Abstract
The kinetics of initiator transfer RNA (tRNA) interaction with the messenger RNA (mRNA)-programmed 30S subunit and the rate of 50S subunit docking to the 30S preinitiation complex were measured for different combinations of initiation factors in a cell-free Escherichia coli system for protein synthesis with components of high purity. The major results are summarized by a Michaelis–Menten scheme for initiation. All three initiation factors are required for maximal efficiency (kcat/KM) of initiation and for maximal in vivo rate of initiation at normal concentration of initiator tRNA. Spontaneous release of IF3 from the 30S preinitiation complex is required for subunit docking. The presence of initiator tRNA on the 30S subunit greatly increases the rate of 70S ribosome formation by increasing the rate of IF3 dissociation from the 30S subunit and the rate of 50S subunit docking to the IF3-free 30S preinitiation complex. The reasons why IF1 and IF3 are essential in E. coli are discussed in the light of the present observations.
Keywords: initiation, initiation factors, initiator tRNA, protein synthesis, ribosome
Introduction
During protein synthesis in the bacterial cell, ribosomal recycling factor RRF and elongation factor G (EF-G) bind to post-termination ribosomes (Janosi et al, 1996), and split them into 30S and 50S subunits in a GTP-dependent reaction (Karimi et al, 1999; Peske et al, 2005; Zavialov et al, 2005). Then, initiation factor IF3 binds rapidly to the 30S subunit, where it stimulates dissociation of deacylated transfer RNA (tRNA) and messenger RNA (mRNA) (Karimi et al, 1999; Peske et al, 2005) and blocks premature subunit docking (Subramanian and Davis, 1970). Subsequent formation of a translation competent 70S ribosome with initiator tRNA (fMet-tRNAfMet) in the peptidyl (P) site takes place in two major steps (Figure 1A). In the first, formation of a 30S preinitiation complex, containing a new mRNA and fMet-tRNAfMet, is promoted by the three initiation factors IF1, IF2 and IF3 (Benne et al, 1973; Hershey, 1987; Gualerzi and Pon, 1990). In the second, IF2 promotes subunit docking (Benne et al, 1973; Fakunding and Hershey, 1973; Grunberg-Manago et al, 1975; Hershey, 1987; Antoun et al, 2003). After GTP hydrolysis on IF2 and subsequent IF2 release, the 70S ribosome is ready to form the first peptide bond in a nascent protein (Benne et al, 1973; Luchin et al, 1999; Antoun et al, 2003).
Figure 1.
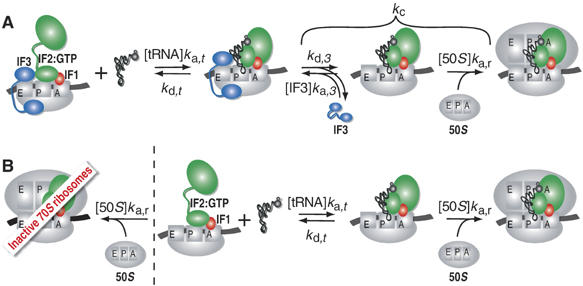
Kinetic scheme for initiation of protein synthesis. (A) Binding of fMet-tRNAfMet to mRNA-programmed 30S subunits equipped with three initiation factors destabilizes IF3 binding. IF3 dissociates then with the rate kd,3 after which it can rebind with the rate ka,3[IF3]. Alternatively, a 50S subunit can dock to an IF3-free 30S preinitiation complex with the rate ka,r[50S]. (B) In the absence of IF3, both 50S subunits and fMet-tRNAfMet can bind with comparable rates to the mRNA-programmed 30S subunits. Binding of 50S subunits gives rise to an empty 70S ribosome inactive in initiation, whereas fMet-tRNAfMet binding results in a subsequent fast docking of 50S subunits and formation of active 70S initiation complexes.
IF1 is essential for Escherichia coli (Cummings and Hershey, 1994) for an unknown reason (Croitoru et al, 2004). It accelerates dissociation of empty 70S ribosomes into subunits (Grunberg-Manago et al, 1975; Naaktgeboren et al, 1977) and stimulates formation of the 30S preinitiation complex (Pon and Gualerzi, 1984; Hershey, 1987).
The presence of IF2 on the 30S subunit promotes the binding of initiator tRNA (Canonaco et al, 1986; Hershey, 1987; Gualerzi and Pon, 1990). It has also been suggested that IF2 forms a complex with initiator tRNA in the cytoplasm and transports it to the 30S subunit (Wu and RajBhandary, 1997). The importance of IF2 and GTP for rapid docking of ribosomal subunits was high-lighted in more recent work (Antoun et al, 2003).
Several roles have been suggested for IF3 in initiation of protein synthesis (Hershey, 1987; Petrelli et al, 2001), for example, (i) prevention of 70S ribosome formation from empty subunits (Subramanian and Davis, 1970; Grunberg-Manago et al, 1975), (ii) acceleration of fMet-tRNAfMet association to the 30S subunit (Gualerzi et al, 1977), (iii) acceleration of dissociation of 30S-bound elongator tRNAs (Risuleo et al, 1976; Hartz et al, 1989) and (iv) acceleration of dissociation of initiator tRNA bound to 30S subunits programmed with other start codons than AUG, GUG or UUG (Petrelli et al, 2001).
The effects of initiation factors on preinitiation complex formation were previously studied with NAc-Phe-tRNAPhe from yeast and poly(U) programmed 30S subunits from E. coli (Wintermeyer and Gualerzi, 1983), but their effects on the kinetics of interaction between fMet-tRNAfMet and 30S subunits programmed with heteropolymeric mRNAs are unknown. There exist but few reports on the rate of subunit docking (Grunberg-Manago et al, 1975; Chaires et al, 1981; Antoun et al, 2003) and there has been no systematic study of how initiation factors and initiator tRNA affect the rate of subunit docking in initiation of protein synthesis.
In this work, we describe the effects of combinations of IF1, IF2 and IF3 on the rates by which fMet-tRNAfMet associates to and dissociates from a 30S subunit, programmed with a heteropolymeric mRNA with a strong Shine–Dalgarno (SD) sequence. We also report how combinations of IF1, IF2, IF3 and initiator tRNA affect the rate of 70S ribosome formation from ribosomal subunits. Our results extend the antiassociation model of IF3 action (Naaktgeboren et al, 1977; Chaires et al, 1981; Hershey, 1987) by showing that spontaneous dissociation of IF3 from the preinitiation complex is also required for docking of the 50S subunit to the canonical 30S preinitiation complex. We demonstrate that the presence of fMet-tRNAfMet on an mRNA-programmed 30S subunit, containing all three initiation factors greatly accelerates subunit docking by increasing (i) the rate of IF3 dissociation from the 30S subunit, (ii) the rate of 50S subunit docking to an IF3-free 30S preinitiation complex.
Results
30S preinitiation complex formation
The kinetics of fMet-tRNAfMet interaction with the 30S·mRNA complex was studied with nitrocellulose filter binding (NC) and stopped-flow with detection of Rayleigh light scattering (Antoun et al, 2003, 2004) for all combinations of initiation factors at 37°C in a modified polymix buffer, LS, containing ATP and GTP (Materials and methods). The polymix buffer has an ionic composition mimicking that in the E. coli cell (Jelenc and Kurland, 1979) and leads to rapid and accurate protein synthesis (Ehrenberg et al, 1990). All kinetic and equilibrium data are summarized in Table I, and a scheme for the whole initiation process with definitions of rate constants is shown in Figure 1.
Table 1.
