Abstract
Major structural changes occur in the spliceosome during its catalytic activation, which immediately precedes the splicing of pre-mRNA. Whereas changes in snRNA conformation are well documented at the level of secondary RNA–RNA interactions, little is known about the tertiary structure of this RNA–RNA network, which comprises the spliceosome's catalytic core. Here, we have used the hydroxyl-radical probe Fe-BABE, tethered to the tenth nucleotide (U+10) of the 5′ end of a pre-mRNA intron, to map RNA–RNA proximities in spliceosomes. These studies revealed that several conserved snRNA regions are close to U+10 in activated spliceosomes, namely (i) the U6 snRNA ACAGAG-box region, (ii) portions of the U6 intramolecular stem-loop (U6-ISL) including a nucleotide implicated in the first catalytic step (U74), and (iii) the region of U2 that interacts with the branch point. These data constrain the relative orientation of these structural elements with respect to U+10 in the activated spliceosome. Upon conversion of the activated spliceosome to complex C, the accessibility of U6-ISL to hydroxyl-radical cleavage is altered, suggesting rearrangements after the first catalytic step.
Keywords: pre-mRNA splicing, site-directed hydroxyl-radical probing, spliceosome
Introduction
Nuclear pre-mRNA splicing proceeds by two transesterification reactions. In the first step, the 2′ hydroxyl group of the branch-point adenosine attacks the 5′ splice site (5′ss), resulting in a cleaved 5′ exon and an intron-3′-exon lariat. During the second step, the 3′ end of the 5′ exon attacks the 3′ splice site (3′ss), resulting in mature mRNA and excised intron lariat. The human spliceosome forms by association of small nuclear ribonucleoprotein particles (U1, U2, U4/U6.U5 snRNPs) and non-snRNP proteins with pre-mRNA, and attains its catalytic conformation by undergoing a series of protein and RNA rearrangements that are highly conserved between yeast and human (Burge et al, 1999; Brow, 2002). First, a pre-spliceosome (complex A) is formed, in which the U2 snRNA interacts with the branch point region and the U1 snRNA with the 5′ss of the pre-mRNA. Then the U4/U6.U5 tri-snRNP binds to this, producing the precatalytic spliceosome (complex B). In this complex, the U4 and U6 snRNAs are base-paired, forming two intermolecular stems. In the next step, this interaction is disrupted and U6 enters into new base-pairing interactions with the 5′ss of the intron and with U2 snRNA. During this rearrangement, U6 is re-folded such that the region previously forming the U4 base-paired stem II now forms an intramolecular stem-loop (U6-ISL). At the same time, the U1 and U4 snRNPs are released, resulting in activated, catalytically competent spliceosomes (complex B*) in which the intron's branch point and 5′ss are juxtaposed for the first transesterification reaction (Burge et al, 1999; Brow, 2002). After the first step, the 5′ and 3′ exon are aligned with the help of U5 snRNA (Sontheimer and Steitz, 1993), leading to complex C and subsequently to the second step of splicing (Umen and Guthrie, 1995).
The nature of the U2/U6 interaction has been a matter of debate in recent years. In mammals, U6 is believed to form helices I and II with U2, while U2 maintains parts of the U2-ISL (Sun and Manley, 1995; Figure 1A). Evidence for this is provided by NMR studies of yeast U2 and U6 snRNAs (Sashital et al, 2004) and by mutational studies in yeast (McPheeters and Abelson, 1992). In an alternative model, U6 is predicted to form more extensive base pairs with U2, leading to helices Ia and Ib (Madhani and Guthrie, 1992; Hilliker and Staley, 2004; Figure 1B). However, these two possibilities could reflect different states of spliceosome activation, as discussed elsewhere (Sun and Manley, 1995; Sashital et al, 2004) and need therefore not be mutually contradictory.
Figure 1.
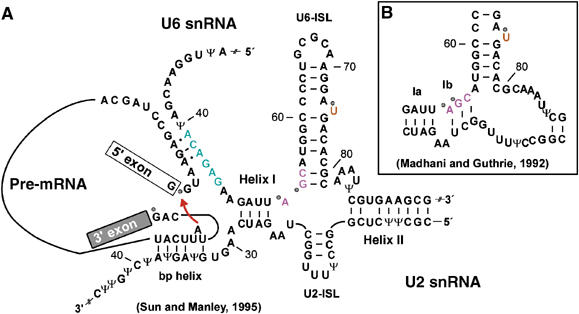
Secondary-structure models of the RNA–RNA network of the spliceosome. (A) Secondary structure of the human U2/U6/pre-mRNA complex in the activated spliceosome just before the first catalytic step of splicing (according to Sun and Manley, 1995). (B) Secondary structure of the alternative human U2/U6 interaction mode, drawn according to the yeast structure (Madhani and Guthrie, 1992). In both panels, the conserved ACAGAG box (green), the AGC triad (pink) and the bulged U (orange) are highlighted. Mg2+-binding sites at the 5′ and 3′ss and at the analogous yeast nts U80, A53, G54 (Fabrizio and Abelson, 1992; Yu et al, 1995; Sontheimer et al, 1997; Gordon et al, 2000; Yean et al, 2000) are indicated by dots. Ψ denotes the modified nucleotide pseudouridine. The red arrow schematically shows the attack of the bulged-out A (branch point) on the 5′ss during the first catalytic step of splicing.
Although activation of the spliceosome is well documented at the level of changes in these base-pairing interactions, much less is known about higher-order interactions and their dynamics. Knowledge of these interactions is essential for mapping the tertiary structure of the catalytic core—in which the two steps of splicing occur—and for understanding the overall mechanism of splicing.
Much evidence supports the idea that splicing catalysis is mediated by the RNA components of the spliceosome (discussed by Nilsen, 1998; Villa et al, 2002). First of all, a reaction similar to the first step of splicing can be catalysed by protein-free U2 and U6 snRNA in vitro (Valadkhan and Manley, 2001). Secondly, spliceosomes are metalloenzymes; that is, the phosphates at each splice site (5′ss or 3′ss) bind essential catalytic divalent ion at both active sites of the spliceosome (Moore and Sharp, 1993; Sontheimer et al, 1997). Additional catalytic factors in spliceosomes have yet to be identified. An interesting candidate for direct involvement in catalysis is the U6-ISL. In yeast, replacement of a nonbridging phosphate oxygen by sulphur at nucleotide U80, which is equivalent to human U74, revealed the existence of essential Mg2+–phosphate interactions (Fabrizio and Abelson, 1992; Yean et al, 2000). Furthermore, for U80, a possible contribution of the magnesium ion to catalysis of the first step has been suggested (Yean et al, 2000). However, no such information is at present available for the human spliceosome.
To investigate the RNA neighbourhood of the 5′ss, we chose hydroxyl-radical probing, using the EDTA derivative Fe-BABE (Joseph and Noller, 2000; Figure 2A). Hydroxyl radicals generated from Fe-BABE upon addition of H2O2 are useful for probing local RNA structure, as they induce cleavage of nearby ribose moieties, predominantly within a distance of 10 Å (Wang and Cech, 1992; Han and Dervan, 1994), regardless of how they are integrated into the overall structure. This method has been used to investigate tertiary interactions in ribosomes, by tethering Fe-BABE to RNA or protein (Culver and Noller, 2000; Joseph and Noller, 2000, and references therein), and more recently, the pre-spliceosome, where a close proximity of the branch point and 3′ss to the 5′ss was revealed (Kent and MacMillan, 2002).
Figure 2.
