Abstract
Hsp70 molecular chaperones function in protein folding in a manner dependent on regulation by co-chaperones. Hsp40s increase the low intrinsic ATPase activity of Hsp70, and nucleotide exchange factors (NEFs) remove ADP after ATP hydrolysis, enabling a new Hsp70 interaction cycle with non-native protein substrate. Here, we show that members of the Hsp70-related Hsp110 family cooperate with Hsp70 in protein folding in the eukaryotic cytosol. Mammalian Hsp110 and the yeast homologues Sse1p/2p catalyze efficient nucleotide exchange on Hsp70 and its orthologue in Saccharomyces cerevisiae, Ssa1p, respectively. Moreover, Sse1p has the same effect on Ssb1p, a ribosome-associated isoform of Hsp70 in yeast. Mutational analysis revealed that the N-terminal ATPase domain and the ultimate C-terminus of Sse1p are required for nucleotide exchange activity. The Hsp110 homologues significantly increase the rate and yield of Hsp70-mediated re-folding of thermally denatured firefly luciferase in vitro. Similarly, deletion of SSE1 causes a firefly luciferase folding defect in yeast cells under heat stress in vivo. Our data indicate that Hsp110 proteins are important components of the eukaryotic Hsp70 machinery of protein folding.
Keywords: chaperones, Hsp70, Hsp110, Ssa1p, Sse1p
Introduction
Heat shock proteins of 70 kDa (Hsp70) comprise a major family of molecular chaperones involved in the cellular folding of newly synthesized proteins and the prevention of protein aggregation under stress conditions (Bukau and Horwich, 1998; Frydman, 2001; Hartl and Hayer-Hartl, 2002). Canonical Hsp70s, like the bacterial DnaK and the eukaryotic Hsc70, consist of a highly conserved ∼44 kDa N-terminal ATPase domain of regulatory function and a ∼27 kDa peptide-binding domain (PBD) with affinity to hydrophobic peptide segments exposed in non-native proteins (DeLuca-Flaherty et al, 1990; Zhu et al, 1996). In the ATP-bound state, peptide associates and dissociates rapidly from Hsp70, but remains tightly bound in the ADP-bound state. Repeated binding and release is thought to drive the folding of substrate proteins in an iterative process until the native state is achieved and hydrophobic segments are no longer exposed. For their physiological function, canonical Hsp70s require two auxiliary proteins, a J-domain protein and a nucleotide exchange factor (NEF). J-domain proteins trigger ATP hydrolysis by Hsp70 and induce tight peptide binding. NEFs (GrpE in bacteria, HspBP1/Fes1p and BAG-1 in eukaryotes) catalyze the release of ADP from Hsp70, allowing the binding of ATP, and thereby drive the folding cycle forward. A major difference between the bacterial and eukaryotic isoforms of Hsp70 lies in the kinetics of these reactions. Whereas the bacterial Hsp70 homologue DnaK is locked in the ADP-bound state and cannot function efficiently in the absence of the NEF GrpE, nucleotide exchange of eukaryotic Hsp70 becomes rate-limiting only under strong stimulation of ATP hydrolysis (Liberek et al, 1991; Brehmer et al, 2001).
Eukaryotes contain a cytosolic protein class related to Hsp70 named after the mammalian Hsp110, which differs from Hsp70 by a more divergent ATPase domain and insertions of varying lengths in the C-terminal domain (Lee-Yoon et al, 1995; Easton et al, 2000). Various cytosolic Hsp110 isoforms have been described including Hsp110 (also known as Hsp105), Apg-1, and Apg-2 in mammalian cytosol; and Sse1p and Sse2p in the cytosol of yeast (Yasuda et al, 1995; Chen et al, 1996; Craven et al, 1996; Kaneko et al, 1997). The yeast genes encoding Sse1p and Sse2p are not individually essential, but deletion of both SSE1 and SSE2 results in synthetic lethality, while deletion of SSE1 alone confers a growth defect (Mukai et al, 1993; Shaner et al, 2004). Initially, Hsp110 family proteins were characterized as ‘holdases' that keep denatured proteins in solution, but no endogenous client proteins have been described so far (Oh et al, 1997, 1999). More recent evidence suggests a cooperation between yeast Hsp110 Sse1p and the Hsp90 chaperone machinery (Lu and Cyr, 1998; Liu et al, 1999; Goeckeler et al, 2002; Zhao et al, 2005b). Another group of Hsp70-related proteins with C-terminal extensions resides in the ER, including mammalian Grp170 and yeast Lhs1p. The C-terminal segments of these Hsp170s differ from those of the Hsp110s (Chen et al, 1996; Craven et al, 1996).
Association of Hsp110 with Hsp70 family members has also been reported, but only recently have studies established the interaction between mammalian Hsp105α and Hsp70; plant Hsp101 and Hsp70; Sse1p and Ssa1p; Sse1p and Ssb1p, a ribosome-associated yeast Hsp70 (Wang et al, 2000; Steel et al, 2004; Yamagishi et al, 2004; Shaner et al, 2005; Yam et al, 2005; Zhang and Guy, 2005). In the mammalian complex, only the ultimate C-terminus and a charged loop in the PBD of Hsp105α are dispensable for complex formation (Yamagishi et al, 2004). Hsp105α was additionally shown to synergistically increase the ATPase rate of Hsp70 in the presence of the J-domain protein Hsp40, but to inhibit folding of the model substrate firefly luciferase. Acceleration of ATP hydrolysis was also reported for the Sse1p–Ssa1p–Ydj1p system (Shaner et al, 2005).
Perhaps the most intriguing clue as to how Hsp110 family proteins may regulate Hsp70 function came from a recent analysis of the ER-lumenal Lhs1p in yeast (Steel et al, 2004). Similar to the ER-resident NEF Sls1p/Sil1p, Lhs1p catalyzes nucleotide exchange on the ER-homologue of Hsp70, Kar2p, which results in an overall enhancement of Kar2p ATPase activity. On the other hand, Kar2p triggers the ATPase activity of Lhs1p.