Association rate constants ka,t, dissociation rate constants kd,t and equilibrium binding constants Kd,t for fMet-tRNAfMet interaction with mRNA-programmed 30S subunits in the presence of different combinations of initiation factors
| IF1 | IF2 | IF3 | ka,t (μM−1 s−1) | kd,t (s−1) | Kd,t (nM) |
|---|---|---|---|---|---|
| + | + | + | 12.5 | 0.034 | 2.7 |
| + | − | + | 0.24 | 0.11 | 460 |
| − | + | + | 1.7 | 0.0065 | 3.8 |
| − | − | + | 0.15 | 0.013 | 87 |
| + | + | − | 1.35 | 1.5 × 10−4 | 0.11 |
| + | − | − | 0.035 | 3.5 × 10−4 | 10 |
| − | + | − | 0.34 | 1.8 × 10−4 | 0.53 |
| − | − | − | 0.031 | 1.1 × 10−4 | 3.6 |
| Rate constants ka,t, and kd,t and their s.d.'s were calculated from experimental data with the nonlinear regression program Origin 7 (OriginLab Corp.). S.d. for ka,t were in most cases around 10% and never exceeded 20%, whereas those for kd,t were around 5% and never exceeded 10%. |
Kinetics of initiator tRNA binding to the 30S subunit in the absence of IF2. From NC experiments, the association rate constant (ka,t) for the binding of [3H]fMet-tRNAfMet to either 30S·mRNA or 30S·mRNA·IF1 was estimated as 0.031 or 0.035 μM−1 s−1, respectively (Figure 2A). The association rates depended linearly on the fMet-tRNAfMet concentration (Figure 2A, insert), implying second-order reactions. From NC experiments, in which [3H]fMet-tRNAfMet prebound to the 30S subunit was chased with unlabelled initiator tRNA, the corresponding dissociation rate constants (kd,t) were estimated as 0.00011 or 0.00035 s−1, respectively (Figure 2B). From similar NC chase experiments, kd,t values for the dissociation of fMet-tRNAfMet from a 30S·mRNA·fMet-tRNAfMet·IF3 or a 30S·mRNA·fMet-tRNAfMet·IF3·IF1 complex were estimated as 0.013 or 0.11 s−1, respectively (Figure 2C). Accordingly, kd,t increased about hundred- or thousand-fold by addition of IF3 or IF3 and IF1. From NC experiments, in which the fMet-tRNAfMet concentration was titrated at a fixed concentration of 30S·mRNA·IF3 or 30S·mRNA·IF3·IF1, the corresponding equilibrium dissociation constants (Kd,t=kd,t/ka,t) were estimated as 87 or 460 nM, respectively (Figure 2D). Using directly measured kd,t and Kd,t values, the corresponding association rate constants ka,t=kd,t/Kd,t were calculated as 0.15 or 0.24 μM−1 s−1, respectively.
Figure 2.
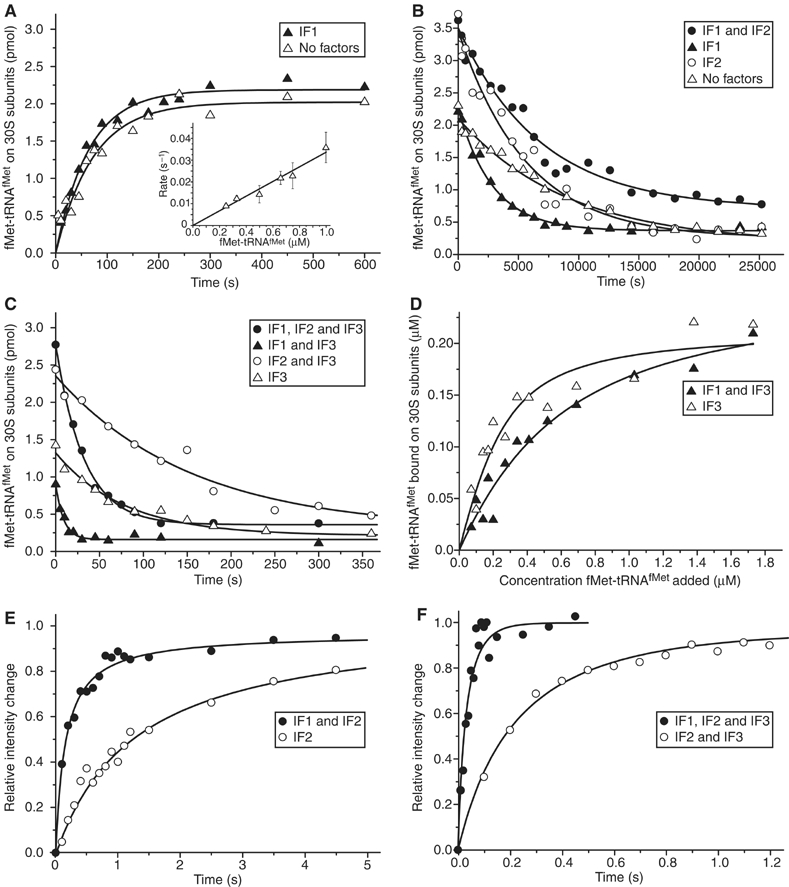
Effect of initiation factors on fMet-tRNAfMet binding to 30S ribosomal subunits. (A) [3H]fMet-tRNAfMet association to mRNA-programmed 30S subunits was measured by NC filtration technique in the absence of initiation factors or with only IF1 present. Insert: dependence of fMet-tRNAfMet association rate on the concentration of fMet-tRNAfMet in the absence of initiation factors. (B) [3H]fMet-tRNAfMet dissociation from 30S preinitiation complexes assembled with different combinations of IF1 and IF2 was measured by NC filtration in the presence of a 10-fold excess of nonlabeled chasing fMet-tRNAfMet. (C) The same as in (B) except that IF3 was also present in the 30S preinitiation complex. (D) Dependence of the 30S·mRNA·IF3·fMet-tRNAfMet complex formation on the concentration of fMet-tRNAfMet in the presence or absence of IF1 measured by NC filtration. (E) Kinetics of fMet-tRNAfMet binding to 30S·mRNA·IF2 or to 30S·mRNA·IF1·IF2 complexes measured by light scattering. (F) The same as in (E) but in the presence of IF3.
Kinetics of initiator tRNA binding to the 30S subunit in the presence of IF2. The ka,t for the binding of initiator tRNA to 30S·mRNA·IF2 or 30S·mRNA·IF2·IF1 was estimated from stopped-flow experiments with light scattering as 0.34 or 1.35 μM−1 s−1, respectively (Figure 2E). The corresponding kd,t, obtained from NC chase experiments, was estimated as 0.00018 or 0.00015 s−1, respectively (Figure 2B).
From similar light scattering experiments, we estimated ka,t for the binding of initiator tRNA to 30S·mRNA·IF2·IF3 or 30S·mRNA·IF2·IF3·IF1 as 1.7 or 12.5 μM−1 s−1, respectively (Figure 2F). The corresponding kd,t value obtained from NC chase experiments was 0.0065 or 0.034 s−1, respectively (Figure 2C).
Kd,t for the binding of initiator tRNA to the mRNA-programmed 30S subunit in the absence of any initiation factor or in the presence of all initiation factors was estimated as 3.6 or 2.7 nM, respectively (Table I). At the same time, ka,t and kd,t increased 400- and 300-fold, respectively, when all three initiation factors were added to the 30S subunit (Table I).
70S ribosome formation from subunits
Stopped-flow techniques with detection of Rayleigh-scattered light were used to estimate the rate constant, kc, for docking of ribosomal subunits in the presence of combinations of IF1, IF2, IF3 and fMet-tRNAfMet on the 30S subunit (Table II).
Table 2.
