Abstract
Escherichia coli and other enterobacteria exploit the H+-consuming reaction catalysed by glutamate decarboxylase to survive the stomach acidity before reaching the intestine. Here we show that chloride, extremely abundant in gastric secretions, is an allosteric activator producing a 10-fold increase in the decarboxylase activity at pH 5.6. Cooperativity and sensitivity to chloride were lost when the N-terminal 14 residues, involved in the formation of two triple-helix bundles, were deleted by mutagenesis. X-ray structures, obtained in the presence of the substrate analogue acetate, identified halide-binding sites at the base of each N-terminal helix, showed how halide binding is responsible for bundle stability and demonstrated that the interconversion between active and inactive forms of the enzyme is a stepwise process. We also discovered an entirely novel structure of the cofactor pyridoxal 5′-phosphate (aldamine) to be responsible for the reversibly inactivated enzyme. Our results link the entry of chloride ions, via the H+/Cl− exchange activities of ClC-ec1, to the trigger of the acid stress response in the cell when the intracellular proton concentration has not yet reached fatal values.
Keywords: autoinhibition, bacterial acid resistance, chloride binding, glutamate decarboxylase, pyridoxal 5′-phosphate
Introduction
In the span of a human lifetime, the immense amount of food that passes through the gastrointestinal tract, the largest body surface, is inevitably accompanied by the entry of bacteria. Some are beneficial to the host because, following intestinal colonization, they release vitamins, stimulate the immune system and limit colonization by pathogens (Bengmark, 1998). The gastrointestinal tract provides barriers to invading bacteria in the form of gastric acidity, lysozyme production, rapid turnover of epithelial cells and the constant presence of immune system cells. However, everyday experience shows that, periodically, pathogenic bacteria successfully colonize the intestine having overcome all the barriers, including the primary bactericidal barrier of the gastrointestinal tract, namely the stomach (Giannella et al, 1972).
Ingested bacteria, both pathogenic and commensal, must endure the strongly acidic stomach (pH <2.5) for approximately 2 h on their way to the more hospitable intestine. Among several mechanisms whereby enteric bacteria prevent their intracellular contents from reaching fatal levels of acidity, the most efficient is that based on the H+-consuming conversion of glutamate to γ-aminobutyrate (GABA). This mechanism is found in bacteria as different as Escherichia coli, Listeria monocytogenes and Lactococcus lactis (Sanders et al, 1998; Castanie-Cornet et al, 1999; De Biase et al, 1999; Cotter et al, 2001). The enzyme catalysing this reaction is the pyridoxal 5′-phosphate (PLP)-dependent glutamate decarboxylase (Gad; EC 4.1.1.15), a component of the glutamate-based acid resistance (AR) system (Castanie-Cornet et al., 1999; De Biase et al., 1999). In Escherichia coli, the other known component of this system is the glutamate/GABA antiporter GadC (Hersh et al, 1996; Castanie-Cornet et al, 1999; De Biase et al, 1999). In this same microorganism, a similar but less-efficient AR system employs the biodegradative arginine decarboxylase (AdiA, converting arginine into agmatine; Lin et al, 1995) and the arginine/agmatine antiporter AdiC (Gong et al, 2003; Iyer et al, 2003). The mechanism of action of the glutamate- and arginine-based AR systems is simple: the decarboxylase replaces the leaving α-carboxyl group of the corresponding amino-acid substrate with H+, while the cognate inner-membrane antiporter exports the decarboxylation product in exchange for new amino-acid substrate. In this way, two needs are satisfied: protons, which leak into the cell when the extracellular pH becomes very acidic, are consumed and the local environment is made less acid because the exported product is less acidic than the imported substrate. Recent studies of E. coli cells in vivo have shown that at external pH 2.5, in the absence of amino-acid supplementation, the internal pH falls to 3.6, but when glutamate or arginine are present in the medium, internal pH does not drop below 4.2 and 4.7, respectively (Richard and Foster, 2004). Moreover, the two amino-acid-based AR systems not only contribute to pH homeostasis but also counteract illicit entry of protons. Removal of one intracellular negative charge per decarboxylation step (i.e. Glu-1 to GABA0, Arg+1 to Agm+2) helps to repel incoming protons by inversion of the membrane potential (Richard and Foster, 2004), a strategy also adopted by extreme acidophiles (Matin, 1999). A crucial role in the E. coli AR is also played by the CIC chloride channels. E. coli cells lacking these channels fail to survive an extreme acidic shock (Iyer et al, 2002). Electrophysiological techniques (Accardi and Miller, 2004; Accardi et al, 2004) and the high-resolution X-ray structures of EcClC (Dutzler et al, 2002; Dutzler et al, 2003) identified this protein as a 1H+–2Cl− exchanger. Thus, the biological role of the EcClC antiporter in the amino-acid-based AR systems is likely to be the import of Cl− anions in exchange for protons (Foster, 2004). This process promotes proton extrusion from the cell and controls transmembrane potential by avoiding the excessive positive ΔΨ values, which would be generated by inleaking H+ and by the accumulation of decarboxylation products.
Structural studies on one of the two E. coli glutamate decarboxylase isoforms, GadB, have demonstrated that the enzyme undergoes pH-dependent changes in conformation, cellular localization and enzymatic activity, which combine to regulate the intracellular pH (Capitani et al, 2003). Recently, a crystallographic study of the other isoform, GadA, unveiled the structure of the enzyme in complex with the noncovalent inhibitor glutarate (Dutyshev et al, 2005).
It has long been known that Gad interconverts between active and inactive forms in a process that, since it involves the simultaneous association or dissociation of several protons, makes the transition between these forms more abrupt than it would be if a single proton were involved (O'Leary and Brummund, 1974). Monovalent anions such as I−, Br− and Cl− were reported to increase the rate of GadB conversion from the neutral-pH inactive form into the low-pH active form and to decrease the opposite. It was thus concluded that these anions bind only to the low-pH conformation of the enzyme (O'Leary and Brummund, 1974). The crystal structures of GadB, a 315-kDa homohexameric enzyme, at neutral and at low pH (PDB entries 1PMO and 1PMM, respectively (Capitani et al, 2003)) identified the structural elements that undergo the pH-dependent conformational change that leads to enzyme activation. GadB was deduced to be inactive at neutral pH (7.6) because each active site funnel is blocked by the C-terminus of the same subunit and by a β-hairpin from the neighbouring subunit. At the same pH, the N-termini of all six subunits are disordered. The X-ray structure at low pH (4.6), very close to the pH-optimum of the enzyme (Shukuya and Schwert, 1960a), reveals an opposite scenario: disordered C-termini and shifted β-hairpins allow full access of the substrate to the six active sites, while the N-termini are ordered and form two triple α-helical bundles, parallel to the three-fold axis of the hexamer. Membrane association studies of wild-type GadB and of its deletion mutant, GadBΔ1–14, conducted at acid and neutral pH showed the bundles to be crucial for recruitment of the protein, under acidic conditions, to the inner E. coli membrane, the place thought to be most affected by inleaking protons (Capitani et al, 2003).