Here we report that human Hsp110 and the yeast homologues Sse1p and Sse2p function generally as NEFs for cytosolic Hsp70s. Mutational analysis revealed that a functional ATPase domain and the C-terminal region are important for NEF activity. Hsp110 releases peptide substrate from Hsp70 in an ATP-dependent manner. Equimolar addition of the yeast Hsp110, Sse1p, to the refolding reaction of thermally denatured firefly luciferase with yeast Hsp70 and Hsp40 considerably increased not only the yield but also the rate of refolding. Excess Sse1p, however, inhibited luciferase refolding. Consistently, deletion or overexpression of SSE1 resulted in a firefly luciferase folding defect in yeast cells subjected to heat shock. Taken together, the data indicate that Hsp110 proteins constitute an important additional element of the Hsp70 machinery of protein folding in eukaryotic cells.
Results
Hsp110s enhance nucleotide exchange on Hsp70s
We set out to determine whether cytosolic Hsp110 class chaperones could act as NEFs for Hsp70 class chaperones. To this end, we used the fluorescent ADP analog, N8-(4-N′-methylanthraniloylaminobut yl)-8-aminoadenosine 5′-diphosphate (MABA-ADP). The fluorescence of MABA-ADP decreases upon release from Hsp70 and can be monitored using stopped flow fluorimetry (Theyssen et al, 1996; Packschies et al, 1997). MABA-ADP is an ideal spectroscopic probe for this purpose, as it has been shown to be similar to ADP with respect to Hsp70 in all kinetic and thermodynamic parameters tested to date.
We first determined whether Sse1p, a yeast Hsp110 homologue, could stimulate nucleotide release from Ssa1p, a binding partner of Sse1p in the yeast cytosol (Shaner et al, 2005; Yam et al, 2005). Equimolar amounts of MABA-ADP and recombinant Ssa1p were incubated to form MABA-ADP/Ssa1p complexes, which were then mixed with an excess of ADP in the presence or absence of Sse1p (Figure 1A). Both resulting time-dependent MABA-ADP fluorescence signals exhibited a single-exponential decay, indicating a first-order reaction with a dissociation rate constant (koff) of 0.47±0.01 s−1 in the absence of Sse1p, and 5.73±0.06 s−1 in the presence of Sse1p (Supplementary Figure S1). The ∼12-fold acceleration of MABA-ADP dissociation in the presence of Sse1p indicates that this protein can indeed function as an NEF for Ssa1p and probably also for its close relatives, Ssa2p and the stress inducible isoforms, Ssa3p and Ssa4p. Next, we compared the nucleotide exchange efficiency of Sse1p with that of Fes1p, the only other known yeast cytosolic NEF (Figure 1B). In both cases, the time-dependent fluorescence signal exhibited a single exponential decay, but Fes1p increased the koff only up to 2.07±0.02 s−1, which was more than two times slower than the release obtained with Sse1p. These results suggest that Sse1p is a more efficient NEF than Fes1p.
Figure 1.
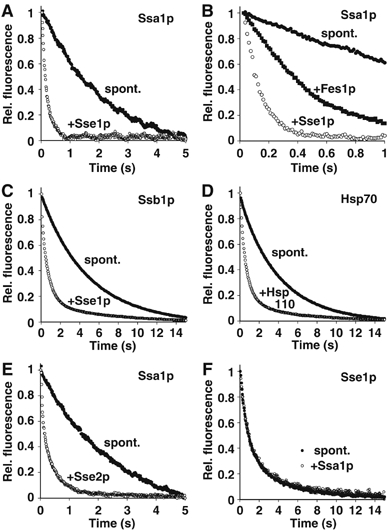
Hsp110 accelerates nucleotide exchange on Hsp70. (A) Accelerated dissociation of MABA-ADP from Ssa1p as monitored by stopped-flow fluorescence spectroscopy. Equimolar amounts of MABA-ADP and the indicated Hsp70 homologue were preincubated to form a complex, which was then mixed with an excess of ADP either in presence or absence of Sse1p. (B) Comparison of the efficiencies of nucleotide exchange in the presence of Sse1p and the canonical NEF, Fes1p. When applied at the same concentration (0.5 μM), Sse1p accelerates nucleotide release more efficiently than Fes1p. (C) Accelerated dissociation of MABA-ADP from Ssb1p. (D) The effect of human Hsp110 on the displacement of MABA-ADP from human Hsp70. (E) Release of MABA-ADP from Ssa1p by Sse2p. (F) Ssa1p does not trigger displacement of MABA-ADP from Sse1p. In all panels, effector-triggered and spontaneous dissociation is indicated by open and closed circles, respectively. Fes1p-triggered dissociation is indicated by closed squares. The molar ratio of effector to Hsp70 was 1:2 in all experiments.
Yeast cytosol furthermore contains the highly similar, ribosome-associated proteins Ssb1p and Ssb2p, which belong to a more distantly related class of canonical Hsp70s. We therefore tested the effect of Sse1p on Ssb1p/MABA-ADP complexes and observed a ∼5-fold acceleration of MABA-ADP dissociation (Figure 1C), indicating that a conserved mechanism applies to the Sse1p interaction with both yeast Hsp70 homologues, Ssa1p and Ssb1p. Analogous experiments were also performed using the human homologues, Hsp70 and Hsp110 (HspH1) (Figure 1D). In this case, the dissociation rate constant of MABA-ADP was increased ∼5-fold in the presence of human Hsp110 (Supplementary Figure S1), suggesting that the nucleotide exchange function of Hsp110s on Hsp70s is conserved from yeast to humans.
The cytosol of Saccharomyces cerevisiae contains another isoform of Sse1p, Sse2p. Sse1p and Sse2p have a sequence identity of 76%, but under normal growth conditions, the expression level of Sse2p stays far below that of its relative (Mukai et al, 1993; Ghaemmaghami et al, 2003). With this in mind, we tested whether Sse2p can compensate for its low concentration by increased nucleotide exchange activity on Ssa1p (Figure 1E), but found a similar ∼7-fold increase in the nucleotide exchange rate of MABA-ADP (Supplementary Figure S1), suggesting that the role of Sse2p may be to provide extra assistance to Sse1p during conditions of stress.
