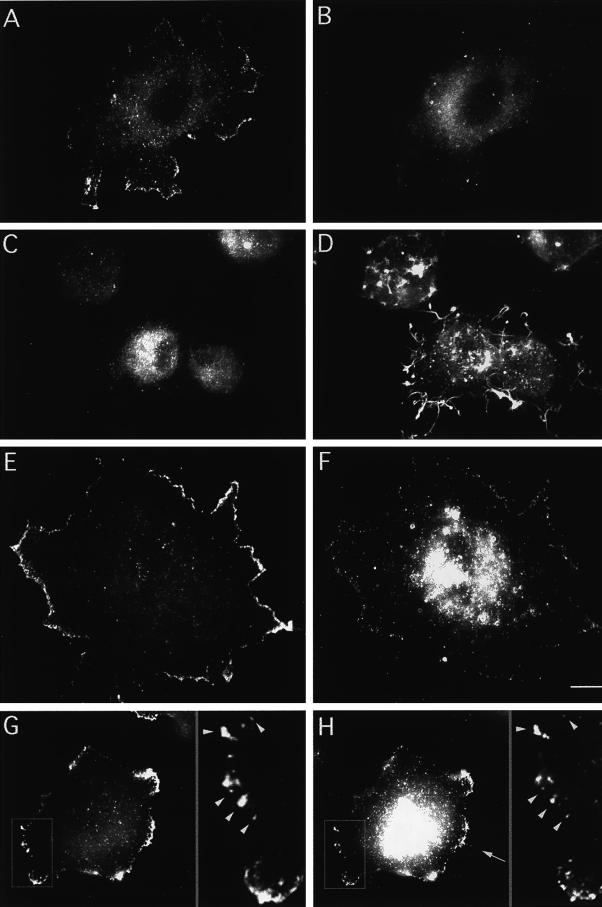
Figure 4.
Indirect immunolabelling of endogenous mAbp1 reveals a punctate distribution colocalizing with the cortical actin cytoskeleton at sites of cellular growth. (A, C, D, G, and H) Indirect immunolabeling of mouse Abp1. F-actin staining by rhodamine-phalloidin is shown in B, D, and E. Rab8 labeling of exocytic vesicles is shown in F and G. (A and B) NIH 3T3 cells. Bar, 10 μm. (C–E) Protrusion of a fetal human IMR90 fibroblast; the insets show a 2.25× higher magnification of the protrusion tip. Abp1 immunolocalization is shown in green (C), and F-actin stained with rhodamine-phalloidin is in red (E); areas of colocalization appear as different shades of yellow in the merged image (D). Bar, 10 μm. (F–H) Rab8 labeling (green; F), Abp1 labeling (red; H) in NIH 3T3 fibroblasts; areas of colocalization appear as different shades of yellow in the merged image (G). Bar, 10 μm.
In general, the mAbp1-positive dots we detected were noncortically distributed, as observed by confocal microscopy (our unpublished results). The density of mAbp1-positive dots often appeared slightly greater in the perinuclear region. Intense accumulations of mAbp1 were often detected in round protrusive structures of fibroblasts (Figure 4A). Some of these structures were stained brightly by rhodamine-phalloidin (Figure 4B), whereas others exhibited only a very faint F-actin staining (our unpublished results). Only a partial colocalization of mAbp1 dots to F-actin fibers was observed in the cell body. However, a striking colocalization of mAbp1 and F-actin structures occurred in cortical areas, such as the actin-rich protrusion tips of fibroblasts (Figure 4, C–E). These results are consistent with our biochemical demonstration that mAbp1 is an F-actin-binding protein.
Because yeast Abp1 plays a role in spatial regulation of cell growth, we next determined whether the peripheral structures enriched for mAbp1 in fibroblasts represent sites of cellular growth. We used Rab8 as a marker protein (Figure 4, F–H). Rab8 is a small GTPase that associates specifically with constitutive secretory vesicles, which are targeted to the plasma membrane for fusion (reviewed in Novick and Zerial, 1997). Although we observed no mAbp1 colocalization with Rab8-containing secretory vesicles within the cell body, sites of mAbp1 accumulation at the cell periphery clearly corresponded to apparent sites of exocytosis and cell growth marked by accumulation of Rab8 at these sites (Figure 4, F–H). Because the areas heavily stained for mAbp1 were of about the same thickness as other parts of the cell extentions (≤3 μm) and the thickness of the cell body easily reached 15 μm, volume effects do not contribute to the observed enrichment. Instead, it appears that mAbp1 specifically accumulates at regions of high membrane dynamics and cellular growth, regions where the actin cytoskeleton may also show high dynamics.
We made use of the fact that the actin cytoskeleton is resistant to detergent extraction to further characterize the cytosolic and cytoskeletal pools of mAbp1. As shown in Figure 5, brief detergent extraction before fixation (Figure 5, C–F) extracted the cytosolic pool of mAbp1, but the lamellipodial fraction remained associated with the actin-rich cortex. In contrast, no colocalization with actin stress fibers was observed, showing that mAbp1 associates specifically with lamellipodial actin. Thus, in resting cells, a large pool of mAbp1 is cytosolic and readily extractable, whereas a smaller pool is nonextractable and is associated with actin-rich lamellipodia.