The recent finding that the EcClC acts as a H+/Cl− antiporter (Accardi and Miller, 2004) and the biochemical observation that halides positively affect the activation of Gad (O'Leary and Brummund, 1974) suggest that the entry of chloride (103 mEq/l in gastric juice) in the bacterial cells satisfies two needs, namely prevention of dangerous membrane hyperpolarization (to more positive ΔΨ) and promotion of Gad activation.
Prompted by the above considerations, we studied GadB activation by chloride and other halides using a combination of structural, mutagenesis and kinetic approaches. In this work, we describe not only the structural basis for chloride-dependent GadB activation and its biological implications, but provide also ‘crystallographic snapshots' of the pH-dependent conformational changes that the enzyme undergoes in the transition from active to inactive state. In addition, our analysis reveals a novel mode of GadB autoinhibition that may be of general significance for the superfamily of PLP-dependent enzymes.
Results
The H+-dependent activation of GadB is a cooperative process positively affected by halides and by N-terminal helix formation
Gad switches between inactive and active forms over a narrow pH range. The switch is accompanied by a change in absorption of the PLP cofactor between the yellow species, absorbing maximally at 420 nm and corresponding to the active low-pH form of the enzyme, and the colourless species, absorbing maximally at 340 nm and corresponding to the inactive high-pH form of the enzyme (Shukuya and Schwert, 1960b). The titration curve, generated by plotting the 420 (or 340) nm absorbance versus pH, fits a process in which the enzyme binds more than one proton, with the number of protons involved having been estimated as 4 (Shukuya and Schwert, 1960b) and 5 (Tramonti et al, 2002).
![]()
In this work, we have chosen to present our results by expressing the controlled variable as [H+] rather than pH because the observed effects are more clearly seen and because, in illustrating cooperativity of ligand binding, it is conventional to express the ligand concentration linearly rather than logarithmically. We fitted our results (Figure 1) to the Hill equation (Hill, 1910). All of our observations made with the wild-type enzyme showed sigmoid dependence and fitted well to the Hill equation. In the absence of added anions, the best-fit values returned for n and K were 9.9±1.3 and 4.2 μM, respectively. In the presence of 50 mM NaCl, Hill analysis returned values of n=7.3±1.0 and K=1.8 μM. In the presence of 50 mM NaI (or NaBr), Hill analysis gave n>7 and K=1 μM (data not shown). Similar experiments were performed with the GadBΔ1–14 mutant, lacking one of the protein parts that changes conformation during enzyme activation and that is required for the recruitment of GadB to the E. coli inner membrane under acidic conditions (Capitani et al, 2003). The results showed dependence on [H+] to be much less sigmoidal. Best fits to the Hill equation gave n=2.0±0.2 and K=3.6 μM in the absence of chloride and n=1.7±0.2 and K=2.9 μM in the presence of 50 mM NaCl.
Figure 1.
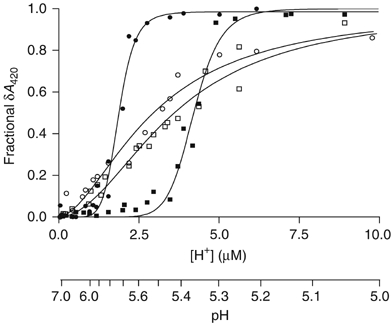
Effect of chloride and removal of N-terminal 14 residues on activation of GadB by H+. GadB wild type, ▪; GadB wild type+50 mM NaCl, •; GadBΔ1–14, □; GadBΔ1–14+50 mM NaCl, ○. Continuous lines through the data points are those of best fit to the Hill equation (Equation (1) in Supplementary data). The experimental points for each condition are results of several absorbance measurements at equilibrium (see Supplementary data) conducted at different enzyme concentrations, all in the range 6–9 μM, and in 0.1 M Na acetate buffer. Data are presented as fraction of the maximal cofactor absorbance change observed in each experiment.
This equilibrium analysis shows that halides act as positive effectors of a highly cooperative process. In the presence of Cl−, the enzyme switches on sharply at pH ∼5.7 (Figure 1). The analysis also shows that when the N-terminal 14 residues are removed, much of the cooperativity of the activation process is lost. In separate experiments, we compared the ability of wild-type and Δ1–14 GadB to catalyse decarboxylation of glutamate at pH 4.6. Both forms of the enzyme catalyse the reaction with similar kcat and Km values (data not shown). Thus, we conclude that the triple helix bundles play no part in the catalytic reaction but that their reversible formation is the major source both of the cooperativity itself and of its dependence on halides.
Chloride binding to allosteric sites stabilizes the triple-helix bundle
To address the issue of where halides bind, two different crystal forms of GadB, both obtained in the presence of the substrate analogue acetate, were analysed in the presence of various anions. The P1 crystal form (form A) was obtained at pH 4.6 (Capitani et al, 2003) and used for soaking experiments with KI. Co-crystallization experiments with KBr at pH 5.5 yielded a new GadB crystal form belonging to the space group P6322 (form B), which was also used for chloride soaks. Crystals of this latter form were also obtained in KCl co-crystallization experiments.
The structure of GadB (form A) obtained by soaking crystals in 100 mM KI shows how iodide binds to the enzyme. A Bijvoet difference-Fourier map was calculated after data collection (λ=1.3 Å), processing and scaling. The initial map, based on phases from the active low-pH form of GadB (1PMM), revealed six clear peaks (height>8σ) corresponding to one iodide ion per GadB monomer. These iodide-binding sites were located at the bottom of each N-terminal helix in both triple-helix bundles (Figure 2B). Each iodide ion is strongly bound to a site (S1) in a buried cleft that exists only in the low-pH form of the enzyme. The ion interacts with residues Ser16, Arg17 and Phe18. At low pH, but not at neutral pH, these residues, together with Gly19, form a characteristic loop that acts as a platform for the formation of an α-helix contributed by residues 3–15 of each monomer. The side chains of Trp67, Leu279, Leu334, Val342 and Arg427, from a neighbouring subunit, provide a hydrophobic environment and interact with the ion from the other side. The hydroxyl group of Ser16 and the Cδ and guanidinium group of Arg427 interact most closely with iodide (3.2 and 3.5 Å, respectively). Since the iodide ion is sandwiched between residues Ser16 and Arg427, the aforementioned Ser16–Gly19 loop is strongly stabilized and properly orients the N-terminal helix so that it forms a bundle with the corresponding N-terminal helices from two other subunits. A superposition of the low-pH GadB structure (PDB: 1PMM) onto the GadB–iodide complex structure (both crystallized at the same pH and belonging to the same space group) did not reveal any major main or side chain movements upon iodide binding.