Since Sse1p and Ssa1p might form a pseudo-symmetrical dimer that exerts similar effects on both of its partners, we also tested whether Ssa1p might conversely act as an NEF for Sse1p. For this purpose, we loaded Sse1p with MABA-ADP and monitored release triggered by mixing with Ssa1p and an excess of ADP (Figure 1F). Here, however, no accelerated dissociation of the nucleotide analog was observed, indicating the absence of mutuality in the Sse1p–Ssa1p complex with respect to nucleotide exchange. Similarly, Sse1p did not exert nucleotide exchange activity on Sse1p–MABA-ADP (data not shown). In summary, these data corroborate the clearly asymmetrical roles of Sse1p and Ssa1p in the heterodimer, despite their assumed overall structural similarity.
Titration of NEFs
To estimate the maximal nucleotide release rates caused by Sse1p versus Fes1p, we tested the effect of increasing concentrations of these NEFs on Ssa1p/MABA-ADP complexes. Already at 6 μM of Sse1p, a nucleotide release rate of 95±16 s−1 was obtained, reaching the upper limit of detection for our stopped-flow instrument (Figure 2A). After plotting the obtained koff values versus Sse1p concentration, it was clear that the titration curve did not reach saturation (Figure 2C). The best fit for the data points was a sigmoidal curve with a maximal release rate of 114±3 s−1. As saturation was not obtained, it remains unclear if the presented fit is the best description of the Sse1p–Ssa1p interaction. In contrast, saturation was reached in the titration experiment with Fes1p, and a maximal release rate of 29±3 s−1 with Kd of 5.1±0.9 μM was determined, based on a hyperbolic curve fit (Figure 2B and C). Taken together, these data confirm that Sse1p is a more efficient NEF than Fes1p.
Figure 2.
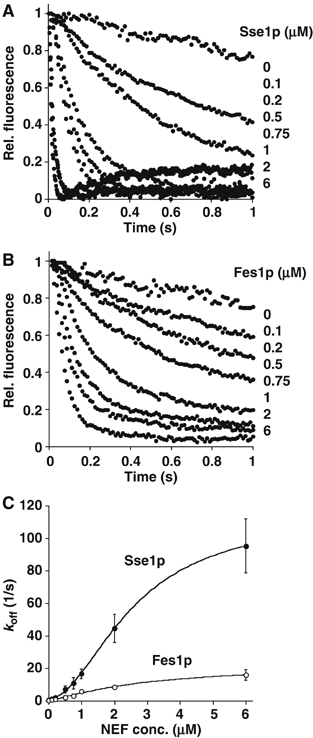
Dependence of nucleotide dissociation rate constant on NEF concentration. (A) Time course of displacement of MABA-ADP from Ssa1p–MABA-ADP complexes in the presence of increasing concentrations (0–6 μM) of Sse1p or (B) the canonical NEF Fes1p. For clarity, only the first second of traces has been shown. In both cases, higher concentrations of NEF lead to faster nucleotide exchange. (C) Observed dissociation rate constants were plotted versus NEF concentration. Each data point represents the mean of three measurements. The maximal nucleotide dissociation constant was estimated to be 114 s−1 for Sse1p and 29 s−1 for Fes1p.
Functional analysis of Sse1p mutants
Since Sse1p is a rather large multidomain protein compared to the conventional NEFs of Hsp70, HspBP1 and BAG-1, we wondered whether Sse1p could be functionally dissected into domains. In order to identify regions of Sse1p critical for nucleotide exchange on Hsp70, truncation mutants of Sse1p were generated and tested for nucleotide exchange activity on Ssa1p and Ssb1p using the stopped-flow assay (Figure 3). Consistent with previous reports, a fragment comprising the N-terminal ATP-binding domain (fragment 3, residues 1–383) could not be expressed as a soluble protein in Escherichia coli, precluding further analysis (Shaner et al, 2004). The remaining C-terminal fragment containing the PBD (fragment 4, residues 393–693) was soluble, but exhibited no NEF activity. Truncation mutants, in which a short unstructured region and two α-helical segments at the C-terminus of Sse1p were incrementally removed (secondary structure assignment based on prediction by the Jpred server (Cuff et al, 1998)), were successfully purified as soluble proteins, but also exhibited no nucleotide exchange activity (Figure 3A, fragments 5–7 and Figure 3B and C). The loss in NEF activity correlated with a reduced binding affinity of the mutant Sse1p proteins for Hsp70, as revealed in a binding assay with Ssb1 (Supplementary Figure S2). Sse1p mutants were incubated with Ssb1p, and the resulting complexes were immunoprecipitated with α-Ssb1p. Consistent with previous studies, Sse1p stably interacted with Ssb1p (Shaner et al, 2005; Yam et al, 2005). Introduction of the G233D mutation into Sse1p reduced its interaction with Ssb1p (this study and Shaner et al, 2005). Sse1p 1–674 (fragment 5) also interacted weakly with Ssb1p, while Sse1p 1–649 (fragment 6) and Sse1p 1–636 (fragment 7) did not detectably bind. Taken together, these findings suggest that both the ATP-binding domain and the less-conserved C-terminus of Sse1p are important for physical as well as functional interactions with Hsp70.
Figure 3.