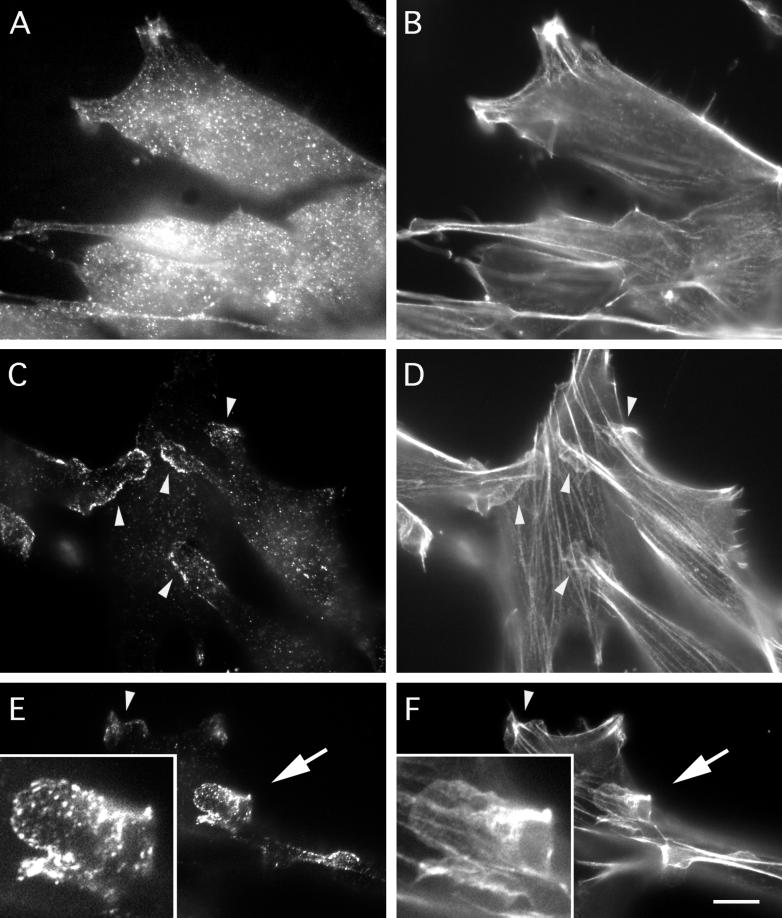
Figure 5.
mAbp1 localization in extracted and unextracted NIH 3T3 cells. (A and B) Immunolocalization of mAbp1 (A) and rhodamine-phalloidin localization of F-actin (B) in cells fixed before extraction. (C–F) Immunolocalization of mAbp1 (C and E) and rhodamine-phalloidin localization of F-actin (D and F) in cells briefly extracted with Triton X-100 before fixation. Arrowheads identify examples of areas showing association of mAbp1 with lamellipodial actin. Insets in E and F are 2.5× enlargements of a large lamellipodium (arrow) at the end of a cell protrusion that appears to be crawling over another cell. Bar, 10 μm.
The Punctate mAbp1 Staining within the Cell Body Seems Not to Represent Vesicles of the Receptor-mediated Endocytosis Pathway
Because the mAbp1 immunostaining displayed a punctate subcellular pattern, and the related yeast protein Abp1p has been shown to play a role in receptor-mediated endocytosis, we labeled different endosomal compartments in Cos-7 cells via transferrin-Texas Red uptake and tested for colocalization with mAbp1 (Figure 6). Neither the individual endosomal compartments labeled by pulse–chase experiments (Figure 6, A–C) nor those marked by continuous uptake of transferrin-Texas Red at 15°C (Figure 6D) or 37°C (our unpublished results) displayed any colocalization with mAbp1. Only in the more densely labeled perinuclear area was a low level of overlap observed. Thus, the mAbp1-positive dots do not represent early, late, fast-recycling, or perinuclear-recycling endosomes of the receptor-mediated endocytosis pathway.
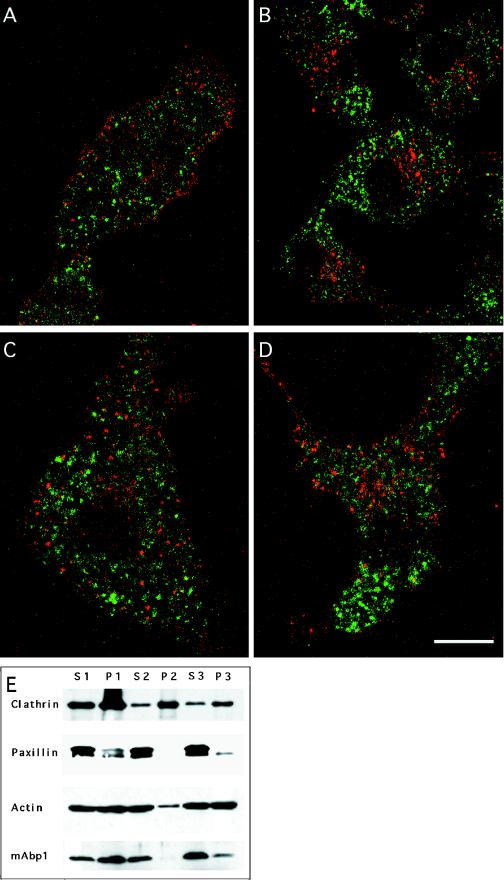
Figure 6.
The punctate mAbp1 immunostaining does not represent vesicular compartments of the receptor-mediated endocytosis pathway. (A–C) Individual labeling of endosomal compartments of the receptor-mediated endocytosis pathway by a pulse–chase experiment in Cos-7 cells. Transferrin-Texas Red conjugates (red) were prebound to transferrin membrane receptors in the cold; internalization was allowed by raising the temperature to 37°C; and the transferrin marker was followed for different times: A, 5 min; B, 30 min; and C, 60 min. No colocalization of mAbp1 (green) was observed by confocal microscopy. (D) Continuous uptake of transferrin at 15°C for 2 h. Bar, 15 μm. (E) Subcellular fractionation of mAbp1. Different fractions of mouse spleen homogenate were examined by SDS-PAGE and immunoblotting for the indicated proteins (see MATERIALS AND METHODS). The same amount of total protein was loaded in each lane.
To determine whether the punctate, subcellular mAbp1 distribution detected by our antibodies represented vesicular compartments, we also performed subcellular fractionations. As shown in Figure 5E, mAbp1 behaved like a soluble cytosolic protein and was not enriched in vesicular compartment fractions (P2 and P3). In contrast, clathrin, used as a marker for membrane compartments, was enriched in the vesicular fractions P2 and P3. The vast majority of the focal adhesion protein paxillin was soluble in these studies, providing evidence that cell disruption and homogenization were relatively effective. The detection of some mAbp1 in P1 may reflect association of mAbp1 with large pieces of plasma membrane via interactions with the cortical actin cytoskeleton, consistent with the observation that actin was also readily detected in P1 and consistent with the results of our extraction experiments (see Figure 5). Summarized, our analysis indicates that mAbp1 does not copurify with vesicles but is mostly cytosolic and possibly associated with the actin cytoskeleton. The observed, punctate mAbp1 immunostaining within the cell body seems not to represent vesicular compartments.
mAbp1 Is Recruited to the Leading Edge of Migrating Cells
The actin cytoskeleton becomes polarized toward the leading edge of cells induced to move into a wound. Because the leading edge of fibroblasts is characterized by directed exocytosis and high dynamics of the actin cytoskeleton, moving fibroblasts represent an attractive system in which to test our hypothesis that mAbp1 accumulates at sites of high plasma membrane and actin cytoskeleton dynamics. By wounding a confluent layer of quiescent fibroblasts, we were able to test whether mAbp1 would colocalize with actin and sites of enhanced exocytosis at the leading edge.