Figure 2.
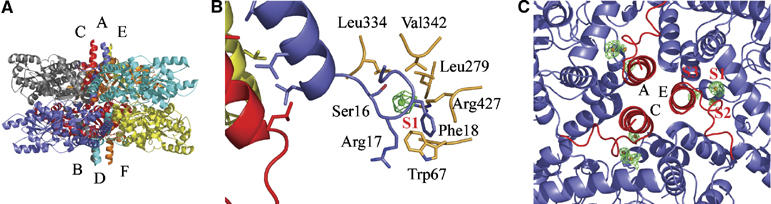
Halide binding to the N-terminal region of GadB. (A) Side view of GadB in its active (low pH) conformation (1PMM) with the helix bundles protruding from top and bottom of the hexamer. (B) Detailed view of the iodide-binding site S1 with one helix bundle partly visible. Residues interacting with the ion are depicted in blue and orange. Chloride anomalous difference density contoured at 2.8σ is superimposed on the GadB–I− structure. (C) View along the three-fold axis of GadB onto the N-terminal three helical bundle (in red) with bound bromide ions (S1, S2, S3). Bromide ions appear as brown spheres with Bijvoet difference-Fourier electron density in green contoured at 3.3σ.
A second iodide site (S2) per GadB subunit is found in the 2mFo-DFc electron density map. It is located at the subunit interface, also near the N-termini in the central cavity of the protein. The ion binds to the side chains of His73 and Asn81, to the amide nitrogen of Asp68 and to the side chain of Trp67 from another subunit. The protein backbone as well as the side chain conformation of residues Thr66, Trp67, Asp68 and Asp69 in the GadB–iodide structure are similar to those in the low-pH active form of GadB (1PMM), which in this region differs significantly from the neutral-pH inactive form (1.1 Å r.m.s.d. for the Cα atoms of residues 65–70 between 1PMM and 1PMO). Asn81 and Trp67, both involved in binding of the S2 iodide ion, are only five residues away from Asp86 and Thr62, residues known to interact with the distal carboxylate of the substrate glutamate in the low-pH crystal structure (Capitani et al, 2003). This was confirmed in the low-pH crystal structure by the orientation of the carboxylate groups of the substrate analogues acetate and glutarate (Capitani et al, 2003; Dutyshev et al, 2005).
Co-crystallization of GadB with 180 mM KBr at pH 5.5 yielded crystals of form B (space group P6322). Diffraction data, collected at a wavelength near the bromide absorption edge (λ=0.9 Å), produced a very clear Bijvoet difference-Fourier map featuring three strong peaks per GadB subunit (S1–S3 in Figure 2C). In each GadB monomer, two bromide ions occupy sites S1 and S2, in the same fashion as iodide ions do. The third bromide site (S3), found only in the GadB–Br− complexes, is provided by the amide nitrogen and the side chain of Arg17 and by the Cβ of Asp69 from a neighbouring subunit.
Minor iodide- and bromide-binding sites, exhibiting lower occupancies (39–74%), were also identified and are described in detail in Supplementary data.
As a next step, we analysed the binding to GadB of the physiologically relevant chloride ion. A two-fold approach was followed to detect chloride binding. First, a model of GadB with bound chloride ions was obtained by soaking crystals of the GadB–bromide complex (form B) in a solution containing 180 mM KCl and collecting diffraction data. Upon KCl soak, chloride displaces bromide, as demonstrated by the disappearance of the anomalous bromide signal when data collection was carried out near the bromide absorption edge (0.9 Å). Calculation of an mFo−DFc difference electron density map immediately showed three peaks of positive density with maximal heights of 6.5σ (S1), 6.9σ (S2) and 7.3σ (S3) per GadB monomer near the N-terminus. The three peaks coincide with the three positions known to be the bromide sites (S1, S2 and S3). Calculation of a Bijvoet difference-Fourier electron density map revealed no residual bromide anomalous signal, indicating that the bromide ions were completely replaced by chloride ions.
In a confirmatory experiment, aimed at the direct detection of chloride, GadB was co-crystallized with 180 mM KCl. The GadB–chloride complexes also crystallized in space group P6322. The wavelength for data collection was set to 1.7 Å, a compromise between maximizing the very weak anomalous signal of chloride (f″=0.84 e− at λ=1.7 Å) and minimizing absorption effects and radiation damage. Calculation of Bijvoet difference-Fourier electron density maps at low resolution (6.2 Å) produced very clear peaks for most of the bound chloride ions and even peaks for many sulphur atoms (f″=0.67 at λ=1.7 Å) of GadB were present. Owing to the weak affinity of chloride (KD=8 mM, O'Leary and Brummund, 1974), the flexible binding mode of the easily polarizable Cl− ion, the very low chloride anomalous scattering contribution of <1% (Hendrickson and Ogata, 1997) and the low resolution of the data, anomalous peaks (>2.8σ) were present only for six out of 12 expected sites (S1 and S2 cannot be distinguished at a resolution of 6.2 Å). The major site underneath the triple-helix bundle known from experiments with bromide and iodide (S1 and Figure 2B) exhibits a strong peak in five subunits out of six with a maximal height of 5.0σ. The binding of Cl− was additionally investigated using  isomorphous difference-Fourier maps at 4.2 Å resolution, which showed clear and distinct peaks for chloride binding sites S1 and S2 (see Supplementary data).
isomorphous difference-Fourier maps at 4.2 Å resolution, which showed clear and distinct peaks for chloride binding sites S1 and S2 (see Supplementary data).