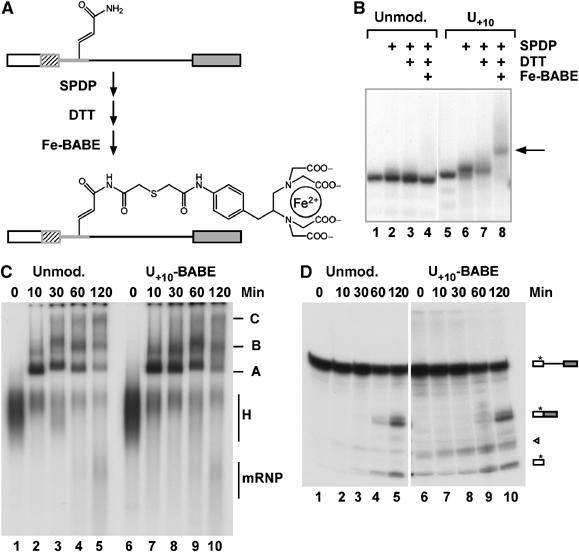
Generation and characterisation of pre-mRNA with Fe-BABE introduced at intron position +10. (A) Site-directed chemical modification of pre-mRNA containing a single reactive amido group. The amido group was first modified with the disulphide-containing reagent SPDP and then reduced with DTT, leading to an SH group that could readily be modified with Fe-BABE. The maximum length of the linker is ∼18 Å. (B) Unmodified and amide-modified 5′-[32P] synthetic oligomers used in the ligation (see Materials and methods) were treated as in (A) and the resulting products at each step were analysed by denaturing PAGE. The arrow (right) shows the product Fe-BABE–RNA. (C) Time course of splicing complex formation. Spliceosomal complex formation was analysed on a native agarose gel. The identity of the complexes is indicated on the right. (D) Time course of splicing of unmodified and Fe-BABE-modified pre-mRNA. Pre-mRNA was radiolabelled in the 5′ exon (asterisk), and RNAs were analysed by denaturing PAGE. The substrate and products are shown schematically on the right (arrowhead, RNA degradation product). The concentrations of pre-mRNA and nuclear extract were 4 nM and 30%, respectively. Splicing efficiencies were ∼40% for both pre-mRNAs, as assessed by comparing the levels of mRNA and pre-mRNA after 120 min.
The results presented here demonstrate that in activated B* spliceosomes, U+10 is close to regions of U6-ISL (including U74) and also to the nucleotides of U2 that base-pair with the branch point region. The proximity relationships are only partially maintained after the first step of splicing (i.e. in complex C). These studies provide important information regarding the higher-order structure of the RNA network in the B* (activated) and C spliceosomal complexes.
Results
Experimental strategy for determining RNA–RNA proximities
To investigate RNA neighbourhoods in the spliceosome by site-directed hydroxyl-radical probing, we devised a strategy in which a pre-mRNA containing an amide or a primary amine is first synthesised by RNA-ligation of a synthetic oligonucleotide with flanking pieces of RNA (Rhode et al, 2003). Subsequently, Fe-BABE is introduced by modification of the amide or primary amine in a two-step conjugation reaction. This general strategy allows attachment of the Fe-BABE to the base through a 5-acrylamido group on a uridine (Figure 2A) or to the ribose through a 2′-amino group (not shown).
For our analysis, we targeted Fe-BABE to the uridine base at position +10 of the intron. This position is probably constrained in the spliceosome, for two reasons: first, it is adjacent to nucleotides +4 to +6, which bind the conserved ACAGAG-box (nucleotides 41–46) of human U6 snRNA, and second, it is on the 3′ side of a psoralen crosslink observed between C37 of U6 and the pre-mRNA, suggesting that base-pairing between U6 and the pre-mRNA can be extended to nucleotide +9 of the pre-mRNA (Wassarman and Steitz, 1992; Kandels-Lewis and Séraphin, 1993; Chan et al, 2003). A modification at intron position +10 should therefore not interfere with pre-mRNA binding to U6 snRNA or with spliceosome function.
The conjugation procedure yielded Fe-BABE modification of U+10 in excess of 90%, as was assayed with the amide-containing oligomer (Figure 2B, lane 8) used in the ligation. We therefore prepared unmodified and Fe-BABE-modified pre-mRNAs and compared their properties in splicing. Analysis of the kinetics of spliceosomal complex formation revealed that the two pre-mRNAs behave almost identically, as shown by the equal amounts of individual spliceosomal complexes formed on each pre-mRNA in a given time (Figure 2C, compare left and right panels). In addition, splicing efficiency was similar, as can be seen from the amounts of mRNA produced from each pre-mRNA (Figure 2D, compare left and right panels). These analyses revealed that introduction of Fe-BABE at position U+10 had no effect on the efficiency or kinetics of spliceosomal complex formation or splicing. However, when Fe-BABE was attached through the 5-acrylamido derivatives at position −2 or +2 relative to the 5′ss, complete or partial inhibition of splicing, respectively, was observed (Supplementary Figure S1). For the +2 position, this inhibition could only partly be overcome by attaching Fe-BABE to the ribose (Supplementary Figure S1). Therefore, we used the Fe-BABE attached to U+10 in subsequent experiments.
Site-directed hydroxyl-radical probing of spliceosomes separated by glycerol-gradient centrifugation
To investigate RNA in the proximity of intron position +10 before the first step of splicing, a splicing reaction was performed under conditions in which spliceosomes were fully assembled but had not yet catalysed splicing. This was achieved by kinetic control of the splicing reaction and by increasing the pre-mRNA concentration (for details, see legend to Figure 3). Fully assembled spliceosomes (migrating as 50S particles) were separated from pre-spliceosomes (25S) by glycerol-gradient centrifugation (Frendewey and Keller, 1985; Lamond et al, 1988). Native gel analysis confirmed that the 25S peak contains pre-spliceosomes and unspecific complex H, whereas the 50S peak contains a complex with mobility lower than that of the pre-spliceosome and lacks contaminating complexes from the 25S peak (not shown). RNA analysis across the gradient showed that the 50S peaks obtained from incubations with both the unmodified and Fe-BABE-modified pre-mRNAs contained predominantly (>90%) unspliced pre-mRNA (Figure 3A).
Figure 3.
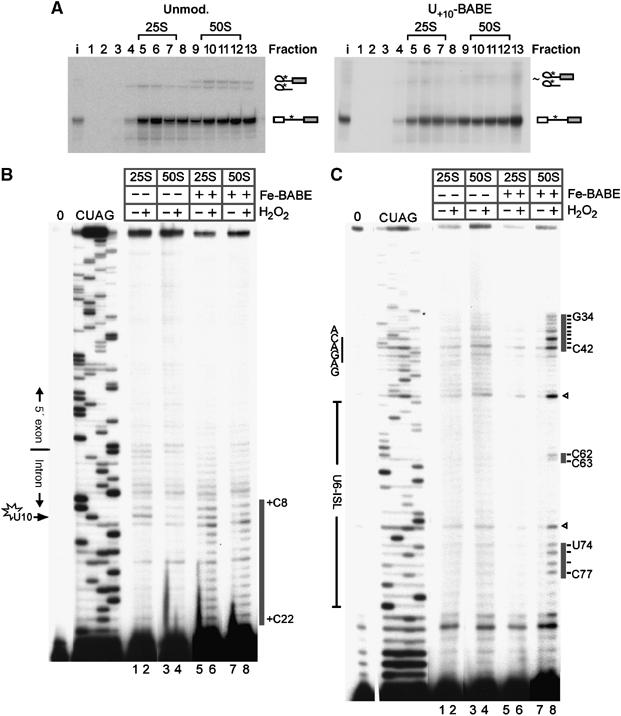
Site-directed hydroxyl-radical probing of gradient-purified spliceosomes. (A) A 30-min splicing reaction containing 20 nM unmodified (left panel) or Fe-BABE modified (right panel) pre-mRNA was fractionated by glycerol-gradient centrifugation, and the RNA in each fraction was analysed by denaturing PAGE. The pre-mRNA was radioactively labelled at intron position +12 (asterisk); RNA species are identified on the right. A higher pre-mRNA concentration than in Figures 2C and D was used, and therefore almost no splicing intermediates are observed after 30 min. (B) Analysis of hydroxyl-radical cleavage of the pre-mRNA close to the site of Fe-BABE modification. Cleavage sites on the pre-mRNA are indicated by a grey bar on the right. 0: input RNA; C, U, A, G: dideoxy sequencing reactions on input RNA. Lanes 1–4: background cleavage of pre-mRNA not modified with Fe-BABE; lanes 5–8: cleavage of spliceosomal complexes containing Fe-BABE pre-mRNA. (C) Analysis of hydroxyl-radical cleavage of the U6 snRNA. Cleavage sites in U6 snRNA are indicated by grey bars on the right. Cleavages at U6 nucleotides G72 and U52 (open arrowheads) were observed in only two out of five experiments. In addition, these positions are prone to erratic and spontaneous reverse transcriptase pauses (compare the dideoxy sequences and ‘0' controls). They were therefore considered as background unrelated to hydroxyl-radical cleavage.