Effects of initiation factors and fMet-tRNAfMet on the first-order rate constant, kc, of 50S subunit docking to 30S·mRNA preinitiation complexes and on KM and kcat/KM parameters of the whole initiation process
| IF1 | IF2 | IF3 |
ka,r (μM−1 s−1) |
kc (s−1) | kcat=kc (s−1) | KM (μM) | kcat/KM (μM−1 s−1) | |
|---|---|---|---|---|---|---|---|---|
| − | fMet | − | fMet | fMet | fMet | |||
| + | + | + | — | — | 0 | 2.9 | 0.23 | 12.4 |
| + | − | + | — | — | 0 | 0.0098 | 0.48 | 0.020 |
| − | + | + | — | — | 0.020 | 8.7 | 5.1 | 1.7 |
| − | − | + | — | — | 0.020 | 0.038 | 0.33 | 0.11 |
| + | + | − | 4.7 | 120 | 1.4 | 42 | 31 | 1.4 |
| + | − | − | 1.4 | 0.80 | 0.43 | 0.24 | 6.9 | 0.035 |
| − | + | − | 2.1 | 140 | 0.63 | 36 | 110 | 0.34 |
| − | − | − | 1.8 | 1.0 | 0.56 | 0.30 | 9.7 | 0.031 |
| The first-order rate constant kc corresponds to the rate of 70S formation measured at 0.3 μM concentration of 30S and 50S subunits in the reaction mixture. The second-order rate constants ka,r in the absence of IF3 were calculated as ka,r=kc/[50S] or obtained from 50S subunit titration experiments (Figure 4A). kcat/KM, kcat and KM parameters were calculated according to Equation (1) and Equation (3) or Equation (4) using kc and ka,r data from this table and ka,t and kd,t rate constants from Table I. Rate constant kc and its s.d.'s were calculated from original light-scattering data with the nonlinear regression program Origin 7 (OriginLab Corp.). S.d.'s for kc were in most cases around 3% and never exceeded 10%. | ||||||||
Subunit docking in the absence of initiator tRNA. Addition of IF1 to 30S subunits decreased kc from 0.56 to 0.43 s−1 in the absence and increased kc from 0.63 to 1.4 s−1 in the presence of IF2 (Figure 3A). When also IF3 was present, kc was small in the absence and almost zero in the presence of IF1 independently of the presence of IF2 (Figure 3B). Furthermore, the extent of 70S ribosome formation was small, as the equilibrium was strongly shifted towards free subunits. This was confirmed in ‘reverse experiments', where one syringe of the stopped-flow instrument contained 70S ribosomes and mRNA and the other various combinations of initiation factors (Figure 3B). After mixing, the light intensity decreased in each case to the same equilibrium value as in the docking experiment. Subunit separation in the presence of IF3 and IF1 was complete and five times faster than in the presence of just IF3 (Figure 3B).
Figure 3.
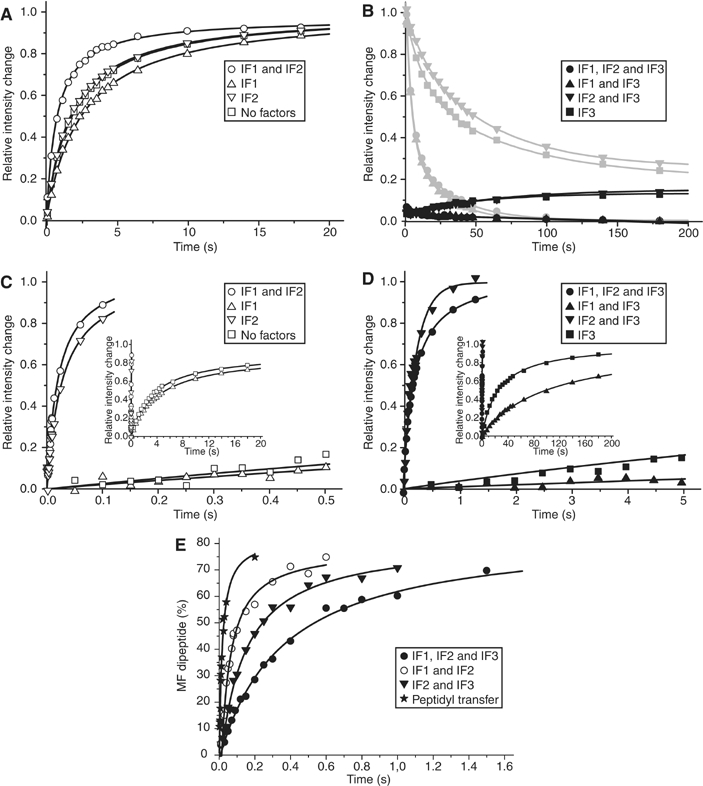
Effect of initiation factors on the association of ribosomal subunits. (A) 50S subunit docking to mRNA-programmed 30S complexes containing different combinations of IF1 and IF2 in the absence of fMet-tRNAfMet measured by light scattering. (B) The same as in (A) but all 30S complexes contained IF3. The gray curves show dissociation of the corresponding 70S complexes assembled without IF3 after IF3 addition. (C) 50S subunit docking to 30S complexes containing fMet-tRNAfMet and different combinations of IF1 and IF2 measured by light scattering. Insert: extended time range. (D) The same as in (C) but 30S complexes contained IF3. Insert: extended time range. (E) Rate of 50S subunit docking to 30S preinitiation complexes monitored as the rate of dipeptide formation in a quench-flow experiment. One syringe of the quench-flow instrument contained 50S subunits and ternary complexes, whereas the other contained 30S preinitiation complexes assembled with all three initiation factors, or in the absence of either IF1 or IF3. The time course of the dipeptide formation reaction (peptidyl transfer) with preformed 70S initiation complexes is shown for comparison.
Subunit docking in the presence of initiator tRNA. With initiator fMet-tRNAfMet present on the 30S subunit, kc was 0.3 s−1 without initiation factors, 0.24 s−1 in the presence of only IF1, 0.038 s−1 in the presence of only IF3 and 0.0098 s−1 in the presence of IF1 and IF3 (Figure 3C).
Addition of IF2 to a 30S subunit containing fMet-tRNAfMet increased kc from 0.3 to 36 s−1 in the absence, and from 0.24 to 42 s−1 in the presence of IF1 (Figure 3C). Addition of IF2 to a 30S subunit containing both fMet-tRNAfMet and IF3 increased kc from 0.038 to 8.7 s−1 in the absence and from 0.0098 to 2.9 s−1 in the presence of IF1 (Figure 3D, Table II). This means that in the presence of IF3, rapid subunit docking required the presence of both fMet-tRNAfMet and IF2.
Quench-flow experiments (Figure 3E) show that 70S initiation complexes, rapidly formed by 50S subunits docking to 30S preinitiation complexes lacking either IF1 or IF3, were fully competent in peptide elongation (Figure 3E). The rate constant, kPT, of dipeptide formation was estimated as 3.6 s−1 in the presence of IF1, IF2 and IF3. It increased to 27 or 7.1 s−1 by removal of IF3 or IF1, respectively. These kPT values were similar to the corresponding kc values for subunit docking as estimated from stopped-flow experiments (Table II). However, in the presence of IF1 and IF2, kc was estimated as 42 s−1, a value significantly greater than the corresponding kPT value of 27 s−1. This difference arises because dipeptide formation requires, in addition to subunit docking, dissociation of IF2 from the 70S ribosome followed by formation of the first peptide bond on IF2-free ribosomes, where the latter reaction occurs at a rate of ∼60 s−1 (Figure 3E).
Subunit docking in the presence of IF3. Stopped-flow experiments with varying concentration of 50S subunits in one and constant concentration of empty 30S subunits in the other syringe (Figure 4A, insert) show that the rate, kc, of 70S ribosome formation obeyed second-order kinetics (Wishnia et al, 1975; Antoun et al, 2004). From these data (Figure 4A, insert), the association rate constant ka,r was estimated as ∼4.0 μM−1 s−1, a value significantly smaller than the ka,r value of ∼10 μM−1 s−1 obtained in the absence of phosphoenolpyruvate (PEP) in the reaction buffer (Antoun et al, 2004). This difference is owing to the previously observed sensitivity of the rate of subunit docking to the concentrations of Mg2+ ions and PEP in the buffer (Wishnia et al, 1975; Antoun et al, 2004).
Figure 4.
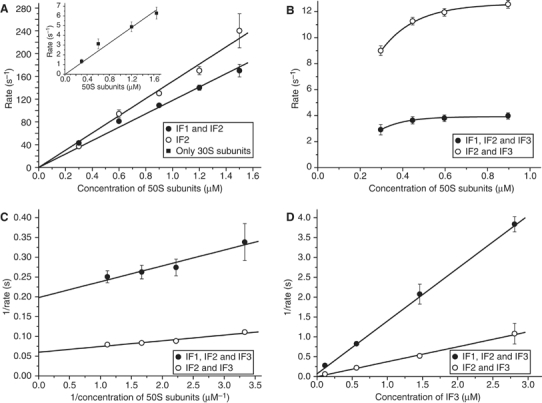
Formation of 70S ribosomes requires IF3 dissociation from 30S subunits. (A) Dependence of the rate of the 50S subunit docking to the 30S·mRNA·IF2·fMet-tRNAfMet complex or to the same complex containing IF1 on the concentration of 50S subunits. Insert: the rate of 70S formation from empty 30S and 50S subunits plotted versus the concentration of 50S subunits. Error bars smaller than symbol size are not visible. (B) The same as in (A) but 30S complexes also contained IF3. (C) The rate dependences in (B) replotted as reciprocal of the rate versus the reciprocal of the concentration of 50S subunits. Error bars smaller than symbol size are not visible. (D) The reciprocal of the rate of the 50S subunit docking to the 30S·mRNA·IF3·IF2·fMet-tRNAfMet complex or to the same complex containing IF1 plotted versus the concentration of IF3 in the reaction mixture. Error bars smaller than symbol size are not visible.
