Abstract
Polarization of naïve CD4 T cells into T helper type 2 (TH2) cells is characterized by expression of IL-4 and silencing of IFN-γ. Here we show that during TH2 polarization, the DNA methyltransferase Dnmt3a is recruited to the IFN-γ promoter and correspondingly the promoter undergoes progressive de novo methylation. Notably, the CpG located at the −53 position becomes methylated rapidly and this methylation inhibits ATF2/c-Jun and CREB transcription factor binding in vitro. In vivo, the same factors bind to the unmethylated IFN-γ promoter in T helper type 1 (TH1) cells but not the methylated IFN-γ promoter in TH2 cells. Furthermore, methylation at the −53 CpG alone is sufficient to inhibit the IFN-γ promoter-driven reporter gene expression in a TH1 cell line. These findings suggest that rapid methylation of the evolutionarily conserved −53 CpG by Dnmt3a may suppress IFN-γ transcription in developing TH2 cells by directly inhibiting transcription factor binding.
Keywords: factor binding, IFN-γ promoter, methylation, T helper cells, transcription
Introduction
One of the most important aspects of the adaptive immune system is the ability to adjust its response in order to more effectively combat different forms of pathogenic infection. At the center of this adaptive process lies the polarization of CD4+ helper T cells. When a naïve CD4 T cell becomes activated, it can polarize into either a T helper type 1 (TH1) phenotype or a T helper type 2 (TH2) phenotype (Murphy and Reiner, 2002). The defining difference between the two helper T-cell subsets is the cytokines the cells express. Whereas TH2 cells express IL-4, IL-5 and IL-13, the primary cytokine expressed by TH1 cells is interferon-γ (IFN-γ) (Murphy and Reiner, 2002). Studies have shown that the key factor that determines the outcome of TH1 versus TH2 cell differentiation is the induction of transcription factors T-bet for TH1 cells and GATA3 for TH2 cells (Zhang et al, 1997; Lee et al, 2000; Szabo et al, 2000). Expression of T-bet is sufficient to induce TH1 polarization of naïve CD4 T cells (Szabo et al, 2000), whereas expression of GATA3 is sufficient to induce a TH2 phenotype in CD4 T cells (Lee et al, 2000). Furthermore, T-bet suppresses expression of TH2 cytokines (Hwang et al, 2005), whereas GATA3 suppresses transcription of IFN-γ (Usui et al, 2003). Therefore, once a sufficient level of T-bet or GATA3 is induced, a CD4 T cell tends to polarize to one of the two subsets.
In addition to the induction of T-bet or GATA3, proper polarization of helper T cells also requires the suppression of inappropriate cytokine gene expression. Under both TH1 and TH2 polarizing conditions, both IFN-γ and IL-4 transcription are initiated within 1 h of naïve CD4 T cell activation (Grogan et al, 2001). In contrast, T-bet and GATA3 mRNA levels are not significantly increased until 72 h postactivation (Grogan et al, 2001). These observations indicate that IFN-γ and IL-4 can be expressed without upregulation of T-bet or GATA3, respectively. Furthermore, factors sufficient for transcription of IFN-γ and IL-4 pre-exist in naïve CD4 T cells and, upon activation, conditions temporarily become conducive to the expression of both cytokines. A sustained expression of IFN-γ and IL-4 following CD4 T cell activation could interfere with TH cell polarization because IFN-γ promotes TH1 polarization but inhibits TH2 polarization (Seder et al, 1992; Afkarian et al, 2002), whereas IL-4 promotes TH2 polarization but inhibits TH1 polarization (Hsieh et al, 1992; Seder et al, 1992). Thus, proper TH cell polarization likely requires a mechanism to suppress the inappropriate cytokine gene expression. Consistent with this notion, as CD4 T cell polarization progresses, transcription of IFN-γ is repressed in cells polarized under TH2 conditions, whereas transcription of IL-4 is repressed in cells polarized under TH1 conditions (Grogan et al, 2001).
One mechanism that has been implicated in long-term suppression of IFN-γ expression is DNA methylation (Young et al, 1994; Yano et al, 2003). CpG methylation in the promoter region often correlates with gene expression in that hypomethylated loci are transcribed whereas hypermethylated loci tend to be silent (Richards and Elgin, 2002). The IFN-γ locus is hypomethylated in IFN-γ-expressing TH1 cell lines but hypermethylated in TH2 cell lines, which do not express IFN-γ (Young et al, 1994). Although there are conflicting reports as to the methylation status of the IFN-γ promoter in naïve CD4 T cells (Melvin et al, 1995; White et al, 2002; Yano et al, 2003; Winders et al, 2004), a recent study shows that, in murine T cells, at least some of the CpGs in the promoter are hypomethylated and undergo de novo methylation under TH2 polarizing conditions (Winders et al, 2004). Because hypermethylation can induce chromatin remodeling through a variety of mechanisms, including the recruitment of histone deacetylases (Richards and Elgin, 2002), it provides a possible mechanism for the chromatin-mediated suppression of the IFN-γ locus that has been observed in long-term polarized TH2 cells (Grogan et al, 2001; Avni et al, 2002).
Because establishment of chromatin-mediated transcriptional suppression is usually a slow process, the question that arises is whether the de novo methylation at the IFN-γ promoter directly and immediately inhibits IFN-γ transcription during the course of TH2 polarization. There are at least two possible binding sites for AP1 and CREB/ATF family members in the IFN-γ promoter, one located at −90 to −78 bp (distal) and the other at −70 to −44 bp (proximal) relative to the transcription start site (Supplementary Figure S1) (Penix et al, 1996; Aune et al, 1997; Zhang et al, 1998), with both sites capable of driving some level of reporter gene expression in cells that express IFN-γ (Penix et al, 1993; Aune et al, 1997). Interestingly, one of the two CpGs that are conserved in the IFN-γ promoter among mouse, rat, dog, chimpanzee, and human resides in the proximal AP1-binding site (the −53 CpG) (Supplementary Figure S1) and methylation of this CpG results in a change in factor binding to the site (Young et al, 1994). Considering that CpG methylation can inhibit the binding of some AP1 family members to the promoter sequences of TrkA and neurotensin/neuromedin N genes (Dong et al, 2000; Fujimoto et al, 2005), these observations point to the possibility of a direct role of CpG methylation in regulating IFN-γ transcription. However, the kinetics and the exact nature and consequences of CpG methylation in the promoter on IFN-γ transcription during TH2 polarization are yet to be elucidated.
We have here examined in detail the methylation changes that occur in the IFN-γ promoter during helper T-cell polarization and how these changes affect transcription factor binding and promoter function. We show that −53 CpG in the IFN-γ promoter becomes rapidly methylated during TH2 polarization, resulting in inhibition of CREB and ATF2/c-Jun binding to the proximal AP1 site. Importantly, methylation of the −53 CpG alone is sufficient to inhibit the IFN-γ promoter-driven reporter gene expression in a TH1 cell line. These findings suggest that rapid and site-specific methylation of the IFN-γ promoter can immediately suppress IFN-γ transcription during TH2 cell polarization by directly inhibiting transcription factor binding.
Results
The IFN-γ promoter is already hypomethylated in precursor T cells
In mice, the IFN-γ proximal promoter contains six CpG dinucleotides (Figure 1A and Supplementary Figure S1), two of which, one at −53 position and the other at −190 position, are also conserved in rat, dog, chimpanzee, and human (Supplementary Figure S1). Studies have shown that the IFN-γ proximal promoter is hypomethylated in IFN-γ-expressing TH1 cells and hypermethylated in TH2 cells, which do not express IFN-γ (Young et al, 1994). However, in naïve CD4 T cells, from which the TH1 and TH2 populations arise, the methylation status of IFN-γ promoter has been reported as hypermethylated (Melvin et al, 1995; White et al, 2002) as well as hypomethylated (Yano et al, 2003; Winders et al, 2004). To resolve this inconsistency, we measured the extent of methylation of all six CpGs in the proximal promoter as well as two CpGs in the transcribed region immediately adjacent to the promoter in various cell types and tissues. To make quantitative measurement, we used sodium bisulfite conversion of genomic DNA, followed by PCR amplification, cloning, and sequencing. The frequency of conversion of C to T following the sodium bisulfite treatment and PCR in our studies was greater than 99% at non-CpG sites (data not shown), indicating adequacy of the approach.