After wounding the cell layer, cells adjacent to the wound reorganized their actin cytoskeleton toward the wound and developed lamellipodia that were decorated by short filopodia (Figure 7, A and C). The Golgi apparatus, from which constitutive exocytic vesicles presumably stream toward the leading edge, was reoriented toward the lamellipodia in these cells (our unpublished results) as described by Bergmann et al. (1983). The mAbp1 distribution in these cells shifted dramatically to the induced lamellipodia. mAbp1 either outlined the rim (Figure 7B) or localized to the entire lamellipodial area (Figure 7D). In both cases, intense mAbp1 staining was detected at the leading edge, accompanied by a marked reduction in cell body staining (Figure 7, B and D). Comparing the overall distribution of F-actin and mAbp1 within these cells, it became clear that mAbp1 does not simply localize to areas rich in F-actin but specifically accumulates in lamellipodial structures directed forward (Figure 7, A and C).
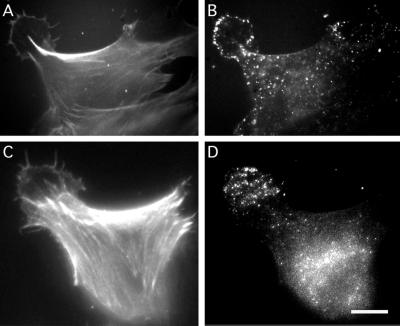
Figure 7.
Abp1 is recruited to the leading edge or the entire lamellipodium in NIH 3T3 fibroblasts moving into a wound. Direction of movement, i.e., the position of the wound, upper left corner. F-actin (A and C) detected by rhodamine-phalloidin and mAbp1 (B and D) detected by anti-mAbp1 antibody and FITC anti-guinea pig secondary antibody in two moving NIH 3T3 cells 3 h after wounding. Bar, 10 μm.
mAbp1 Rapidly Accumulates at the Cell Periphery in Response to Growth Factors
Next, we attempted to identify the signals that lead to the relocation of mAbp1 to the leading edge of motile fibroblasts by activating pathways that specifically lead to activation of Cdc42, Rac1, and RhoA, GTPases that induce rapid actin rearrangements (reviewed in Hall, 1998; Aspenström, 1999). We hoped that these conditions might allow us to uncouple exocytic events from cytoskeletal rearrangements.
We first examined the distribution of F-actin and mAbp1 in quiescent and serum-starved NIH 3T3 fibroblasts. The cells displayed actin-rich retraction spikes and nonfibrous, phalloidin-stainable F-actin structures that were often found accumulated in the perinuclear region. The stress fiber content of the cells appeared low (Figure 8A). mAbp1 was observed throughout the cytosol and also often appeared enriched in the perinuclear area (Figure 8B).
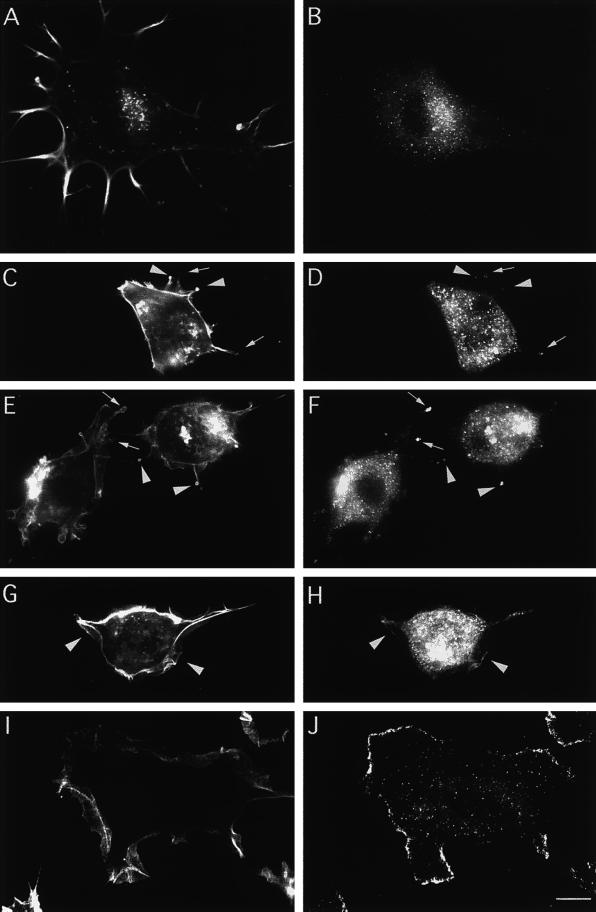
Figure 8.
Abp1 distribution is rapidly shifted toward the periphery of NIH 3T3 cells in response to growth factors that lead to Rac1 activation (here addition of 5 ng/ml PDGF). F-actin was stained with rhodamine-phalloidin (A, C, E, G, and I), and mAbp1 was detected by anti-mAbp1 antibody GP5 and FITC-conjugated secondary antibody (B, D, F, H, and J). (A and B) Quiescent cells starved for serum overnight. (C–J) Different time points of the response to growth factor: 1 min (C and D), 2 min (E and F), 4 min (G and H), and 10 min after PDGF addition (I and J). Arrowheads indicate protrusive areas with high F-actin content (C and E) at early time points at which mAbp1 was detected and accumulated (D and F). Arrows in C–F identify areas characterized by high mAbp1 accumulation with lower cortical actin accumulation. Arrowheads in G and H mark areas where the first continuous leading edge mAbp1 labeling at newly forming lamellipodia occurred at 4 min. Bars, 10 μm.