The 25S (fraction 6 or 7) and 50S (fraction 11) peaks were subjected to hydroxyl radicals from the Fe-BABE label and consequent cleavage of pre-mRNA, and the U6, U2 and U5 snRNAs were investigated by primer extension. The existence of a cleavage site implies proximity of the cleaved nucleotide to the Fe-BABE label. RNA proximities to U+10 of the intron could thus be identified and compared between the two spliceosomal complexes. Sites of cleavage induced by hydroxyl radicals are detected as bands that appear in the ‘+BABE +H2O2' lanes but are missing, or less intense, in the corresponding control lanes (+BABE –H2O2 and –BABE +H2O2). Analysis of cleavage of the pre-mRNA revealed that the hydroxyl radicals attack the RNA in the vicinity of the Fe-BABE-modified nucleotide U+10 (Figure 3B). The pre-mRNA cleavage pattern observed with the 25S complexes (pre-spliceosomes) and 50S complexes (fully assembled spliceosomes) was essentially similar: cleavage sites were observed between nucleotides +8 and +22, with strongest cleavage between nucleotides +8 and +13 of the intron (black line in Figure 3B; compare lane 6 with lanes 5 and 2, and lane 8 with lanes 7 and 4). While this confirms that the method itself is working, it also shows that the overall accessibility of the pre-mRNA for hydroxyl radicals produced from U+10 does not differ between the two different spliceosomal complexes.
Analysis of cleavage sites on U6 snRNA was performed with a primer that anneals to its 3′ end (Figure 3C). Distinct cleavage was observed in the 50S fraction only (compare –H2O2 with +H2O2 in Figure 3C, lanes 8–7 and 6–5). This was only observed in the presence of Fe-BABE on the pre-mRNA, and not in fractions containing unmodified pre-mRNA (lanes 2 and 3). It is therefore clear that portions of U6 snRNA are close to U+10 of the pre-mRNA in the spliceosomes that migrate at 50S in the gradient.
One region of cleavages was found at nucleotides G34–C42 of the U6 snRNA, with the strongest cleavages at U40–C42 (Figure 3C, lane 8). Since nucleotides A41 and C42 are part of the conserved ACAGAG box, we infer that this element is in close proximity to U+10 in the 50S spliceosomes. Considering that this element indeed approaches the 5′ end of the intron only upon formation of the 5′ss/U6 snRNA helix (Figure 1), that is upon spliceosome activation, the data imply that the preparation analysed contains activated spliceosomes.
An additional set of cleavage sites was observed at nucleotides C62, C63 and U74–C77 of the U6 snRNA (Figure 3C, lane 8). In the activated spliceosomes, these two regions of cleavages map to a row of base pairs around U74 on the highly conserved U6-ISL (Figure 5A). Since the cleavage pattern is consistent with a helical structure (see below), the data are thus consistent with the existence of the U6-ISL and further corroborate the above finding that the 50S fractions contain activated spliceosomes.
Figure 5.
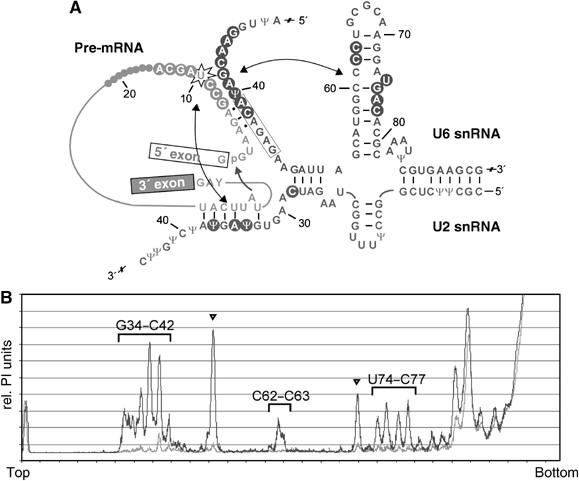
Summary of hydroxyl-radical induced cleavage sites in the secondary-structure model of the activated spliceosome. (A) Secondary-structure model of U2/U6/pre-mRNA before the first step of splicing according to Sun and Manley (1995). The attack of the branch point A at the 5′ss is indicated by an arrow. Hydroxyl-cleavage sites in U6 and the pre-mRNA are indicated by circles. The double-headed arrows indicate proximity relationships found between U+10 of the pre-mRNA and U2 snRNA as well as U6-ISL. (B) Densitometric analysis of cleavage of U6 snRNA from Figure 3C, lanes 7 and 8. The background lane is shown in grey; hydroxyl-induced cleavages are shown in black. Nucleotides on U6 snRNA specifically cleaved by hydroxyl radicals show higher peak values and are labelled with the corresponding nucleotides. Arrowheads indicate enhanced cleavage sites that are not reproducibly observed (compare with Figure 3C).
To allow a classification of the genuine cleavages in Figure 3C, according to Joseph et al (2000), cleavage intensities were quantified by a densitometric analysis of the bands observed in the absence versus the presence of H2O2 (Figure 5B). This quantification revealed ‘very strong' cleavage between U40 and C42 of U6 snRNA, while the remaining cleavages were found to be ‘strong' (intensities about 50% of the ‘very strong' ones). While we have not attempted to calibrate distances in our system, we can nevertheless conclude that, relatively speaking, the nucleotides U40–C42 are closer to U+10 than the nucleotides cleaved on U6-ISL.
An analysis of cleavages on U2 snRNA revealed distinct hydroxyl-radical-induced cleavage at nucleotides Ψ34, A35 and Ψ37 of the branch-point-binding region (Figure 4A, compare lane 4 with lanes 1–3). The cleavage sites detected in U2 are indicated in the secondary structure model of the spliceosome (Figure 5A). No cleavage was found 5′ of C28 of U2 snRNA, indicating that these regions either are not near U+10 or are shielded by protein.
Figure 4.