Also the rate, kc, of docking of 50S subunits to mRNA-programmed 30S subunits containing initiator tRNA and combinations of IF1 and IF2 obeyed the second-order kinetics with kc=ka,r [50S] (Figure 4A). From these data, ka,r was estimated as 120 in the presence of IF1 and IF2 and 140 μM−1 s−1 in the presence of only IF2. When, however, IF3 was added, kc, became a hyperbolic function of [50S] (Figure 4B) with well-defined plateau values (Figure 4C). At fixed concentrations of subunits, mRNA, fMet-tRNAfMet, IF2 and, when present, IF1, increasing concentrations of IF3 strongly inhibited subunit association, with a linear dependence of 1/kc on 1/[IF3] (Figure 4D). This behavior implies mutually exclusive interactions of the 50S subunit and IF3 with the 30S subunit (Naaktgeboren et al, 1977; Chaires et al, 1981). When the concentrations of IF3 and 50S subunits are much larger than that of 30S subunits, then 1/kc is approximated by
![]()
Here, kd,3 is the rate constant for dissociation of IF3 from the 30S preinitiation complex, ka,3 is rate constant for association of IF3 to this complex and ka,r is the rate constant for 50S subunit association to the IF3-free 30S preinitiation complex. Accordingly, 1/kg and 1/(ka,r[50S]) are the average times the 30S preinitiation complex spends in IF3-bound and IF3-free forms, respectively, before binding the 50S subunit (see Supplementary data). The intercepts at the y-axis of the straight lines in Figure 4C estimate the rate constant, kd,3, for IF3 dissociation from the 30S preinitiation complex as 13 s−1 in the absence and as 5.2 s−1 in the presence of IF1. The slope of the straight line in the reciprocal plot in Figure 4D is given by
![]()
It was 0.35 μM−1 s in the absence and 1.2 μM−1 s in the presence of IF1 (Figure 4D). From these estimates in combination with the values of kd,3 and ka,r as determined above and with [50S]=0.3 μM, ka,3 was estimated as 180 μM−1 s−1.
An independent estimate of ka,3 was obtained from a stopped-flow experiment with 30S subunits (0.3 μM) bound to mRNA, fMet-tRNAfMet, IF1 and IF2 in one, and with 50S subunits (0.3 μM) together with IF3 (0.45 μM) in the other syringe. After mixing, there was a fast phase, when 50% of the 70S ribosomes were formed by direct subunit docking, and a slow phase when subunit docking was conditional on IF3 dissociation from the 30S subunit (Figure 5A). This result implies that [IF3]·ka,3≈[50S]·kr,3, so that ka,3≈0.3/0.45·120 μM−1 s−1=80 μM−1 s−1, a value about half of the previous estimate. With kd,3=5.2 s−1, we estimate Kd,3 (=kd,3/ka,3) for IF3 binding to a 30S preinitiation complex containing IF1, IF2 and fMet-tRNAfMet as ∼70 nM.
Figure 5.
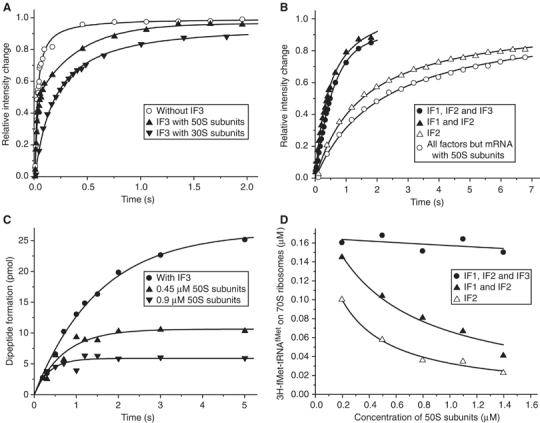
IF3 prevents the formation of abortive 70S initiation complexes. (A) Competition between IF3 and the 50S subunit for binding to the 30S·mRNA·IF2·fMet-tRNAfMet complex; 50S subunits were added to the 30S complexes or to the same complexes containing IF3. Alternatively, a mixture containing 50S subunits and IF3 was added to the 30S complexes. (B) Kinetics of the formation of 70S ribosomes from 30S·mRNA·IF2 complexes containing different combinations of IF1 and IF3 after addition of 50S subunits together with fMet-tRNAfMet. Alternatively, 70S ribosomes were formed by mixing 30S·IF1·IF2·IF3 complexes with a mixture containing mRNA, 50S subunits and fMet-tRNAfMet. (C) fMet-Phe dipeptide formation after the addition of 30S·mRNA·IF1·IF2 complexes to the mixture B containing EF-Tu·GTP·Phe-tRNAPhe ternary complexes, fMet-tRNAfMet and 50S subunits in 0.45 μM or 0.9 μM concentration. The reaction was also repeated with 30S·mRNA·IF1·IF2·IF3 complexes and the same mixture B containing 0.45 μM 50S subunits. (D) Reduction in the concentration of fMet-tRNAfMet containing 70S ribosomes assembled by addition of 30S·mRNA·IF2 complexes to a mixture containing increasing concentrations of 50S subunits and fMet-tRNAfMet at fixed concentration. 30S·mRNA·IF2 complexes contained different combinations of IF1 and IF3. The amount of 70S ribosomes containing [3H]fMet-tRNAfMet was measured by nitrocellulose filtration after 30 s from the start of the assembly reaction.
Michaelis–Menten formulation of initiation of protein synthesis
In the steady state of the living cell, with [30S0] as the total concentration of mRNA-programmed 30S subunits, the rate j of ribosome initiation per cell volume is given by j=v·[30S0], where the rate v per 30S subunit is given by the Michaelis–Menten relation (Supplementary Data):
![]()
Here, [fMet-tRNAfMet] is the concentration of free initiator tRNA, kcat is the maximal value of v at saturating [fMet-tRNAfMet] and KM is the value of [fMet-tRNAfMet] at which v=kcat/2. The kcat and KM parameters in Equation (2) for the initiation scheme in Figure 1A can be calculated from the measured rate constants in Table II according to (see Equation (1) and Supplementary data)
![]()
In the absence of IF3, the initiation proceeds according to the kinetic scheme in Figure 1B for which the Michaelis–Menten parameters in Equation (2) are given by
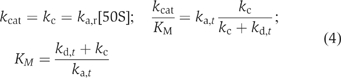
The reason why kg (see Equation (1)), rather than kc, appears in kcat/KM in Equation (3) is explained in Supplementary data. Table II shows that the catalytic efficiency parameter kcat/KM (Fersht, 1999, p. 362) was maximized by the presence of all three initiation factors. The kcat value, in contrast, was maximal ∼40 s−1 with IF2 alone or with IF1 and IF2 and decreased to ∼9 or ∼3 s−1, respectively, by the addition of IF3.
IF3 prevents formation of 70S ribosomes lacking initiator tRNA
To study the effect of IF3 on 70S ribosome formation under more in vivo like conditions, we designed a series of stopped-flow experiments in which one syringe was loaded with the 50S subunit and initiator tRNA and the other with 30S·mRNA·IF2, 30S·mRNA·IF2·IF1 or 30S·mRNA·IF2·IF1·IF3 complex. Similar rates of subunit docking were obtained for the IF1/IF2 (1.9 s−1) and IF1/IF2/IF3 (1.8 s−1) combinations and a significantly smaller rate was estimated for IF2 alone (0.65 s−1) (Figure 5B).
