Abstract
The zinc-finger transcription factor Krox20 constitutes a key regulator of hindbrain development, essential for the formation and specification of rhombomeres (r) 3 and 5. It is in particular responsible for the respective activation and repression of odd- and even-numbered rhombomere-specific genes, which include Hox genes. In this study, we have identified PIASxβ as a novel direct interactor of Krox20. In addition, we found that PIASxβ is able to activate the r4-specific gene Hoxb1. Binding of Krox20 prevents this activation, providing a molecular basis for the repression of Hoxb1 by Krox20. The same domain in the Krox20 protein, the zinc-fingers, is involved in DNA binding for transcriptional activation and in interaction with PIASxβ for transcriptional repression, although the actual precise contacts are different. Our findings add an additional level in the complexity of Hox gene regulation and provide an example of how a single regulator can coordinate the activation and repression of a set of genes by very different mechanisms, acting as a molecular switch to specify cell identity and fate.
Keywords: hindbrain segmentation, Hox gene, pattern formation, transcription control
Introduction
The development of the vertebrate hindbrain involves a transient segmentation process along the anterior–posterior (AP) axis leading to the formation of 7–8 transversal morphological units, called rhombomeres (r) (reviewed in Lumsden and Krumlauf, 1996). The rhombomeres behave as compartments, constituting units of cell lineage restriction (Fraser et al, 1990) and domains of specific gene expression (reviewed in Lumsden and Krumlauf, 1996). This subdivision presages the metameric pattern of neuronal specification in the hindbrain (Clarke et al, 1998), underlies the pathways of neural crest cell migration into the branchial arches and participates in its patterning (Birgbauer et al, 1995; Trainor and Krumlauf, 2000), thus playing an essential role in craniofacial morphogenesis.
Numerous genes have been implicated at different levels of the segmentation process, including the initial formation of segmental territories (Frohman et al, 1993; Schneider-Maunoury et al, 1993; Barrow et al, 2000; Choe and Sagerstrom, 2004; McNulty et al, 2005 and references therein), the specification of their AP identities (Rijli et al, 1993; Studer et al, 1996; Seitanidou et al, 1997; Rossel and Capecchi, 1999), the stabilization of the pattern by restriction of cell intermingling between adjacent rhombomeres (reviewed in Pasini and Wilkinson, 2002) and the development of specific cell populations at boundaries (Cheng et al, 2004). Segment formation and specification are highly intricated processes in the hindbrain (Gavalas et al, 1998; Rossel and Capecchi, 1999; Voiculescu et al, 2001), relying on a complex network of transcription factors. Hox proteins play a key role in this network, which receives inputs from several signalling cascades, including the FGF and retinoid pathways, acting both intrinsically and extrinsically to the neuroepithelium (Marin and Charnay, 2000; Dupe and Lumsden, 2001; Walshe et al, 2002; Serpente et al, 2005). Deciphering Hox gene regulation and function therefore appears as an essential step in our understanding of the molecular mechanisms controlling hindbrain segmentation.
A key regulator of Hox genes in the hindbrain is Krox20, which encodes a zinc-finger transcription factor expressed in r3 and r5 and is essential for the development and specification of these segments (Schneider-Maunoury et al, 1993; Seitanidou et al, 1997; Voiculescu et al, 2001). Krox20 has been shown to directly activate the transcription of several Hox genes (Hoxb2, Hoxa2 and Hoxb3) by binding to nearby transcriptional enhancers (Sham et al, 1993; Nonchev et al, 1996; Manzanares et al, 2002). Krox20 was also shown to repress the expression of another Hox gene, Hoxb1, although the mechanisms underlying this second type of transcriptional regulation mediated by Krox20 have not been elucidated (Seitanidou et al, 1997; Giudicelli et al, 2001; Voiculescu et al, 2001). A likely possibility is that Krox20 exerts positive and negative activities on transcription by interacting with different cofactors. Several Krox20 interactors have been identified, including NAB1 and NAB2, which modulate its transcriptional activity (Russo et al, 1995; Svaren et al, 1996). So far, however, these factors cannot account for the dual transcriptional activity of Krox20 in the hindbrain.
In the present work, we have performed a two-hybrid screening designed to identify novel Krox20 interactors that might be involved in the different aspects of Hox gene regulation by Krox20. Among the positive clones, we have identified PIASxβ. PIAS (protein inactivator of activated STAT) proteins were first reported as inhibitors of the DNA-binding and transcription activation by STAT (signal transducer and activator of transcription) (Shuai, 2000). The PIAS mammalian family includes at least five members: PIAS1, PIAS3, PIASx (with two isoforms, PIASxα or ARIP3, and PIASxβ or Miz1) and PIASγ (Shuai, 2000; Johnson, 2004). In addition to their role in the modulation of transcription, they have been shown to carry an E3 ligase activity for the small ubiquitin-related modifier proteins (SUMO) (Johnson, 2004). We have found that PIASxβ is expressed in the developing neural tube, that its overexpression leads to ectopic Hoxb1 activation and that a specific PIASxβ deletion mutant represses Hoxb1 expression, presumably acting as a dominant-negative molecule. Our findings identify PIASxβ as a novel positive regulator of Hoxb1 in the hindbrain. We also show that Krox20 antagonizes this novel activity of PIASxβ and that its ability to repress Hoxb1 does not require its DNA-binding activity, but its capacity to interact with PIASxβ. Together, these data reveal a novel mechanism of control of Hox gene expression by Krox20, in which repression is mediated by antagonizing a positive regulatory factor.
Results
Krox20 interacts with PIASxβ
In an attempt to identify factor(s) interacting with Krox20 during hindbrain development, we performed a yeast two-hybrid screening. As full-length Krox20 fused to the Gal4 DNA-binding domain was able alone to activate the transcription of a β-galactosidase reporter linked to Gal4 binding sites (data not shown), we decided to use as bait a Krox20 deletion (Krox20(184–470)), lacking the N-terminal part containing the known transcriptional activation domains (Figure 1A; Vesque and Charnay, 1992). Yeast cells containing the expression plasmid for the fusion protein between Krox20(184–470) and the Gal4 DNA-binding domain were transfected with a library of 8.5 dpc mouse embryo cDNAs fused to the Gal4 activation domain coding sequence. Approximately 2.5 × 106 transformants were screened on the basis of transcriptional activation of survival factors through Gal4 binding sites (see Materials and methods). Selected clones were further checked for their level of expression of the β-galactosidase reporter mentioned above. A total of 26 clones were finally retained at this stage and nucleotide sequence analysis of the cDNAs revealed that they corresponded to 12 different genes. Two of them corresponded to already known Krox20 cofactors, Nab1 and Nab2 (Russo et al, 1995; Svaren et al, 1996) (two and six clones, respectively), indicating that the two-hybrid selection was performed appropriately. The 10 newly identified putative Krox20 partners included PIASxβ (represented by a single clone), which we selected for further analysis on the basis of preliminary investigations (data not shown).
Figure 1.