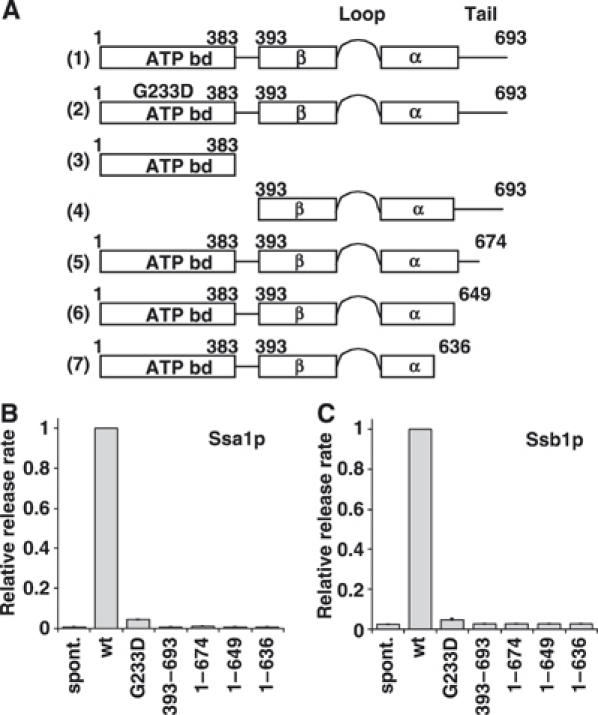
Mutational analysis of Sse1p function. (A) Schematic representation of a set of Sse1p constructs assayed for their ability to accelerate nucleotide exchange on Ssa1p and Ssb1p. (B, C) Nucleotide exchange activity of the Sse1p constructs on Ssa1p and Ssb1p, respectively. Sse1p variants and Ssa1p or Ssb1p were mixed at a molar ratio of 5:1. In the bar graphs, the average nucleotide exchange activity of wt Sse1p was set to 1. The ATP binding mutant G233D retained only about 5% of wt activity, that is, koff is slightly above the spontaneous release rate.
We also tested the influence of the Sse1p-G233D point mutant on the displacement of MABA-ADP from Ssa1p and Ssb1p (Figure 3B and C). In order to resolve rather subtle effects, a five-fold excess of Sse1p mutants over Ssa1p was used in these experiments. Surprisingly, the release rate of MABA-ADP was reduced more than 95% relative to wild-type (wt) Sse1p in the presence of Sse1p-G223D, suggesting that the nucleotide exchange activity of Sse1p requires a functional ATP binding domain. In a companion study, similar experiments were performed using another Sse1p mutant, K69M, which binds ATP (or ADP), but cannot hydrolyze ATP (Raviol et al, 2006). Surprisingly, the rate of MABA-ADP release from Ssa1p in the presence of the Sse1p K69M mutant was approximately the same as in the presence of wt Sse1p. However, the K69M mutant retains the capability to bind Sse1p (Shaner et al, 2005).
In a limited proteolysis experiment probing for conformational changes of Sse1p in the absence and presence of nucleotide, no major changes in the fragmentation pattern were observed, suggesting only limited conformational rearrangements in Sse1p upon nucleotide binding (data not shown). Nucleotide-induced conformational shifts may however only become apparent in the complex with Ssa1p. On the other hand, we cannot exclude a specific effect of the G223D mutation in Sse1p.
Effect of Hsp110 on the interaction of Hsp70 with peptide substrate
In canonical Hsp70 proteins, ATP binding after nucleotide exchange triggers dissociation of the bound peptide substrate, allowing further cycles of protein folding. Accelerated substrate release in the presence of ATP is therefore another hallmark of NEF activity. In order to determine how Hsp110 influences substrate release from Hsp70, we utilized a fluorescent dye-labeled peptide probe, Dansyl-NRLLLTGC (D-NR). The peptide D-NR binds with high affinity and specificity to Hsp70, and its release from Hsp70 is characterized by a decrease in its fluorescence, which can be monitored using stopped-flow fluorimetry (Gragerov et al, 1994; Gässler et al, 2001). We first determined whether Hsp110 could influence the displacement of D-NR from nucleotide free Hsp70-D-NR complexes. Preformed nucleotide free Hsp70-D-NR complex was mixed with the indicated combinations of excess NR peptide, ATP, and Hsp110 (Figure 4A). Upon mixing of nucleotide free Hsp70-D-NR with NR, or with both NR and Hsp110, D-NR remained stably bound to Hsp70. Upon mixing the complex with NR and ATP, the half-time (t1/2) of release of D-NR from Hsp70 was estimated to be ∼16.9 s (Figure 4A and D). However, a substoichiometric amount of Hsp110 (Hsp110:Hsp70 ratio=1:2) together with NR and ATP accelerated D-NR release by four-fold, corresponding to a half-time of 3.9 s. These data indicate that Hsp110 enhances substrate release in an ATP-dependent manner. For comparison, similar experiments were performed in the presence of HspBP1 to analyze the influence of a conventional NEF on the displacement of peptide from nucleotide free Hsp70–D-NR complex (Figure 4B and D). Consistent with previous reports, HspBP1 also accelerated substrate release from Hsp70 in an ATP-dependent manner (Shomura et al, 2005). Another identical set of experiments using ADP–Hsp70–D-NR complex indicated that the presence of ADP did not alter the rate of D-NR release achieved by Hsp110 (Figure 4C and D). Taken together, these data suggest that Hsp110 releases substrates from Hsp70 like a conventional NEF.
Figure 4.
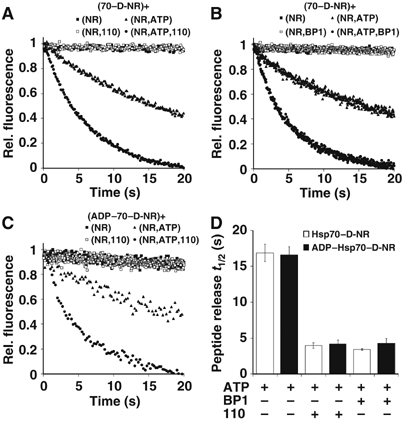
Hsp110 accelerates substrate release from Hsp70. (A) Hsp110 accelerates displacement of fluorescent dye-labeled peptide (D-NR) from nucleotide free Hsp70–D-NR complex in an ATP-dependent manner. Hsp70–D-NR was mixed with the indicated combination of unlabeled peptide (NR), Hsp110 (110), and ATP, and release of D-NR was monitored by stopped-flow fluorimetry. Components shown in parentheses indicate content of stopped-flow syringes, and ‘+' indicates the mixing event. (B) Acceleration of D-NR release from nucleotide free Hsp70–D-NR complex triggered by the NEF HspBP1. (C) Hsp110-mediated acceleration of D-NR release from an ADP–Hsp70–D-NR complex in an ATP-dependent manner. Preformed ADP–Hsp70–D-NR complex was mixed with the indicated components and analyzed as in (A). (D) Bar graph representation of the peptide (D-NR) release half times from Hsp70–D-NR and ADP–Hsp70–D-NR complexes under the conditions tested.