Figure 1.
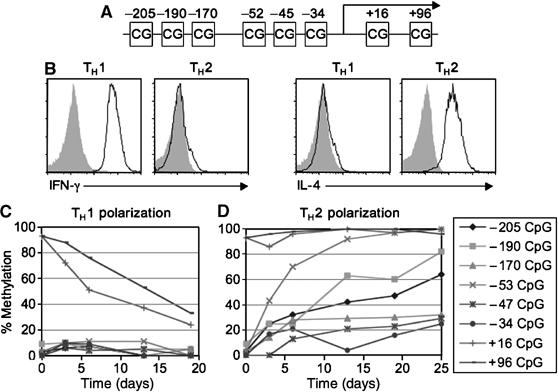
Methylation changes in the IFN-γ locus during TH1 and TH2 cell polarization. (A) CpG locations in the IFN-γ promoter and the contiguous transcribed region. (B) IFN-γ and IL-4 expression by polarized TH1 and TH2 cells. Day 6-polarized TH1 and TH2 cells were stimulated with PMA and ionomycin for 3 h and cytokine expression was measured. Open histogram, anti-IFN-γ or anti-IL-4 staining; shaded histograms, isotype control. (C and D) Changes in the levels of methylation at different CpGs in the IFN-γ locus during the course of TH1 (C) and TH2 (D) polarization. Polarized TH cells were stimulated every 6 days and, with the exception of day 3, were harvested for methylation assay just before restimulation. The same data are shown in part of Table II.
As summarized in Table I, all six CpGs in the IFN-γ promoter and the two CpGs in the transcribed region were almost completely methylated (>94%) in DNA isolated from kidney and heart tissues, which do not transcribe IFN-γ. In contrast, in DNA isolated from freshly purified naïve CD4 T cells, which also do not transcribe IFN-γ, the IFN-γ promoter was hypomethylated, whereas the transcribed region of the gene was hypermethylated (Table I). The methylation status on the reverse strand was similar to those on the forward strand, with the promoter region hypomethylated (<10%) and the coding region hypermethylated (>86%). Furthermore, the IFN-γ promoter was hypomethylated and the transcribed region was hypermethylated in naïve CD8T cells. These results are consistent with and further extend those reported by Winders et al (2004).
Table 1.
Percentages of CpG methylation at the IFN-γ locus in various cell types and tissues
| Cell type/tissue | N | −205 | −190 | −170 | −53 | −45 | −34 | 16 | 96 |
|---|---|---|---|---|---|---|---|---|---|
| Kidney | 50 | 96 | 96 | 100 | 98 | 96 | 98 | 98 | 98 |
| Heart | 18 | 100 | 94 | 100 | 94 | 94 | 100 | 100 | 100 |
| B cells | 15 | 80 | 87 | 73 | 93 | 87 | 100 | 93 | 93 |
| NK cells | 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| DN Thymocytes | 20 | 0 | 0 | 5 | 0 | 0 | 5 | 100 | 100 |
| DP Thymocytes | 29 | 0 | 0 | 0 | 0 | 3 | 3 | 97 | 100 |
| CD4 Thymocytes | 25 | 0 | 4 | 4 | 0 | 0 | 4 | 96 | 100 |
| CD8 Thymocytes | 23 | 0 | 0 | 0 | 4 | 0 | 4 | 100 | 100 |
| Naïve CD4 T cells | 34 | 3 | 9 | 3 | 3 | 0 | 0 | 85 | 97 |
| Naïve CD4 rev. strand | 21 | ND | 5 | 5 | 10 | 5 | 10 | 86 | ND |
| TH0 cells | 31 | 0 | 10 | 10 | 13 | 0 | 7 | 87 | 87 |
| TH1 cells | 45 | 7 | 4 | 7 | 11 | 4 | 9 | 51 | 76 |
| TH2 cells | 47 | 32 | 26 | 28 | 70 | 13 | 21 | 96 | 98 |
| Naïve CD8 T cells | 26 | 4 | 0 | 4 | 4 | 4 | 0 | 92 | 100 |
| Effector CD8 T cells | 23 | 0 | 0 | 0 | 0 | 0 | 0 | 65 | 91 |
| Memory CD8 T cells | 16 | 6 | 6 | 6 | 0 | 0 | 0 | 31 | 69 |
| Cell populations were obtained as described in the Materials and methods section. The number in the N column represents the number of clones sequenced. With the exception of memory CD8 T cell and reverse strand values, sequenced clones are from at least two different PCR amplifications with two independently isolated DNA samples. The other columns represent the percentages of CpG methylation at the specific position at the IFN-γ locus in the indicated cell types or tissues. ND, not determined. P-values were calculated by the χ2 test. |
We investigated the stages during T-cell development in which the IFN-γ promoter first becomes hypomethylated. Thus, CD4−CD8− (double negative or DN), CD4+CD8+ (double positive or DP), CD4+ (single positive or SP), and CD8+ SP thymocytes were purified and assayed for methylation at the IFN-γ locus. In all four thymocyte populations, the IFN-γ promoter was hypomethylated whereas the transcribed region was hypermethylated (Table I). To further determine the developmental stages when IFN-γ promoter becomes hypomethylated, we assayed the methylation status of freshly purified natural killer (NK) cells and B cells. Like T cells, NK cells are differentiated from DN thymocytes; but unlike naïve T cells, NK cells constitutively transcribe IFN-γ (Stetson et al, 2003). All CpGs in both the promoter and the transcribed region were completely demethylated in NK cells (Table I). In contrast, in B cells, which are derived from the common lymphoid progenitors but do not express IFN-γ under physiological conditions, the CpGs in both the promoter and the transcribed region were hypermethylated. These results suggest that the IFN-γ promoter becomes hypomethylated after lymphoid lineage commitment but before bifurcation between T- and NK-cell development in the thymus.
Hypomethylation of the IFN-γ promoter is correlated with the cells' potential to express IFN-γ
The above results also indicate that hypomethylation of the IFN-γ promoter is correlated with the ability of a cell to differentiate into an IFN-γ-expressing cell rather than with the cell's actual expression of the gene. To further investigate this possibility, we assayed the methylation status of the IFN-γ locus in CD4 T cells following TH1 and TH2 polarization. Naïve CD4 T cells were purified from BALB/c mice and activated by exposure to plate-bound anti-CD3 and anti-CD28, and recombinant IL-2. TH1 polarization was induced by addition of IL-12 and anti-IL-4 antibody to the culture, whereas TH2 polarization was induced by addition of IL-4 and anti-IFN-γ antibody to the culture. After 6 days, methylation was assayed in polarized cells. Following culture under TH1 conditions, during which IFN-γ expression is induced (Figure 1B), the IFN-γ promoter remained hypomethylated, but the transcribed region underwent some level of demethylation as compared to naïve CD4 T cells (P⩽0.001) (Table I). When cells were cultured under TH2 polarizing conditions, during which IL-4 but not IFN-γ was expressed (Figure 1B), the IFN-γ promoter became hypermethylated as compared to that in naïve CD4 T cells (P⩽0.001) and the transcribed region remained hypermethylated. In particular, the −53 CpG became significantly more methylated than the other CpGs within the promoter (70% versus 13–32%, P⩽0.001) (Table I). When cultured in the absence of polarizing cytokines (TH0), there was no detectable change in methylation of the IFN-γ promoter as compared to naïve CD4 T cells.