Treatments leading to Rac1 activation, including addition of PMA (our unpublished results) or PDGF (Figure 8, C-J), resulted in a dramatic redistribution of mAbp1. Within 10 min, cell body staining was greatly reduced, and mAbp1 accumulated at the periphery of the cells (Figure 8J). Colabeling of F-actin revealed the sites of mAbp1 accumulation to be the membrane-proximal edge of lamellipodia (Figure 8I).
This reorganization of the actin cytoskeleton and mAbp1 localization was rapid. Within the first minute of treatment (Figure 8, C and D), cells rounded up and extended small areas that were either enriched for F-actin (Figure 8C, arrowheads) or were only weakly surrounded by F-actin (Figure 8C, arrows). Some mAbp1 already localized to these sites (Figure 8D). After 2 min, larger protrusive structures were observed; these circular areas contained modest levels of cortical F-actin staining (Figure 8E, arrows) but were now brightly labeled by anti-mAbp1 antibodies (Figure 8F). By 4 min, mAbp1 began to outline small and narrow lamellipodial belts at the cell periphery (Figure 8, G and H, arrowheads). By 10 min, cells had increased dramatically in diameter and further extended their lamellipodia as mAbp1 accumulated at the periphery (Figure 8, I and J).
In contrast to the dramatic change in mAbp1 localization upon activation of Rac1 by PDGF or PMA, the activation of Cdc42 by bradykinin, which leads to filopodia formation (Kozma et al., 1995), resulted in only a barely detectable association of mAbp1 with newly formed filopodia (our unpublished results). In addition, RhoA activation by lysophosphatidic acid, leading to stress fiber assembly and focal adhesion induction (Ridley and Hall, 1992), failed to lead to an altered mAbp1 distribution pattern or accumulation of mAbp1 at focal adhesions (our unpublished results). Thus, two different signaling pathways converging on Rac1, activation by PMA or PDGF to induce lamellipodia formation, both resulted in redistribution of mAbp1. Furthermore, mAbp1 is recruited to lamellipodia as they are forming from small roundish areas protruding from the cell body.
The Dramatic Shift of mAbp1 to the Cell Periphery Coincides with Sites of De Novo F-actin Assembly and Is Dependent on Cortical F-actin Polymerization
We next used the rapid Rac1-inducible mAbp1 relocalization to gain insights into the mechanisms underlying the accumulation of mAbp1 at the leading edge. We first asked whether under Rac1-activating conditions exocytosis and rapid actin assembly would be spatially uncoupled. Indeed, when serum-starved cells were induced with PDGF, Rab8 remained near the Golgi apparatus (Figure 9B), whereas mAbp1 relocated to the leading edge (Figure 9A).
Figure 9.
Abp1 accumulation at the leading edge after 10 min of stimulation with growth factors leading to Rac1 activation is dependent on actin polymerization and coincides with de novo actin nucleation. (A and B) Comparison of the location of mAbp1 (A) and Rab8 (B) in quiescent, serum-starved NIH 3T3 cells treated with PDGF for 10 min. (C and D) Simultaneous addition of 2 μM Latrunculin A suppresses the change of the mAbp1 distribution (C), although some F-actin structures still persist after 10 min of treatment (D). (E–H) Colocalization of mAbp1 (E and G) and the Arp2/3 complex (F and H) at areas of the periphery of NIH 3T3 cells to which both proteins were recruited upon 10 min of stimulation with 5 ng/ml PDGF. Insets in G and H represent 3× enlargements of the area marked in G and H. Examples of mAbp1 and Arp2/3 colocalization at high resolution are marked by arrowheads. An example of Arp2/3 complex localization to filopodia is marked by an arrow in H. Bar, 10 μm.
Next, to test whether the recruitment of mAbp1 is F-actin dependent, we incubated the cells with Latrunculin A, a G-actin-sequestering drug, before addition of growth factor. Despite activation of the PDGF or PMA signaling cascades, no accumulation of mAbp1 at the cell periphery was observed under these conditions (our unpublished results). To dissect the role of the actin cytoskeleton further, we next asked whether de novo actin assembly is required for relocation of mAbp1 to lamellipodia. For this purpose, we incubated cells with PDGF and Latrunculin A simultaneously. Under these conditions, actin monomers are sequestered, and new assembly is therefore inhibited; however, F-actin structures persist at the cell periphery (Figure 9D). Despite the persistence of these F-actin structures, no mAbp1 recruitment occurred upon stimulation with growth factor, and mAbp1 did not colocalize with the remaining F-actin (Figure 9C). Identical results were obtained by adding PMA in the presence of Latrunculin A (our unpublished results). Thus, the redistribution of mAbp1 to the cell periphery appears to be dependent on de novo actin assembly rather than just F-actin integrity.
Finally, we determined whether the Arp2/3 complex, shown to nucleate actin filament assembly (for review, see Machesky and Gould, 1999), displays a similar shift in its localization upon activation of Rac1. If it did, we wanted to know whether the stretches of the cell periphery brightly labeled for mAbp1 would correspond to areas of high Arp2/3 complex concentration. PDGF- and PMA-treated cells indeed showed a relocalization of the Arp2/3 complex to the cell periphery (Figure 9, F and H). The Arp2/3 complex, however, was far less restricted to the leading edge than mAbp1. Even after 10 min of growth factor stimulation, a time point when mAbp1 was highly concentrated at the leading edge, readily detectable cell body staining of the Arp2/3 complex was still observed (Figure 9, F and H). We also noticed that the Arp2/3 complex localized to filopodia (when present) more intensely than mAbp1 (Figure 9H, arrow). Although the Arp2/3 complex relocated to the periphery to a lesser extent than mAbp1, a clear colocalization of mAbp1 and the Arp2/3 complex at the leading edge was observed. Areas of the periphery that displayed a low Arp2/3 abundance were also devoid of mAbp1 immunolabeling, whereas stretches of the periphery showing an accumulation of Arp2/3 complex also exhibited a bright mAbp1 immunolabeling (Figure 9, E–H). Thus, mAbp1 relocation was not only dependent on actin polymerization but also coincided with sites of actin nucleation.