By combining the above results, it is possible to conclude that the halides I−, Br− and Cl− bind strongly to the N-terminal part of GadB in protein cavities present only in the low-pH, active conformation of the enzyme. Halide binding clearly stabilizes the low-pH form of GadB by fixing the turn built by residues 16–19 in the right conformation, thereby stabilizing the triple α-helix bundle that is required for migration of the enzyme towards the membrane (Capitani et al, 2003). I−, Br− and Cl− bind to sites S1 and S2, while Br− and Cl− also bind to site S3. The physico-chemical properties may account for the differential binding of these anions to GadB, as radii decrease from 2.2 Å (I−) to 2.0 Å (Br−) and 1.8 Å (Cl−), with polarizability following the Hofmeister series (I−>Br−>Cl−) (Hofmeister, 1888).
pH-shift experiments in cristallo: monitoring folding and unfolding events in GadB
The positive effect of halides on GadB activation is achieved by shifting the activation curve towards higher pH (Figure 1). Thus, at pH 5.5, the enzyme converts almost completely from inactive to active form (as judged spectroscopically and by activity measurements) when the halide concentration is increased from zero to 50 mM. Our GadB–bromide crystals (form B) were obtained at pH 5.5 in the presence of 180 mM KBr, conditions in which the enzyme is almost fully active. Fortuitously, the packing in the P6322 crystal form allows the N- and C-terminal regions, which undergo conformational changes during enzyme activation, to move freely as these regions are not involved in crystal contacts. To detect possible intermediate structures in the transition described by Capitani et al (2003), pH-shift experiments were performed in cristallo. GadB–bromide crystals, grown at pH 5.5, were transferred into soaking solutions at pH 5.9 and 6.5, respectively. The crystals were cryoprotected and diffraction data collected. The pH-shift affected the crystal lattice (unit cell parameters change significantly in the case of pH 6.5, Supplementary Table I) and diffraction quality, so that only moderate resolution and data quality could be achieved. Careful analysis of the electron density allowed unambiguous assignment of structural details relevant for this study and a sort of ‘crystallographic snapshots' of the pH-driven changes was obtained:
(1) In a first experiment, the pH was changed from 5.5 to 5.9, a pH close to the pK 6.0 for the pH-dependent transition for bromide-complexed GadB. Diffraction data were collected and analysed (Supplementary Table I) by simulated annealing composite omit maps (Hodel et al, 1992). The electron density map shows that the C-termini in all subunits have become ordered up to residue Gln460. Thus, the turn following the last long helix of the small domain is formed upon this pH-change. In subunit D, however, the C-terminus is fully ordered up to the final residue, Thr466. Unexpectedly, electron density in this region hints at a covalent bond between the penultimate residue, His465, and C4′ of the cofactor through its side chain. At this pH, it is likely that a covalent ternary complex (aldamine) has been formed between His465, the PLP-cofactor and Lys276 (see also next paragraph). For the most part, the conformation of the β-hairpin (residues 300–313) closely resembles the neutral-pH conformation, which is seen to close the active site in 1PMO. However, it is more disordered at the β-turn itself. This makes sense since residues at the side of the tip (Leu306, Tyr305) interact with residues 461–463 of the C-terminus and can only be well defined if the C-terminus is also ordered. An unexpected asymmetry is evident in the conformations of the N-termini. The N-termini of chains A, C and E are completely ordered in a triple α-helix bundle and bromide sites S1, S2 and S3 are fully occupied underneath each helix (Figure 3A). This structural arrangement is typical of the low-pH (active) form of the protein (Figure 2A and B). However, the helical bundle on the other side of the molecule, formed by the N-termini of chains B, D and F, is completely unfolded without any electron density for the helices. The turn formed by residues Ser16–Gly19 is in the neutral pH-conformation as described for 1PMO (Capitani et al, 2003) and no bromide ions are found in their sites (Figure 3A). We analysed the above pH-driven conformational changes also by using isomorphous difference-Fourier maps (see Supplementary data).
Figure 3.
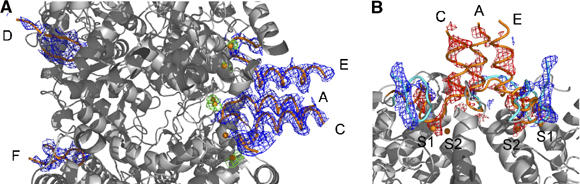
In cristallo pH-shift experiments reveal conformational changes at the N-termini of GadB. (A) Cross-section of the central region of GadB–Br− after pH shift (5.5–5.9). A 2mFo−DFc composite annealed omit map is contoured at 1σ (blue), with residues 3–20 in tube representation (orange). Br− ions are depicted as brown spheres with their Bijvoet difference-Fourier electron density in green (2.2σ). (B) Detailed view of one N-terminal three helical bundle after a pH-shift from pH 5.5 to 6.5. The starting conformation of residues 3–25 in tube representation is depicted in orange while the final conformation at pH 6.5 is shown in cyan. mFo−DFc electron density calculated with phases from the starting model of GadB–Br− at pH 5.5 is displayed in red (−2.4σ) and blue (+2.4σ).
(2) Shifting the pH from 5.5 to 6.5 induced even further significant conformational changes: the C-termini of all six GadB subunits go from a disordered to a completely folded state in the active site funnel, thus blocking active site entry (Supplementary Figure 3). Clear electron density for a covalent bond (see also next paragraph) between the distal side chain nitrogen of His465 and the C4′ atom of the PLP-cofactor is now visible also in subunits B, C, E and F (2mFo–DFc simulated annealing composite omit map contoured at 0.8σ). The β-hairpin (residues 300–313) at pH 6.5 shifts towards the folded C-terminal tail, thereby preventing access of substrate and solvent to the cofactor. Both triple helical bundles are completely unfolded. The turn following the α-helices (residues 16–19) has rearranged from a right-handed to a left-handed turn. All bromide ions, kept in position by this turn, have been released, as confirmed by the absence of any significant peak in a Bijvoet difference-Fourier map for the known bromide sites (Figure 3B).
It is remarkable that not only all conformational changes known to occur in GadB between pH 4.6 and 7.6 can be reproduced in cristallo but also that intermediates of a folding process can be trapped and analysed. In particular, the structure at pH 5.9 suggests that folding/unfolding of the C- and N-termini happens at the same pH but with independent mechanisms. The unfolding of the N-terminal helix bundles seems to be a two-step process, which comes to completion at a slightly different pH for each side of the protein, probably driven by cooperativity effects influencing helical bundle formation (Lupas and Gruber, 2005). Shifting the pH from 5.5 to 6.5, the in cristallo structural transition from the active to the inactive form of the enzyme is complete. This is exactly the same pH range in which titrations in solution show the halide-bound enzyme to convert completely its absorption spectrum from the active 420 nm-absorbing form to the inactive 340 nm-absorbing form (Figure 1).
Capturing the aldamine: a novel mode of GadB autoinhibition revealed by the X-ray structure of GadBΔ1–14
The N-terminal deletion mutant GadBΔ1–14 lacking the first 14 amino acids was crystallized in space group P212121 at pH 5.5. From the titration curve, it is evident that at this pH the GadBΔ1–14 mutant is mostly in the inactive conformation (Figure 1). Even though the refined structure exhibits only moderate r.m.s. deviations of 0.3 Å if compared with 1PMO (chain A, residues 30–452, Cα atoms), detailed analysis revealed interesting structural features. No electron density is visible for residues 15–28. Thus, the presence of the first 14 residues seems to be necessary for ordering also the following part of each GadB polypeptide chain. Since no significant domain rearrangement is detected in the mutant structure, the inability of GadBΔ1–14 to migrate to the E. coli membrane at low pH (Capitani et al, 2003) can only be attributed to the missing N-terminal α-helical bundles.