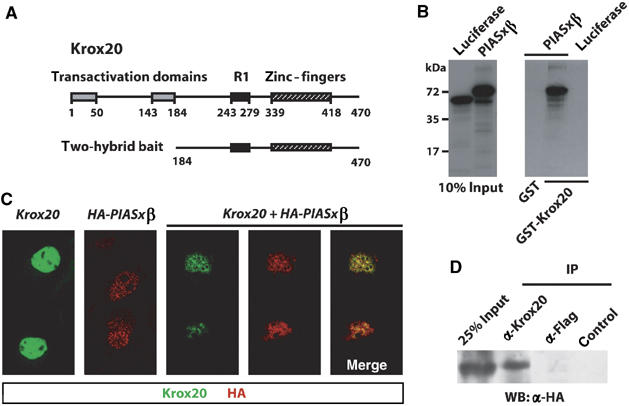
Krox20 interacts with PIASxβ. (A) Schematic structure of the Krox20 protein showing the location of the transactivation domains, NAB interaction domain (R1) and DNA-binding domain (zinc-fingers). Numbers below the line indicate amino-acid positions. The 287 C-terminal amino acids were used as bait in the two-hybrid screening. (B) In vitro binding of Krox20 and PIASxβ. In vitro-translated, 35S-labelled luciferase and PIASxβ were resolved by SDS–PAGE and detected by fluorography. Left: 10% of the input was directly deposited on gel; right: luciferase or PIASxβ retained on GST or GST-Krox20 beads was analysed. PIASxβ specifically binds to GST-Krox20. (C) Colocalization of Krox20 and PIASxβ in the cell nucleus. COS7 cells were transfected with expression vectors encoding Krox20 and/or HA-PIASxβ and 36 h later the proteins were revealed by immunofluorescence analysis (Krox20 is labelled with FITC (green) and HA-PIASxβ with Cy3 (red)). When transfected alone, Krox20 appeared homogenously distributed within the nucleus, whereas Piasxβ localized to nuclear bodies. Cotransfection led to a re-distribution of Krox20, with colocalization of both proteins. (D) Co-immunoprecipitation of Krox20 and PIASxβ. COS7 cells were cotransfected with expression vectors encoding Krox20 and HA-PIASxβ. Cell lysates were subjected to immunoprecipitation with antibodies directed against Krox20 or Flag, or with no antibody (control) and the precipitates were subsequently analysed by Western blotting using an anti-HA antibody.
Comparison of the selected PIASxβ clone with available mouse ESTs revealed that it lacked the sequence encoding the three N-terminal amino acids and the cDNA was therefore completed. In a first series of experiments, we attempted to confirm the Krox20–PIASxβ interaction by GST pull-down experiments. For this purpose, we prepared GST and GST-Krox20 fusion proteins that were incubated with radiolabelled, in vitro-translated PIASxβ or luciferase as a control. As shown in Figure 1B, GST-Krox20, but not GST alone, specifically retained PIASxβ. These data establish that Krox20 and PIASxβ can interact in vitro and that the interaction is direct.
To determine whether Krox20 and PIASxβ also interact in mammalian cells, we transfected COS7 cells with expression vectors for Krox20 and an N-terminally HA-tagged version of PIASxβ, alone or in combination. The cellular localization of the proteins was established by immunofluorescence performed with antibodies directed against Krox20 and the HA epitope (Figure 1C). When the expression vectors were transfected alone, Krox20 appeared distributed relatively homogeneously within the nucleus, whereas PIASxβ presented a punctuated nuclear pattern, consistent with its previous localization to nuclear bodies (Kotaja et al, 2002). When both expression vectors were cotransfected, the nuclear distribution of Krox20 was modified towards a punctuated pattern, largely overlapping with that of PIASxβ (Figure 1C). These data suggest that Krox20 and PIASxβ colocalize when they are coexpressed, owing to sequestering of Krox20 by PIASxβ in nuclear bodies. We also performed immunoprecipitation analyses on cotransfected COS7 cells, which revealed the presence of PIASxβ among the proteins precipitated with an antibody directed against Krox20 (Figure 1D). Together, these data demonstrate that Krox20 and PIASxβ can interact in vivo.
Identification of the PIASxβ/Krox20 interaction domains
Several domains have been predicted within PIASxβ, on the basis of amino-acid sequence analysis and comparison with protein sequence databases (Figure 2A): an N-terminal SAP domain, containing a putative DNA-binding bi-helical motif (Aravind and Koonin, 2000); a proline-rich, putative SH3-binding domain (Wu et al, 1997); a core region containing an SP-RING, related to RING zinc-fingers present in many E3 ubiquitin ligases and known to be required for Ubc9 binding and SUMO ligase activity of PIAS family members (Kahyo et al, 2001; Sachdev et al, 2001); a SIM domain for non-covalent SUMO binding (Minty et al, 2000); a nuclear localization signal (NLS); a C-terminal serine/threonine-rich domain, which has been shown to be required for transcription activation in Gal4 fusions in yeast (Wu et al, 1997).
Figure 2.
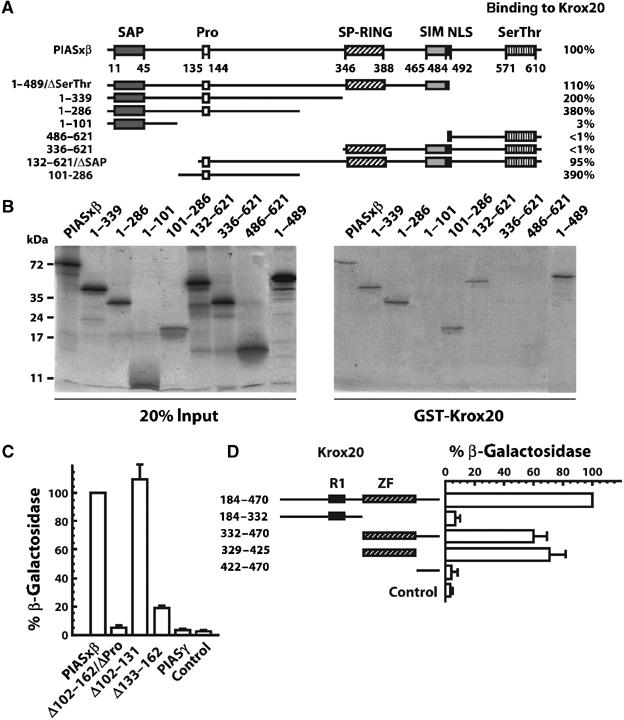
Identification of the domains involved in PIASxβ/Krox20 interaction. (A) Schematic representation of the PIASxβ structural domains and of the deletion mutants. PIASxβ contains SAP, proline-rich (Pro), SP-RING, SIM domains, an NLS and a C-terminal serine/threonine-rich (Ser/Thr) domain. Numbers below the line indicate amino-acid positions. The amino-acid positions of the extremities of truncated PIASxβ proteins are indicated on the left. The efficiency of the binding to Krox20, indicated in percentage on the right, was estimated from the recovery of each deleted protein after GST-Krox20 pull-down assay (see part B), normalized with wild-type PIASxβ. The PIASxβ interacting region was localized between amino acids 132 and 286. (B) GST-Krox20 pull-down assay performed on deleted PIASxβ. In vitro-translated, 35S-labelled PIASxβ truncated proteins were resolved by SDS–PAGE and detected by fluorography. Left: deposit of 20% of the input proteins; right: PIASxβ proteins recovered after binding to immobilized GST-Krox20 protein. (C) Quantitative yeast two-hybrid assay performed on PIASxβ deletions. PIASγ and a series of PIASxβ deletions fused to the GAL4 activation domain were evaluated for their capacity to bind to a Krox20-Gal4 DNA-binding domain bait by measuring the expression of a lacZ reporter. The β-galactosidase activity was normalized by the level obtained with the wild-type PIASxβ construct. The PIASxβ proline-rich domain appears necessary for the interaction with Krox20. Control corresponds to no PIASxβ insert. (D) Identification of the Krox20 domain required for the interaction with PIASxβ, using the quantitative yeast two-hybrid assay. In this case, a series of Krox20 deletions fused to the Gal4 DNA-binding domain were confronted to the wild-type PIASxβ-Gal4 activation domain fusion and the interaction was recorded by measuring the expression of the lacZ reporter gene. The β-galactosidase activity was normalized by the level obtained with the 184–470 Krox20 deletion construct. The region of interaction with PIASxβ was localized within the zinc-finger domain. Control corresponds to no Krox20 insert.