Hsp110 enhances the refolding activity of Hsc70 in an Hsp40-dependent manner
Hsp110 class chaperones are thought to prevent aggregation by binding protein substrates during thermal denaturation and holding them in a folding-competent state (Oh et al, 1997, 1999). It is possible however, that Hsp110 has an additional role in the refolding process that may be mediated by its nucleotide exchange activity. We therefore asked whether Hsp110 influences the refolding of a heat-denatured substrate, in a manner that is independent of its holdase function during denaturation. Firefly luciferase was denatured at 42°C in the presence or absence of Hsc70, and refolding was initiated at 30°C by addition of ATP in the presence or absence of Hsp40 and/or Hsp110 (Figure 5A). When luciferase was denatured in the presence of Hsc70 to prevent aggregation, followed by refolding in the absence of other chaperones, it was reactivated to less than 10% of its prior activity level. The reactivation of luciferase was only slightly improved when either Hsp40 or Hsp110 was added to the refolding reaction, but was greatly improved when both Hsp40 and Hsp110 were added, with luciferase activity reaching more than 50% of its previous level. This is in line with an NEF function of Hsp110 because NEF activity is dispensable for eukaryotic Hsc70 without additional ATPase stimulation by Hsp40. The folding yield remained unchanged when the sequence of chaperones was reversed, that is, when Hsp110 was incubated with luciferase during unfolding (Figure 5B). Thus, Hsc70 was as effective as Hsp110 in prevention of protein aggregation, arguing against a predominant holdase function of Hsp110. Our data instead suggest that Hsp110, acting together with Hsc70 and Hsp40, promotes the refolding of denatured protein substrates.
Figure 5.
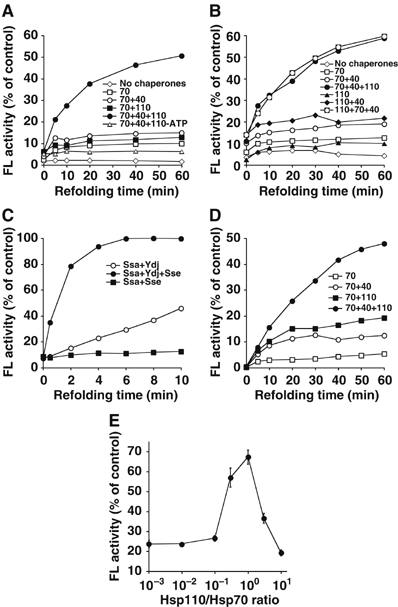
Hsp110 and Hsp70 mediate luciferase refolding in an Hsp40-dependent manner. (A) Luciferase refolding after preincubation with Hsc70. Firefly luciferase was heat-denatured at 42°C in the presence of Hsc70. Refolding was performed at 30°C in the presence of either Hsp110, Hsp40, or both, and ATP. In total, 100% activity corresponds to the enzyme activity of native luciferase in refolding buffer. (B) Luciferase refolding after preincubation with Hsp110 or Hsc70. Here, luciferase was denatured at 42°C in presence of Hsp110 or Hsc70 and folded at 30°C by adding ATP and different combinations of Hsc70, Hsp110, and Hsp40. Folding yields were similar irrespective of the order of Hsc70 and Hsp110 addition, indicating that the prevention of aggregation activity of Hsp110 is not a critical determinant for efficient protein refolding by the Hsp70–Hsp110 system. (C) Luciferase refolding by Ssa1p, Sse1p, and Ydj1p (yeast Hsp70/Hsp110/Hsp40). Luciferase was denatured in the presence of Ssa1p as described in (A). (D) Refolding of chemically denatured luciferase by Hsp70/Hsp110/Hsp40. (E) Hsp110 concentration dependence of luciferase refolding by the Hsp70/Hsp110/Hsp40 system. The concentrations of all other components were unchanged.
We next tested whether Sse1p had the same effect as mammalian Hsp110 on Hsp70-assisted refolding. Firefly luciferase was heat-denatured in the presence of Ssa1p and refolding was initiated at 30°C in the presence or absence of Ydj1p and/or Sse1p and ATP (Figure 5C). Within 10 min of incubation, luciferase activity reached ∼40% of its prior activity when Ydj1p was added to the refolding reaction, but regained full activity when Sse1p was also present. Strikingly, addition of Sse1p accelerated the rate of refolding by more than 10-fold, an effect that was not observed with Fes1p as NEF (data not shown). The overall efficiency of refolding in the presence of Ssa1p, Sse1p and Ydj1p exceeded that observed with their mammalian counterparts. While this difference between the two systems might reflect different physiological folding demands, the ability to enhance refolding of substrates after denaturation is apparently conserved among Hsp110s.
During heat denaturation, luciferase loses its activity rapidly but probably becomes only partially unfolded. To test whether the Hsp70/40/110 system can handle a completely unfolded protein resembling a nascent protein at the ribosome, we have performed similar experiments with luciferase denatured in guanidinium chloride. Hsc70 alone mediated refolding of such a substrate with a yield of up to 5% of its prior activity (Figure 5D). Addition of either Hsp40 or Hsp110 increased the yield of the refolded luciferase to 15 and 20%, respectively. However, when Hsp40 and Hsp110 were added together with Hsc70, 50% of luciferase was refolded. This agrees well with the data obtained for heat-denatured luciferase, suggesting that the Hsp70/40/110 folding system can handle both thermally and chemically denatured protein substrates.