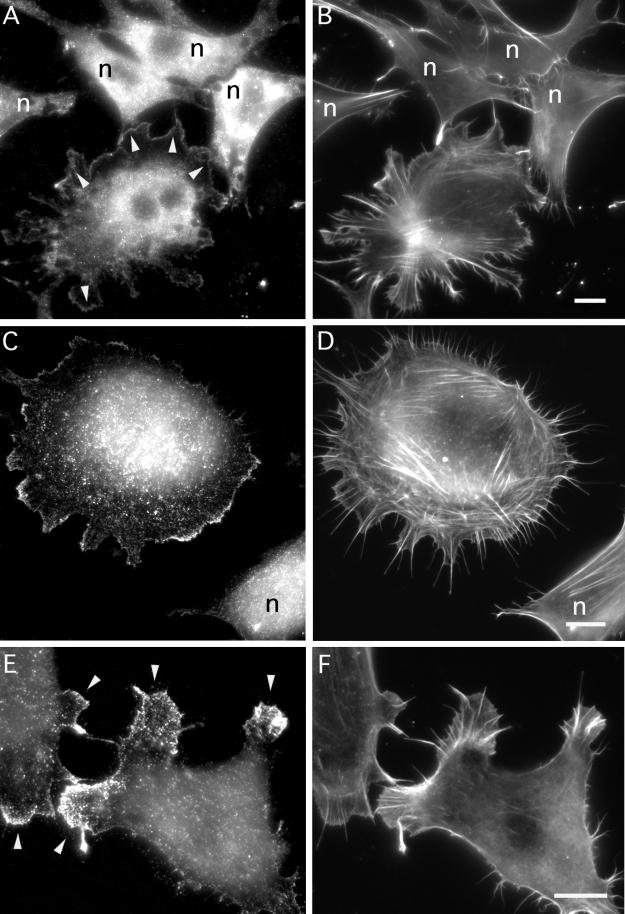
Mouse Abp1 Localization Does Not Depend on Src Activation but Is Responsive to Rac1 Activation
mAbp1/SH3P7 is a substrate of Src family tyrosine kinases (Lock et al. 1998; Larbolette et al. 1999). These kinases are activated via a signaling cascade activated by PDGF and also linked in various ways to the cytoskeleton-controlling GTPases acting further downstream. Therefore, we decided to test whether Src activation is required for translocation of mAbp1 to dynamic regions of the actin cytoskeleton at a cell's leading edge. Thus, we studied mAbp1 localization under conditions known to render Src family kinases and thus their downstream effectors inactive (reviewed by Brown and Cooper, 1996); i.e., we activated Rac1 under conditions of serum starvation and a lack of integrin activation. The Rac signaling pathway was selectively activated by transfecting serum-starved fibroblasts plated on glass with a dominant-positive Rac1 mutant (L61). As shown in Figure 10, introduction of Rac1L61 into NIH 3T3 cells is sufficient for lamellipodia formation (Figure 10, B, D, and F) and recruitment of mAbp1 to the cell periphery (Figure 10, A, C, and E, some examples marked by arrowheads). As shown in Figure 10E, this accumulation of mAbp1 in some cases correlates with a marked reduction in the amount of mAbp1 detected in the cell body (also see Figures 8J and 9, E and G). In untransfected cells (labeled n throughout Figure 10), no accumulation of mAbp1 in actin-rich areas of the cell periphery (Figure 10, A and C) and no formation of lamellipodia (Figure 10, B and D) were observed. In control transfections in which the pRK5 mycRac1L61plasmid was left out, cells appeared as untransfected cells (our unpublished results).
Figure 10.
Evidence that translocation of mAbp1 to the cell periphery is Rac1 mediated but Src independent. Serum-starved NIH 3T3 cells were transfected with mycRac1L61. (A and B) Field of nontransfected cells (marked by n throughout) and one Rac1L61-positive cell, which shows lamellipodia formation and mAbp1 translocation (some such areas are marked by arrowheads in A) despite prolonged serum starvation. (C–F) Examples of Rac1L61-transfected cells displaying accumulation of mAbp1 at the leading edge of lamellipodial areas (labeled by arrowheads in E) at higher magnifications. (A, C, and E) mAbp1 immunolabeling; (B, D, and F) F-actin staining with rhodamine-phalloidin. Bars, 10 μm.
DISCUSSION
mAbp1 Is Part of an Ancient Line of F-actin-binding Proteins
Genetic studies in yeast have suggested that Abp1 links the actin cytoskeleton to the membrane dynamics that underlie polarized cell growth. Here, we report the identification and initial characterization of mAbp1, a mouse homologue of the yeast Abp1p. To evaluate the function of mAbp1, we focused first on its interaction with actin and second on how its localization is affected by Rac, Cdc42, and Rho GTPases.
mAbp1 shows overall sequence homology to Abp1s from different yeasts and to the product of a C. elegans ORF. Furthermore, mAbp1 shows high N-terminal similarity to drebrins, brain-specific F-actin-binding proteins that have been identified in a variety of vertebrates (Shirao and Obata, 1985; Toda et al., 1993) and are thought to mediate neuronal morphogenesis by an unknown mechanism (Shirao, 1995). Despite the high N-terminal sequence similarity between mAbp1 and drebrins, mAbp1 is not a drebrin isoform. The SH3 domains of Abp1s from phylogenetically distinct organisms (e.g., yeast, C. elegans, and mouse) form a distinct SH3 subfamily (Figure 1A) and therefore are likely to interact with a distinct subset of SH3 domain-binding proteins. In contrast, drebrins seem not to contain SH3 domains. Furthermore, our analysis of ADF-H domains indicates that the gene duplication that separated drebrins from Abp1s was a relatively ancient event that occurred when the neuronal system became more complex. In support of this conclusion, we did not find a drebrin in the now fully sequenced C. elegans genome. Instead we identified an Abp1 sequence.