To determine which region(s) of PIASxβ are important for physical interaction with Krox20, we first generated a series of external deletions. The interactions with Krox20 were then analysed by GST pull-down as described above (Figure 2A). Quantitative analysis of the retention of deleted proteins as compared to full-length PIASxβ indicated that amino acids upstream to position 132 or downstream to position 286 were not essential for efficient interaction with Krox20 (Figure 2A and B). Consistently, a region corresponding to amino-acid sequence 101–286 and encompassing the proline-rich region was sufficient for binding (Figure 2A and B). To confirm these data and localize the region of interaction more precisely, we analysed internal PIASxβ deletion mutants in the yeast two-hybrid system, using the Gal4 site-driven β-galactosidase reporter. Consistent with the external deletion analysis, deletion of amino acids 102–162 (ΔPro) completely abolished the interaction with Krox20 (Figure 2C). The interface domain was more precisely localized to the 133–162 region, which contains the proline-rich domain, as its deletion severely impaired the interaction, whereas deletion of region 102–131 did not affect binding (Figure 2C). In agreement with these data, another PIAS family member, PIASγ, which shows a general high sequence similarity to PIASxβ but totally differs from it at the level of the proline-rich domain, did not interact with Krox20 (Figure 2C).
The region of the Krox20 protein required for the interaction with PIASxβ was subsequently localized in a similar manner, using a series of external deletions and monitoring β-galactosidase reporter activity in the yeast two-hybrid system. We found that the zinc-finger region, corresponding to the DNA-binding domain, was necessary and sufficient for effective interaction with PIASxβ (Figure 2D). Attempts to further delimit the region of interaction were unsuccessful, as elimination of the first or the third zinc-finger led to loss of Krox20/PIASxβ interaction (data not shown). This suggests that the domain of interaction with PIASxβ overlaps with the entire Krox20 DNA-binding domain.
PIASxβ can antagonize Krox20 transcriptional activity
The physical interaction observed between Krox20 and PIASxβ, both in vitro and in vivo, raised the possibility of a functional interaction between the two proteins in the developing hindbrain. To investigate this possibility, we turned to the chick embryo in which gain-of-function experiments can easily be performed by in ovo electroporation. We first analysed the expression of PIASxβ by in situ hybridization to determine whether Krox20 and PIASxβ are coexpressed during hindbrain segmentation. We found that PIASxβ expression is induced in the anterior part of the neural tube around the 3–4 somite stage (ss) and that this expression rapidly extends caudally to form an AP-decreasing gradient at the 5–6 ss (Figure 3A and data not shown). Krox20 is expressed in r3 from 4 to 5 ss and in r5 from 7 to 8 ss (Irving et al, 1996; Giudicelli et al, 2001) and therefore the two genes are coexpressed in these rhombomeres. Around the 12–16 ss, PIASxβ is downregulated posterior to r2, except at the level of r4 (Figure 3B). Around the 24–30 ss stage, PIASxβ expression appeared generally decreased in the neural tube (data not shown).
Figure 3.
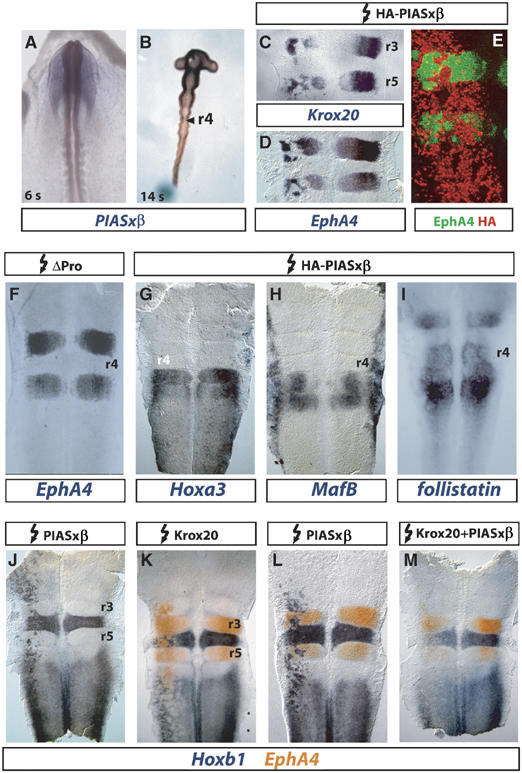
Ectopic PIASxβ expression antagonizes Krox20 activity and leads to Hoxb1 activation. (A, B) Analysis of PIASxβ expression in chick embryos. Whole-mount in situ hybridization on 6 ss (A) or 14 ss (B) embryos. At 6 ss, PIASxβ is expressed in the neural tube in a decreasing AP gradient. At 14 ss, higher levels of expression are maintained in the anterior CNS up to r2 and in r4. (C–M) Flat-mounted hindbrains from chick embryos electroporated between stages 3 and 8 ss with expression plasmids for the proteins indicated above. The embryos were collected 24 h after electroporation and expression of the markers indicated below was analysed by in situ hybridization (C, D, F–M) or immunochemistry (E). (C, D) Krox20 and EphA4 are repressed in r3 and r5. (E) Double immunohistochemistry with anti-EphA4 (green) and anti-HA (red) antibodies. EphA4-negative patches in r3 and r5 are positive for HA-PIASxβ. Only the left (electroporated) side of the embryo is shown. (F) ΔPro, which has lost the ability to interact with Krox20, does not repress EphA4. (G–I) Hoxa3, MafB and follistatin are not affected by PIASxβ. (J–M) Hoxb1 is ectopically activated by PIASxβ and this activity is antagonized by co-electroporated Krox20 (M). In r3 and r5, Hoxb1 expression domains coincide with the EphA4-negative patches (L). Electroporations were always performed on the left side.
We then investigated the consequences of PIASxβ ectopic expression in the developing hindbrain. The HA-tagged PIASxβ expression plasmid was unilaterally electroporated in the chick neural tube at the 4–8 ss. Electroporated embryos were recovered at 20 ss and we analysed the patterns of expression of different segmentally expressed genes by in situ hybridization, first focusing on genes that are directly, positively regulated by Krox20: Krox20 itself and EphA4 (Theil et al, 1998; Giudicelli et al, 2001). Krox20 and EphA4 expressions, which are restricted to r3 and r5, were severely affected on the electroporated side, with the appearance of large patches of non-expressing cells in these rhombomeres (Figure 3C and D), indicating that PIASxβ can repress these genes. Double immunolabelling experiments, designed to detect both the EphA4 protein and the electroporated, tagged PIASxβ, revealed that the loss of EphA4 in a cell is correlated with the presence of exogenous PIASxβ (Figure 3E), indicating that the repression occurs in a cell-autonomous manner. The formation of Krox20- or EphA4-negative patches could therefore be due to a phenomenon of segregation, as previously reported (Giudicelli et al, 2001). No repression of EphA4 expression was observed when the hindbrain was electroporated with the ΔPro mutant, which has lost the ability to interact with Krox20 (Figure 3F). This suggests that the repression activity of PIASxβ involves antagonizing Krox20-positive transcriptional activity. This latter possibility was reinforced by the observation that Hoxa2, another positive target of Krox20, was also repressed by PIASxβ (data not shown). In contrast, the expression patterns of two genes whose regulation is known to be independent of Krox20, Hoxa3 and MafB (Giudicelli et al, 2001; Manzanares et al, 2002), were not affected by ectopic PIASxβ expression (Figure 3G and H). Finally, taken together, our data indicate that PIASxβ appears capable of preventing the transcriptional activation of all the genes tested that are positively regulated by Krox20, presumably by directly antagonizing Krox20.