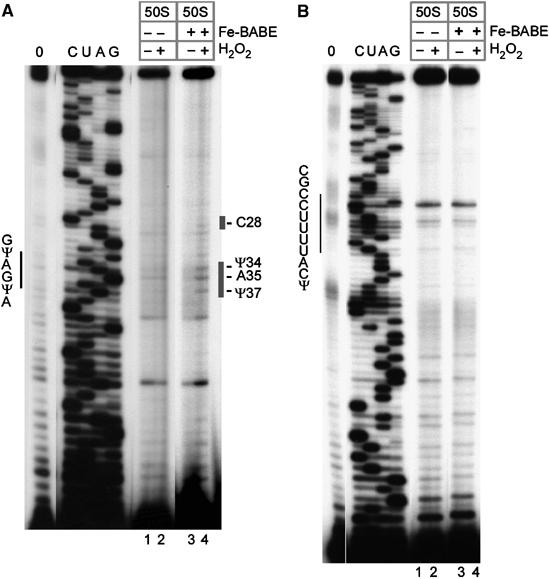
Site-directed hydroxyl-radical probing of gradient-purified spliceosomes. (A) Analysis of hydroxyl-radical cleavage of the U2 snRNA. Cleavage sites on the U2 snRNA are indicated by a bar on the right. Lanes 1 and 2: background cleavage of U2 snRNA not modified with Fe-BABE; lanes 3 and 4, cleavages in spliceosomal complexes containing Fe-BABE pre-mRNA. (B) Analysis of hydroxyl-radical cleavage of the U5 snRNA. The position of the U5 loop (nt 36–46) is indicated on the left. Lanes are as in (A). Further labelling is as in Figure 3B.
As the U2 snRNA cleavages mapped to the branch-point-binding region, we also scanned the 3′ portion of the pre-mRNA for cleavages. This analysis revealed no additional specific cleavages in the branch-point region, the polypyrimidine tract, the 3′ss, or the 3′ exon (data not shown). Therefore, either these regions of the pre-mRNA are not close to nucleotide U+10 or, alternatively, they are protected. These protections may be by direct contact or by indirect shielding through intervening protein or RNA. In addition, the analysis was extended to the U5 snRNA, which is known to undergo numerous interactions with the 5′ss and the 3′ss (Wyatt et al, 1992; Sontheimer and Steitz, 1993; McConnell and Steitz, 2001). This revealed no specific hydroxyl-radical-induced cleavages of the U5 snRNA in the region examined (Figure 4B, lanes 3 and 4).
Site-directed hydroxyl-radical probing of affinity-purified activated spliceosomes
The gradient-purified spliceosomes analysed so far could potentially contain a mixture of different spliceosomal complexes, and the cleavages observed could belong to different assembly states. Consequently, a direct correlation of all the observed cleavages with the activated state cannot be inferred from these experiments. To prove that the cleavages indeed occurred in activated spliceosomes, we isolated homogenous activated spliceosome according to the method of Makarov et al (2002) using the Fe-BABE-modified pre-mRNA. In this procedure, activated spliceosomes (B* complexes) are immunoaffinity purified by using an antibody against the SKIP protein, which associates stably with spliceosomes at the time of their catalytic activation (Makarov et al, 2002; Makarova et al, 2004). Previous characterisation of such spliceosomes by psoralen crosslinking confirmed the existence of the U6/pre-mRNA interaction and the U2/U6 interaction in the activated spliceosome, while the U4/U6 interaction was largely disrupted (Makarova et al, 2004).
The affinity-purified B* complexes, which were confirmed to contain stoichiometric amounts of pre-mRNA along with U2, U5 and U6 snRNAs (data not shown), were subjected to hydroxyl-radical probing as above. The pre-mRNA and the U6 and U2 snRNAs were analysed for cleavage (Figure 6). The cleavage pattern of the pre-mRNA was comparable to that observed with the 50S gradient fractions, in that it extended from nts +9 to +20 of the intron (compare lane 2 of Figure 6A with lane 8 of Figure 3B). Strikingly, the cleavage pattern of U6 snRNA was identical to that observed with the 50S gradient fractions (compare lane 2 of Figure 6B with lane 8 of Figure 3C). As with the 50S spliceosome, cleavage sites were detected at nucleotides G34–C42 and at nucleotides C62, C63 and U74–C77 of U6 snRNA, corroborating the results obtained above. Likewise, an analysis of U2 snRNA revealed cleavages at nucleotides Ψ34, A35 and Ψ37 (Figure 6C, lane 2), identical to those observed in the gradient-purified 50S spliceosomes (Figure 4B).
Figure 6.
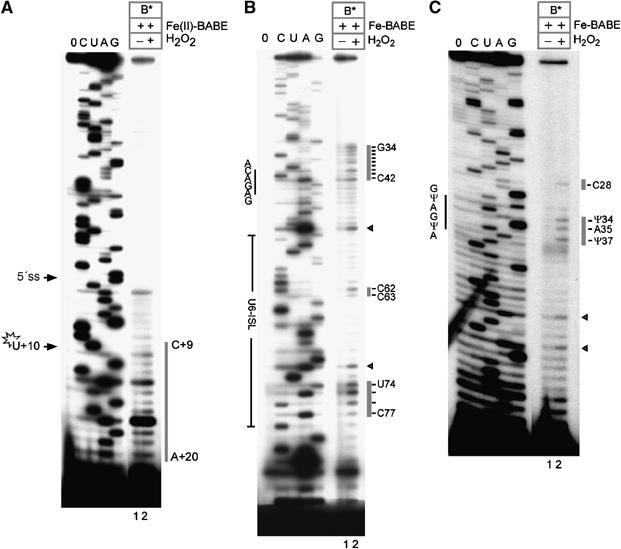
Site-directed hydroxyl-radical probing in the affinity-purified B* complex. (A) Analysis of hydroxyl-radical cleavage of pre-mRNA. Cleavage sites in the pre-mRNA are indicated by a grey bar on the right. (B) Analysis of hydroxyl-radical cleavage of U6 snRNA. Cleavage sites in U6 snRNA are indicated by grey bars on the right. Arrowheads indicate sites of increased background cleavage not observed in every experiment. (C) Analysis of hydroxyl-radical cleavage of U2 snRNA. Cleavage sites in U2 snRNA are indicated by the grey bars on the right. Further labelling is as in Figure 3B. See also densitometric analysis in Supplementary Figure S2.
Taken together, our data demonstrate that all the specific cleavages described so far can be assigned to the activated spliceosomes. Thus, the cleavage patterns can indeed be interpreted in the context of the 5′ss/U6 snRNA interactions and the U6-ISL, as was suggested above. Moreover, since U+10 is an integral part of the 5′ss/U6 snRNA interaction helix, this structural element as a whole must be close to the U6-ISL, such that U+10 is oriented towards the U6-ISL. A third element that is likewise close to U+10 at this stage of the spliceosomal cycle is the U2 snRNA/branch point helix.
Mapping the cleavages onto known RNA structural elements
To investigate the relative orientation of the U6-ISL and the U2/branch point helix to U+10, we initially mapped the cleavage sites onto the known homologous 3D structures from yeast. The yeast U6-ISL (Sashital et al, 2004) and U2/bp-helix (Newby and Greenbaum, 2002) should both fold in a manner very similar, if not identical, to that in which their human counterparts fold.
When the U6 cleavage sites at C62, C63, and U74–C77 are mapped to their homologous residues of the structure of the isolated yeast U6-ISL as determined by NMR, it becomes apparent that they form a continuous stretch of nucleotides around U74 (yeast U80; Figure 7A). This is consistent with the presence of the U6-ISL in our activated spliceosomes (see above). Moreover, the only way to explain this cleavage pattern with hydroxyl radicals arising from U+10 is to assert that the U6-ISL is oriented in the spliceosome in such a way that these nucleotides face U+10. The data thus constrain the possibilities for orientation of U6-ISL with respect to U+10 in the activated spliceosome.
Figure 7.