Experiments on wild-type GadB at pH 6.5 provided evidence for the existence of a ternary complex (aldamine), arising from reaction of the distal nitrogen of the imidazole ring of His465 with the PLP-Lys276 Schiff base. Since the GadBΔ1–14 mutant was crystallized in the inactive conformation, a model lacking residues 453–466 was used for electron density calculations in order not to introduce any model bias in this region: a clear conformation of the C-termini in the active site became visible. Evident positive mFo–DFc difference electron density was present in every GadB subunit, revealing a covalent bond between the C4′ atom of PLP, already involved in a covalent bond with Lys276, and the penultimate residue in the polypeptide chain, His465 (Figure 4). Residues 453–464 extend into the active site in a similar way as in 1PMO, while His465 is in a completely new position with its distal side chain nitrogen covalently attached to the PLP-cofactor (C4′). A sharp turn of the main chain follows, so that the C-terminal residue, Thr466, points towards the solvent. The C4′ atom of the cofactor is sp3-hybridized and shares two covalent bonds with the ɛ-nitrogen of Lys276 and with the distal side chain nitrogen of His465 in an R-configuration (Figure 4). The best fit of this substituted aldamine moiety to the electron density was achieved with a slightly distorted tetraehedral geometry around the C4′ centre. In the neutral pH (inactive) structure (1PMO) described earlier by Capitani et al. (2003), no electron density was detectable for a covalent bond between the cofactor and the side chain amino group of Lys276. The gap was attributed to reduction of the Schiff base by NaBH3CN for sample preparation, combined with radiation damage upon data collection at the synchrotron. A protein–protein BLAST (http://us.expasy.org/tools/blast) using the sequence of E. coli GadB as query against the subsection bacteria of the UniProt database identified 48 sequences of highly similar proteins (E-value <1e-100). His465 (GadB) turns out to be fully conserved in the multiple alignments of these homologous sequences. In 15 sequences this histidine is found at the penultimate position as in GadB, in 29 sequences at the ultimate position and in four sequences it is followed by 9–13 residues. In the crystal structure of GadBΔ1–14, the protein was not treated with NaBH3CN and the new chemical species is clearly present in the autoinhibited inactive form of GadB. The nature of the 340 nm-absorbing chromophore was attributed either to the deprotonated enolimine form of the PLP-Schiff base (Shukuya and Schwert, 1960b; Tramonti et al, 2002) or to a substituted aldamine formed by covalent bonding of lysine 276 and of an unidentified group X to the PLP-cofactor (O'Leary, 1971; O'Leary and Brummund, 1974). Our structural proof for the existence of a substituted aldamine in form of a geminal diamine with a protein side chain (His465) clearly identifies the 340 nm-absorbing species and ends a long period of speculation about the chemical state of PLP present in the catalytically inactive form of GadB.
Figure 4.
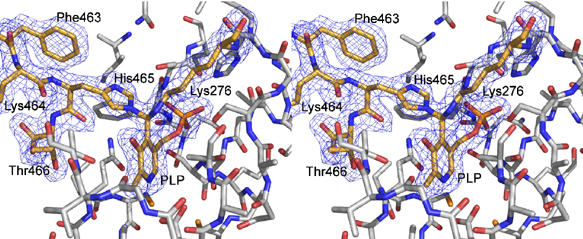
Stereo view of the substituted aldamine in the active site of GadBΔ1–14. The C-terminal residues 463–466, the PLP-cofactor and Lys276 appear as sticks (orange and atom colours, while other residues are in white and atom colours) with the corresponding 2mFo−DFc electron density (1σ). Both the ɛ-nitrogen of Lys276 and the distal side chain nitrogen of His465 form a covalent bond with the C4′ atom (hybridized sp3) of the PLP-cofactor.
Effect of anions on the kinetics of transitions between active and inactive forms of GadB wild type and GadBΔ1–14
We analysed by stopped-flow spectrophotometry the effect of anions on the kinetics of transitions from the low-pH form to the high-pH form of GadB wild type and Δ1–14. We found that the absence of the N-terminal 14 residues decreases by 20-fold the rate at which inactivation of GadB occurs upon a pH jump from 4.6 to 5.9 and makes the rate of inactivation insensitive to chloride (Figure 5), thus confirming the major role of this structural determinant in controlling cooperativity, sensitivity to chloride and inactivation rates. Our analysis also showed that the conversion of GadB wild type from the low-pH to the high-pH form follows a kinetic profile which can only be fitted by a three-step process (Figure 5A and details in Supplementary data). These results are in agreement with the crystallographic snapshots analysis showing that two consecutive steps (sequential unfolding of the bundles), not associated with 420-nm absorbance decrease (aldamine formation), must occur before the major spectroscopic changes are detected.
Figure 5.

Effect of Cl− on decrease in A420 after pH jump-up. The enzyme dissolved in 0.1 M sodium acetate pH 4.6 was mixed with 0.2 M Na2HPO4 containing various concentrations of NaCl to give a final pH of 5.9. (A) WT-GadB (8 μM) at [NaCl]=(a) 0; (b) 12.5 mM; (c) 25 mM; (d) 50 mM and (e) 75 mM. Experimental results (dots) were fitted (thin black line) to the three-step model described in detail in Supplementary data. (B) Δ1–14 GadB (8 μM) at (NaCl)=0; 25; 50 and 100 mM.
Discussion
We undertook structural and functional studies of chloride binding to GadB. Our data indicate that both components of the host ‘chemical weapon' HCl, H+ and Cl−, are used by some enteric bacteria for triggering a highly efficient response that enables survival and, in the case of pathogenic bacteria, turns against the host. We believe that our results provide a novel picture of the importance of chloride in the glutamate-based AR system, because they extend the biochemical and physiological effects of this halide well beyond the proposed model in which the import of Cl− by the ClC antiporter serves to control transmembrane potential (Foster, 2004). Our observation that binding of Cl− near the N-terminus of each monomer of the GadB enzyme anticipates the formation of and stabilizes the two triple-helix bundles, typical of the active form of the enzyme, provides a molecular explanation for additional roles of chloride in E. coli AR. The influence of different sodium chloride concentrations and pH values on the growth of stressed and unstressed E. coli cells (O157:H7) was studied by Jordan and Davies (2001). They found that addition of sodium chloride to the medium at pH 5 increased the growth rate of acid-stressed cells dramatically and also decreased lag times for bacterial growth. Herein, we demonstrate that the hypersensitivity of the enzyme to pH resides in its ability to form the above-mentioned bundles and Cl− significantly raises the pH at which this activating conformational change occurs. Thus, not only are the pH-correcting properties of glutamate decarboxylase turned on closer to neutrality but also, as a result of the same structural change, the enzyme would be expected to transfer to the membrane (Capitani et al, 2003) where its pH-correcting properties would be most advantageous.