PIASxβ act as an activator of Hoxb1 expression
We next investigated whether PIASxβ could also interfere with the expression of genes that are downregulated by Krox20. The follistatin gene is expressed in r2 and r4–r6 around the 20 ss in the chick (Figure 3I), and is known to be repressed by Krox20 in r3 (Seitanidou et al, 1997). Its expression was not modified by PIASxβ electroporation (Figure 3I), suggesting that PIASxβ is not able to interfere with Krox20 repressive activity in this case.
Another gene subjected to repression by Krox20 is Hoxb1, a major determinant of r4 identity: in particular, we have previously demonstrated that ectopic Krox20 expression leads to Hoxb1 repression in r4, r6 and the spinal cord (Giudicelli et al, 2001) and that Krox20 loss-of-function mutation allows rostral extension of Hoxb1 expression in r3 (Voiculescu et al, 2001). Ectopic PIASxβ expression resulted in Hoxb1 activation outside of its territories of normal expression, an increase in its level of expression in r6 and the spinal cord and an enlargement of the r4 domain (Figure 3J and L). In r3 and r5, the domains of activation of Hoxb1 precisely corresponded to the patches of repression of EphA4 (Figure 3L and data not shown). Surprisingly, this ectopic activation of Hoxb1 expression was observed at least from the midbrain to the spinal cord and was therefore not restricted to the territories of Krox20 expression. This indicates that the capacity of PIASxβ to activate Hoxb1 is not dependent on Krox20. Nevertheless, co-electroporation of Krox20 and PIASxβ expression vectors indicated that the two proteins can antagonize each other: PIASxβ could prevent activation of EphA4 by Krox20 (compare Figure 3K and M) and reciprocally Krox20 could prevent activation of Hoxb1 by PIASxβ (compare Figure 3L and M).
In conclusion, we unexpectedly found that PIASxβ acts as an activator of Hoxb1 expression, independently of Krox20. As Hoxb1 is under Krox20 negative control and Krox20 can antagonize PIASxβ-inducing activity, this raises the possibility that Krox20 may actually repress Hoxb1 by antagonizing PIASxβ in vivo.
PIASxβ is involved in Hoxb1 activation
To further investigate the involvement of PIASxβ in the control of Hoxb1 expression, we generated a series of HA-tagged, external and internal deletion mutants that were tested by electroporation in the chick hindbrain (Figure 4A). In order to normalize our analysis, the relative amount of protein produced by these different constructs was estimated by anti-HA immunohistochemistry on electroporated embryos. No major variations were observed (data not shown).
Figure 4.
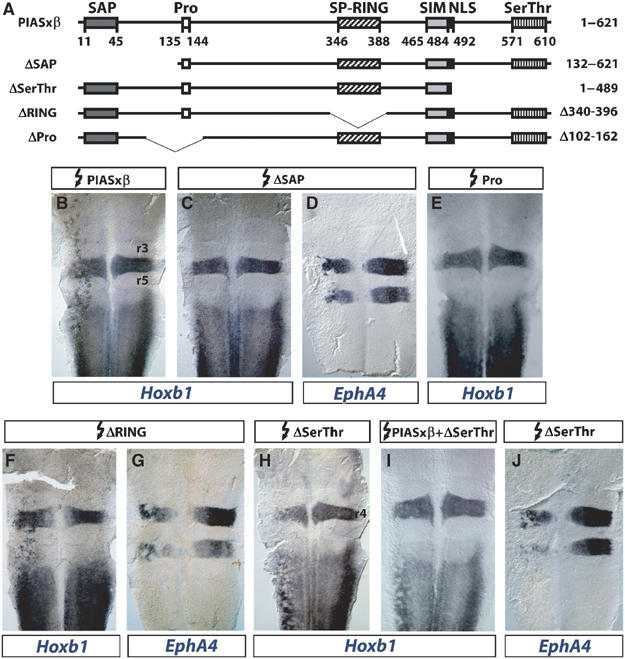
Identification of PIASxβ domains required for Hoxb1 activation. (A) Schematic representation of PIASxβ and deletion mutants (see Figure 2 for details). (B–J) Flat-mounted hindbrains from chick embryos electroporated on the left side with the constructs indicated above and in situ hybridized with the probes indicated below. Whereas deletion of the SP-RING domain does not affect the activity of PIASxβ (B, F, G), elimination of the SAP domain prevents activation of Hoxb1 without affecting the repression of EphA4 (C, D), and deletion of the proline-rich domain prevents activation of Hoxb1 (E). Deletion of the serine/threonine-rich domain does not affect EphA4 repression (J), but transforms PIASxβ into a repressor of Hoxb1 (H), able to antagonize the wild-type protein (I).
Deletion of the N-terminal SAP domain abrogated Hoxb1 activation but preserved EphA4 repression (Figure 4B–D). In contrast, the SP-RING domain was surprisingly not required for either activity (Figure 4F and G). Deletion of the proline-rich domain prevented Hoxb1 activation (Figure 4E). Finally, the deletion of the C-terminal serine/threonine-rich region turned out to be particularly interesting. This mutant had not only lost the capacity to activate Hoxb1, but it actually appeared to repress endogenous Hoxb1 expression (Figure 4H) and to antagonize wild-type PIASxβ in this respect (Figure 4I). The ΔSerThr mutant kept its capacity to repress EphA4 (Figure 4J), consistent with the maintenance of the interaction with Krox20. It did not interfere with the expression of MafB (data not shown), a gene that we have shown previously not to be affected by the ectopic expression of wild-type PIASxβ (Figure 3H). This latter observation indicates that the acquisition of an Hoxb1 repression capacity by the ΔSerThr mutant is specific and does not reflect a general repressive activity of the construct.
In conclusion, these data indicate that the SAP and proline-rich domains are required for Hoxb1 transcriptional activation, whereas the SP-RING domain is not. The specific repression of Hoxb1 by the ΔSerThr mutant suggests that this mutant acts as a dominant-negative molecule and competes with endogenous PIASxβ, supporting the notion that PIASxβ is involved in the normal transcriptional activation of Hoxb1.
Krox20 does not require DNA binding to repress Hoxb1
The data presented above suggest that the repression of Hoxb1 by Krox20 is mediated by the interaction of Krox20 with PIASxβ, an activator of Hoxb1. However, we have previously shown that, when a point mutation preventing DNA binding was introduced into the third zinc-finger of Krox20, corresponding to an arginine to tryptophan substitution (R409W) observed in a human peripheral myelinopathy (Warner et al, 1998, 1999), Hoxb1 repression by Krox20 was abolished (Giudicelli et al, 2001; Figure 5B). The latter experiment rather suggested that Krox20 repression of Hoxb1 required DNA binding by Krox20. Therefore, we have further examined the involvement of DNA binding in Krox20 repression of Hoxb1. For this purpose, we have compared the activities of the wild-type Krox20 protein on Hoxb1 and EphA4 expression with R409W and another human peripheral myelinopathy mutant, S382R/D383Y, which corresponds to serine and aspartic acid to arginine and tyrosine substitutions in the second zinc-finger (Warner et al, 1998) and is also defective in DNA binding (Warner et al, 1999).