We also assayed the methylation status of the IFN-γ locus in effector and memory CD8T cells, in which IFN-γ is transcribed (Grayson et al, 2001). The effector CD8T cells were generated in vitro whereas memory CD8T cells were isolated from immunized mice (see Materials and methods). As shown in Table I, the IFN-γ promoter remained hypomethylated in these CD8 T cells (Table I). Notably, the two CpGs in the transcribed region became more hypomethylated in memory CD8T cells than in naïve CD8T cells (P⩽0.001). These results further support the notion that hypomethylation of the IFN-γ promoter is correlated with the cells' potential to express IFN-γ.
The −53 CpG in the IFN-γ promoter is rapidly methylated during TH2 cell polarization
Increased level of methylation of the IFN-γ promoter in day 6-polarized TH2 cells suggest that IFN-γ promoter can become de novo methylated during TH2 cell polarization. To unequivocally demonstrate this event, we examined the methylation changes at the IFN-γ locus during TH1 and TH2 polarization over 25 days. In T cells cultured under TH1 conditions, the promoter remained hypomethylated, and the transcribed region was gradually demethylated from day 3 to day 19 (Figure 1C and Table II). In T cells polarized under TH2 conditions, in contrast, the promoter became increasingly methylated from day 3 to day 25, whereas the transcribed region remained hypermethylated (Figure 1D and Table II). These patterns in methylation change are consistent with the observation that the long-term TH1 polarized cell line AE7 is almost completely hypomethylated at the IFN-γ locus, whereas the long-term TH2 polarized cell line D10 is almost completely hypermethylated at the IFN-γ locus (Table II). It is notable that the −53 CpG was methylated much faster and more completely than other CpGs in the promoter during TH2 polarization, with the −190 CpG ranked second in de novo methylation rate. As both −53 and −190 CpGs are evolutionarily conserved (Supplementary Figure S1) and the −53 CpG resides in the proximal AP1-binding site of the IFN-γ promoter (Penix et al, 1993; Penix et al, 1996; Aune et al, 1997), the rapid de novo methylation of the −53 CpGs may help to promote TH2 cell polarization by inhibiting IFN-γ transcription.
Table 2.
Percentages of CpG methylation at the IFN-γ locus during the course of TH1 and TH2 cell polarization
| Cell type | N | −205 | −190 | −170 | −53 | −45 | −34 | 16 | 96 |
|---|---|---|---|---|---|---|---|---|---|
| Naïve CD4 T cells | 34 | 3 | 9 | 3 | 3 | 0 | 0 | 85 | 97 |
| TH1 day 3 | 32 | 6 | 9 | 9 | 9 | 6 | 9 | 72 | 88 |
| TH1 day 6 | 45 | 7 | 4 | 7 | 11 | 4 | 9 | 51 | 76 |
| TH1 day 13 | 19 | 0 | 5 | 0 | 11 | 5 | 0 | 37 | 53 |
| TH1 day 19 | 21 | 0 | 5 | 5 | 0 | 0 | 0 | 24 | 33 |
| TH2 day 3 | 28 | 25 | 25 | 14 | 43 | 0 | 17 | 86 | 96 |
| TH2 day 6 | 47 | 32 | 26 | 28 | 70 | 13 | 21 | 96 | 98 |
| TH2 day 13 | 29 | 42 | 63 | 29 | 92 | 21 | 4 | 100 | 100 |
| TH2 day 19 | 30 | 47 | 60 | 30 | 97 | 23 | 17 | 97 | 100 |
| TH2 day 25 | 28 | 64 | 82 | 32 | 100 | 29 | 25 | 100 | 96 |
| TH1 (AE7) | 18 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 11 |
| TH2 (D10) | 18 | 94 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Cell populations were obtained as described in the Materials and methods section. The number in the N column represents the number of clones sequenced. Except for the day 3 values, sequenced clones are from at least three different PCR amplifications with two independently isolated DNA. The other columns represent the percents of CpG methylation at the specific position at the IFN-γ locus in the indicated cell types. P-values were calculated by the χ2 test. |
Methylation of the −53 CpG impairs ATF2/c-Jun and CREB binding in vitro
We next investigated the effect of the −53 CpG methylation on transcription factor binding by electrophoretic mobility shift assays (EMSA). For these studies, we used nuclear extracts from the mouse TH1 cell line AE7 and oligonucleotide probes corresponding to the proximal AP1 site (−62 through −32) of the IFN-γ promoter, which were either unmethylated, or methylated at the −53 CpG, or the −47 CpG, or the −34 CpG, or methylated at all three CpGs simultaneously. When the EMSA was carried out with the unmethylated oligonucleotide probe, there were two major shifted bands (Figure 2A), indicating factor binding. Methylation at either the −47 CpG or the −34 CpG had no effect on the formation of these bands. In contrast, the same sequence methylated at only the −53 position or at all three positions abolished the formation of the EMSA complexes.
Figure 2.
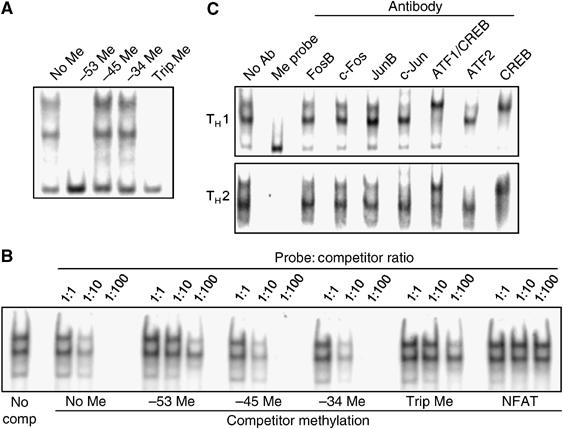
Inhibition of ATF2/c-Jun and CREB binding by the −53 CpG methylation. (A) EMSA using probes corresponding to the proximal AP1 site of the IFN-γ promoter methylated at the indicated CpGs and nuclear extracts from the AE7 (TH1) cells. (B) EMSA in the presence of competitor oligonucleotides. The assays were carried out with AE7 nuclear extract and labeled, unmethylated AP1 probe in the presence of increasing amount of unlabeled, methylated or unmethylated, or nonspecific (NFAT) competitor oligonucleotides. The first lane was from the incubation without competitor oligonucleotide. The remaining lanes were from incubations with the indicated unlabeled competitor oligonucleotides. (C) EMSA in the presence of specific antibodies. The assays were carried out with nuclear extracts from either AE7 or D10 (TH2) cells and labeled, unmethylated AP1 probe in the absence (No Ab) or presence of the indicated antibodies. The EMSA with labeled, methylated AP1 probe serves as controls. The anti-ATF1 antibody crossreacts with CREB. Data shown are representative from one of the three to five experiments.
To confirm the observed results, we carried out competition assays in which increasing amounts of unlabeled competitor oligonucleotides were added to a fixed amount of labeled, unmethylated probe during the incubation. As expected, formation of the two complexes was effectively inhibited by an excess of the unlabeled, unmethylated probe itself (Figure 2B). Oligonucleotides that were methylated at either the −45 or −34 CpG also inhibited the formation of the two complexes to the same degree. In contrast, oligonucleotides that were methylated at the −53 position, or at all three CpGs were much less effective, but still more effective than nonspecific NFAT oligonucleotide, in inhibiting the complex formation (Figure 2B). Based on the relative intensities of the shifted bands, −53 CpG methylation reduced factor binding by 10-fold or more. These results show that methylation at the −47 or −34 CpGs does not affect factor binding, whereas methylation at the −53 CpG reduces factor binding significantly in vitro.