Titration of Hsp110 with the other components held at constant concentration revealed that Hsp110 is most effective for folding at an equimolar ratio to Hsc70 (Figure 5E). This could mean that Hsp110 operates as a stoichiometric component of a catalytic complex, but the details of the Hsp70/40/110 folding mechanism remain to be analyzed. We have also observed that Hsp110, when added in 10-fold excess, can have a negative effect on the refolding process, perhaps by accelerating substrate cycling to an extent where the reaction becomes unproductive for folding.
Does Hsp110 also improve luciferase folding in vivo? To this end, we compared the activity of newly synthesized luciferase expressed in wt yeast, an SSE1 deletion strain (sse1Δ), and an SSE1 overexpressing strain (SSE1↑). Western blot analysis of extracts from these strains revealed that overexpression increased Sse1p levels ∼4-fold (data not shown). Equal amounts of yeast cells were transferred into media that induced luciferase synthesis and grown at 37°C. At various time points after induction, luciferase activity was measured in vivo and found to be significantly lower in the sse1Δ and SSE1↑ strains relative to wt (Figure 6A). Parallel analyses indicated that the kinetics of luciferase synthesis, as well as the solubility of luciferase, were similar among all three yeast strains (Figure 6B and data not shown), thus ruling out that Sse1p influences luciferase synthesis or solubility. Specific luciferase activity (SLA) determined 6 h postinduction revealed that deletion or overexpression of SSE1 reduced the SLA to 27 or 54% of wt levels, respectively (Figure 6A), suggesting the accumulation of substantial amounts of misfolded but soluble protein. These results indicate that Sse1p is an important component of the folding machinery in vivo, in line with findings from a companion study that used yeast strains with a different genetic background (Raviol et al, 2006). Furthermore, the fes1Δ mutant has similarly been shown to exhibit a defect in luciferase folding at 37°C (Shomura et al, 2005).
Figure 6.
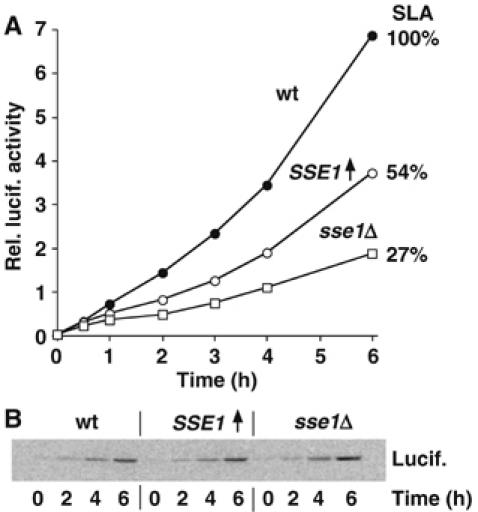
De novo folding of luciferase under thermal stress conditions is dependent on Sse1p levels. (A) Luciferase activity measurements in wt yeast, an SSE1 deletion strain (sse1Δ), and an SSE1 overexpression strain (SSE1↑). Equal numbers of cells were transferred into media that induces luciferase synthesis and grown at 37°C. Aliquots of cells were taken at the indicated time points after induction, and luciferase activity was measured in vivo. SLA was determined 6 h postinduction, revealing that deletion and overexpression of SSE1 reduced the SLA to 27 and 54% of wt levels, respectively. (B) Luciferase protein levels are similar in analyzed yeast strains. Aliquots of cells from (A) were lysed at the indicated time points, and luciferase protein levels were analyzed via SDS–PAGE and Western blotting.
Discussion
In this study, we have investigated the functional interaction between Hsp110 class molecular chaperones and Hsp70s involved in de novo folding and in re-folding of stress-denatured proteins. Hsp110s function as NEFs for Hsp70s, and consistently, Hsp110 regulates the kinetics of Hsp70-substrate interactions. Mutational analyses indicate that both the ATPase domain and the C-terminal peptide binding domain are necessary for the NEF function of Sse1p, the S. cerevisiae Hsp110 homologue. Additional experiments suggest that Sse1p modulates the efficiency of firefly luciferase folding in vivo, and refolding in vitro. Together with recent data reporting stable complex formation between Sse1p and Ssa1p (Shaner et al, 2005; Yam et al, 2005), our findings suggest that this complex, together with Ydj1p, is a functional folding unit in the yeast cytosol, incorporating both ATPase stimulation and nucleotide exchange activity.
Nucleotide exchange activity of Hsp110s
Both Hsp110s and Hsp170s are evolutionarily divergent families of Hsp70s, which reside in the cytosol and the ER-lumen, respectively. Interestingly, recent studies have shown that Lhs1p, a yeast member of the Hsp170 family, acts as an NEF for the major ER-lumenal Hsp70, Kar2p, which in turn stimulates the ATPase activity of Lhs1p (Steel et al, 2004). Since the folding pathways and the respective machineries in the ER lumen and cytoplasm differ substantially, it seemed possible that such a regulatory mechanism may be restricted to the ER. Our results demonstrate that the Hsp110s have NEF activity for cytosolic Hsp70s. We found that Sse1p catalyzed nucleotide exchange on both Ssa1p and Ssb1p (Figure 1), and thus presumably on the four remaining isoforms of Ssa and Ssb present in the cytoplasm of S. cerevisiae. However, we were unable to study NEF activity on Ssz1p, a noncanonical ribosome-associated Hsp70, or its stable complex with the J-domain protein zuotin, RAC (ribosome-associated complex), with our stopped-flow assay, since MABA-ADP did not bind efficiently to these components. Both yeast Hsp110 isoforms, Sse1p and Sse2p, were equally efficient in their nucleotide exchange activity assayed on Ssa1p. Furthermore, we found NEF activity on Hsp70 for human Hsp110, which is more distantly related to Sse1p (39% identity). Thus, Hsp70 NEF activity appears to be a generally conserved property of Hsp110 homologues in eukaryotes.