Figure 5.
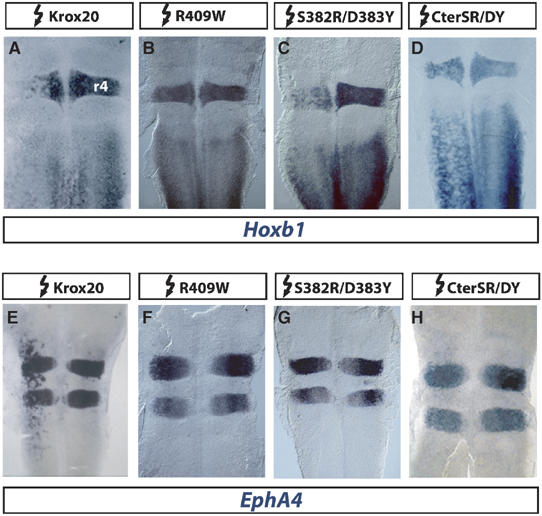
Hoxb1 repression by Krox20 does not require DNA binding. (A–H) Flat-mounted hindbrains from chick embryos electroporated on the left side with wild-type and mutant Krox20 constructs as indicated above and in situ hybridized with Hoxb1 or EphA4 probes. (A–D) Whereas the R409W mutation prevents repression of Hoxb1, S382R/D383Y and CterSR/DY conserve this activity, despite their loss of DNA binding. (E–H) In the three mutants, loss of DNA binding correlates with the inability to activate EphA4.
As indicated above, R409W has lost the ability to repress Hoxb1 (Figure 5A and B). It is also unable to induce ectopic EphA4 expression (Figure 5E and F), consistent with the fact that EphA4 has been shown to constitute a direct transcriptional target of Krox20 (Theil et al, 1998). S382R/D383Y presented a strikingly different behaviour: whereas it was unable to activate EphA4 (Figure 5G), it repressed Hoxb1 (Figure 5C). Furthermore, a deleted Krox20 protein containing only the 160 C-terminal amino acids, including the zinc-fingers, and carrying the double mutation S382RD383Y (CterSR/DY) was also able to repress Hoxb1 and unable to activate EphA4 (Figure 5D and H). Finally, these data were confirmed using co-electroporation of constructs carrying a lacZ reporter under the control of a β-globin minimal promoter and EphA4 and Hoxb1 cis-acting regulatory sequences (see Supplementary data). The effects on the reporter constructs were fully consistent with those observed on the endogenous EphA4 and Hoxb1 genes (Supplementary Figure 1).
In conclusion, this analysis clearly establishes that the DNA-binding activity of Krox20, although absolutely required for EphA4 induction, is not necessary to repress Hoxb1. This latter conclusion is in sharp contrast with what could have been deduced from the sole examination of the R409W mutant.
Hoxb1 repression by Krox20 correlates with binding to PIASxβ
As the two examined point mutants in Krox20 zinc-fingers differed drastically in their capacity to repress Hoxb1, we investigated whether this behaviour could be correlated with their ability to interact with PIASxβ. This latter property was estimated in the yeast two-hybrid system using the lacZ reporter driven by the PIASxβ-Gal4 hybrid as indicated above. We found that the R409W mutant was highly impaired in its activation of the reporter, whereas the behaviour of the S382R/D383Y mutant was close to that of the wild-type protein (Figure 6). As controls, we performed the same assay, but using a Par4-Gal4 hybrid as lacZ driver. Par4 is another interactor of the zinc-finger domain of Krox20 identified in the two-hybrid screening (data not shown). The two Krox20 mutants appeared to perform very similarly and slightly better than the wild type in this latter assay, indicating that the differences observed with the PIASxβ-Gal4 driver are not due to differences in the synthesis or stability of the two mutant proteins, but rather in the capacity to interact with PIASxβ (Figure 6).
Figure 6.
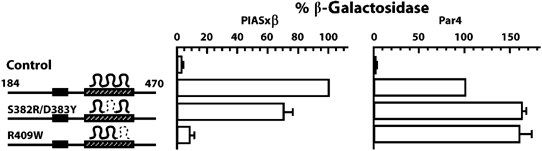
The ability of Krox20 to repress Hoxb1 correlates with its capacity to interact with PIASxβ. The 184–470 Krox20 deletions, in their wild-type version or carrying mutations in the DNA-binding domain, fused to the Gal4 DNA-binding domain were confronted to the Gal4 activation domain fused to PIASxβ or Par4 in the yeast two-hybrid system. The interaction was recorded by measuring the expression of the lacZ reporter gene. The β-galactosidase activities were normalized with the levels obtained with the wild-type construct. Mutant S382R/D383Y, which has retained Hoxb1 repression activity, has also maintained its capacity to interact with PIASxβ, whereas R409W has lost both activities. As a control, interaction with Par4 is not seriously affected in either mutant. Control corresponds to no Krox20 insert.
In conclusion, these results establish that the R409W is strongly hampered in its interaction with PIASxβ, whereas S382R/D383Y behaves similarly to the wild-type protein. This indicates that although DNA-binding and PIASxβ interaction domains overlap within the Krox20 protein, they are distinct and at least DNA binding can be abolished without seriously affecting PIASxβ binding. In addition, we observed that in these two mutants, the capacity of Krox20 to repress Hoxb1 correlates with its ability to interact with PIASxβ, supporting the idea that Krox20 represses Hoxb1 by antagonizing one of its activators, PIASxβ. This conclusion was further supported by the observation that co-electroporation with PIASxβ can release Krox20-mediated repression of Hoxb1, whereas this is not the case with ΔPro, which cannot interact with Krox20 (data not shown).
Discussion
In this study, we have identified PIASxβ as a novel interactor of Krox20, which plays an important role in hindbrain segmentation, regulating the expression of several Hox genes. We unexpectedly found that PIASxβ is able to activate Hoxb1 expression and that its interaction with Krox20 is likely to constitute the molecular basis of Hoxb1 repression by Krox20 in the hindbrain. Our findings reveal an additional level of complexity in the mechanisms controlling Hox gene regulation and raise the possibility that PIAS family members might be involved in novel and important developmental regulatory processes.
Direct interaction between Krox20 and PIASxβ
We have accumulated several lines of evidence supporting a direct biochemical and functional interaction between Krox20 and PIASxβ: (i) the existence of the interaction was initially established in a yeast two-hybrid assay, which also allowed, together with GST pull-down experiments, to define the interaction interfaces on each proteins; (ii) GST pull-down data also established that binding was direct (Figure 1B); (iii) the existence of an in vivo interaction was supported by the coexpression of the two genes in the developing hindbrain, by the subcellular relocalization of Krox20 upon forced expression of PIASxβ in mammalian cells (Figure 1C) and by co-immunoprecipitation of the two proteins (Figure 1D). Colocalization of Krox20 with the different PIASxβ deletion mutants (Supplementary Figure 2) adds support to this conclusion; (iv) the functionality of the interaction is supported by the crossregulatory activities of the two proteins.