Various members of the AP1 and CREB/ATF families of transcription factors have been shown to bind the proximal AP1 site (Cippitelli et al, 1995; Penix et al, 1996; Yano et al, 2003). We determined the composition of transcription factors in the two complexes in the TH1 nuclear extracts by inclusion of specific antibodies in EMSA. Antibodies specific for FosB, c-Fos, and JunB did not significantly affect the two shifted bands (Figure 2C, upper panel). In contrast, antibodies specific for c-Jun and ATF2 diminished the formation of the upper complex, whereas antibodies specific for CREB or both ATF1 and CREB supershifted the lower complex. Furthermore, when the EMSA was carried out with nuclear extracts from the mouse TH2 cell line D10, the same two complexes were detected and the same effects were observed with the various antibodies (Figure 2C, lower panel). Thus, the upper complex contains ATF2/c-Jun and the lower complex contains CREB (and possibly ATF1) and these factors are present in both TH1 and TH2 cells.
CREB, ATF2, and c-Jun are bound to the IFN-γ promoter in TH1 but not in TH2 cells
To assess CREB and ATF2/c-Jun binding to the IFN-γ promoter in vivo, we performed chromatin immunoprecipitation (ChIP) assays using the AE7 and D10 cells. The IFN-γ promoter is nearly completely unmethylated in AE7 cells and nearly completely methylated in D10 cells (Table II), and therefore should give a clear indication as to any correlation between promoter methylation and factor binding in vivo. Cells were treated with formaldehyde to crosslink proteins to DNA, lysed, and sonicated, followed by immunoprecipitation with various antibodies. The presence of the IFN-γ promoter sequence in the precipitates was determined by PCR, which amplifies a 175 bp fragment containing both the proximal and distal AP-1 sites. PCR product at the expected size was detected in the input DNA, but not in precipitates where no antibody was added (Figure 3A). In DNA from TH1 precipitates, the same size PCR product was detected when antibodies specific for FosB, JunB, c-Jun, CREB/ATF1, ATF2, and CREB were used, but not when anti-c-Fos was used (Figure 3A, upper panel). In DNA from TH2 precipitates, the PCR product was detected when antibodies specific for FosB, c-Fos, JunB, and CREB/ATF1 were used, but not when antibodies specific for c-Jun, ATF2, and CREB alone were used (Figure 3B, lower panel). The observed factor binding is likely specific for the IFN-γ promoter because PCR product assaying for a portion of IFN-γ intron-1, located 1 kb downstream of the promoter, was detected only in input DNA, but not in any antibody-precipitated DNA samples. These results suggest that the unmethylated IFN-γ promoter is bound by CREB, ATF2, and c-Jun in the TH1 cell line, whereas the methylated IFN-γ promoter in the TH2 cell line is not. Co-precipitation of the IFN-γ promoter sequence with CREB/ATF1 in TH2 cells likely reflects CREB/ATF1 binding to the distal AP-1 site, because CREB/ATF1 do not bind to the proximal AP-1 site when the −53 CpG is methylated (Figure 2).
Figure 3.
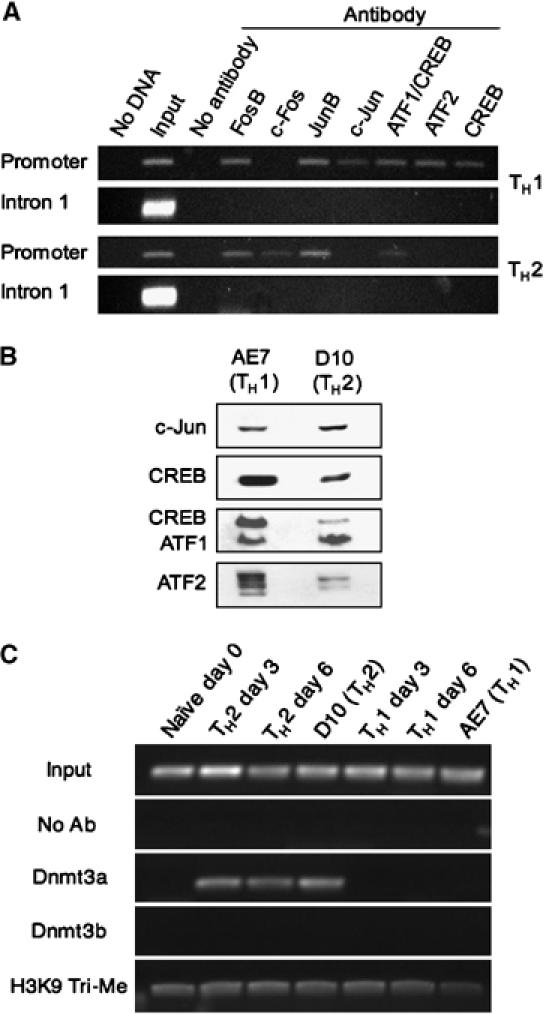
Transcription factor binding to the IFN-γ promoter in TH1 and TH2 cells. (A) ChIP assays of the indicated AP1 and CREB/ATF family members binding within the IFN-γ promoter in the AE7 or the D10 cell lines. PCR assays of IFN-γ intron 1 were used to demonstrate the specificity of immunoprecipitation. Representative data from one of two independent experiments is shown. (B) Western blots demonstrating the relative nuclear protein levels of c-Jun, CREB, ATF1/CREB, and ATF2 in AE7 and D10 cells. (C) ChIP assays of Dnmt3a and Dnmt3b binding and tri-methyl H3K9 levels within the IFN-γ promoter in the indicated cell populations.
To verify the differences in factor binding of the IFN-γ promoter between TH1 and TH2 cells, we assayed for c-Jun, CREB, ATF1/CREB, and ATF2 in nuclear extracts by Western blotting. As shown in Figure 3B, the relative levels of c-Jun and ATF1 were similar in TH1 and TH2 nuclear extracts, whereas the relative levels of ATF2 and CREB were significantly higher in TH1 than in TH2 extracts. Thus, nuclear protein levels, in addition to methylation status of the IFN-γ promoter, may contribute to the observed differences in factor binding between the TH1 and TH2 cells.
Methylation of the −53 CpG inhibits the IFN-γ promoter activity
We further investigated the effect of CpG methylation on IFN-γ promoter activity. For these studies, we constructed a vector, referred to as γ-luc, in which the firefly luciferase gene was under the control of the proximal 250 bp of the IFN-γ promoter. Transfections were carried out in either the TH1 cell line AE7, which expressed IFN-γ following PMA plus ionomycin (P/I) stimulation (Figure 4A), or the TH2 cell line D10, which did not express IFN-γ. Compared to the vector without the IFN-γ promoter (pGL3), significant luciferase activities were observed when the γ-luc vector was transiently transfected into the AE7 and the D10 cell lines, especially following PMA plus ionomycin stimulation (Figure 4B). When the proximal AP1 site was deleted from the promoter, luciferase activity was reduced to a level similar to that observed with the control pGL3 vector (Figure 4C). These results suggest that the 250 bp IFN-γ promoter is sufficient to drive the reporter gene expression, and that the proximal AP1 site is one of the critical elements. The presence of luciferase activities in the TH2 cell line is consistent with the presence of transcription factors known to be involved in IFN-γ transcription in D10 cells (Figure 3) (Zhang et al, 1998; Yano et al, 2003). Nevertheless, the 250 bp IFN-γ promoter is significantly more active in AE7 cells than in D10 cells, both with and without PMA plus ionomycin stimulation (P<0.02 for both).
Figure 4.