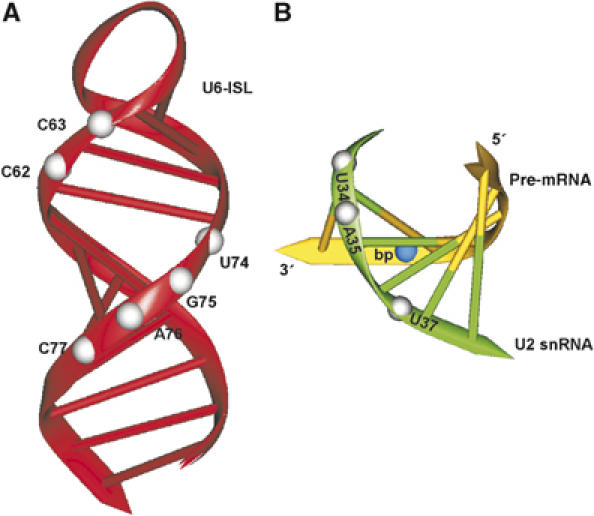
Mapping the cleavages onto RNA structural elements. (A) Model showing the position of cleavage sites of U6 snRNA observed in the activated spliceosome in the NMR structure of the yeast U6-ISL (Sashital et al, 2004; PDB code 1XHP). Corresponding human nucleotides at hydroxyl-radical cleavage sites are shown by white balls. (B) Model showing the position of cleavage sites of U2 snRNA observed in the activated spliceosome in the NMR structure of the yeast U2/bp-helix (Newby and Greenbaum, 2002; PDB code 1LPW). Corresponding human nucleotides at positions of hydroxyl-radical cleavage sites are shown by white balls. The branch site is shown by a blue ball. The structural views were prepared with the program Drawna 2.1 (Westhof et al, 1985).
With the NMR structure of the yeast U2/branch-point helix as a basis for mapping our observed U2 cleavage sites, it becomes readily apparent that the pre-mRNA strand has to be oriented away from U+10 to account for the positions of cleavage observed (Figure 7B). Only then can hydroxyl radicals produced at U+10 cleave the nucleotides Ψ34, A35 and Ψ37. Our data thus provide a second constraint for localising the catalytic centre in the activated spliceosome, in that the U2 snRNA strand of the U2/branch-point helix is most probably located between U+10 and the branch-point adenosine. Consistently with such a configuration, we do not find cleavage at the branch point.
The third structural element that is attacked by hydroxyl radicals in the activated spliceosome is the 5′ss/U6 snRNA helix. In an ideal double-stranded RNA helix, cleavage sites three base pairs apart on opposite strands would be positioned on the same side and within the major groove of the helix. This is indeed what we observe, since the strongest cleavages of U6 (Ψ40–C42) and the strongest cleavages around the Fe-BABE at U+10 (+8 to +13) are separated by three intervening bases (Figure 3B and C; see also Figure 5A). These observations are therefore consistent with the existence of an extended 5′ss/U6 snRNA helix, as outlined above.
Site-directed hydroxyl-radical probing in spliceosomal complex C
Spliceosomes have been suggested to undergo rearrangements between the first and the second step of splicing (Moore and Sharp, 1993). We therefore finally investigated whether the RNA cleavage pattern observed with the activated spliceosome is maintained after the first step of splicing. As C complexes are difficult to isolate, we made use of a well-described mutation, that is a GG mutation introduced at the 3′ss, to stall splicing before the second step (Gozani et al, 1994). After 150 min of splicing, accumulated C complexes were fractionated by glycerol-gradient centrifugation. An RNA analysis of the gradient fractions showed that in the 50S gradient peak more than 80% of the complexes contained intron-lariat joined to the 3′ exon, consistent with the presence of C complexes (Figure 8A). These fractions were subjected to hydroxyl-radical probing. The pre-mRNA cleavage pattern was essentially identical to that in the activated spliceosome, in that nucleotides +7 to +18 were cleaved (compare Figures 8B and 3C). In contrast, the cleavage pattern of U6 in complex C differed from that obtained with activated spliceosomes, in two respects. First, fewer nucleotides were cleaved around the ACAGAG-box region (i.e. only G38 to C42; Figure 8C, lane 2), but A41 and C42 of the box were still cleaved. These results show that the Fe-BABE reagent is still close to the ACAGAG box and is consistent with the persistence of the 5′ss/U6 snRNA base-pairing interaction in complex C (Sontheimer and Steitz, 1993). Second, cleavage at U74, G75 and A76 disappeared, whereas cleavage at C62, C63 and C77 (which was observed in the activated spliceosome) was still detected. Therefore, several nucleotides of U6-ISL (including U74) are no longer accessible to site-directed hydroxyl-radical cleavage in complex C. An analysis of U2 snRNA cleavage was not attempted, because of the small amounts of splicing complexes routinely obtained per fraction (less than 1/4 when compared with the 120 fmol of activated spliceosomes in Figures 3 and 4) and the inherently lower efficiency of primer extension on U2 snRNA (not shown). An overview of the U6 snRNA cleavage pattern is shown in Figure 8D. In summary, these data suggest that upon transition to C complex either (i) protection of nucleotides U74 to A76, or (ii) a relative repositioning of the U6-ISL with respect to U+10 has occurred. The latter may reflect the requirement to remodel the spliceosome before the second step (Moore and Sharp, 1993). In contrast, the 5′ss/U6 snRNA binding region was still largely helical at this stage in the C complexes analysed.
Figure 8.
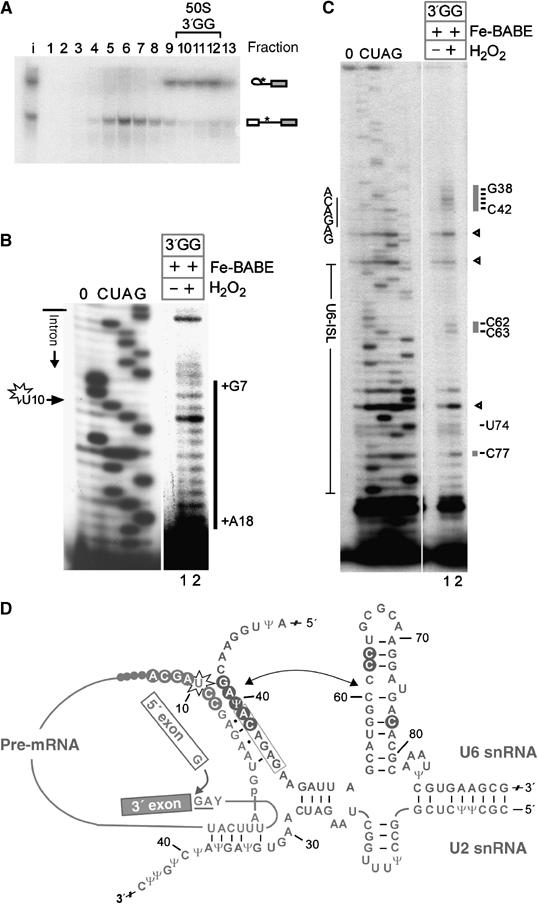
Additional U6-ISL nucleotides are protected from hydroxyl radicals after the first step of splicing. (A) Gradient centrifugation of a 150-min splicing incubation containing 50% nuclear extract and Fe-BABE-pre-mRNA-3′GG (9 nM), analysed as in Figure 2. The pre-mRNA was radioactively labelled at intron position +12 (asterisk); RNA species are identified on the right. Hydroxyl-radical probing of pre-mRNA (B) and U6 snRNA (C) in complex C. Cleavage sites are indicated by grey bars on the right. The further labelling of the figure is as in Figure 6A and B. See also densitometric analysis in Supplementary Figure S2. (D) Secondary-structure model of U2/U6/pre-mRNA after the first step of splicing. The attack of the 5′ exon at the 3′ss is indicated by an arrow. Hydroxyl-radical cleavage sites in U6 and the pre-mRNA are indicated by circles. The double-headed arrow indicates the proximity found between U6-ISL and U+10 of the pre-mRNA. Further labelling is as in Figure 3B.