When the main pathway to 70S ribosome formation was via an fMet-tRNAfMet containing 30S preinitiation complex (Figure 1A), the rate, v, of 70S ribosome formation could be calculated from Equation (2) and the data in Table II. When IF3 was present, this condition was fulfilled, as then subunit docking was very slow in the absence of fMet-tRNAfMet on the 30S subunit (Figure 3B, Table II). For the IF1/IF2/IF3 combination, this calculation gives a v value of 1.9 s−1, close to the v value estimated from light scattering as 1.8 s−1 (Figure 5B). When IF3 was absent, this condition could be violated, as then direct binding of a 50S subunit to a tRNA-less 30S preinitiation complex was quite fast (Figure 3A, Table II). For the IF1/IF2 and IF2 combinations, the v values calculated from Equation (2) and Table II were 0.62 and 0.15 s−1, respectively, and much smaller than the corresponding measured v values of 1.9 and 0.65 s−1, respectively (Figure 5B). When also direct subunit docking involving 30S subunits lacking initiator tRNA (Table II) was taken into account (Supplementary data), the calculated rates of 70S ribosome formation for the IF1/IF2 and IF2 cases, 2.0 and 0.78 s−1, respectively, were in much better correspondence with the measured ones. This suggests that large fractions of 70S ribosomes formed in the absence of IF3 (Figure 5B) lacked initiator tRNA. This was confirmed by quench-flow results, showing that the extent of [3H]fMet-Phe formation in the absence of IF3 was only 40% of that in the presence of IF3. When the 50S subunit concentration was doubled, this fraction dropped to about 20% (Figure 5C), owing to increased competition between 50S subunits and the initiator tRNA for 30S subunit binding. Formation of 70S ribosomes lacking initiator tRNA in the absence of IF3 was also directly observed in NC experiments, where 30S·mRNA·IF2, 30S·mRNA·IF2·IF1 or 30S·mRNA·IF2·IF1·IF3 complexes were mixed with 50S subunits at a varying and initiator tRNA at a constant concentration. For the IF1/IF2/IF3 combination, the fraction of initiator tRNA-containing 70S ribosomes was large and did not change with increasing [50S] (Figure 5D). For IF3-less combinations, in contrast, the fraction of initiator tRNA-bound 70S ribosomes decreased markedly with increasing [50S] and the decrease was more pronounced for the IF2 than for the IF1/IF2 combination (Figure 5D).
Interestingly, when mRNA, 50S subunits and fMet-tRNAfMet were rapidly mixed with the 30S·IF1·IF2·IF3 complex, the rate of 70S formation was only 0.45 s−1 (Figure 5B), showing that here the association of mRNA to the 30S subunit was rate limiting for initiation (see Discussion).
Discussion
In the first part of this study, we document how the kinetics of initiator tRNA interaction with the 30S subunit in complex with an mRNA is affected by combinations of IF1, IF2 and IF3 (Table I). In the second part, we show how the rate of subunit docking is tuned by combinations of IF1, IF2, IF3 and initiator tRNA (Table II). We have derived Michaelis–Menten parameters (kcat, KM, kcat/KM) for initiator tRNA binding to the 30S subunit and subsequent formation of 70S initiation complex (Equation (2), Table II). To clarify the essential nature of IF1 and IF3, we have studied the conditions for formation of 70S ribosomes lacking initiator tRNA (Table II).
Interaction of initiator tRNA with the 30S subunit
Initiator tRNA has similar affinity to the mRNA-programmed 30S subunit in the presence (Kd=2.7 nM) and absence (Kd=3.6 nM) of all three initiation factors, meaning that the roles of the factors are to be found in their kinetic, rather than equilibrium, effects on initiation. The rate of initiator tRNA association to the 30S subunit is maximized by the simultaneous presence of IF1, IF2 and IF3 and the association as well as the dissociation rate constants for initiator tRNA are accelerated about 400-fold by the supply of the three initiation factors (Table I).
Initiation factor IF3 greatly reduces the affinity of initiator tRNA to the 30S subunit by increasing the association rate constant about five-fold and the dissociation rate constant about 100-fold (Table I). This finding is at variance with the previous suggestion that IF3 specifically stabilizes the binding of initiator tRNA, but not other tRNAs, to the mRNA-programmed 30S subunit (Risuleo et al, 1976; Gualerzi et al, 1979; Hartz et al, 1989), but fully in line with recent observations that IF3 destabilizes the binding of fMet-tRNAfMet in the authentic 30S preinitiation complex (Lancaster and Noller, 2005; Lomakin et al, 2006). We have, in addition, found that the IF3-dependent decrease in the affinity of initiator tRNA to the 30S subunit is even more pronounced in the presence of IF1 and that the highest Kd value (460 nM) for initiator tRNA binding to the 30S subunit occurs in the presence of IF1 and IF3 (Table I).
IF2 enhances the affinity of initiator tRNA to the 30S subunit by a 10-fold increase of the association rate constant and, interestingly, the presence of IF1 makes this affinity enhancing effect of IF2 even more pronounced resulting in a Kd value of 0.11 nM (Table I). This implies that IF1 amplifies both the affinity-reducing effect of IF3 and the affinity-enhancing effect of IF2 on initiator tRNA binding to the 30S subunit.
The present data allow for comparison with a previous study under different buffer conditions, where NAc-Phe-tRNAPhe from yeast and poly(U) substituted fMet-tRNAfMet and heteropolymeric mRNA, respectively (Wintermeyer and Gualerzi, 1983). They used stopped-flow with fluorescence detection to estimate the rate constant, ka,t, for NAc-Phe-tRNAPhe association to the 30S subunit for combinations of initiation factors. The results are qualitatively similar to our ka,t values (Table I). For instance, they estimated ka,t for NAc-Phe-tRNAPhe binding to the poly(U)-programmed 30S subunit as 0.3 μM−1 s−1 in the presence of only IF3 and as 5 μM−1 s−1 in the presence of all initiation factors, whereas we estimated the corresponding ka,t values as 0.15 and 12.5 μM−1 s−1, respectively. The kinetic correspondence suggests that NAcPhe may interact with IF2 very much like fMet and that the codon–anticodon complexes for tRNAfMet and tRNAPhe affect the association kinetics of these different tRNAs in quite similar ways.
Initiation factors and subunit docking
In the absence of IF3, the rate of subunit docking increases about 60-fold upon addition of initiator tRNA and IF2, whereas the presence of either one of these molecules alone has little effect on the docking rate (Table II). Therefore, rapid docking of the two ribosomal subunits results from the synergistic action of fMet-tRNAfMet and IF2 (Table II). Addition of IF3 to a 30S subunit in complex with fMet-tRNAfMet and IF2 greatly decreases the rate of subunit docking and the IF3-dependent decrease is even more pronounced when IF1 is also in the 30S complex. The reason for the rate decrease is that IF3 must dissociate spontaneously from the 30S preinitiation complex before subunit docking can occur. This follows from our observation that the rate of subunit docking reaches plateau values when the 50S subunit concentration is titrated to large values (Figure 4B and C). If, in contrast, the 50S subunit could dock to an IF3-containing 30S preinitiation complex and form a 70S ribosome complex from which IF3 were rapidly ejected, the straight lines in the inverse plots in Figure 4C would go through the origin, which clearly is not the case. The rate of subunit docking would, furthermore, be insensitive to the free concentration of IF3, once the 30S subunit had been saturated with the factor. What we observe, in contrast, is that the rate of subunit docking goes to zero when the IF3 concentration increases beyond saturation of the 30S subunit (Figure 4D). These conclusions extend the ‘antiassociation' model for the effect of IF3 on the docking of 50S to the empty 30S subunit (Grunberg-Manago et al, 1975; Naaktgeboren et al, 1977; Chaires et al, 1981; Hershey, 1987), to the docking of the 50S subunit also to the canonical 30S preinitiation complex.
These conclusions are also in line with foot-printing experiments, which position the C-domain of IF3 near the platform of the 30S subunit in direct contact with the 16S rRNA helices 23, 24 and 45. These data suggest blockage of the B2 bridges between rRNA helix 69 of the 50S subunit and 16S rRNA of the 30S subunit by IF3, which effectively inhibits 70S ribosome formation (Dallas and Noller, 2001).
The reason why the plateau value for the rate of subunit association in Figure 4B is smaller in the presence than in the absence of IF1 is that the rate constant for dissociation of IF3 from the 30S subunit is reduced by the presence of IF1. This interpretation is supported by the observation that the affinity of IF1 to the 30S subunit is enhanced by the presence of IF3 (Zucker and Hershey, 1986), since it follows from detailed balance considerations (Fersht, 1999, pp 125–127) that the affinity of IF3 to the 30S subunit must be also enhanced by the presence of IF1.