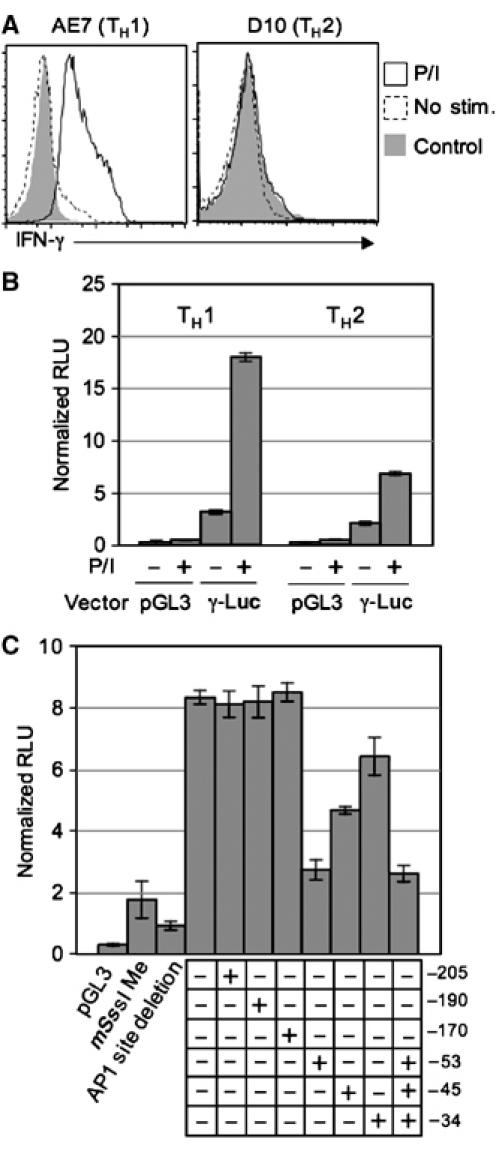
Inhibition of IFN-γ promoter activity by site-specific methylation. (A) IFN-γ expression by AE7 and D10 cells. IFN-γ expression in AE7 and D10 cells with or without P/I stimulation (for 2 h) was detected by cytokine recapture assay. Stimulated cells were also stained with an isotype control antibody. (B) The 250 bp IFN-γ promoter is capable of driving reporter gene expression in both TH1 and TH2 cells. Data shown are firefly luciferase activities normalized to renilla luciferase activities for γ-luc and pGL3 vectors in transfected TH1 and TH2 cells with or without P/I stimulation. The error bars indicate standard deviations from triplicate samples in each experiment. Representative data from one of the two independent experiments are shown. (C) Effects of CpG methylation on the IFN-γ promoter activity. The γ-luc vector with the indicated methylation(s), or methylated at all CpG sites by mSss I, or deleted of the proximal AP1-binding site, and the pGL3 vector were transfected together with the renilla luciferase vector into AE7 cells. After 6 h, the transfected cells were stimulated with P/I for 2 h and luciferase activities were measured. (+) Indicates a methylated CpG at the indicated position, whereas (−) indicates an unmethylated CpG at the indicated position. The error bars indicate standard deviations from triplicate samples in each experiment. P-values were calculated by Student's t-test. Representative data from one of the three to five independent experiments are shown.
We modified the γ-luc vector so that each of the promoter's six CpGs was individually methylated or the three CpGs at −53, −45, and −34 positions were methylated simultaneously by a PCR-mediated mutagenesis (see Materials and methods) (Martinowich et al, 2003). As controls, we generated unmethylated γ-luc vector by the same PCR technique, and a completely methylated vector in which every CpG was methylated by mSss I methyltransferase. These vectors were cotransfected into AE7 cells with a reference renilla luciferase plasmid. The transfected cells were stimulated with PMA and ionomycin and assayed for luciferase activities. The firefly luciferase activities (normalized to the renilla luciferase activities) were virtually the same in cells transfected with the unmethylated vector or vector methylated at the −205, −190, or −170 position (Figure 4C). In contrast, methylation at the −53 site reduced the luciferase activity significantly (P<0.001) to a level (33%) that was not significantly different from the reduction observed with the completely methylated vector (P=0.113) (mSss I, Figure 4C). Methylation at the −45 or −34 sites also reduced luciferase activity, although by a lesser, but still significant, amount (P=0.003 and P=0.04, respectively). However, methylation of the −53 site along with the −45 and −34 sites in the same vector did not reduce luciferase activities more than methylation of the −53 site alone. These results show that the methylation of the −53 CpG alone has the most pronounced effect on IFN-γ promoter activity.
Dnmt3a is rapidly recruited to the IFN-γ promoter in polarizing TH2 cells
To elucidate the mechanisms underlying the observed rapid de novo methylation, we examined the interaction between DNA methyltransferase and IFN-γ promoter during the course of TH2 polarization. In mammals, there are three known DNA methyltransferases, Dnmt1, Dnmt3a, and Dnmt3b (Bestor et al, 1988; Okano et al, 1998; Okano et al, 1999). Dnmt1 preferentially methylates hemimethylated DNA (Bestor, 1992; Li et al, 1992) and is responsible for the maintenance of established methylation patterns during cell division. In contrast, Dnmt3a and Dnmt3b exhibit an enzymatic preference for unmethylated DNA over hemimethylated DNA (Okano et al, 1998), suggesting that these enzymes function as de novo methyltransferases. We performed ChIP assays with antibodies specific for either Dnmt3a or Dnmt3b in naïve CD4 T cells, day 3- and day 6-polarized T cells, AE7 cells, and D10 cells. As expected, IFN-γ promoter-specific PCR product was detected in input DNA, but not in precipitates when antibody was not added during the precipitation reaction. When anti-Dnmt3a was used, PCR product of the expected size was detected in precipitates of both day 3- and day 6-TH2 polarized T cells and D10 cells, but not in precipitates of naïve CD4 T cells, TH1 polarized T cells, or AE7 cells (Figure 3C). In contrast, no PCR product was detected in any precipitates when anti-Dnmt3b was used. Thus, Dnmt3a interacts with the IFN-γ promoter during TH2 polarization and likely mediates the de novo methylation of the promoter during the process.
Tri-methylation of lysine 9 on histone H3 (H3K9) has been implicated in the recruitment of DNA methyltransferases (Lehnertz et al, 2003). To investigate the role of tri-methyl H3K9 in the de novo methylation of the IFN-γ promoter during TH2 polarization, we performed ChIP assays on the above cell lysates using antibodies specific for tri-methyl H3K9. Similar levels of IFN-γ-specific PCR product was detected in all the precipitates (Figure 3C), suggesting that tri-methyl H3K9 levels at the IFN-γ promoter do not significantly change during T helper cell polarization.
Discussion
Dynamic relationships between methylation status and transcription at the IFN-γ locus
The methylation status of the IFN-γ promoter in naïve CD4 T cells has been controversial (Melvin et al, 1995; White et al, 2002; Yano et al, 2003; Winders et al, 2004). Although a recent report suggests that the promoter is hypomethylated in naïve CD4 T cells, that study examined only two CpG sites in the proximal promoter by semiquantitative Southern blotting. Our quantitative analyses of all six CpGs in the proximal promoter have now unequivocally demonstrated that the IFN-γ promoter is hypomethylated in naïve CD4 T cells. Our results also show that the IFN-γ promoter is hypomethylated in precursor thymocytes before T-cell and NK-cell bifurcation but after common lymphocyte progenitor commitment. Despite hypomethylation in the promoter during precursor T-cell development, the transcribed region of the IFN-γ locus remains hypermethylated, indicating site specificity of this methylation process. In addition, our analyses of the methylation status of the IFN-γ locus during the course of T helper cell differentiation reveal that the IFN-γ promoter becomes de novo methylated during TH2 cell polarization, whereas the transcribed region becomes demethylated during TH1 cell polarization and memory CD8T cell development. Together, these results demonstrate dynamic changes in DNA methylation at the IFN-γ locus during T-cell, and especially T helper cell, development.