The discovery of this novel interaction raises a number of interesting points. It first adds to the list of Krox20 interactors, illustrating the importance and the complexity of the regulatory activities of Krox20. As PIASxβ carries an E3 ligase activity, SUMOylation of Krox20 by PIASxβ might have been envisaged. However, several pieces of data are against this possibility: (i) Krox20 does not contain any SUMOylation consensus site; (ii) preliminary in vitro SUMOylation assays failed to reveal any SUMOylated form of Krox20 (data not shown); (iii) the deletion of the ΔRING domain is known to inactivate the E3 ligase activity and nevertheless it does not prevent PIASxβ from antagonizing Krox20 (Figure 4). The precise localization of the interaction interfaces between Krox20 and PIASxβ revealed a large overlap with the DNA-binding domain in Krox20, as both zinc-fingers 1 and 3 appear to be required. However, overlap does not mean identity, as we have found a mutation that abolishes DNA binding without significantly affecting PIASxβ binding (Figure 6). Such an overlap has implications at both structural and functional (binding to each target is likely to be exclusive) levels. In PIASxβ, the interaction domain has been shown to include a proline-rich region whose structure has not yet been established but resembles the SH3 domain-binding region of the GAP protein 3BP-1 (Wu et al, 1997).
Both Krox20 and PIASxβ belong to highly conserved multigene families. The interaction between Krox20 and PIASxβ raises the possibility of the existence of similar interactions between other members of the families. The zinc-fingers are extremely conserved between Krox20 and the three other members of the Krox20/Egr family (see O'Donovan et al, 1999 for a review), suggesting that these proteins are likely to bind PIAS as well. PIASxβ and PIASxα are splice variants that are identical in the binding domain. PIASxα should therefore also bind to Krox20. We showed that the sequence between positions 133 and 162 in PIASxβ is absolutely required for binding. This sequence is not present in PIASγ and accordingly this protein does not bind to Krox20 (Figure 2C). This sequence is also poorly conserved between PIASxβ and PIAS1 and PIAS3, with the exception of the hexapeptide Pro-Val-His-Pro-Asp-Val/Ile. Consistently, binding of Krox20 to PIAS3 is poor and interaction with PIAS1 is significant, but much lower than with PIASxβ (Supplementary Figure 3). Accordingly, we did not observe a repression of Krox20 targets upon electroporation of PIAS1 or PIAS3 expression vectors in chick embryos (data not shown).
Hoxb1 regulation by PIASxβ
The patterns of expression of Hoxb1 and PIASxβ in the developing nervous system only partially overlap and in particular Hoxb1 is expressed caudally up to the prospective r3/r4 boundary at the 2–3 ss stage in the chick, before PIASxβ induction (Giudicelli et al, 2001 and this paper), excluding the possibility of a role of PIASxβ in Hoxb1 early activation. At later stages, however, PIASxβ is likely to be implicated in the positive regulation of Hoxb1 in the hindbrain and the spinal cord. This is based both on gain-of-function experiments in which we ectopically expressed PIASxβ in the chick embryo neural tube and on loss-of-function experiments involving expression of a PIASxβ mutant derivative that presumably acts as a dominant-negative molecule.
Our experiments did not address at which level PIASxβ is acting on Hoxb1 expression. Transcription initiation nevertheless constitutes the most likely possibility. This is consistent with several observations: (i) PIASxβ is able to induce accumulation of Hoxb1 mRNA in territories that normally do not express the gene. (ii) Although PIAS family members have been mostly described as transcriptional corepressors (Schmidt and Muller, 2003), roles of transcriptional coactivators have also been reported (Yang and Sharrocks, 2005). Hence, PIASxβ interacts with a histone deacetylase (HDAC) to alleviate transcriptional repression on TFII-I (Tussié-Luna et al, 2002). It is interesting to note that the SAP domain, which is required for Hoxb1 activation, has been shown to be involved in the interaction with HDACs (Gross et al, 2004). (iii) The C-terminus of PIASxβ, which is also required for Hoxb1 activation, is rich in serine and threonine like some transcriptional activation domains and has been shown to activate transcription in Gal4 fusions in yeast (Wu et al, 1997).
Definitive proof for the requirement of PIASxβ in some aspects of Hoxb1 transcription will await the analysis of the knockout mutants. However, owing to the close conservation of the different family members, this is likely to necessitate the combination of mutations in several of them.
Krox20, a molecular switch for coordinated Hox gene regulation
Hoxb1 transcriptional regulation in the developing central nervous system (CNS) is a very complex process. It has been shown to involve an initiation period involving induction by Hoxa1 and the retinoic acid pathway followed by a maintenance and refinement period. This second phase involves both positive regulators, in particular, Pbx, Meis and Hox factors acting through auto- and crossregulatory pathways, and repressors that lead to restriction of the expression to r4. Krox20 belongs to these negative regulators, as indicated by its capacity to repress Hoxb1 expression in gain-of-function experiments in the chick and by the extension of the Hoxb1-positive territory into r3 in the mouse Krox20 null mutant (Giudicelli et al, 2001; Voiculescu et al, 2001). The present work provides a molecular basis for the genetic interaction between Krox20 and Hoxb1. We propose that PIASxβ is required for the second phase of Hoxb1 expression in the hindbrain and that Krox20 represses Hoxb1 by directly binding to PIASxβ and antagonizing its activity (Figure 7). In contrast, the early pattern of expression of PIASxβ in the CNS, with no rhombomere-specific domains (Figure 3A), is not in favour of an implication of PIASxβ in the control of Krox20 activity. The mechanism of repression by Krox20 is very different from its mode of activation of gene expression, which involves specific DNA binding, although the same domain, the zinc-fingers, is involved in both types of interactions (Figure 7). Our interpretation is strongly supported by the fact that Hoxb1 repression does not require the ability of Krox20 to bind DNA, and that it correlates with the capacity of Krox20 to interact with PIASxβ. According to our model, odd- (r3/r5) or even- (r4) numbered rhombomere-specific expression relies on the balance between Krox20 and PIASxβ: when in excess, Krox20 both antagonizes PIASxβ, leading to Hoxb1 repression, and binds to its DNA target sites, directly activating the transcription of odd-numbered rhombomere-specific genes, including Hox genes (Figure 7A). In contrast, in the absence of Krox20, PIASxβ is free to activate Hoxb1 expression and odd-numbered genes are not activated (Figure 7B). Therefore, Krox20 appears as a molecular switch that coordinates gene expression to establish odd-numbered rhombomere identity.
Figure 7.
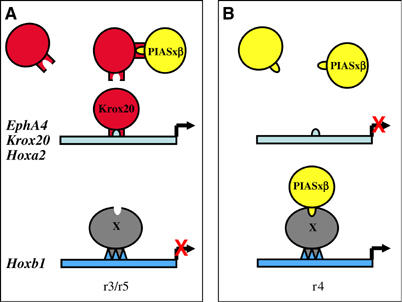
Schematic representation of the possible crossregulatory interactions between Krox20 and PIASxβ. The relative levels of the two proteins determine the expression of r3/r5- and r4-specific genes (see the Discussion section for details of the model). The circles represent the different proteins and the rectangle the cis-acting elements. (A) Situation in r3/r5. (B) Situation in r4. The interaction of Piasxβ with the Hoxb1 locus may be direct or indirect, via a putative protein (X). The domain of PIASxβ involved in this latter interaction has not been characterized. It is arbitrarily represented as the domain overlapping with the Krox20-binding domain. This representation does not take into account the possibility that PIASxβ might be acting by sequestering a repressor of Hoxb1.