The rate of subunit docking when initiator tRNA and IF3 or only IF3 are in complex with the 30S subunit is very small or virtually zero, respectively (Figure 3D, Table II). Fast subunit docking in the presence of both initiator tRNA and IF2 (Figure 3D, Table II) can now be interpreted in terms of three types of effects. Firstly, addition of initiator tRNA to the 30S subunit enhances the rate of dissociation of IF3, which follows from the observation that IF3 reduces the affinity of fMet-tRNAfMet to the 30S subunit (Table I) and from detailed balance considerations (Fersht, 1999, pp. 125–127). Secondly, in the presence of fMet-tRNAfMet, IF2 greatly stimulates the rate of subunit association after release of IF3 (Figure 3C, Table II), which, thirdly, reduces the probability that IF3 rebinds to the 30S subunit before subunit docking.
The pivotal role played by IF2 in synergy with initiator tRNA for subunit docking is highlighted by two recent cryo-EM reconstructions of the initiation 70S ribosome complex with IF2 in the GTP form; one with E. coli components (Allen et al, 2005) and the other with components from Thermus thermophilus (Myasnikov et al, 2005). In both cases, the contact area between IF2 and the 50S subunit is remarkably large, which we deem to be the structural corollary to the large acceleration of subunit docking by IF2 as observed here (Figure 3C, Table II).
Initiation factors and the efficiency of initiation
Inspection of the Michaelis–Menten parameters (kcat, KM, kcat/KM) in Table II for combinations of initiation factors shows that the efficiency of initiation of protein synthesis (kcat/KM) is maximized by the simultaneous presence of IF1, IF2 and IF3 (Table II). Removal of IF1, IF3 or IF2 reduces the initiation efficiency seven-fold, nine-fold or 600-fold, respectively (Table II). At the in vivo concentration of fMet-tRNAfMet (∼1 μM) (Hershey, 1987; Gualerzi and Pon, 1990), the rate of initiation appears to be maximized by the three initiation factors (Equation (2), Table II), but at an elevated concentration of fMet-tRNAfMet the rate of initiation as defined in Equation (2) would, in fact, increase by deletion of either IF1 or IF3. These data suggest that the essential nature of IF1 (Cummings and Hershey, 1994) and IF3 (Olsson et al, 1996; Laursen et al, 2005) must have different causes than their stimulatory effects on the rate of initiation according to Equation (2). To understand why these factors are essential, one must therefore take other observations into account. We have found that a large fraction of 70S ribosomes, formed when fMet-tRNAfMet and 50S subunits are mixed with 30S·mRNA·IF1·IF2 but not with 30S·mRNA·IF1·IF2·IF3 complexes, lacks initiator tRNA (Figure 5B–D). This is because, firstly, the rate of subunit docking in the absence of initiator tRNA is quite large in the absence but very small in the presence of IF3 and virtually zero in the presence of both IF3 and IF1 (Figure 3A and B; Table II). Secondly, the rate of initiator tRNA association to the 30S subunit is very small in the absence of IF1 and IF3, whereas it is large in their presence (Table I). From this follows that IF1 and IF3 effectively prevent formation of initiator tRNA-less 70S ribosomes by greatly accelerating the binding of fMet-tRNAfMet to the 30S subunit and by greatly inhibiting docking of 50S subunits to initiator tRNA-less 30S subunits. These observations place the early conclusion that IF1 and IF3 maintain the pool of empty 30S subunits (Naaktgeboren et al, 1977; Hershey, 1987) in its functional context. In addition, IF3 and IF1 at high in vivo like concentrations can rapidly dissolve such inactive ribosome complexes (Figure 3B). From this we suggest that the essential nature of IF1 and of IF3 are to be found in their ability to minimize the fraction of ribosomes sequestered in an initiator tRNA lacking state, which allows for the large fraction of ribosomes in the elongation phase (∼80%) observed in vivo (Farewell and Neidhardt, 1998).
The time to initiate protein synthesis in vivo, including the times for binding of mRNA and initiator tRNA to the 30S subunit and for subunit docking, has been estimated as about 4 s (Ingraham et al, 1983; Farewell and Neidhardt, 1998). Here, we have estimated the time for initiator tRNA binding to the mRNA containing 30S subunit followed by subunit docking as about 0.4 s. However, in a pilot experiment where initiation was started from mRNA lacking 30S subunits (Figure 5B), the time for 70S ribosome formation was more than 2 s, showing mRNA binding to the 30S subunit to be rate limiting for initiation in this particular experiment and, by inference, possibly also in vivo.
The present data, finally, allow us to update the order of events in initiation of protein synthesis in the E. coli cell. After termination, the ribosome is split into subunits by the joint action of RRF and EF-G (Karimi et al, 1999; Peske et al, 2005; Zavialov et al, 2005). The released 30S subunit rapidly binds IF3, which ejects the deacylated tRNA from the partial P site of the 30S subunit, and leads to rapid release of the mRNA (Karimi et al, 1999; Peske et al, 2005). Thereafter, IF1, IF2 and a new mRNA associate with the 30S subunit. This is followed by rapid binding of initiator tRNA (Table I, Figure 1), which destabilizes IF3 in the 30S complex and induces release of the factor (Table II), allowing for rapid subunit docking and formation of a 70S ribosome containing IF2 in the GTP form and IF1 or, alternatively, rebinding of IF3 to the 30S subunit. In the former case, GTP is rapidly hydrolyzed and this induces release of IF2 and possibly IF1. In the latter case, IF3 will rapidly dissociate again, thereby giving the 50S subunit another chance to associate with the 30S subunit. These association and dissociation events involving IF3 will continue until, finally, the 50S subunit wins the race and unites with the 30S subunit.
Materials and methods
Chemicals and buffers
ATP, UTP, CTP, GTP and radioactive amino acids were from Amersham Biosciences (Sweden). tRNAPhe, PEP, myokinase (MK), pyruvate kinase (PK), putrescine, spermidine and nonradioactive amino acids were from Sigma (USA). All other chemicals were of analytical grade from Merck (Germany). All experiments were carried out in a polymix-like LS buffer containing (95 mM KCl, 5 mM NH4Cl, 5 mM Mg(OAc)2, 0.5 mM CaCl2, 8 mM putrescine, 1 mM spermidine, 5 mM potassium phosphate, 1 mM DTE, 1 mM GTP, 1 mM ATP and 2 mM PEP) at a pH of 7.5 (Jelenc and Kurland, 1979; Antoun et al, 2004).
Components of the translation system
Synthetic mMFTI mRNA with the sequence (gggAATTCGGGCCCTTGTTAACAATTAAGGAGGTA TACTatgtttacgatttaaTTGCAGaaaaaaaaaa aaaaaaaaaaa) encoding the tetra-peptide Met-Phe-Thr-Ile was prepared according to Pavlov et al (1997). 70S ribosomes, 50S and 30S subunits were prepared from the E. coli strain MRE 600 according to Rodnina and Wintermeyer (1995). Elongation factors, [3H]fMet-tRNAfMet and Phenylalanine tRNA synthetase (PheRS) were prepared according to Antoun et al (2004). One unit (U) of PheRS is the amount of PheRS that aminoacylates 1 pmol of tRNAPhe per second in LS buffer under standard conditions with phenylalanine at 0.1 mM and tRNAPhe at 1 μM concentration. N-terminally His-tagged initiation factors were prepared as described for His-tagged IF2 (Antoun et al, 2004).
Light scattering assays
Subunit docking and binding of fMet-tRNAfMet to the 30S subunit were monitored with light scattering after rapid mixing in a stopped-flow instrument (Bio-sequential SX-18MV, Applied Photophysics, Leatherhead, UK) as described (Antoun et al, 2003, 2004).
Effects of IF1 and IF3 on the binding of fMet-tRNAfMet to 30S·mRNA·IF2 complexes. Two mixtures, A and B, were prepared in LS buffer. A contained 4 μM mMFTI mRNA, 2 μM 30S subunits, 4 μM IF2 and different combinations of IF1 (4 μM) and IF3 (3 μM). B contained 4 μM of [3H]fMet-tRNAfMet.