Targeted methylation at the IFN-γ locus
It is notable that de novo methylation does not occur with the same kinetics at the six CpGs within the IFN-γ proximal promoter during TH2 cell polarization (Figure 1D). The more rapid and complete methylation of the −53 CpG suggests that the methylation machinery is targeted to some CpGs more readily than others. This and other observations raise the questions as to what signal initiates the de novo methylation event, which methyltransferase mediates the de novo methylation, and how methylation machinery is targeted to particular CpG. Although a thorough understanding of the underlying molecular mechanisms requires further investigations, findings presented here already shed light on aspects of these questions.
First, we found that the IFN-γ promoter remains hypomethylated under TH0 polarizing conditions, suggesting that the same signals that induce methylation of the IFN-γ promoter also induce other aspects of TH2 cell development. Because the difference between TH0 and TH2 polarizing conditions is the presence of IL-4 in the TH2 culture, cytokine signaling would appear to trigger the de novo methylation event. Second, we found that Dnmt3a but not Dnmt3b interacts with the IFN-γ promoter in both day 3- and day 6-polarized TH2 cells, but not in naïve CD4 T cells or polarized TH1 cells. These findings suggest that Dnmt3a is rapidly recruited to the IFN-γ promoter under TH2 polarizing conditions and therefore may mediate the observed de novo methylation event. Interestingly, Dnmt3a remains associated with the IFN-γ promoter in long-term polarized TH2 (D10) cells, indicating that Dnmt3a may be part of a larger repressor complex and may be involved in the maintenance of the hypermethylated state of the IFN-γ locus. Third, although tri-methylation of histone H3 (H3K9) has been implicated in the recruitment of DNA methyltransferases (Lehnertz et al, 2003), we were unable to detect any change in H3K9 methylation status during the first 6 days of TH polarization, indicating other mechanisms are involved in the recruitment of Dnmt3a to the IFN-γ promoter. In this respect, we found that several AP1 transcription factor family members associate with the IFN-γ promoter in TH2 cells (Figure 3). It is possible that these and other transcription factors that bind the IFN-γ promoter following TH2 polarization recruit Dnmt3a and target methylation to particular CpGs.
Methylation of the −53 CpG inhibits transcription factor binding to the IFN-γ promoter
The rapid methylation at the conserved −53 CpG during the course of TH2 cell polarization may decrease factor binding to the CpG-containing proximal AP1 site. In support of this hypothesis, our EMSA studies demonstrate that two protein complexes from the nuclear extract of the TH1 cell line AE7 can bind to the proximal AP1 site of the IFN-γ promoter in vitro (Figure 2). One complex contains CREB, whereas the other contains ATF2 and c-Jun, consistent with previous reports of AP1 and CREB/ATF binding at this site (Cippitelli et al, 1995; Penix et al, 1996; Zhang et al, 1998). Importantly, we show that methylation of the −53 CpG alone, but not the −45 or −34 CpGs, is sufficient to reduce the levels of factor binding. These findings are consistent with a previous observation showing that methylation of the −53 CpG results in a change in factor binding to the site although the early study did not identify the specific transcription factors involved (Young et al, 1994). Complementing the in vitro binding results, we found that the same factors are bound to the hypomethylated IFN-γ promoter in a TH1 cell line but not the hypermethylated IFN-γ promoter in a TH2 cell line (Figure 3). While the ChIP assay is unable to determine exactly where in the promoter the factors are bound, the ChIP data are in accord with the EMSA data. Together, these findings show that methylation of the −53 CpG inhibits transcription factor binding to the IFN-γ promoter both in vitro and in vivo.
Methylation of the −53 CpG inhibits IFN-γ promoter activity
The effect of −53 CpG methylation on IFN-γ promoter activity was directly examined by reporter assays in a TH1 cell line. We showed that a 250 bp fragment of the IFN-γ promoter is capable of driving significant reporter gene expression in the TH1 cell line, especially after PMA plus ionomycin stimulation (Figure 4). The proximal AP1 site is critical for the promoter activity because its deletion from the vector results in a reduction of luciferase activity to a level similar to that observed with the control vector lacking any known promoter element. Most importantly, methylation of the −53 CpG alone in the reporter vector results in a significant reduction of luciferase activity, reaching a level not significantly different from that obtained with a completely methylated vector.
Surprisingly, methylation of the proximal promoter at either the −45 or the −34 CpG also results in a significant reduction of luciferase activity, although methylation at these sites do not affect the observed factor binding to the proximal AP1 site in vitro. It is possible that the inhibition of transcription by methylation of the −45 or −34 CpG in vivo is a result of recruitment of 5-methylcytosine-binding proteins that interfere with binding of activating transcription factors to the proximal AP1-binding site. Alternatively, there are other factors that contribute to induction of transcription of the reporter construct but that are unable to exert their effect unless the −53 CpG site is also occupied. In support of the latter interpretation, methylation of the −53, −45, and −34 CpGs simultaneously does not reduce luciferase activities more than methylation of the −53 CpG alone. Together, these results suggest a critical role of methylation of the conserved −53 CpG in inhibiting IFN-γ promoter activity. They further suggest that methylation of other CpGs in the vicinity helps to suppress this promoter's activity.
Targeted CpG methylation and T helper cell development
Following activation of naïve CD4 T cells, both IFN-γ and IL-4 transcriptions are activated. Because IFN-γ inhibits TH2 polarization, IFN-γ expression has to be rapidly suppressed in polarizing TH2 cells. Our findings suggest that the rapid and site-specific methylation of the promoter likely contributes to this regulation. Studies have shown that a dominant negative variant of c-Jun inhibits IFN-γ expression (Cippitelli et al, 1995; Penix et al, 1996), suggesting that this transcription factor normally activates IFN-γ transcription. However, the role of CREB in IFN-γ expression is less clear, as some studies suggest that it plays a repressing role (Penix et al, 1993; Zhang et al, 1998), whereas others suggest that it plays an activating role (Samten et al, 2005). Both our and other studies (Yano et al, 2003) show that CREB and c-Jun are present in both TH1 and TH2 cells. The conflicting observations with regard to the role of CREB in IFN-γ expression may be a result of CREB being a component of both repressive and activating complexes.
We propose a general model that integrates the contributions of both promoter CpG methylation and ATF2/c-Jun in IFN-γ transcriptional regulation in CD4 T cells. In naïve CD4 T cells, although the promoter is hypomethylated IFN-γ is not transcribed, perhaps because activating complexes, such as c-Jun/ATF2, do not bind to the promoter owing to the chromatin structure of the promoter (Avni et al, 2002) (Figure 5A). Upon activation of CD4 T cells, there are significant changes of the promoter chromatin structure owing to histone modifications. As a result, the activating complexes, such as c-Jun/ATF2, can bind the promoter and activate IFN-γ transcription (Figure 5B). Under TH2 polarizing conditions, however, Dnmt3a is recruited to the IFN-γ promoter and catalyzes a rapid methylation of the −53 CpG, which directly inhibits c-Jun/ATF2 binding and therefore suppresses IFN-γ transcription in polarizing TH2 cells (Figure 5C). As the TH2 polarization progresses, the remaining CpGs in the IFN-γ promoter gradually become hypermethylated, contributing to chromatin remodeling and the permanent suppression of the locus (Figure 5D).
Figure 5.