The precise mode of action of Krox20 on PIASxβ activity is not known. However, we have shown that on PIASxβ the interaction occurs at the level of a region that contains an SH3-binding domain (proline-rich region). Therefore, binding of Krox20 to the proline-rich region may prevent the interaction of PIASxβ with an SH3 factor and its incorporation into a transcription activation complex. In support of this idea, the deletion of the proline-rich domain not only prevents interaction with Krox20, but also abrogates Hoxb1 activation. Identification of this putative SH3 factor will constitute a further step in the understanding of Hoxb1 regulation.
Materials and methods
Yeast two-hybrid screening and yeast β-galactosidase assays
For all constructions, see Supplementary data. Yeast two-hybrid screening was performed with the ProQuest Two-Hybrid System (Invitrogen) in the MaV203 strain, according to the manufacturer's protocol. We used a ProQuest two-hybrid, 8.5 dpc mouse embryo cDNA library cloned in the pPC86 vector (Invitrogen). Quantification of β-galactosidase activity was performed using o-nitrophenyl-β-D-galactopyranoside as substrate.
Protein expression, purification, immunoprecipitation and pull-down assays
GST and GST fusion constructs were transformed in Escherichia coli BL21(DE3) and GST fusions were prepared and purified on glutathione-Sepharose beads (Amersham Biosciences Inc.) according to the manufacturer's protocol. In vitro transcription/translation reactions were performed with the TNT Quick Coupled Transcription/Translation System (Promega) in the presence of [35S]methionine (Amersham Biosciences Inc.) using 1 μg of template plasmid. Pull-down experiments and protein immunoprecipitation are detailed in Supplementary data.
Cell culture, transfections and indirect immunofluorescence
COS7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal bovine serum (Invitrogen). Transient transfections were performed with Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen). Cells were analysed 36 h after transfection. For immunofluorescence analysis, cells were permeabilized for 2 min at room temperature with 0.5% Triton X-100 in PBS, fixed for 10 min in 4% paraformaldehyde (PFA) in PBS and blocked with 10% fetal bovine serum in PBS containing 0.1% Triton X-100. Dissected hindbrains were fixed for 2 h in 4% PFA in PBS and blocked with 5% donkey serum in PBS containing 0.25% Triton X-100. Antibodies and dilutions were as follows: Krox20, rabbit polyclonal (1:100; Babco); HA, rat monoclonal (1:400; Roche); EphA4, mouse monoclonal (1:50; Hirano et al, 1998); FITC- and Cy3-conjugated secondary antibodies (1:200 and 1:800, respectively; Jackson Immuno Research). Immunofluorescence pictures were acquired on a Leica TCS 4D confocal microscope and assembled with Adobe Photoshop.
In ovo electroporation and whole-mount in situ hybridization
Electroporation, preparation of the embryos for immunochemistry and in situ hybridization were carried out as described previously (Giudicelli et al, 2001). To evaluate the efficiency of electroporation, a GFP expression vector (pEGFP-N1; Clontech) was systematically co-electroporated. The chick probes for in situ hybridization were as follows: Krox20 (Giudicelli et al, 2001), EphA4 (Sajjadi and Pasquale, 1993), MafB (Kataoka et al, 1994), Hoxa3 (Grapin-Botton et al, 1995), Hoxb1 (Guthrie et al, 1992), follistatin (Graham and Lumsden, 1996), PIASxβ (BBSRC chick EST ChEST604b10, ChEST350g13 and ChEST289m24).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Information
Acknowledgments
We thank J Ghislain for critical reading of the manuscript and A Dejean and J Seeler for help in the Somoylation assay. MG-D was supported by Marie Curie (European Union) and Ramon y Cajal (MEC, Spain) contracts. This work was supported by grants from INSERM, MENRT, EC, ARC and AFM to PC.
References
- Aravind L, Koonin EV (2000) SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci 25: 112–114 [DOI] [PubMed] [Google Scholar]
- Barrow JR, Stadler HS, Capecchi MR (2000) Roles of Hoxa1 and Hoxa2 in patterning the early hindbrain of the mouse. Development 127: 933–944 [DOI] [PubMed] [Google Scholar]
- Birgbauer E, Sechrist J, Bronner-Fraser M, Fraser S (1995) Rhombomeric origin and rostrocaudal reassortment of neural crest cells revealed by intravital microscopy. Development 121: 935–945 [DOI] [PubMed] [Google Scholar]
- Cheng YC, Amoyel M, Qiu X, Jiang YJ, Xu Q, Wilkinson DG (2004) Notch activation regulates the segregation and differentiation of rhombomere boundary cells in the zebrafish hindbrain. Dev Cell 6: 539–550 [DOI] [PubMed] [Google Scholar]
- Choe SK, Sagerstrom CG (2004) Paralog group 1 hox genes regulate rhombomere 5/6 expression of vhnf1, a repressor of rostral hindbrain fates, in a meis-dependent manner. Dev Biol 271: 350–361 [DOI] [PubMed] [Google Scholar]
- Clarke JD, Erskine L, Lumsden A (1998) Differential progenitor dispersal and the spatial origin of early neurons can explain the predominance of single-phenotype clones in the chick hindbrain. Dev Dyn 212: 14–26 [DOI] [PubMed] [Google Scholar]
- Dupe V, Lumsden A (2001) Hindbrain patterning involves graded responses to retinoic acid signalling. Development 128: 2199–2208 [DOI] [PubMed] [Google Scholar]
- Fraser S, Keynes R, Lumsden A (1990) Segmentation in the chick embryo hindbrain is defined by cell lineage restrictions. Nature 344: 431–435 [DOI] [PubMed] [Google Scholar]
- Frohman MA, Martin GR, Cordes SP, Halamek LP, Barsh GS (1993) Altered rhombomere-specific gene expression and hyoid bone differentiation in the mouse segmentation mutant, kreisler (kr). Development 117: 925–936 [DOI] [PubMed] [Google Scholar]
- Gavalas A, Studer M, Lumsden A, Rijli FM, Krumlauf R, Chambon P (1998) Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch. Development 125: 1123–1136 [DOI] [PubMed] [Google Scholar]
- Giudicelli F, Taillebourg E, Charnay P, Gilardi-Hebenstreit P (2001) Krox-20 patterns the hindbrain through both cell-autonomous and non cell-autonomous mechanisms. Genes Dev 15: 567–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A, Lumsden A (1996) Interactions between rhombomeres modulate Krox-20 and follistatin expression in the chick embryo hindbrain. Development 122: 473–480 [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A, Bonnin MA, McNaughton LA, Krumlauf R, Le Douarin NM (1995) Plasticity of transposed rhombomeres: Hox gene induction is correlated with phenotypic modifications. Development 121: 2707–2721 [DOI] [PubMed] [Google Scholar]
- Gross M, Yang R, Top I, Gasper C, Shuai K (2004) PIASy-mediated repression of the androgen-receptor is independent of sumoylation. Oncogene 23: 3059–3066 [DOI] [PubMed] [Google Scholar]
- Guthrie S, Muchamore I, Kuroiwa A, Marshall H, Krumlauf R, Lumsden A (1992) Neuroectodermal autonomy of Hox-2.9 expression revealed by rhombomere transpositions. Nature 356: 157–159 [DOI] [PubMed] [Google Scholar]
- Hirano S, Tanaka H, Ohta K, Norita M, Hoshino K, Meguro R, Kase M (1998) Normal ontogenic observations on the expression of Eph receptor tyrosine kinase, Cek8, in chick embryos. Anat Embryol (Berl) 197: 187–197 [DOI] [PubMed] [Google Scholar]