The multiple roles of chloride in AR explain why eliminating the ClC antiporters in E. coli impairs the ability to withstand an extreme acidic shock (Iyer et al, 2002). It has recently been shown that also eukaryotic members of the ClC family involved in acidifying compartments of the endosomal/lysosomal pathway (ClC-4 and ClC-5) function as electrogenic Cl−/H+ exchangers (Picollo and Pusch, 2005; Scheel et al, 2005). This makes it very likely that H+-coupled Cl− transport over a membrane is a general principle used when a proton gradient has to be altered or controlled.
Sensitivity to ligand concentration as high as that observed for GadB is rarely observed. The abrupt nature of the H+-dependent transition between active and inactive forms means that the enzyme behaves almost as a switch that is either on or off. Precise measurement of number of protons (n) in this situation is difficult because of the need to obtain experimental points in the narrow range where the transition occurs. Nevertheless, the values we measured were consistently in the range 7–10 considerably higher for instance than the value of 4.5 observed for activation of the adenylyl cyclase responsible for sensing pH in Mycobacterium tuberculosis (Tews et al, 2005). We observed that most of the cooperativity leading to the hypersensitivity of GadB to [H+] is lost when the N-terminal 14 residues are removed. This strongly suggests that the formation of the triple helix that accompanies lowering of pH is somehow linked to the activating changes occurring at the active site. Such a link has not been easy to identify. However, the formation of each three-helix bundle is expected to be a strongly cooperative process since all four carboxylic acids in each aminoterminal pentadecamer would be protonated in the active helical form and unprotonated in the inactive random form. Our observation that the allosteric activator Cl− binds to protein cavities that do not exist when the helices are not formed reinforces the identification of this helix-coil transition as the source of the allosteric behaviour.
It is still unclear how the migration of GadB to the inner E. coli membrane under acidic conditions can take place. A direct insertion of the triple-helix bundle into the lipid bilayer is not very likely due to the hydrophilic surface properties of the bundle. An alternative possibility would be that the tip of the bundle, containing the sequence MDKKQ, with a net positive charge, electrostatically interacts with the membrane phospholipid heads. A third possibility is docking of GadB to an integral membrane protein either in a direct mode via the helix bundle or in an indirect way via an adaptor protein. To elucidate the mechanism of GadB membrane affinity and to identify possible binding partners will be a very challenging task for future structural studies.
The ‘crystallographic snapshots', taken at pH values covering the range over which the activating transitions occur, demonstrate that, unlike classical examples of allostery such as haemoglobin and aspartate transcarbamylase, the symmetry of this hexameric enzyme is not conserved. At pH 5.9, the enzyme was seen to have a triple-helix bundle on one side of the hexamer, but the N-termini on the other side remained disordered. At the same pH, all six carboxy termini were ordered as far as the last six residues but only one of the six monomers had the C-terminus fully ordered with His465 covalently bound to the cofactor. This monomer was one of the three with disordered N-termini. At pH 6.5, all N-termini were disordered, and symmetrically on either side, five of the six C-termini were fully ordered with covalent bonds between His465 and the cofactor. The kinetic analysis of the pH jump-up also detected multiple steps, consistent with a model in which the triple-helix bundles unfold independently without change in spectrum, but unfolding of both was required before the covalent reaction (aldamine formation) resulting in loss of absorbance at 420 nm and inactivation occurred. Thus, the snapshots of the conformational changes of GadB obtained by pH-shift experiments in cristallo as well as stopped flow experiments are both suggestive of an unexpected protein behaviour: the formation of the bundles appears to be sequential instead of simultaneous.
Interestingly, in our in cristallo pH-shift experiments, the estimated pK for the formation of the triple-helix bundle (one bundle formed on one side and the other unfolded on the other side) is approximately 5.9 in the presence of bromide, in agreement with the K=1 μM (pK=6) obtained by fitting the in solution GadB titration curve in the presence of bromide. Combining the in cristallo pH jump-up experiments with the kinetic pH jump-up data, it can be concluded that the conversion of GadB from the active form, absorbing at 420 nm, into the inactive form, absorbing at 340 nm, is the sum of two events: the first, spectroscopically undetectable and rate limiting, consists in the unfolding of the bundles, a process that is negatively affected by chloride; the second, spectroscopically visible and much slower than the first, consists in the closure of the active site with the final formation of an aldamine structure.
A finding of particular relevance concerns the ternary adduct (aldamine) through which GadB autoinhibition takes place. The structural identification of such an adduct is an absolute novelty and its importance might extend well beyond GadB to other PLP-dependent enzymes like tryptophanase (Ikushiro et al, 1998), cystalysin from Treponema denticola (Bertoldi et al, 2002) or 5-aminolevulinate synthase (Zhang et al, 2005). Our observation that the 340 nm-absorbing chromophore, typical of the inactive form of the enzyme, is a covalent adduct formed between the imidazole ring of His465 and the Lys276-PLP imine, ends a long period of uncertainty about the chemical nature of the cofactor when the enzyme's activity is reversibly switched off by increasing pH. Our crystallographic demonstration of this structure suggests that histidine is not only the unknown substituent X (O'Leary, 1971; O'Leary and Brummund, 1974) in GadB but also a possible substituent in other PLP-dependent enzymes (other possible substituents are lysines and tyrosines).
In conclusion, considerable interest has been shown in the importance of chloride to bacterial acid resistance. It has been demonstrated that the presence of chloride is essential for bacteria to survive low pH (Jordan and Davies, 2001) and that an antiporter protein in the inner membrane, EcClC, exchanges H+ for Cl− (Accardi and Miller, 2004). On the basis of these and other observations, it has been proposed that Cl− is driven into the bacterial cell to counteract the increased polarization of the membrane resulting from accumulation of the products of amino-acid decarboxylation (Foster, 2004). We propose that a major beneficial consequence of the entry of Cl− is that it ensures that the pH-correcting properties of Gad are turned on at pH values sufficiently near neutrality so that the lethal consequences of low intracellular pH are avoided.