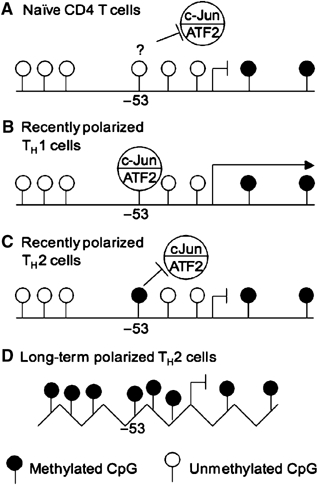
IFN-γ promoter methylation and transcriptional repression. (A) In naïve CD4 T cells, the promoter is hypomethylated but IFN-γ is not transcribed possibly because activating complexes, such as c-Jun/ATF2, do not bind to the promoter. (B) Activation under TH1 polarizing conditions allows the c-Jun/ATF2 complex to bind to the promoter and contribute to IFN-γ transcription. (C) During activation under TH2 polarizing conditions, rapid methylation of the −53 CpG prevents c-Jun/ATF2 from binding to the promoter, and thus contributes to the suppression of IFN-γ transcription. (D) In long-term polarized TH2 cells, the remaining CpGs in the IFN-γ promoter are methylated, contributing to chromatin remodeling and the permanent suppression of the locus.
In conclusion, our results show that the IFN-γ promoter is hypomethylated in naïve CD4 T cells and becomes progressively methylated during TH2 cell polarization. The rapid methylation of the −53 CpG probably prevents IFN-γ transcription by directly inhibiting transcription factor binding to the promoter. As the −53 CpG is conserved in the IFN-γ promoter among mouse, rat, dog, chimpanzee, and human, the rapid methylation of the −53 CpG is likely a general mechanism for repressing IFN-γ transcription during TH2 cell polarization.
Materials and methods
Cell purification, culture, and cytokine recapture assay
Primary naïve CD4 T cells and B cells were purified from lymph nodes and spleens of BALB/c mice by FACS. DP and SP thymocytes were sorted from BALB/c thymus. DN thymocytes and NK cells were sorted from RAG1-deficient mice. Naïve CD8T cells were sorted from spleens and lymph nodes of C57BL/6 mice. Purity of the various cell populations was >95%. All antibodies used in flow cytometry were obtained from BD Pharmingen (San Diego, CA).
Sorted naïve CD4 T cells were stimulated with plate-bound anti-CD3 and anti-CD28. TH1 polarized cultures contained recombinant IL-12 and neutralizing anti-IL-4 antibody (BD Pharmingen), while TH2 polarized cultures contained recombinant IL-4 and neutralizing anti-IFN-γ antibody (BD Pharmingen). TH0 cells contained both anti-IL-4 antibody and anti IFN-γ antibody. After 24 h, 40 μ/ml IL-2 was added to all cultures.
Effector CD8T cells were generated by stimulating naïve CD8 T cells with plate-bound anti-CD3 and anti-CD28 for 3 days. Memory phenotype CD8T cells were generated by sorting naïve CD8T cells from the spleen and lymph nodes of 2C TCR transgenic mice on an RAG1-deficient background. These cells were adoptively transferred intravenously into RAG1-deficient mice, which were then immunized with SIYRYYGL peptide in CFA. Approximately 6 months later, CD8+CD44hi 2C T cells were sorted from spleens and lymph nodes of these mice.
Cytokine recapture staining was carried out using either the Mouse IFN-γ Secretion Assay kit or the Mouse IL-4 Secretion Assay kit (Miltenyi Biotec, Auburn, CA) according to the manufacturer's instructions. When restimulations were carried out beforestaining, cells were restimulated with 5 ng/ml PMA and 100 ng/ml ionomycin for either 2 or 3 h.
Methylation analysis
Methylation analysis was carried out by bisulfite conversion of genomic DNA using the CpGenome Universal DNA Modification Kit (Chemicon International, Temecula, CA) according to the manufacturer's instructions. The IFN-γ promoter region was PCR amplified using the following primers: Forward strand: 5′: TAGAGAATTTTATAAGAATGGTATAGGTGGGTAT and 3′: CCATAAAAAAAAACTACAAAACCAAAATACAATA Reverse strand: 5′: ACAATTTCCAACCCCCACCCCAAATAATATAAAA 3′: TAGGAGGAGAAGTTTAGAATTTTTGTTTTAAGTT. The PCR product was cloned using the TOPO TA Cloning kit (Invitrogen).
EMSA, Western blotting, and ChIP
Nuclear extracts of AE7 and D10 cell lines were generated as previously described (Schreiber et al, 1989), except that a Complete Mini protease inhibitor cocktail tablet (Roche, Mannheim, Germany) was included in buffers A and C. DNA-binding reactions were performed as described (Shen and Stavnezer, 2001). For competition experiments, 1-, 10- or 100-fold molar excess of the unlabeled competitor oligonucleotide was added to the reaction. For antibody supershift experiments, 200 ng of the indicated antibodies were added to the reactions. The following oligonucleotide sequence corresponding to the proximal AP1 site of the IFN-γ promoter and its complement were used with the three CpGs either methylated or unmethylated: GTGAAAATACGTAATCCCGAGGAGCCTTCGA. Methylated CpG oligonucleotides were prepared by incorporating the methyl-cytosine during the chemical synthesis (IDT, Coralville, IA). The NFAT consensus oligonucleotide used had the following sequence: TATGAAACAAATTTTCCTCTTTGGGCG. The antibodies specific for Fos B, Jun B, c-Fos, c-Jun, ATF1/CREB, and ATF2 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA) and the CREB antibody was obtained from Zymed (San Francisco, CA).
For Western blotting, 50 μg/lane of the nuclear extracts were fractionated by SDS–PAGE and transferred to a nitrocellulose membrane. The membranes were probed with specific primary antibodies. Blots were visualized using Western Lightning chemiluminescence reagent (PerkinElmer, Boston, MA).
The ChIP assay was carried out using the ChIP Assay Kit from Upstate Biotech (Lake Placid, NY). The same antibodies used for the EMSA experiments were used for ChIP. Antibodies specific to Dnmt3a and Dnmt3b were from Imgenex (San Diego, CA) and anti-tri-methyl H3K9 antibody was from Upstate Biotech. Presence of the IFN-γ promoter sequences was determined using the following PCR primers: 5′: GCTGTCTCATCGTCAGAGAGCCCA and 3′: TGATCGAAGGCTCCTCGGGATTACG. The following primers were used to amplify a region of intron 1: 5′: CAGTAACAGTGTTTGGCTACATGC and 3′: ACCTGCCCTGAAAATATCTATCA.
Vector construction and methylation
The sequences between −250 and +100 relative to the IFN-γ transcriptional start site was amplified by PCR and inserted into the SacI and XhoI sites of the pGL3 basic reporter vector (Promega). The resulting vector is referred to as γ-luc. The Quickchange Site-Directed Mutagenesis Kit (Stratagene) was used to delete the proximal AP1 site (−47 through −57) in the γ-luc vector. Methylation of the γ-luc vector was carried out using either CpG Methylase (mSss I) (New England Biolabs) or the Quickchange Site-Directed Mutagenesis Kit using a protocol adapted from Martinowich et al (2003). Briefly, the γ-luc vector with methylation at the −205, −190, −170, −53, −45, or −34 CpG of the IFN-γ proximal promoter was generated using site-specific methylated oligonucleotides purchased from IDT. The following primers and their reverse complements, containing either methylated CpGs or unmethylated CpGs, were used: −53, −45, and −34 CpGs: GTGAAAATACGTAATCCCGAGGAGCCTTCGATCAGGTATAAAAC; −205 and −190 CpGs: GGGCACAGCGGGGCTGTCTCATCGTCAGAGAGCCCAAGG; −170 CpG: CGTCAGAGAGCCCAAGGAGTCGAAAGGAAACTCTAAC. PCR was carried out according to the manufacturer's protocol with 100 ng of γ-luc vector DNA per reaction. Following the PCR, the reaction was treated with DpnI for 2 h to remove the template DNA. The product was purified using the Qia-Quick method from Qiagen, reannealed, and precipitated. DNA concentration was estimated by gel electrophoresis.