Association of empty subunits. Two mixtures, A and B, were prepared in LS buffer. A contained 0.6 μM 30S subunits, whereas B contained different concentrations of 50S subunits.
Effects of initiation factors on the rate of 70S ribosome formation. Two mixtures, A and B, were prepared in LS buffer. A contained 0.6 μM 30S subunits, 1.2 μM mMFTI mRNA with different combinations of 4 μM IF1, 0.9 μM IF3, 0.9 μM IF2 and 0.9 μM of [3H]fMet-tRNAfMet. B contained 0.6 μM 50S subunits.
Docking of the 30S preinitiation complex to the 50S subunit at varying concentration. Two mixtures, A and B, were prepared in LS buffer. A contained 0.6 μM 30S subunits, 1.2 μM mMFTI mRNA, 0.9 μM IF2, 0.9 μM of [3H]fMet-tRNAfMet and 4 μM IF1, and also 0.9 μM IF3 when specified. B contained different concentrations of 50S subunits.
Docking of the 30S preinitiation complex to the 50S subunit at varying IF3 concentration. Two mixtures, A and B, were prepared in LS buffer. A contained 0.6 μM 30S subunits, 1.2 μM mMFTI mRNA, 4 μM IF1, 0.9 μM IF2, 0.9 μM of [3H]fMet-tRNAfMet and different concentrations of IF3 as specified. B contained 0.6 μM 50S subunits.
Rate of IF3 binding to the 30S preinitiation complex. To study the binding of IF3 to 30S preinitiation complexes in competition with 50S subunits, two mixtures A and B were prepared in LS buffer. A contained 0.6 μM 30S subunits, 1.2 μM mMFTI mRNA, 4 μM IF1, 0.9 μM IF2, 0.9 μM of [3H]fMet-tRNAfMet and 0.9 μM of IF3 when indicated. B contained 0.6 μM 50S subunits and 0.9 μM IF3 when indicated. Kinetics of 50S subunit binding was measured for the cases when IF3 was present only in A, only in B, and in the absence of IF3 in both mixtures.
Effects of IF1 and IF3 in the 30S·mRNA·IF2 complex on the rate of 70S formation upon addition of 50S subunits together with fMet-tRNAfMet. Two mixtures, A and B, were prepared in LS buffer. A contained 0.6 μM 30S subunits, 0.9 μM mMFTI mRNA, 0.9 μM IF2 and, when indicated, 4 μM IF1 and/or 0.9 μM IF3. B contained 0.9 μM of [3H]fMet-tRNAfMet and 0.6 μM 50S subunits. Alternatively, mMFTI mRNA was added in mixture B, instead of mixture A.
Dipeptide formation assay
Two mixtures A and B were prepared in LS buffer as described below. After preincubation for 10 min at 37°C, equal volumes of A and B were mixed in a quench-flow instrument (KinTek Corp., USA), incubated for specified times and quenched by 20% TCA. The samples were centrifuged and the amount of formed fMet-Phe-tRNAPhe in the pellet was determined by HPLC as described (Pavlov et al, 1997).
Effects of IF1 and IF3 in the 30S preinitiation complex on the rate of 50S subunit docking and subsequent dipeptide formation. Mixture A contained 0.9 μM 30S subunits, 1.6 μM mMFTI mRNA, 2 μM [3H]fMet-tRNAfMet, 1.6 μM IF2, 3 μM IF1 and 1.4 μM IF3. To study the effects of IF1 or IF3, the corresponding initiation factor was omitted from mixture A. Mixture B contained 0.9 μM 50S subunits, 5 μM EF-Tu, 0.2 μM EF-Ts, 2 μM tRNAPhe (Sigma, USA), 200 μM phenylalanine, 1 U/ml PheRS, 1 μg/ml PK and 0.1 μg/ml MK.
Effects of IF3 on the extent of elongation competent 70S ribosome formation. Mixture A contained 0.6 μM 30S subunits, 1.2 μM mMFTI mRNA, 0.9 μM IF2, 4 μM IF1 and 0.9 μM IF3 when specified. Mixture B contained 0.9 μM [3H]fMet-tRNAfMet and different concentrations of 50S subunits in the presence of 5 μM EF-Tu, 7.5 μM tRNAPhe (Sigma, USA), 30 μM phenylalanine, 1 U/ml PheRS, 1 μg/ml PK and 0.1 μg/ml MK.
Nitrocellulose assays
Two mixtures, A and B, were prepared in LS buffer. After preincubation for 10 min at 37°C, equal volumes (10 μl) of A and B were mixed and incubated for indicated times. Then, 15 μl of each reaction mixture was diluted in 6 ml ice-cold Polymix buffer (Jelenc and Kurland, 1979), rapidly filtered through BA85 nitrocellulose (NC) filters (Shleicher & Schuell, Dassel, Germany), put into vials with 5 ml Filter Safe (Zinsser Analytic, Germany) scintillation cocktail and counted in an LC6500 scintillation counter (Beckman, USA) to determine the amount of ribosome bound [3H]fMet-tRNAfMet.
The association rate of [3H]fMet-tRNAfMet to 30S·mRNA complexes. Mixture A contained 0.6 μM 30S subunits, 1.2 μM mMFTI mRNA and different combinations of IF1 (4 μM), IF2 (0.9 μM) and IF3 (0.9 μM). Mixture B contained 0.9 μM [3H]fMet-tRNAfMet.
The dissociation rate of [3H]fMet-tRNAfMet from 30S·mRNA complexes in the presence of different combinations of initiation factors. Mixture A contained 0.6 μM 30S subunits, 1.2 μM mMFTI mRNA, 0.9 μM [3H]fMet-tRNAfMet and different combinations of IF1 (4 μM), IF2 (0.9 μM) and IF3 (0.9 μM). Mixture B contained a 10-fold molar excess (9 μM) nonlabeled fMet-tRNAfMet.
The equilibrium constant for [3H]fMet-tRNAfMet binding to 30S·mRNA complexes. NC filtration was carried out here with one mixture, containing 0.3 μM 30S subunits, 0.6 μM mMFTI mRNA, specified combinations of IF1 (2 μM), IF2 (0.45 μM), IF3 (0.45 μM) and different concentrations of [3H]fMet-tRNAfMet. This mixture was incubated at 37°C for 1 h before quenching with ice-cold Polymix buffer and filtration.
Effects of IF3 on abortive 70S initiation complex formation. Mixture A contained 0.6 μM 30S subunits, 1.2 μM mMFTI mRNA, 0.9 μM IF2 and different combinations of IF1 (4 μM) and IF3 (0.9 μM). Mixture B contained 1.5 μM [3H]fMet-tRNAfMet and varying 50S subunit concentration. The incubation time was 30 s.
Data treatment
Kinetic parameters from light-scattering data. To estimate the first-order rate constant kc for subunit association from light-scattering data, the following relation was used:
![]()
Here, I0 is the initial, I(t) the current and Ieq the equilibrium intensity of the scattered light (Antoun et al, 2004). Relation (Equation (5)) is valid at equal initial concentrations of 30S and 50S subunits (see Supplementary data for details), which was the case for most experiments reported here. In the presence of fMet-tRNAfMet, IF3 and IF2 (with or without IF1) on the 30S subunit, kc is related to elemental rate constants (Figure 1A) according to Equation (1) (Supplementary data). In the absence of IF3, kc is given by
![]()
which allowed for estimation of the association rate constant ka,r for subunit docking (Antoun et al, 2004). When the concentration of 50S was much larger than the concentration of 30S subunits or when the binding of fMet-tRNAfMet to the 30S subunit was studied, the relation
![]()
was used to calculate kc from light-scattering data. The treatment of light-scattering data in more complex (and rare) cases of moderate excess of 50S subunits over 30S subunits is described in Supplementary data.
Presentation of light-scattering data. The original stopped-flow traces were used to calculate all kinetic parameters in Tables I and II. For clarity of presentation, we used a digital filter to reduce the noise and the numbers of data points in the figures.
Supplementary Material
Supplementary data
Acknowledgments
This work was supported by the Swedish Research Council and the Egyptian Mission Department.