- Irving C, Nieto A, DasGupta R, Charnay P, Wilkinson DG (1996) Progressive spatial restriction of Sek-1 and Krox-20 gene expression during hindbrain segmentation. Dev Biol 173: 26–38 [DOI] [PubMed] [Google Scholar]
- Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 [DOI] [PubMed] [Google Scholar]
- Kahyo T, Nishida T, Yasuda H (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell 8: 713–718 [DOI] [PubMed] [Google Scholar]
- Kataoka K, Fujiwara KT, Noda M, Nishizawa M (1994) MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol Cell Biol 14: 7581–7591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Karvonen U, Janne OA, Palvimo JJ (2002) PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol Cell Biol 22: 5222–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R (1996) Patterning the vertebrate neuraxis. Science 274: 1109–1115 [DOI] [PubMed] [Google Scholar]
- Manzanares M, Nardelli J, Gilardi-Hebenstreit P, Marshall H, Giudicelli F, Martinez-Pastor MT, Krumlauf R, Charnay P (2002) Krox20 and kreisler co-operate in the transcriptional control of Hoxb3 hindbrain segmental expression. EMBO J 21: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin F, Charnay P (2000) Hindbrain patterning: FGFs regulate Krox20 and mafB/kr expression in the otic/preotic region. Development 127: 4925–4935 [DOI] [PubMed] [Google Scholar]
- McNulty CL, Peres JN, Bardine N, van den Akker WM, Durston AJ (2005) Knockdown of the complete Hox paralogous group 1 leads to dramatic hindbrain and neural crest defects. Development 132: 2861–2871 [DOI] [PubMed] [Google Scholar]
- Minty A, Dumont X, Kaghad M, Caput D (2000) Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem 275: 36316–36323 [DOI] [PubMed] [Google Scholar]
- Nonchev S, Vesque C, Maconochie M, Seitanidou T, Ariza-McNaughton L, Frain M, Marshall H, Sham MH, Krumlauf R, Charnay P (1996) Segmental expression of Hoxa-2 in the hindbrain is directly regulated by Krox-20. Development 122: 543–554 [DOI] [PubMed] [Google Scholar]
- O'Donovan KJ, Tourtellotte WG, Millbrandt J, Baraban JM (1999) The EGR family of transcription-regulatory factors: progress at the interface of molecular and systems neuroscience. Trends Neurosci 22: 167–173 [DOI] [PubMed] [Google Scholar]
- Pasini A, Wilkinson DG (2002) Stabilizing the regionalisation of the developing vertebrate central nervous system. BioEssays 24: 427–438 [DOI] [PubMed] [Google Scholar]
- Rijli FM, Mark M, Lakkaraju S, Dierich A, Dolle P, Chambon P (1993) A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell 75: 1333–1349 [DOI] [PubMed] [Google Scholar]
- Rossel M, Capecchi MR (1999) Mice mutant for both Hoxa1 and Hoxb1 show extensive remodeling of the hindbrain and defects in craniofacial development. Development 126: 5027–5040 [DOI] [PubMed] [Google Scholar]
- Russo MW, Sevetson BR, Milbrandt J (1995) Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc Natl Acad Sci USA 92: 6873–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev 15: 3088–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjadi FG, Pasquale EB (1993) Five novel avian Eph-related tyrosine kinases are differentially expressed. Oncogene 8: 1807–1813 [PubMed] [Google Scholar]
- Schmidt D, Muller S (2003) PIAS/SUMO: new partners in transcriptional regulation. Cell Mol Life Sci 60: 2561–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Maunoury S, Topilko P, Seitandou T, Levi G, Cohen-Tannoudji M, Pournin S, Babinet C, Charnay P (1993) Disruption of Krox-20 results in alteration of rhombomeres 3 and 5 in the developing hindbrain. Cell 75: 1199–1214 [DOI] [PubMed] [Google Scholar]
- Seitanidou T, Schneider-Maunoury S, Desmarquet C, Wilkinson DG, Charnay P (1997) Krox-20 is a key regulator of rhombomere-specific gene expression in the developing hindbrain. Mech Dev 65: 31–42 [DOI] [PubMed] [Google Scholar]
- Serpente P, Tumpel S, Ghyselinck NB, Niederreither K, Wiedemann LM, Dolle P, Chambon P, Krumlauf R, Gould AP (2005) Direct crossregulation between retinoic acid receptor {beta} and Hox genes during hindbrain segmentation. Development 132: 503–513 [DOI] [PubMed] [Google Scholar]
- Sham MH, Vesque C, Nonchev S, Marshall H, Frain M, Gupta RD, Whiting J, Wilkinson D, Charnay P, Krumlauf R (1993) The zinc finger gene Krox20 regulates HoxB2 (Hox2.8) during hindbrain segmentation. Cell 72: 183–196 [DOI] [PubMed] [Google Scholar]
- Shuai K (2000) Modulation of STAT signaling by STAT-interacting proteins. Oncogene 19: 2638–2644 [DOI] [PubMed] [Google Scholar]
- Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R (1996) Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature 384: 630–634 [DOI] [PubMed] [Google Scholar]
- Svaren J, Sevetson BR, Apel ED, Zimonjic DB, Popescu NC, Milbrandt J (1996) NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol Cell Biol 16: 3545–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil T, Frain M, Gilardi-Hebenstreit P, Flenniken A, Charnay P, Wilkinson DG (1998) Segmental expression of the EphA4 (Sek-1) receptor tyrosine kinase in the hindbrain is under direct transcriptional control of Krox-20. Development 125: 443–452 [DOI] [PubMed] [Google Scholar]
- Trainor PA, Krumlauf R (2000) Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nat Rev Neurosci 1: 116–124 [DOI] [PubMed] [Google Scholar]
- Tussié-Luna MI, Michel B, Hakre S, Roy AL (2002) The SUMO ubiquitin-protein isopeptide ligase family member Miz1/PLASxB/Siz2 is a transcription cofactor for TFII-I. J Biol Chem 277: 43185–43193 [DOI] [PubMed] [Google Scholar]
- Vesque C, Charnay P (1992) Mapping functional regions of the segment-specific transcription factor Krox-20. Nucleic Acids Res 20: 2485–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiculescu O, Taillebourg E, Pujades C, Kress C, Buart S, Charnay P, Schneider-Maunoury S (2001) Hindbrain patterning: Krox20 couples segmentation and specification of regional identity. Development 128: 4967–4978 [DOI] [PubMed] [Google Scholar]
- Walshe J, Maroon H, McGonnell IM, Dickson C, Mason I (2002) Establishment of hindbrain segmental identity requires signaling by FGF3 and FGF8. Curr Biol 12: 1117–1123 [DOI] [PubMed] [Google Scholar]
- Warner LE, Mancias P, Butler IJ, McDonald CM, Keppen L, Koob KG, Lupski JR (1998) Mutations in the early growth response 2 (EGR2) gene are associated with hereditary myelinopathies. Nat Genet 18: 382–384 [DOI] [PubMed] [Google Scholar]
- Warner LE, Svaren J, Milbrandt J, Lupski JR (1999) Functional consequences of mutations in the early growth response 2 gene (EGR2) correlate with severity of human myelinopathies. Hum Mol Genet 8: 1245–1251 [DOI] [PubMed] [Google Scholar]
- Wu L, Wu H, Ma L, Sangiorgi F, Wu N, Bell JR, Lyons GE, Maxson R (1997) Miz1, a novel zinc finger transcription factor that interacts with Msx2 and enhances its affinity for DNA. Mech Dev 65: 3–17 [DOI] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD (2005) PIASx acts as an Elk-1 coactivator by facilitating derepression. EMBO J 24: 2161–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Information