Cell transfection and luciferase detection
AE7 and D10 cells were transfected using the Amaxa Nucleofector Device and the Amaxa Mouse T-Cell Nucleofector Kit (Amaxa, Koelin, Germany). Briefly, 2 × 106 AE7 or D10 cells were resuspended in 100 μl nucleofector solution in which 2 μg of the test vector and 100 ng of a control CMV promoter-driven renilla luciferase vector were added. After electroporation, the cells were cultured in 1.5 ml of the Mouse T-Cell Nucleofector Medium for 6 h in 37 degrees. The cells were then restimulated with 5 ng/ml PMA and 100 ng/ml ionomycin for 2 h, following which cells were harvested and lysed. Luciferase activities were determined using the Dual Reporter Luciferase Assay System (Promega).
Supplementary Material
Supplementary Figure S1
Acknowledgments
We thank Drs Anjana Rao and Herman N Eisen for critique of the manuscript, Dr Laurie Glimcher for the AE7 and D10 cell lines, Dr Qing Ge for assistance in the generation of memory CD8T cells, and Dr Ching-Hung Shen for help with the EMSA assays. This work was supported in part by grants from the National Institutes of Health AI40416 and AI50631 (to JC) and a Cancer Center Core Grant (to Tyler Jacks).
References
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM (2002) T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol 3: 549–557 [DOI] [PubMed] [Google Scholar]
- Aune TM, Penix LA, Rincon MR, Flavell RA (1997) Differential transcription directed by discrete gamma interferon promoter elements in naive and memory (effector) CD4T cells and CD8T cells. Mol Cell Biol 17: 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A (2002) T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol 3: 643–651 [DOI] [PubMed] [Google Scholar]
- Bestor T, Laudano A, Mattaliano R, Ingram V (1988) Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol 203: 971–983 [DOI] [PubMed] [Google Scholar]
- Bestor TH (1992) Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J 11: 2611–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cippitelli M, Sica A, Viggiano V, Ye J, Ghosh P, Birrer MJ, Young HA (1995) Negative transcriptional regulation of the interferon-gamma promoter by glucocorticoids and dominant negative mutants of c-Jun. J Biol Chem 270: 12548–12556 [DOI] [PubMed] [Google Scholar]
- Dong Z, Wang X, Evers BM (2000) Site-specific DNA methylation contributes to neurotensin/neuromedin N expression in colon cancers. Am J Physiol Gastrointest Liver Physiol 279: G1139–G1147 [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Kitazawa R, Maeda S, Kitazawa S (2005) Methylation adjacent to negatively regulating AP-1 site reactivates TrkA gene expression during cancer progression. Oncogene 24: 5108–5118 [DOI] [PubMed] [Google Scholar]
- Grayson JM, Murali-Krishna K, Altman JD, Ahmed R (2001) Gene expression in antigen-specific CD8+ T cells during viral infection. J Immunol 166: 795–799 [DOI] [PubMed] [Google Scholar]
- Grogan JL, Mohrs M, Harmon B, Lacy DA, Sedat JW, Locksley RM (2001) Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity 14: 205–215 [DOI] [PubMed] [Google Scholar]
- Hsieh CS, Heimberger AB, Gold JS, O'Garra A, Murphy KM (1992) Differential regulation of T helper phenotype development by interleukins 4 and 10 in an alpha beta T-cell-receptor transgenic system. Proc Natl Acad Sci USA 89: 6065–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH (2005) T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science 307: 430–433 [DOI] [PubMed] [Google Scholar]
- Lee HJ, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, O'Garra A, Arai N (2000) GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med 192: 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R (1992) Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE (2003) DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302: 890–893 [DOI] [PubMed] [Google Scholar]
- Melvin AJ, McGurn ME, Bort SJ, Gibson C, Lewis DB (1995) Hypomethylation of the interferon-gamma gene correlates with its expression by primary T-lineage cells. Eur J Immunol 25: 426–430 [DOI] [PubMed] [Google Scholar]
- Murphy KM, Reiner SL (2002) The lineage decisions of helper T cells. Nat Rev Immunol 2: 933–944 [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E (1999) DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E (1998) Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 19: 219–220 [DOI] [PubMed] [Google Scholar]
- Penix L, Weaver WM, Pang Y, Young HA, Wilson CB (1993) Two essential regulatory elements in the human interferon gamma promoter confer activation specific expression in T cells. J Exp Med 178: 1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penix LA, Sweetser MT, Weaver WM, Hoeffler JP, Kerppola TK, Wilson CB (1996) The proximal regulatory element of the interferon-gamma promoter mediates selective expression in T cells. J Biol Chem 271: 31964–31972 [DOI] [PubMed] [Google Scholar]
- Richards EJ, Elgin SC (2002) Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108: 489–500 [DOI] [PubMed] [Google Scholar]
- Samten B, Howard ST, Weis SE, Wu S, Shams H, Townsend JC, Safi H, Barnes PF (2005) Cyclic AMP response element-binding protein positively regulates production of IFN-g by T cells in response to a microbial pathogen. J Immunol 174: 6357–6363 [DOI] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Muller MM, Schaffner W (1989) Rapid detection of octamer binding proteins with ‘mini-extracts', prepared from a small number of cells. Nucleic Acids Res 17: 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seder RA, Paul WE, Davis MM, Fazekas de St Groth B (1992) The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J Exp Med 176: 1091–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CH, Stavnezer J (2001) Activation of the mouse Ig germline epsilon promoter by IL-4 is dependent on AP-1 transcription factors. J Immunol 166: 411–423 [DOI] [PubMed] [Google Scholar]
- Stetson DB, Mohrs M, Reinhardt RL, Baron JL, Wang ZE, Gapin L, Kronenberg M, Locksley RM (2003) Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J Exp Med 198: 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH (2000) A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100: 655–669 [DOI] [PubMed] [Google Scholar]
- Usui T, Nishikomori R, Kitani A, Strober W (2003) GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity 18: 415–428 [DOI] [PubMed] [Google Scholar]
- White GP, Watt PM, Holt BJ, Holt PG (2002) Differential patterns of methylation of the IFN-gamma promoter at CpG and non-CpG sites underlie differences in IFN-gamma gene expression between human neonatal and adult CD45RO- T cells. J Immunol 168: 2820–2827 [DOI] [PubMed] [Google Scholar]
- Winders BR, Schwartz RH, Bruniquel D (2004) A distinct region of the murine IFN-gamma promoter is hypomethylated from early T cell development through mature naive and Th1 cell differentiation, but is hypermethylated in Th2 cells. J Immunol 173: 7377–7384 [DOI] [PubMed] [Google Scholar]
- Yano S, Ghosh P, Kusaba H, Buchholz M, Longo DL (2003) Effect of promoter methylation on the regulation of IFN-gamma gene during in vitro differentiation of human peripheral blood T cells into a Th2 population. J Immunol 171: 2510–2516 [DOI] [PubMed] [Google Scholar]
- Young HA, Ghosh P, Ye J, Lederer J, Lichtman A, Gerard JR, Penix L, Wilson CB, Melvin AJ, McGurn ME, Lewis DB, Taub DD (1994) Differentiation of the T helper phenotypes by analysis of the methylation state of the IFN-gamma gene. J Immunol 153: 3603–3610 [PubMed] [Google Scholar]
- Zhang DH, Cohn L, Ray P, Bottomly K, Ray A (1997) Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls. Th2-specific expression of the interleukin-5 gene. J Biol Chem 272: 21597–21603 [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang DZ, Boothby M, Penix L, Flavell RA, Aune TM (1998) Regulation of the activity of IFN-gamma promoter elements during Th cell differentiation. J Immunol 161: 6105–6112 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1