References
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J (2005) The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121: 703–712 [DOI] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Andersson K, Tenson T, Ehrenberg M (2003) The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J 22: 5593–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Tenson T, Ehrenberg MM (2004) Ribosome formation from subunits studied by stopped-flow and Rayleigh light scattering. Biol Proc Online 6: 35–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne R, Ebes F, Voorma HO (1973) Sequence of events in initiation of protein synthesis. Eur J Biochem 38: 265–273 [DOI] [PubMed] [Google Scholar]
- Canonaco MA, Calogero RA, Gualerzi CO (1986) Mechanism of translational initiation in prokaryotes. Evidence for a direct effect of IF2 on the activity of the 30 S ribosomal subunit. FEBS Lett 207: 198–204 [DOI] [PubMed] [Google Scholar]
- Chaires JB, Pande C, Wishnia A (1981) The effect of initiation factor IF-3 on Escherichia coli ribosomal subunit association kinetics. J Biol Chem 256: 6600–6607 [PubMed] [Google Scholar]
- Croitoru V, Bucheli-Witschel M, Hagg P, Abdulkarim F, Isaksson LA (2004) Generation and characterization of functional mutants in the translation initiation factor IF1 of Escherichia coli. Eur J Biochem 271: 534–544 [DOI] [PubMed] [Google Scholar]
- Cummings HS, Hershey JW (1994) Translation initiation factor IF1 is essential for cell viability in Escherichia coli. J Bacteriol 176: 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas A, Noller HF (2001) Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell 8: 855–864 [DOI] [PubMed] [Google Scholar]
- Ehrenberg M, Bilgin N, Kurland C (1990) Design and use of a fast and accurate in vitro translation system. In Ribosomes and Protein Synthesis. A Practical Approach, Spedding G (ed), pp 101–128. Oxford, UK: IRL Press at Oxford University Press [Google Scholar]
- Fakunding JL, Hershey JW (1973) The interaction of radioactive initiation factor IF-2 with ribosomes during initiation of protein synthesis. J Biol Chem 248: 4206–4212 [PubMed] [Google Scholar]
- Farewell A, Neidhardt FC (1998) Effect of temperature on in vivo protein synthetic capacity in Escherichia coli. J Bacteriol 180: 4704–4710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A (1999) Structure and Mechanism in Protein Science: a Guide to Enzyme Catalysis and Protein Folding. New York: W.H. Freeman and Company [Google Scholar]
- Grunberg-Manago M, Dessen P, Pantaloni D, Godefroy-Colburn T, Wolfe AD, Dondon J (1975) Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. J Mol Biol 94: 461–478 [DOI] [PubMed] [Google Scholar]
- Gualerzi C, Risuleo G, Pon CL (1977) Initial rate kinetic analysis of the mechanism of initiation complex formation and the role of initiation factor IF-3. Biochemistry 16: 1684–1689 [DOI] [PubMed] [Google Scholar]
- Gualerzi C, Risuleo G, Pon C (1979) Mechanism of the spontaneous and initiation factor 3-induced dissociation of 30S.aminoacyl-tRNA.polynucleotide ternary complexes. J Biol Chem 254: 44–49 [PubMed] [Google Scholar]
- Gualerzi CO, Pon CL (1990) Initiation of mRNA translation in prokaryotes. Biochemistry 29: 5881–5889 [DOI] [PubMed] [Google Scholar]
- Hartz D, McPheeters DS, Gold L (1989) Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev 3: 1899–1912 [DOI] [PubMed] [Google Scholar]
- Hershey JW (1987) Protein synthesis. In Escherichia coli and Salmonella Typhimurium: Cellular and Molecular Biology, Neidhardt FC, Ingraham J, Low K, Magasanik B, Schaechter M, Umbarger H (eds), Washington, DC: American Society for Microbiology [Google Scholar]
- Ingraham JL, Maaloe O, Neidhardt FC (1983) Growth of Bacterial Cell. Sunderland, MA 01375: Sinauer Associates Inc. [Google Scholar]
- Janosi L, Hara H, Zhang S, Kaji A (1996) Ribosome recycling by ribosome recycling factor (RRF)—an important but overlooked step of protein biosynthesis. Adv Biophys 32: 121–201 [DOI] [PubMed] [Google Scholar]
- Jelenc PC, Kurland CG (1979) Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc Natl Acad Sci USA 76: 3174–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R, Pavlov MY, Buckingham RH, Ehrenberg M (1999) Novel roles for classical factors at the interface between translation termination and initiation. Mol Cell 3: 601–609 [DOI] [PubMed] [Google Scholar]
- Lancaster L, Noller HF (2005) Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol Cell 20: 623–632 [DOI] [PubMed] [Google Scholar]
- Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU (2005) Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev 69: 101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV (2006) The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J 25: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchin S, Putzer H, Hershey JW, Cenatiempo Y, Grunberg-Manago M, Laalami S (1999) In vitro study of two dominant inhibitory GTPase mutants of Escherichia coli translation initiation factor IF2. Direct evidence that GTP hydrolysis is necessary for factor recycling. J Biol Chem 274: 6074–6079 [DOI] [PubMed] [Google Scholar]
- Myasnikov AG, Marzi S, Simonetti A, Giuliodori AM, Gualerzi CO, Yusupova G, Yusupov M, Klaholz BP (2005) Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat Struct Mol Biol 12: 1145–1149 [DOI] [PubMed] [Google Scholar]
- Naaktgeboren N, Roobol K, Voorma HO (1977) The effect of initiation factor IF-1 on the dissociation of 70-S ribosomes of Escherichia coli. Eur J Biochem 72: 49–56 [DOI] [PubMed] [Google Scholar]
- Olsson CL, Graffe M, Springer M, Hershey JW (1996) Physiological effects of translation initiation factor IF3 and ribosomal protein L20 limitation in Escherichia coli. Mol Gen Genet 250: 705–714 [DOI] [PubMed] [Google Scholar]
- Pavlov MY, Freistroffer DV, MacDougall J, Buckingham RH, Ehrenberg M (1997) Fast recycling of Escherichia coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. EMBO J 16: 4134–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Rodnina MV, Wintermeyer W (2005) Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell 18: 403–412 [DOI] [PubMed] [Google Scholar]
- Petrelli D, LaTeana A, Garofalo C, Spurio R, Pon CL, Gualerzi CO (2001) Translation initiation factor IF3: two domains, five functions, one mechanism? EMBO J 20: 4560–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon CL, Gualerzi CO (1984) Mechanism of protein biosynthesis in prokaryotic cells. Effect of initiation factor IF1 on the initial rate of 30S initiation complex formation. FEBS Lett 175: 203–207 [DOI] [PubMed] [Google Scholar]
- Risuleo G, Gualerzi C, Pon C (1976) Specificity and properties of the destabilization, induced by initiation factor IF-3, of ternary complexes of the 30-S ribosomal subunit, aminoacyl-tRNA and polynucleotides. Eur J Biochem 67: 603–613 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Wintermeyer W (1995) GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc Natl Acad Sci USA 92: 1945–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian AR, Davis BD (1970) Activity of initiation factor F3 in dissociating Escherichia coli ribosomes. Nature 228: 1273–1275 [DOI] [PubMed] [Google Scholar]
- Wintermeyer W, Gualerzi C (1983) Effect of Escherichia coli initiation factors on the kinetics of N-Acphe-tRNAPhe binding to 30S ribosomal subunits. A fluorescence stopped-flow study. Biochemistry 22: 690–694 [DOI] [PubMed] [Google Scholar]
- Wishnia A, Boussert A, Graffe M, Dessen PH, Grunberg-Manago M (1975) Kinetics of the reversible association of ribosomal subunits: stopped-flow studies of the rate law and of the effect of Mg2+. J Mol Biol 93: 499–515 [DOI] [PubMed] [Google Scholar]
- Wu XQ, RajBhandary UL (1997) Effect of the amino acid attached to Escherichia coli initiator tRNA on its affinity for the initiation factor IF2 and on the IF2 dependence of its binding to the ribosome. J Biol Chem 272: 1891–1895 [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Hauryliuk VV, Ehrenberg M (2005) Splitting of the posttermination ribosome into subunits by the concerted action of RRF and EF-G. Mol Cell 18: 675–686 [DOI] [PubMed] [Google Scholar]
- Zucker FH, Hershey JW (1986) Binding of Escherichia coli protein synthesis initiation factor IF1 to 30S ribosomal subunits measured by fluorescence polarization. Biochemistry 25: 3682–3690 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