As a biological consequence of this study, we highlight the important physiological role of chloride in E. coli AR and provide for the first time insight into its molecular mechanism of action.
Materials and methods
Structural studies
All experiments with GadB wild-type, purified as described by (De Biase et al, 1996), were carried out on two crystal forms obtained by vapour diffusion. Form A (space group P1) was obtained by mixing 1 μl of protein solution (5 mg/ml GadB, 100 mM Na acetate pH 4.6) with 1 μl of reservoir (135 mM Na acetate pH 4.6, 700 mM Na formate, 13% PEG 4000). Form B (space group P6322) was obtained by mixing 3 μl of protein solution (5 mg/ml GadB, 100 mM Na acetate pH 4.6) with 1 μl reservoir solution (19.8% PEG 2000 mME, 180 mM KBr and 100 mM Na acetate pH 5.5).
The GadB–iodide complex was prepared by transferring form A crystals from the mother liquor into 300 μl stabilizing solution (100 mM Na citrate pH 5.15, 800 mM Na formate and 15% PEG 4000). Three 100 μl aliquots of KI-soaking solution (100 mM Na acetate pH 4.6, 800 mM Na formate, 15% PEG 4000 and 200 mM KI) were added. After overnight equilibration, the crystals were cryoprotected by a solution similar to the soaking solution but containing in addition 17% ethylene glycol (w/v) and 100 mM KI.
Crystals of the GadB–bromide complex, of the GadB–chloride complex (soak and co-crystals) as well as crystals used in the pH-shift experiments (pH 5.9 and 6.5) belong to form B. For the GadB–chloride complex (soak), GadB–Br− crystals were transferred into 600 μl soaking solution (19.8% PEG 2000 MME, 180 mM KCl and 100 mM Na acetate pH 5.5). Cryoprotection was achieved using a similar solution containing also 180 mM KCl and additionally 10% ethylene glycol. GadB–chloride co-crystals were obtained by mixing 3 μl of protein solution (5 mg/ml GadB, 100 mM Na acetate pH 4.6) with 1 μl reservoir solution containing 21.6% PEG 2000 MME, 180 mM KCl and 100 mM Na acetate pH 5.5. These crystals were cryoprotected by addition of 18% ethylene glycol to the reservoir solution. In the pH-shift experiments, GadB–Br− form B crystals were directly transferred into solutions (400 μl each) similar to the reservoir condition but containing 100 mM Na cacodylate pH 5.9 and 100 mM Na cacodylate pH 6.5, respectively. The crystals were cryoprotected using solutions similar to the pH-shift solutions but containing 15% ethylene glycol in addition.
Crystals of GadBΔ1–14, purified as previously described (Capitani et al, 2003), were obtained by mixing 1 μl of protein solution (5 mg/ml GadBΔ1–14, 100 mM Na acetate pH 4.6) with 3 ml reservoir solution (22.5% PEG 2000 MME, 180 mM Li2SO4 and 100 mM Na acetate pH 5.5). The crystals were cryoprotected by adding 20 % ethylene glycol to the reservoir solution.
Diffraction data at 100 K were collected at beamline X06SA of the Swiss Light Source and were processed with DENZO, HKL2000 (Otwinowski and Minor, 1996) or XDS (Kabsch, 1993). Data collection statistics are given in Supplementary Table I.
The structure of GadB in crystal form B (space group P6322, one hexamer per a.u.) was solved with Phaser 1.2 (Storoni et al, 2004) using PDB entry 1PMM as search model. The structure of GadBΔ1–14 (space group P212121, one hexamer per a.u.) was solved with AMoRe (Navaza, 1994) with a modified version of PDB entry 1PMO as search model. All GadB structures were refined with CNS (Brünger et al, 1998) using NCS restraints, except for the 1.95 Å GadB–iodide and the 1.9 Å GadBΔ1–14 structure. Refinement statistics for the GadB–iodide, GadB–bromide and GadBΔ1–14 structures (PDB entry codes 2DGM, 2DGL, 2DGK, respectively) are given in Table I. Structural figures were prepared using PyMOL (http://www.pymol.org).
Table 1.
Refinement statistics
| GadB–I− | GadB–Br− | GadB-Δ1–14 | |
|---|---|---|---|
| Space group | P1 | P6322 | P212121 |
| Resolution range (Å) | 40–1.95 | 40–3.15 | 30–1.90 |
| No. of reflections (test) | 191 251 (5338) | 104 053 (2067) | 230 716 (2308) |
| R-factor/Rfree (%) | 22.7/26.7 | 21.8/24.8 | 21.8/24.6 |
| Protein atoms | 21 640 | 21 503 | 20 971 |
| Cofactor/ligand atoms | 90/32 | 90/24 | 90/0 |
| Solvent+buffer atoms | 1973 | 158 | 1806 |
| Rmsd bond lengths (Å) | 0.006 | 0.008 | 0.006 |
| Rmsd bond angles (deg) | 1.26 | 1.27 | 1.23 |
| Wilson/mean B-fac (Å2) | 20.9/21.0 | 39.4/37.7 | 32.0/28.9 |
| Ramachandran plot regions | |||
| Most favoured (%) | 90.7 | 87.1 | 89.3 |
| Additionally/generously allowed (%) | 9.0/0.3 | 12.7/0.3 | 10.5/0.2 |
Equilibrium and kinetic analyses
All experiments related to equilibrium and kinetic analyses are described in Supplementary data.
Supplementary Material
Supplementary Movie
Supplementary information
Acknowledgments
Financial support from the Swiss NCCR Structural Biology to MGG, from the Istituto Pasteur-Fondazione Cenci Bolognetti to DDB and from the Italian MIUR (PRIN) to FB is gratefully acknowledged. We thank the staff of beamline X06SA of the Swiss Light Source, Villigen, Switzerland for excellent technical assistance.