Discussion
In this work, we performed site-directed hydroxyl-radical footprinting with the aim of gaining insight into the structuring of RNA neighbourhoods in the spliceosome. We provide evidence that in activated spliceosomes, a number of highly conserved RNA structural elements are in the direct vicinity of nucleotide U+10 of the intron. These elements include (i) the U6-ISL, which forms as a result of activation of the spliceosome and which positions the important nucleotide U74 of U6 snRNA; (ii) the U2/branch point helix, which holds the branch point in position for the first step of splicing; and (iii) the highly conserved ACAGAG box, which tethers the 5′ss region to the spliceosome, thereby positioning the 5′ss. In addition to providing simple proximity data, our results constrain the possibilities for the relative orientations of these RNA structural elements with respect to U+10, thus providing higher-order constraints for reconstructing the architecture of the spliceosome. By tracking these proximity relationships into a C complex, stalled just before the second step of splicing, we find the 5′ss/U6 snRNA helix unchanged, but changes in the accessibility of nucleotides on U6-ISL. These issues, and their implications for organisation of the catalytic centre of the spliceosome, are discussed in turn.
The Fe-BABE reagent was tethered to nucleotide U+10 for a number of reasons. First of all, U+10 is next to a highly conserved RNA structural element, the 5′ss/U6 snRNA helix (Figure 1), which only forms upon spliceosome activation. In addition, crosslinking experiments in human (Wassarman and Steitz, 1992) and yeast spliceosomes (Chan et al, 2003) suggested that the helix can be extended at least to nucleotide +9 of the intron. Therefore, the U+10 position is likely constrained in the spliceosome. Second, of all positions tested (Supplementary Figure S1), modifications at position U+10 were least detrimental to spliceosome assembly and splicing. For this reason, we used it as an entry point for a site-directed hydroxyl-radical investigation of spliceosomes at defined assembly stages.
The affinity-purified activated spliceosome used in our studies contained the snRNPs U2, U5 and U6, and the pre-mRNA, while it lacked U1 and U4 (Makarov et al, 2002). In this complex, the major structural rearrangements leading to spliceosome activation have occurred, as shown by the presence of U2/U6 helix II and the 5′ss/U6 interaction (Makarova et al, 2004). Consistent with the latter interaction, we now find close proximity of U+10 with the ACAGAG-box in this complex (Figure 5A). The cleavage pattern that we observe is in agreement with a helical structure as outlined above.
We provide strong evidence that before the first step of splicing, C62, C63 and U74–C77 of U6-ISL are accessible to hydroxyl radicals generated at U+10. Since these cleavages are fully compatible with the helical structure proposed for the U6-ISL, our site-directed hydroxyl-radical probing from position U+10 of the intron of the pre-mRNA therefore provides a first direct and straightforward experimental approach for investigation of this important structural element in a native activated spliceosome.
Our data imply that the stretch of nucleotides cleaved on the U6-ISL must be positioned in such a way that they face U+10, the source of the hydroxyl radicals. In addition, these nucleotides are not protected by proteins. Thus, whatever the exact positioning of the U6-ISL relative to U+10 of the 5′ss/U6 snRNA helix, the space between U+10 and the cleaved nucleotides is not occluded by RNA or protein.
Of particular interest is U74. Its homologue in yeast, U80, was shown to bind a magnesium ion at the pro-Sp nonbridging phosphoryl oxygen, which was found to be critically required for the first step of splicing (Yean et al, 2000). Whether the essential magnesium identified by Yean et al (2000) has a structural or catalytic role is at present an unsettled issue (discussed by Nilsen, 2000; DeRose, 2003). For a magnesium ion to perform a catalytic task at U74, the 5′ss or the branch-point adenosine would have to be brought into close proximity (at least 4 Å; Steitz and Steitz, 1993) of the magnesium bound to U80 (or U74 in our case). As far as the 5′ss (i.e. nt +1 of the intron) is concerned, the close proximity of nucleotide +10 to U74 probably precludes the simultaneous proximity of nt +1 to U74. In fact, cleavage of nucleotide +1 or neighbouring nucleotides was never observed in any of the spliceosomal complexes investigated (complexes A, B* and C). Therefore, the 5′ss region of the pre-mRNA is probably not in the vicinity of U+10, which in turn implies that a folding-back of the 5′ss towards the intron nucleotide U+10 is not very likely. In further corroboration of this, nt +2 of the pre-mRNA and U40 of the U6 snRNA were found to be in close proximity before the first step of splicing in yeast (Kim and Abelson, 1996). However, we cannot exclude the possibility that the 5′ss is shielded by protein, or is otherwise unfavourably oriented for attack by hydroxyl radicals from U+10. A proximal position of the branch-point adenosine to U74 is even less likely. The clear hydroxyl-radical-induced cleavage of U2 snRNA implied that the branch-point is oriented away from U+10 (Figure 7B). To allow cleavage of U2 snRNA, and at the same time to maintain proximity of the branch-point to U74, would require the complete U2/branch-point helix to lie between U+10 and U74. This possibility, however, can be excluded by the data, which show that U74 is in the direct ‘line of sight' of U+10. Our data therefore suggest that a structural, rather than catalytic, role should be considered for this nucleotide. For example, a magnesium ion bound to U74 could help tether the intron part of the 5′ss helix to U6 snRNA. This could be important for maintaining the overall fold of the spliceosome's catalytic core. Consistent with a structural role, yeast U80 was found crosslinked in spliceosomes to an intron position 209 nucleotides away from the 5′ss (Ryan et al, 2004).
It is interesting to compare the situation at U74 with recent data from self-splicing group II introns, which are mechanistically related to spliceosomes (Villa et al, 2002). U6-ISL is functionally equivalent to domain V of these introns (Shukla and Padgett, 2002), with C839 being the homologue of the magnesium-binding U80 of yeast. Crosslinking studies revealed a close contact between C839 and intron nucleotides 4–6 (de Lencastre et al, 2005). This makes a proximity of C839 to the 5′ss improbable and, thus, a catalytic role for C839 unlikely. More recently, an NMR study of the closely related domain V RNA from Pylaiella littoralis, combined with detailed magnesium titration experiments, revealed two additional magnesium-binding regions (Seetharaman et al, 2006). By comparison of electrostatic surface potentials between domain V and the U6-ISL structure, the homologous magnesium-binding sites of yeast U6-ISL were identified as the penta-loop closing the stem, the internal bulge with the U80 nucleotide, and the AGC triad of the lower stem (see Figure 1). Thus, at least two further candidate regions, in addition to U80, may potentially be involved in coordinating essential magnesium ions. Further, the distance between neighbouring sites of about 20 Å would preclude a simultaneous involvement of any two of them in a catalytic scheme.
Taken together, our data suggest that U74 is placed in a structurally, rather than catalytically, important position in the activated spliceosomes that we investigated. However, we cannot exclude the possibility that rearrangements placing U74 in a position favourable for participation in catalysis occur.