When Sse1p functions as an efficient NEF for Ssa1p, why is there a need for an additional NEF, Fes1p, in the yeast cytosol? Perhaps the most likely explanation is that the functions of these two structurally unrelated NEFs overlap only partially (Shomura et al, 2005). For instance, we failed to detect a significant acceleration of Ssa1p/Ydj1p-mediated firefly luciferase refolding in presence of Fes1p (unpublished observations), while Sse1p increased both folding rate and yield in this model system (Figure 5C). Furthermore, while Sse1p was found associated with various Hsp90–Hsp70–substrate complexes in a genome-wide screen, Fes1p was notably absent in these complexes, suggesting competition between Fes1p-binding and interaction of Ssa1p with Hsp90 and its co-chaperones and substrates (Zhao et al, 2005b). Sse1p was also shown to be involved in the Hsp90-mediated clearance of the model protein VHL tumor suppressor in S. cerevisiae (McClellan et al, 2005). Interestingly, overexpression of Sse1p in yeast is toxic and results in a slow-growth phenotype, suggesting a potential blockage of Ssa1p function by increased complex formation with Sse1p (Shaner et al, 2004). NEFs may also play differential roles in protein-folding triage, a mechanism mediated by molecular chaperones to determine whether a misfolded protein is destined for further folding attempts or proteasomal degradation (Connell et al, 2001).
Similar to the situation in yeast cytosol, multiple structurally unrelated NEFs are present in the mammalian ER, where the Hsp170 NEF, Grp170, coexists with the Fes1p-related NEFs called BAP in mammals and Sil1p in yeast. Mutations in the mammalian Sil1 gene that potentially inactivate its NEF function cause Marinesco–Sjögren syndrome, an autosomal recessive neurodegenerative disorder in humans, and the ataxia phenotype of woozy mice (Anttonen et al, 2005; Senderek et al, 2005; Zhao et al, 2005a). In both cases, Purkinje cells selectively degenerate while other secretory tissues are much less affected, indicating differential expression levels and/or varying folding demands in different types of secretory cells. Grp170 and BAP might therefore be differentially regulated to accommodate these demands, thus accounting for the existence of more than one NEF in mammalian ER.
Molecular basis of nucleotide exchange
An unexpected finding was that a functional ATP-binding domain is a prerequisite for the nucleotide exchange activity of Sse1p (Figure 3). Why must Sse1p be able to bind nucleotide in order to exchange nucleotide in the Hsp70 molecule? An explanation might be that nucleotide binding induces a conformational rearrangement of Sse1p necessary for functional interaction with Ssa1p. Another explanation could be that a nucleotide shuttling mechanism exists between Sse1p and Ssa1p; however, studies on other Hsp110–Hsp70 pairs suggest this explanation is unlikely (Steel et al, 2004; Yamagishi et al, 2004). It was also unexpected that all C-terminal truncation mutants of Sse1p were absolutely devoid of NEF activity, suggesting that the entire Sse1p molecule is required for this function. Long insertions in the PBD are a hallmark of Hsp110 primary structure, and therefore an important role of these elements in nucleotide exchange can be envisioned.
Folding of multi-domain proteins mediated by the Hsp110–Hsp70 complex
We have shown that Hsp110 can enhance Hsp70-mediated folding of eukaryotic firefly luciferase, both with regard to rate and yield (Figure 5). The other Hsp70 NEFs, Fes1p and HspBP1, failed to produce such an effect, although they accelerated peptide release from Hsp70 (Figure 4B and data not shown). Thus, it would appear that Hsp110 enhances protein folding by a mechanism more complex than simply accelerating substrate cycling on and off Hsp70. This mechanism is most likely related to the combined function of Hsp110 as Hsp70 NEF and chaperone, and may be of special importance in the folding of complex multidomain proteins such as firefly luciferase. Protein domains have evolved as independent structural units with cooperative folding behavior. However, when present in a multidomain protein, unfolded domains could interact with each other, resulting in the formation of kinetically trapped, misfolded states that tend to aggregate (Netzer and Hartl, 1997). In order to prevent this from happening, the sequences of multidomain proteins, such as luciferase, may encode suitably spaced sequence signatures for molecular chaperones that inherently guide a domain-wise folding process, thereby avoiding nonproductive interactions between concomitantly folding domains. With this in mind, we hypothesize that a concerted release of Hsp70 and Hsp110, resulting from the NEF activity of Hsp110, may trigger one domain (or sub-domain) of an unfolded polypeptide to fold, while other domains would remain chaperone-bound (Figure 7). In this model, the functional Hsp70 folding machine for multidomain proteins is envisioned to be composed of both Hsp110 and Hsp70, not just Hsp70 alone. Different substrate specificity of Hsp70 and Hsp110 would be a prerequisite for the proposed synergistic effect on protein folding. We have already obtained preliminary indications of differential substrate spectra for the two chaperones: The peptide probe D-NR binds to Ssa1p, but not to Sse1p, whereas Sse1p is known to bind to other unfolded proteins, such as denatured firefly luciferase (Oh et al, 1999; Goeckeler et al, 2002).
Figure 7.
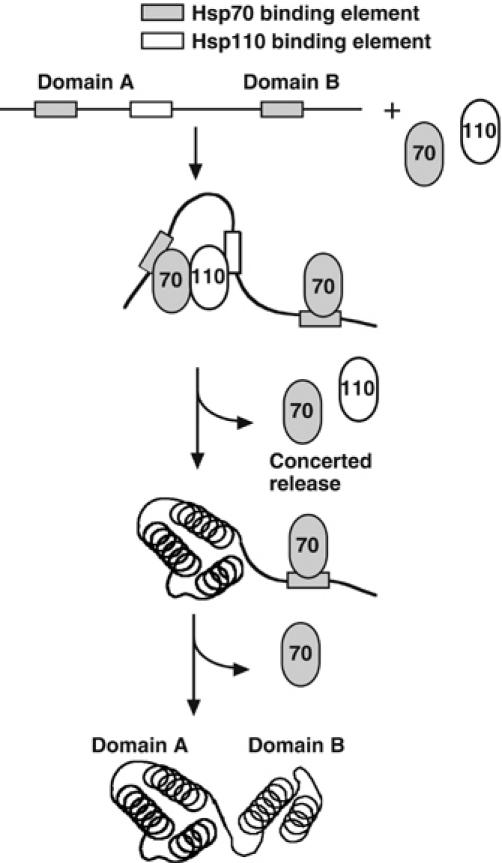
Hypothetical model of cooperative protein folding by Hsp70 and Hsp110. Sequence signatures in an unfolded multidomain protein are recognized by either Hsp70 or Hsp110. Concerted release of Hsp70 and Hsp110 triggers efficient folding of domain A (i.e., a structural unit that folds cooperatively), and subsequent release of Hsp70 triggers folding of domain B.