However, our studies do show that based on functional properties, drebrins and Abp1s belong to the same class of ADF-H domain-containing proteins. Recombinant mAbp1 bound to F-actin with a 1:5 saturation stoichiometry did not show F-actin bundling or severing activity, did not interfere with the nucleotide exchange on actin, and did not bind to G-actin in vitro, properties resembling those of rat drebrin (Ishikawa et al., 1994). The ADF-H domain and the highly charged helical domain were individually able to bind to F-actin. Sequence similarities suggest that this feature might be shared with drebrins. The ADF-H domains of both cofilin/ADF and twinfilin have been shown to bind to actin at a 1:1 ratio (Carlier, 1998; Goode et al., 1998). The low saturation stoichiometry of one mAbp1 or drebrin bound per five actin molecules may be explained by mAbp1 and drebrin binding along filaments and covering a larger surface area of F-actin by the simultaneous interaction of these two F-actin binding domains. This hypothesis is consistent with the apparent lack of bundling activity and is supported by the close proximity of the two domains, probably creating a continuous actin-binding interface.
In Response to Activators of the GTPase Rac, mAbp1 Associates with the Actin Cytoskeleton at Cortical Sites of Cellular Growth
In resting cells, most mAbp1 appears to be cytosolic and is detergent extractable. However, a fraction of the protein is nonextractable and colocalizes with lamellipodial F-actin, particularly at sites of cellular growth as marked by Rab8-containing secretory vesicles. Furthermore, we demonstrated that mAbp1 displays a polarized peripheral distribution in migrating cells. This localization does not reflect the overall F-actin distribution but instead appears to coincide with sites of high F-actin dynamics and membrane insertion at the F-actin-rich leading edge (for review of membrane dynamics in migrating cells, see Bretscher and Aguado-Velasco, 1998). Thus, the functional role for mAbp1 may be related to cell motility and to polarized cell growth, analogous to that suggested for yeast Abp1 (Drubin et al., 1988; Lila and Drubin, 1997). Such a function might also explain why mAbp1 expression levels are highest during early embryonic development and in cell lines.
Because mAbp1 associates with regions enriched in secretory vesicles and the cortical actin cytoskeleton, it was important to dissect the contributions of secretory elements and the cytoskeleton to mAbp1 localization. Treatments resulting in activation of Rac1 altered the subcellular distribution of mAbp1 and did so independently of polarization of the exocytosis machinery. Thus, a polarized exocytosis machinery is not the basis of the polarized distribution of mAbp1 and its accumulation at sites of cellular growth. Instead, activation of signaling pathways leading to polarized actin cytoskeleton rearrangements are responsible for the observed dramatic changes in mAbp1 localization and its association with F-actin within lamellipodia. Furthermore, Latrunculin A blocked the mAbp1 relocalization, demonstrating a dependence of the mAbp1 recruitment to the cell periphery on rapid actin assembly at the cell cortex. We also found that the Arp2/3 complex is rapidly relocated to the cell periphery upon activation of Rac under our conditions, and that mAbp1 and Arp2/3 colocalize at the cell periphery. We thus conclude that the sites at the leading edge to which mAbp1 is recruited are sites of high de novo actin polymerization mediated by the Arp2/3 complex, and that mAbp1 is likely to directly or indirectly participate in rearrangements of the actin cytoskeleton that lead to lamellipodium formation.
The exact molecular function of mAbp1 at the leading edge has not yet been discovered. However, even though recombinant mAbp1 alone had no obvious effects on actin assembly or disassembly in vitro, it is still possible that mAbp1 plays a regulatory role in actin assembly by either affecting it directly or by activating, interfering, or competing with proteins involved in actin dynamics. mAbp1 may also serve as an anchor for proteins that regulate the actin cytoskeleton, that are involved in membrane trafficking, or both.
mAbp1 Shows Sequence and Functional Similarities to the Src Substrate p80/85 Cortactin
Recently, Weed et al. (1998) reported that p80/85 cortactin accumulates at the periphery of serum-starved Swiss 3T3 cells when Rac1 is activated. Cortactin is an F-actin-binding protein and an Src substrate expressed in all tissues (Wu and Parsons, 1993). The gene encoding its human homologue, EMS1, was found to be amplified and overexpressed in certain cancers (Schuuring et al., 1993). Cortactin associates with the Src SH2 domain and colocalizes with v-Src in transformed cells (Okamura and Resh, 1995). Interestingly, cortactin shows a domain organization similar to mAbp1, with an F-actin binding motif at its N terminus and an SH3 domain at its C terminus. Because this SH3 domain is extremely similar to that of mAbp1, cortactin and mAbp1 may bind to the same ligands. Indeed, mAbp1/SH3P7 was isolated by a phage display screen using a cortactin SH3-binding consensus peptide (Sparks et al., 1996). Because we did not find any cortactin homologues in yeast or C. elegans, it seems that the cortactin family may have been formed by combining the ancient SH3 domain of Abp1s with another actin-binding module.
Interestingly, Lock et al. (1998) recently identified mAbp1/SH3P7 in a screen for in vitro substrates of Src kinases. Thus, as cortactin, mAbp1/SH3P7 is a Src family kinase substrate. Larbolette et al. (1999) confirmed this finding, showing that mAbp1/SH3P7 becomes tyrosine phosphorylated in response to lymphocyte activation in vivo and identifying two sites within the flexible domain that are tyrosine phosphorylated, as Lock et al. (1998) predicted.
Src family kinases play an important role in activation of cell proliferation. The subcellular localization of Src family kinases is dynamically regulated, and the relocation of Src to adhesion plaques during cell adhesion, mediated by RhoA, is critical for its growth-promoting activities in fibroblasts (reviewed by Brown and Cooper, 1996; Thomas and Brugge, 1997). Src can be recruited to the cell periphery as early as 30 min after PDGF treatment of NIH 3T3 cells. This relocalization must depend on actin-binding proteins, because it can be blocked by cytochalasin D, and because Src itself is unable to bind to actin directly (Fincham et al., 1996). Because mAbp1 is recruited to lamellipodia by dominant-positive Rac1, under conditions in which Src is rendered inactive, i.e., no integrin activation and serum starvation (Brown and Cooper, 1996), tyrosine phosphorylation of Abp1 by Src seems not to be necessary for mAbp1's accumulation at sites of de novo actin nucleation. This finding is consistent with the observations that E. coli-expressed mAbp1 binds to actin and that PMA treatment, which to our knowledge does not lead to Src activation but acts through activation of protein kinase C, also leads to the actin polymerization-dependent relocation of mAbp1 to the cell periphery. Therefore, the association of mAbp1 with dynamic actin structures appears not to depend on Src activation but depends on lamellipodia formation triggered by activation of Rac and its downstream effectors. Similar results were recently reported for cortactin (Weed et al., 1998). The activation of the signaling cascades that lead to Rac activation, however, is not sufficient to recruit mAbp1 to the cell periphery under conditions in which actin polymerization is inhibited, linking association of mAbp1 at the cell periphery with de novo actin assembly.