References
- Accardi A, Miller C (2004) Secondary active transport mediated by a prokaryotic homologue of ClC Cl− channels. Nature 427: 803–807 [DOI] [PubMed] [Google Scholar]
- Accardi A, Kolmakova-Partensky L, Williams C, Miller C (2004) Ionic currents mediated by a prokaryotic homologue of CLC Cl− channels. J Gen Physiol 123: 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengmark S (1998) Ecological control of the gastrointestinal tract. The role of probiotic flora. Gut 42: 2–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoldi M, Cellini B, Clausen T, Voltattorni CB (2002) Spectroscopic and kinetic analyses reveal the pyridoxal 5′-phosphate binding mode and the catalytic features of Treponema denticola cystalysin. Biochemistry 41: 9153–9164 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Capitani G, De Biase D, Aurizi C, Gut H, Bossa F, Grutter MG (2003) Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J 22: 4027–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW (1999) Control of acid resistance in Escherichia coli. J Bacteriol 181: 3525–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter PD, Gahan CG, Hill C (2001) A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol Microbiol 40: 465–475 [DOI] [PubMed] [Google Scholar]
- De Biase D, Tramonti A, Bossa F, Visca P (1999) The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol Microbiol 32: 1198–1211 [DOI] [PubMed] [Google Scholar]
- De Biase D, Tramonti A, John RA, Bossa F (1996) Isolation, overexpression, and biochemical characterization of the two isoforms of glutamic acid decarboxylase from Escherichia coli. Protein Expr Purif 8: 430–438 [DOI] [PubMed] [Google Scholar]
- Dutyshev DI, Darii EL, Fomenkova NP, Pechik IV, Polyakov KM, Nikonov SV, Andreeva NS, Sukhareva BS (2005) Structure of Escherichia coli glutamate decarboxylase (GADalpha) in complex with glutarate at 2.05 angstroms resolution. Acta Crystallogr D 61: 230–235 [DOI] [PubMed] [Google Scholar]
- Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R (2002) X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature 415: 287–294 [DOI] [PubMed] [Google Scholar]
- Dutzler R, Campbell EB, MacKinnon R (2003) Gating the selectivity filter in ClC chloride channels. Science 300: 108–112 [DOI] [PubMed] [Google Scholar]
- Foster JW (2004) Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol 2: 898–907 [DOI] [PubMed] [Google Scholar]
- Giannella RA, Broitmann SA, Zamcheck N (1972) Gastric acid barrier to ingested microorganisms in man: studies in vivo and in vitro. Gut 13: 251–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Richard H, Foster JW (2003) YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J Bacteriol 185: 4402–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson WA, Ogata CM (1997) Phase determination from multiwavelength anomalous diffraction measurements. In Macromolecular Crystallography, Part A, Vol. 276, pp 494–523 [DOI] [PubMed] [Google Scholar]
- Hersh BM, Farooq FT, Barstad DN, Blankenhorn DL, Slonczewski JL (1996) A glutamate-dependent acid resistance gene in Escherichia coli. J Bacteriol 178: 3978–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AV (1910) The possible effects of the aggregation of the molecules of haemoglobin on its oxygen dissociation curve. J Physiol (London) 40: 4–7 [Google Scholar]
- Hodel A, Kim SH, Brunger AT (1992) Model bias in macromolecular crystal-structures. Acta Crystallographica Section A 48: 851–858 [Google Scholar]
- Hofmeister F (1888) Zur Lehre von der Wirkung der Salze. II. Arch Exp Pathol Pharmakol 24: 247–260 [Google Scholar]
- Ikushiro H, Hayashi H, Kawata Y, Kagamiyama H (1998) Analysis of the pH- and ligand-induced spectral transitions of tryptophanase: activation of the coenzyme at the early steps of the catalytic cycle. Biochemistry 37: 3043–3052 [DOI] [PubMed] [Google Scholar]
- Iyer R, Iverson TM, Accardi A, Miller C (2002) A biological role for prokaryotic ClC chloride channels. Nature 419: 715–718 [DOI] [PubMed] [Google Scholar]
- Iyer R, Williams C, Miller C (2003) Arginine–agmatine antiporter in extreme acid resistance in Escherichia coli. J Bacteriol 185: 6556–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan KN, Davies KW (2001) Sodium chloride enhances recovery and growth of acid-stressed E. coli O157:H7. Lett Appl Microbiol 32: 312–315 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallog 26: 795–800 [Google Scholar]
- Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW (1995) Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol 177: 4097–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas AN, Gruber M (2005) The structure of alpha-helical coiled coils. In Fibrous Proteins: Coiled-Coils, Collagen and Elastomers, Vol. 70, pp 37–78 [DOI] [PubMed] [Google Scholar]
- Matin A (1999) pH homeostasis in acidophiles. Novartis Found Symp 221: 152–163; discussion 163–156 [DOI] [PubMed] [Google Scholar]
- Navaza J (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr A 50: 157 [Google Scholar]
- O'Leary MH (1971) A proposed structure for the 330-nm chromophore of glutamate decarboxylase and other pyridoxal 5′-phosphate dependent enzymes. Biochim Biophys Acta 242: 484–492 [DOI] [PubMed] [Google Scholar]
- O'Leary MH, Brummund W Jr (1974) pH jump studies of glutamate decarboxylase. Evidence for a pH-dependent conformation change. J Biol Chem 249: 3737–3745 [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1996) Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW and Sweet RM (eds) Meth Enzymol Macromolecular Crystallography. Vol. 276, pp 307–326. New York: Academic Press [DOI] [PubMed] [Google Scholar]
- Picollo A, Pusch M (2005) Chloride/proton antiporter activity of mammalian CLC proteins ClC-4 and ClC-5. Nature 436: 420–423 [DOI] [PubMed] [Google Scholar]
- Richard H, Foster JW (2004) Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol 186: 6032–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JW, Leenhouts K, Burghoorn J, Brands JR, Venema G, Kok J (1998) A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol 27: 299–310 [DOI] [PubMed] [Google Scholar]
- Scheel O, Zdebik AA, Lourdel S, Jentsch TJ (2005) Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature 436: 424–427 [DOI] [PubMed] [Google Scholar]
- Shukuya R, Schwert GW (1960a) Glutamic acid decarboxylase. 1. Isolation procedures and properties of the enzyme. J Biol Chem 235: 1649–1652 [PubMed] [Google Scholar]
- Shukuya R, Schwert GW (1960b) Glutamic acid decarboxylase. 2. Spectrum of the enzyme. J Biol Chem 235: 1653–1657 [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ (2004) Likelihood-enhanced fast rotation functions. Acta Crystallogr D 60: 432–438 [DOI] [PubMed] [Google Scholar]
- Tews I, Findeisen F, Sinning I, Schultz A, Schultz JE, Linder JU (2005) The structure of a pH-sensing mycobacterial adenylyl cyclase holoenzyme. Science 308: 1020–1023 [DOI] [PubMed] [Google Scholar]
- Tramonti A, John RA, Bossa F, De Biase D (2002) Contribution of Lys276 to the conformational flexibility of the active site of glutamate decarboxylase from Escherichia coli. Eur J Biochem 269: 4913–4920 [DOI] [PubMed] [Google Scholar]
- Zhang J, Cheltsov AV, Ferreira GC (2005) Conversion of 5-aminolevulinate synthase into a more active enzyme by linking the two subunits: spectroscopic and kinetic properties. Protein Sci 14: 1190–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Movie
Supplementary information