In our stalled C complexes, we find the situation at the 5′ss/U6 snRNA helix essentially unchanged when compared with the activated spliceosome. This suggests that the suspected rearrangement for the second step of splicing does not involve this structural element. In contrast, after the first step of splicing, nucleotides U74 to A76 of U6-ISL are no longer accessible to hydroxyl radicals generated from Fe-BABE at U+10 of the intron, whereas C62, C63 and C77 of U6 snRNA remain accessible. This loss of accessibility could be due to an interaction of proteins with U6-ISL just before the second step of splicing, at which time a number of second step-splicing factors join the spliceosome (Umen and Guthrie, 1995; Schwer and Gross, 1998). Alternatively, it could also reflect a rearrangement within the spliceosome, consistent with the idea that the active site is remodelled just before the second step (Moore and Sharp, 1993; Konarska et al, 2006). To reconcile the different secondary-structure models of the spliceosome, it has been proposed that a four-way junction fold of U2/U6 snRNA is required for the first step, whereas the three-way junction fold is needed for the second step (see Discussion in Sun and Manley, 1995; Sashital et al, 2004). A transition from one type of interaction to another could allow a repositioning of catalytic nucleotides close to the 5′ and 3′ss. As nucleotides of the U6-ISL are still in close proximity to nucleotide U+10 of the intron after step one, a dramatic repositioning of the U6-ISL with respect to the 5′ end of the intron does not appear to occur at this stage. The observed change in the U6-ISL cleavage pattern could possibly arise from a reorientation of the 5′ss helix with respect to U6-ISL. In this event, some nucleotides of U6-ISL would still remain oriented towards the 5′ss helix.
The data presented here demonstrate that hydroxyl-radical probing using tethered Fe-BABE is a valuable tool for detecting RNA proximity relationships at different steps of the splicing cycle. Future work will include introducing Fe-BABE at other positions of the pre-mRNA, and also into the snRNAs or protein, and searching for RNA–RNA proximities in spliceosomes captured at different steps of the spliceosomal cycle. Such studies will provide us with additional constraints for modelling the RNA network of the spliceosome, and will provide further insight into the precise timing of the spliceosome's rearrangements and the catalytic mechanism of splicing.
Materials and methods
Pre-mRNA preparation and Fe-BABE modification
Oligoribonucleotides CCUCCGAACG∣GUAAGAGCCUA and CCUCCGAACG∣GUAAGAGCC(dU*)A were purchased from RNA-Tec, Belgium (the vertical separator denotes the 5′ exon/intron boundary, and dU* is the modified nucleotide 5-acrylamido-deoxyuridine). Preparation of site-specifically modified pre-mRNA was performed as described earlier (Rhode et al, 2003). Briefly, the flanking 5′ and 3′ fragments were generated from MINX pre-mRNA by DNA enzyme digestion (Santoro and Joyce, 1997). Then, 240 pmol each of oligoribonucleotide and 5′ fragment, and 120 pmol of γ-[32P] 3′ fragment, were hybridised to a splint oligodeoxyribonucleotide and ligated with 8 μl T4 DNA ligase (NEB, 2000 U/μl; Moore and Sharp, 1992). This resulted in ∼20 pmol of ligated pre-mRNA. MINX pre-mRNA containing a single AG to GG mutation at the 3′ss was prepared using a site-directed mutagenesis kit (Stratagene). Modification of pre-mRNA with SPDP (N-succinimidyl-3-[2-pyridyld ithio]propionate, Pierce) and subsequent DTT cleavage were performed as described by Cohen and Cech (1997). Fe-BABE was prepared from aminobenzyl-EDTA (Dojindo, Japan; DeRiemer and Meares, 1979). Reaction with the SH was as described (Newcomb and Noller, 1999). Modified pre-mRNA was separated from Fe-BABE by a G50 spin column purification and EtOH precipitation of Fe-BABE-modified pre-mRNA.
Pre-mRNA splicing and complex isolation
Except where otherwise indicated, spliceosomal complexes were prepared by incubation of 2 pmol of pre-mRNA with 40% nuclear extract (Dignam et al, 1983), 1.3 mM ATP, 27 mM creatine phosphate and 2.4 mM MgCl2 in a final volume of 100 μl for different times at 30°C. After incubation, heparin was added to a final concentration of 0.125 mg/ml and the reaction mixtures were applied to linear 10–30% glycerol gradients (1.5 ml, gradient buffer containing 100 mM NaCl, 1.5 mM MgCl2, 0.1 mM EDTA and 20 mM HEPES-KOH, pH 7.9). Centrifugation with a Sorvall S55-S rotor was performed at 55 000 r.p.m. for 105 min at 4°C, and gradients were fractionated into thirteen 110-μl aliquots.
Affinity purification of activated spliceosomes (complex B*) was performed as described (Makarov et al, 2002). Briefly, 8.5 pmol Fe-BABE pre-mRNA was incubated with 40% nuclear extract in a final volume of 800 μl for 14 min under splicing conditions, and the reaction was stopped by adding heparin to a final concentration of 0.5 mg/ml. Immunoprecipitation of activated spliceosomes was performed for 2 h at 4°C with 150 μg anti-SKIP antibody coupled to protein A-Sepharose. Complexes were eluted with cognate SKIP peptide. The eluted material (∼600 μl) contained ∼0.9 pmol B* complex.
Hydroxyl-radical cleavage in isolated complexes
For induction of the hydroxyl-radical reaction, 33 μl of the peak fractions from the gradient or 44 μl of eluted complex B* were diluted to 2 ml with buffer containing 50 mM cacodylic-acid-KOH (pH 7.0), 1.5 mM MgCl2 and 100 mM KCl on ice. A measure of 8 μl ascorbic acid (500 mM) was added and the generation of hydroxyl radicals was initiated with 25 μl H2O2 (0.4% v/v). Dilution to a glycerol concentration well below 0.5% was necessary as glycerol functions as a scavenger of hydroxyl radicals (Tullius and Dombrowski, 1986). The integrity of 50S spliceosomal complexes after dilution was confirmed by glycerol-gradient centrifugation (not shown). The cleavage reaction was allowed to proceed for 10 min on ice and was then stopped by addition of glycerol to a final concentration of 2.5% (v/v). Proteins were removed by digestion with proteinase K and the RNA was recovered. Control reaction mixtures were digested directly with proteinase K.
Primer extension
Primer extension was performed as described (Hartmuth et al, 1999). For the different RNAs, the following oligonucleotide primers (from MWG Biotech) were used (complementary nucleotides are in brackets): MINX intron: GCTTGGGCTGCAGGTAAC (100–83); MINX exon 2: TCTGGAAAGACCGCGAAG (216–199); U6: ATATGGAACGCTTC (103–90); U2: CTCGGATAGAGGACGTATCAG (81–61); U5: GCAAGGCTCAAAAAATTGGGT (103–83). Radioactively labelled pre-mRNA was removed after reverse transcription by adding NaOH to a final concentration of 20 mM and incubating at 60°C for 1 h.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Acknowledgments
We thank E Makarov for helpful discussions and, E Makarov and B Akyildiz for providing affinity-purified anti-SKIP antibody. We also thank CL Will, P Fabrizio and B Kastner for helpful comments on the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (Lu294/12-2) and the Fonds der Chemischen Industrie to RL.