It thus seems possible that mAbp1 serves as a Rac1-activation-dependent anchor for Src in forming lamellipodia. This Src recruitment may subsequently allow Src to perform its role in RhoA-controlled focal adhesion assembly, cell–cell adhesion, and finally cell proliferation (reviewed by Brown and Cooper, 1996; Thomas and Brugge, 1997). Src might also positively or negatively regulate yet-to-be-identified activities of mAbp1 in the cortical actin cytoskeleton, or it might alter mAbp1's F-actin binding activity. The Src-dependent hyperphosphorylation of cortactin appears to down-regulate its actin-binding activity and inhibits its newly discovered actin-bundling activity in vitro (Huang et al., 1997). In addition, cortactin hyperphosphorylation resulted in an inhibition of the entry of the bacterial pathogen Shigella into cells (Duménil et al., 1998), a process that, like lamellipodia formation, is dependent on actin dynamics.
Cortactin and mAbp1 are both F–actin-binding proteins that share a functionally similar domain structure (N-terminal actin binding and C-terminal SH3). Both proteins are Src substrates, both are likely to participate in dynamic actin rearrangements underlying cell growth and membrane trafficking, and both relocate to the cell periphery upon Rac activation, independently of their tyrosine phosphorylation by Src. These observations suggest a commonality in mechanism for the recruitment to sites of rapid actin dynamics. Elucidation of the exact roles of mAbp1 and cortactin in lamellipodia, however, awaits further analysis.
ACKNOWLEDGMENTS
We thank Thomas Lila for initiating this project, Narla Mohandes for generous support of this project since its inception, and Anne Fisher for advice on cell culture. We also thank Brian Kay for generously providing the mAbp1/SH3P7 cDNA, David King for synthesizing the mAbp1 N-terminal peptide, Matthew Welch for sharing his anti-Arp3 antibody with us, and Alan Hall (University College London, London, United Kingdom) for the gift of the pRK5 mycRac1L61 plasmid. We are grateful to Britta Qualmann, Bruce Goode, and Keith Kozminski for helpful comments on the manuscript. This work was supported by Deutsche Forschungsgemeinschaft grant Ke 685/1-1, and by an Otto-Hahn-Research Award for the Max-Planck-Society to M.M.K., and by National Institutes of Health grants GM-50399 and DK-32094 to D.G.D.
REFERENCES
- Aspenström P. Effectors for the Rho GTPases. Curr Opin Cell Biol. 1999;11:95–102. doi: 10.1016/s0955-0674(99)80011-8. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kinston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1990. [Google Scholar]
- Bergmann JE, Kupfer A, Singer SJ. Membrane insertion at the leading edge of motile fibroblasts. Proc Natl Acad Sci USA. 1983;80:1367–1371. doi: 10.1073/pnas.80.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi GQ, Morris RL, Liao G, Alderton JM, Scholey JM, Steinhardt RA. Kinesin- and myosin-driven steps of vesicle recruitment for Ca2+-regulated exocytosis. J Cell Biol. 1997;138:999–1008. doi: 10.1083/jcb.138.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bretscher MS, Aguado-Velasco C. Membrane traffic during cell locomotion. Curr Opin Cell Biol. 1998;10:537–541. doi: 10.1016/s0955-0674(98)80070-7. [DOI] [PubMed] [Google Scholar]
- Brown M, Cooper JA. Regulation, substrates and functions of src. Biochim Biophys Acta. 1996;1287:112–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- Cope MJTV, Yang S, Shang C, Drubin DG. Novel protein kinases Ark1p and Prk1p associate with and regulate the cortical actin cytoskeleton in budding yeast. J Cell Biol. 1999;144:1–17. doi: 10.1083/jcb.144.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHostos EL, Bradke B, Lottspeich F, Gerisch G. Coactosin, a 17 kDa F-actin binding protein from Dictyostelium discoideum. Cell Motil Cytoskeleton. 1993;26:181–191. doi: 10.1002/cm.970260302. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Miller KG, Botstein D. Yeast actin-binding proteins: evidence for a role in morphogenesis. J Cell Biol. 1988;107:2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duménil G, Olivo JC, Pellegrini S, Fellous M, Sansonetti PJ, Tran Van Nhieu G. Interferon α inhibits a Src-mediated pathway necessary for Shigella-induced cytoskeletal rearrangements in epithelial cells. J Cell Biol. 1998;143:1003–1012. doi: 10.1083/jcb.143.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham VJ, Unlu M, Brunton VG, Pitts JD, Wyke JA, Frame MC. Translocation of Src kinase to the cell periphery is mediated by the actin cytoskeleton under the control of the Rho family of small G proteins. J Cell Biol. 1996;135:1551–1564. doi: 10.1083/jcb.135.6.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger FP, Novick P. Spatial regulation of exocytosis: lessons from yeast. J Cell Biol. 1998;142:609–612. doi: 10.1083/jcb.142.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli MI, Riezman H. Endocytic internalization in yeast and animal cells: similar and different. J Cell Sci. 1998;111:1031–1037. doi: 10.1242/jcs.111.8.1031. [DOI] [PubMed] [Google Scholar]
- Goode BL, Drubin DG, Lappalainen P. Regulation of the cortical actin cytoskeleton in budding yeast by twinfilin, a ubiquitous actin sequestering protein. J Cell Biol. 1998;142:723–733. doi: 10.1083/jcb.142.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Wong JJ, Butty A-C, Peter M, McCormack AL, Yates JR, Drubin DG, Barnes G. Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtuble cytoskeleton in yeast. J Cell Biol. 1999;144:83–98. doi: 10.1083/jcb.144.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hartwig JH, Kwiatkowski DJ. Actin-binding proteins. Curr Opin Cell Biol. 1991;3:87–97. doi: 10.1016/0955-0674(91)90170-4. [DOI] [PubMed] [Google Scholar]
- Huang C, Yansong N, Wang T, Gao Y, Haudenschild CC, Zhang X. Down-regulation of the filamentous actin cross-linking activity of cortactin by Src-mediated tyrosine phosphorylation. J Biol Chem. 1997;272:13911–13915. doi: 10.1074/jbc.272.21.13911. [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Hayashi K, Shirao T, Xue Y, Takagi T, Sasaki Y, Kohama K. Drebrin, a development-associated brain protein from rat embryo causes the dissociation of tropomyosin from actin filaments. J Biol Chem. 1994;269:29928–29933. [PubMed] [Google Scholar]
- Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO J. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P, Kessels MM, Cope MJTV, Drubin DG. The ADF homology (ADF-H) domain: a highly exploited actin-binding module. Mol Biol Cell. 1998;9:1951–1959. doi: 10.1091/mbc.9.8.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larbolette O, Wollscheid B, Schweikert J, Nielsen P, Wienands J. SH3P7 is a cytoskeletal adapter protein and is coupled to signal transduction from lymphocyte antigen receptors. Mol Cell Biol. 1999;19:1539–1546. doi: 10.1128/mcb.19.2.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichte B, Veh RW, Meyer H, Kilimann MW. Amphiphysin, a novel protein associated with synaptic vesicles. EMBO J. 1992;11:2521–2530. doi: 10.1002/j.1460-2075.1992.tb05317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lila T, Drubin DG. Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Mol Biol Cell. 1997;8:367–385. doi: 10.1091/mbc.8.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock P, Abram CL, Gibson T, Courtneidge SA. A new method for isolating tyrosine kinase substrates used to identify Fish, an SH3 and PX domain-containing protein, and Src substrate. EMBO J. 1998;17:4346–4357. doi: 10.1093/emboj/17.15.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Gould KL. The Arp2/3 complex: a multifunctional actin organizer. Curr Opin Cell Biol. 1999;11:117–121. doi: 10.1016/s0955-0674(99)80014-3. [DOI] [PubMed] [Google Scholar]
- Mciver SK. How ADF/cofilin depolymerizes actin filaments. Curr Opin Cell Biol. 1998;10:140–144. doi: 10.1016/s0955-0674(98)80097-5. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Cramer LP. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- Moon A, Drubin DG. The ADF-cofilin proteins: stimulus responsive modulators of actin dynamics. Mol Biol Cell. 1995;6:1423–1431. doi: 10.1091/mbc.6.11.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muallem S, Kwiatkowska K, Xu X, Yin HL. Actin filament disassembly is a sufficient trigger for exocytosis in nonexitable cells. J Cell Biol. 1995;128:589–598. doi: 10.1083/jcb.128.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Zerial M. The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- Okamura H, Resh MD. p80/85 cortactin associates with the Src SH2 domain and colocalizes with v-Src in transformed cells. J Biol Chem. 1995;270:26613–26618. doi: 10.1074/jbc.270.44.26613. [DOI] [PubMed] [Google Scholar]
- Peränen J, Rikkonen M, Hyvönen M, Kääräinen L. T7 vectors with a modified T7lac promotor for expression of proteins in Escherichia coli. Anal Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- Puius YA, Mahoney NM, Almo SC. The modular structure of actin regulatory proteins. Curr Opin Cell Biol. 1998;10:23–34. doi: 10.1016/s0955-0674(98)80083-5. [DOI] [PubMed] [Google Scholar]
- Qualmann B, Roos J, DiGregorio PJ, Kelly RB. Syndapin I, a synaptic dynamin-binding protein that associates with the neuronal Wiskott-Aldrich syndrome protein. Mol Biol Cell. 1999;10:501–513. doi: 10.1091/mbc.10.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein Rac regulates growth factor-induced membrane ruffles. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- Rodgers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degradaded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Rost B, Sander C. Combining evolutionary inormation and neuronal networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Safer D. An electrophoretic procedure for detecting proteins that bind actin monomers. Anal Biochem. 1989;178:32–37. doi: 10.1016/0003-2697(89)90351-5. [DOI] [PubMed] [Google Scholar]
- Schuuring E, Verhoeven E, Litvinov S, Michalides RJAM. The product of the EMS1 gene, amplified and overexpressed in human carcinomas, is homologous to a v-src substrate and is located in cell-substratum contact sites. Mol Cell Biol. 1993;13:2891–2898. doi: 10.1128/mcb.13.5.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirao T. The roles of microfilament-associated proteins, drebrins, in brain morphogenesis. J Biochem. 1995;117:231–236. doi: 10.1093/jb/117.2.231. [DOI] [PubMed] [Google Scholar]
- Shirao T, Obata K. Two acidic proteins associated with brain development in chick embryo. J Neurochem. 1985;44:1210–1216. doi: 10.1111/j.1471-4159.1985.tb08745.x. [DOI] [PubMed] [Google Scholar]
- Sparks AB, Hoffman NG, McConnell SJ, Fowlkes DM, Kay BK. Cloning of ligand targets—systematic isolation of SH3 domain-containing proteins. Nat Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- Toda M, Shirao T, Minoshima S, Shimizu N, Toya S, Uyemura K. Molecular cloning of cDNA encoding human drebrin E and chromosomal mapping of its gene. Biochem Biophys Res Commun. 1993;196:468–472. doi: 10.1006/bbrc.1993.2273. [DOI] [PubMed] [Google Scholar]
- Vitale ML, Seward EP, Trifaro JM. Chromaffin cell cortical actin network dynamics control the size of the release-ready vesicle pool and the initial rate of exocytosis. Neuron. 1995;14:353–363. doi: 10.1016/0896-6273(95)90291-0. [DOI] [PubMed] [Google Scholar]
- Weed SA, Du Y, Parsons JT. Translocation of cortactin to the cell periphery is mediated by the small GTPase Rac1. J Cell Sci. 1998;111:2433–2443. doi: 10.1242/jcs.111.16.2433. [DOI] [PubMed] [Google Scholar]
- Wesp A, Hicke L, Palecek J, Lombardi R, Aust T, Munn AL, Riezman H. End4p-SLa2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:2291–2306. doi: 10.1091/mbc.8.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Parsons JT. Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J Cell Biol. 1993;120:1417–1426. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]