References
- Brow DA (2002) Allosteric cascade of spliceosome activation. Annu Rev Genet 36: 333–360 [DOI] [PubMed] [Google Scholar]
- Burge CB, Tuschl T, Sharp PA (1999) Splicing of precursors to mRNAs by the spliceosomes. In The RNA World, Gesteland RF, Cech T, Atkins JF (eds), pp 525–560. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC (2003) The Prp19p-associated complex in spliceosome activation. Science 302: 279–282 [DOI] [PubMed] [Google Scholar]
- Cohen SB, Cech TR (1997) Dynamics of thermal motions within a large catalytic RNA investigated by cross-linking with thiol-disulfide interchange. J Am Chem Soc 119: 6259–6268 [Google Scholar]
- Culver GM, Noller HF (2000) Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol 318: 461–475 [DOI] [PubMed] [Google Scholar]
- de Lencastre A, Hamill S, Pyle AM (2005) A single active-site region for a group II intron. Nat Struct Mol Biol 12: 626–627 [DOI] [PubMed] [Google Scholar]
- DeRiemer LH, Meares CF (1979) BLEDTA: tumor localization by a bleomycin analogue containing a metal-chelating group. J Med Chem 22: 1019–1023 [DOI] [PubMed] [Google Scholar]
- DeRose VJ (2003) Metal ion binding to catalytic RNA molecules. Curr Opin Struct Biol 13: 317–324 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Abelson J (1992) Thiophosphates in yeast U6 snRNA specifically affect pre-mRNA splicing in vitro. Nucleic Acids Res 20: 3659–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendewey D, Keller W (1985) Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell 42: 355–367 [DOI] [PubMed] [Google Scholar]
- Gordon PM, Sontheimer EJ, Piccirilli JA (2000) Metal ion catalysis during the exon-ligation step of nuclear pre-mRNA splicing: extending the parallels between the spliceosome and group II introns. RNA 6: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Patton JG, Reed R (1994) A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J 13: 3356–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Dervan PB (1994) Visualization of RNA tertiary structure by RNA-EDTA.Fe(II) autocleavage: analysis of tRNA(Phe) with uridine-EDTA.Fe(II) at position 47. Proc Natl Acad Sci USA 91: 4955–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmuth K, Raker VA, Huber J, Branlant C, Lührmann R (1999) An unusual chemical reactivity of Sm site adenosines strongly correlates with proper assembly of core U snRNP particles. J Mol Biol 285: 133–147 [DOI] [PubMed] [Google Scholar]
- Hilliker AK, Staley JP (2004) Multiple functions for the invariant AGC triad of U6 snRNA. RNA 10: 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S, Noller HF (2000) Directed hydroxyl radical probing using iron(II) tethered to RNA. Methods Enzymol 318: 175–190 [DOI] [PubMed] [Google Scholar]
- Joseph S, Whirl ML, Kondo D, Noller HF, Altman RB (2000) Calculation of the relative geometry of tRNAs in the ribosome from directed hydroxyl-radical probing data. RNA 6: 220–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandels-Lewis S, Séraphin B (1993) Involvement of U6 snRNA in 5′ splice site selection. Science 262: 2035–2039 [DOI] [PubMed] [Google Scholar]
- Kent OA, MacMillan AM (2002) Early organization of pre-mRNA during spliceosome assembly. Nat Struct Biol 9: 576–581 [DOI] [PubMed] [Google Scholar]
- Kim CH, Abelson J (1996) Site-specific crosslinks of yeast U6 snRNA to the pre-mRNA near the 5′ splice site. RNA 2: 995–1010 [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Vilardell J, Query CC (2006) Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell 21: 543–553 [DOI] [PubMed] [Google Scholar]
- Lamond AI, Konarska MM, Grabowski PJ, Sharp PA (1988) Spliceosome assembly involves the binding and release of U4 small nuclear ribonucleoprotein. Proc Natl Acad Sci USA 85: 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C (1992) A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817 [DOI] [PubMed] [Google Scholar]
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R (2002) Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298: 2205–2208 [DOI] [PubMed] [Google Scholar]
- Makarova OV, Makarov EM, Urlaub H, Will CL, Gentzel M, Wilm M, Lührmann R (2004) A subset of human 35S U5 proteins, including Prp19, function prior to catalytic step 1 of splicing. EMBO J 23: 2381–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell TS, Steitz JA (2001) Proximity of the invariant loop of U5 snRNA to the second intron residue during pre-mRNA splicing. EMBO J 20: 3577–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS, Abelson J (1992) Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell 71: 819–831 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA (1992) Site-specific modification of pre-mRNA: the 2′-hydroxyl groups at the splice sites. Science 256: 992–997 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA (1993) Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature 365: 364–368 [DOI] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL (2002) Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struct Biol 9: 958–965 [DOI] [PubMed] [Google Scholar]
- Newcomb LF, Noller HF (1999) Directed hydroxyl radical probing of 16S ribosomal RNA in the ribosome: spatial proximity of RNA elements of the 3′ and 5′ domains. RNA 5: 849–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen TW (1998) RNA–RNA interactions in nuclear pre-mRNA splicing. In RNA Structure and Function, Simons RW, Grunberg-Manago M (eds), pp 279–307. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Nilsen TW (2000) The case for an RNA enzyme. Nature 408: 782–783 [DOI] [PubMed] [Google Scholar]
- Rhode BM, Hartmuth K, Urlaub H, Lührmann R (2003) Analysis of site-specific protein-RNA cross-links in isolated RNP complexes, combining affinity selection and mass spectrometry. RNA 9: 1542–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DE, Kim CH, Murray JB, Adams CJ, Stockley PG, Abelson J (2004) New tertiary constraints between the RNA components of active yeast spliceosomes: a photo-crosslinking study. RNA 10: 1251–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro SW, Joyce GF (1997) A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci USA 94: 4262–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE (2004) U2–U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat Struct Mol Biol 11: 1237–1242 [DOI] [PubMed] [Google Scholar]
- Schwer B, Gross CH (1998) Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J 17: 2086–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman M, Eldho NV, Padgett RA, Dayie KT (2006) Structure of a self-splicing group II intron catalytic effector domain 5: parallels with spliceosomal U6 RNA. RNA 12: 235–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla GC, Padgett RA (2002) A catalytically active group II intron domain 5 can function in the U12-dependent spliceosome. Mol Cell 9: 1145–1150 [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ, Steitz JA (1993) The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262: 1989–1996 [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ, Sun S, Piccirilli JA (1997) Metal ion catalysis during splicing of premessenger RNA. Nature 388: 801–805 [DOI] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA (1993) A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA 90: 6498–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JS, Manley JL (1995) A novel U2–U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev 9: 843–854 [DOI] [PubMed] [Google Scholar]
- Tullius TD, Dombrowski BA (1986) Hydroxyl radical ‘footprinting': high-resolution information about DNA–protein contacts and application to λ repressor and Cro protein. Proc Natl Acad Sci USA 83: 5469–5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Guthrie C (1995) The second catalytic step of pre-mRNA splicing. RNA 1: 869–885 [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S, Manley JL (2001) Splicing-related catalysis by protein-free snRNAs. Nature 413: 701–707 [DOI] [PubMed] [Google Scholar]
- Villa T, Pleiss JA, Guthrie C (2002) Spliceosomal snRNAs: Mg(2+)-dependent chemistry at the catalytic core? Cell 109: 149–152 [DOI] [PubMed] [Google Scholar]
- Wang JF, Cech TR (1992) Tertiary structure around the guanosine-binding site of the Tetrahymena ribozyme. Science 256: 526–529 [DOI] [PubMed] [Google Scholar]
- Wassarman DA, Steitz JA (1992) Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science 257: 1918–1925 [DOI] [PubMed] [Google Scholar]
- Westhof E, Dumas P, Moras D (1985) Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol 184: 119–145 [DOI] [PubMed] [Google Scholar]
- Wyatt JR, Sontheimer EJ, Steitz JA (1992) Site-specific cross-linking of mammalian U5 snRNP to the 5′ splice site before the first step of pre-mRNA splicing. Genes Dev 6: 2542–2553 [DOI] [PubMed] [Google Scholar]
- Yean SL, Wuenschell G, Termini J, Lin RJ (2000) Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408: 881–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Maroney PA, Darzynkiwicz E, Nilsen TW (1995) U6 snRNA function in nuclear pre-mRNA splicing: a phosphorothioate interference analysis of the U6 phosphate backbone. RNA 1: 46–54 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2