In conclusion, our findings indicate that coordinated regulation of Hsp70 and Hsp110 class molecular chaperones makes a substantial contribution to protein folding in the eukaryotic cytosol. Specialized Hsp70s may have coevolved in eukaryotes as a necessary folding tool for the large number of complex multidomain proteins, which often fail to fold efficiently when expressed in bacterial hosts.
Materials and methods
Reagents
All chemicals were of the highest purity available. Firefly luciferase was from Sigma (L 9009).
Protein expression constructs
S. cerevisiae SSB1, SSE1, and SSE2 were PCR-amplified from yeast genomic DNA and cloned into the expression plasmid pPROEX-HTb (Invitrogen) using the restriction sites for NcoI and XhoI (SSB1) and EheI and XhoI (SSE1, SSE2), respectively. SSE1 was additionally cloned into the yeast expression vector, pCM189 (Gari et al, 1997), using the sites for BamHI and NotI. The expression plasmid for Ssa1p, pPROEX-SSA1, was described previously (Shomura et al, 2005). Human Hsp110 was PCR-amplified from lymphocyte cDNA and cloned into pPROEX-HTb using the EheI and KpnI restriction sites. Deletion and point mutants of SSE1 were produced by using a PCR-based mutagenesis strategy, which relies on amplification of full-length plasmid using primers that contain the required nucleotide alterations in their sequences (Weiner et al, 1994). The correctness of all plasmid inserts was verified by DNA sequencing.
Protein expression and purification
For expression of the His6-tagged fusion protein of Sse1p, the plasmid pPROEX-SSE1 was transformed into E. coli BL21(DE3) cod+ RLI cells (Novagen). His6-Sse1p protein expression was induced in the early log phase with 1 mM IPTG for 4 h at 30°C. Harvested cells were resuspended in lysis buffer (300 mM NaCl, 25 mM HEPES-KOH pH 7.5, 10 mM imidazole, 5 mM β-mercaptoethanol, 1 mM PMSF, Roche Complete protease inhibitors and 0.5 mg/ml Lysozyme) and incubated at RT for 20 min. Cell lysis was completed by repeated freeze–thaw cycles and ultrasonication. After removal of cell debris by centrifugation at 50 000 g for 30 min at 4°C, the resulting supernatant was loaded on Ni-NTA agarose (Qiagen) at 4°C. The column was washed with lysis buffer containing no Lysozyme, and the His6-Sse1p protein was eluted with 250 mM imidazole in the same buffer. For cleavage of the His6-tag, the eluted protein was incubated for 1 h at RT with TEV protease at a molar ratio of 20:1 in the presence of 5 mM DTT. Afterwards, the solution was diluted to a final concentration of 10 mM NaCl in the same buffer and loaded on a MonoQ anion exchange column, and Sse1p was eluted with a linear NaCl-gradient to 500 mM. Fractions containing Sse1p were combined and applied to Ni-NTA agarose to remove residual His6-Sse1p and TEV protease. In a final purification step, the protein was passed through a Superdex-200 size exclusion column equilibrated in 50 mM NaCl, 25 mM HEPES-KOH pH 7.5, and finally the protein was concentrated by ultrafiltration.
Recombinant Sse2p, Ssa1p, Ssb1p, Hsp70 and Hsp110 were expressed and purified similar to Sse1p. TEV protease treatment was done overnight at 4°C for Ssa1p, Ssb1p and Hsp110. Ssa1p and Hsp110 were subjected to hydroxyl apatite chromatography using a linear phosphate gradient (10–500 mM) after TEV protease cleavage, and the anion exchange step was omitted in the case of Ssb1p. Hsc70, which was used in the refolding experiments, was purified from pig brain, as previously described (Minami et al, 1996). All Hsp70 chaperones were determined to be nucleotide free by measuring the absorption spectrum in the range of 240–340 nm after purification.
Nucleotide release measurements
Nucleotide release kinetics of the fluorescent nucleotide analog MABA-ADP (Mo Bi Tec) was followed with an SX.18V stopped-flow instrument (Applied Photophysics, Surrey, UK) as previously described (Theyssen et al, 1996; Brehmer et al, 2001; Gässler et al, 2001). To obtain spontaneous nucleotide release rates, 2 μM Hsp70–MABA-ADP complex was mixed at 30°C with an equal volume of 250 μM ADP (both in HKM buffer, 25 mM HEPES-KOH pH 7.5, 50 mM KCl, 5 mM MgCl2). To assay the nucleotide exchange activity of Sse1p, Sse2p and human Hsp110, the protein samples were added to the ADP solution and subsequently mixed with Hsp70–MABA-ADP complex. Release of labeled nucleotides was monitored by the decrease in fluorescence of the unbound MABA-ADP (excitation 360 nm wavelength, cutoff filter 420 nm).
Substrate dissociation experiments
To determine peptide substrate koff rates, Dansyl-labeled NR peptide (Dansyl-NRLLLTGC, Metabion) was used (Gässler et al, 2001). Nucleotide-free Hsp70 or ADP-Hsp70 preformed complex were preincubated in HKM buffer for 30 min at 30°C with D-NR. Nucleotide-free Hsp70–D-NR complex or ADP–Hsp70–D-NR complex were then mixed (volume ratio 1:1) in the stopped-flow apparatus with 250 μM unlabeled NR and 250 μM ATP in the presence or absence of the NEF. As negative controls, substrate–chaperone complexes were mixed with unlabeled NR in the presence or absence of the NEF, but with no ATP. The molar ratio of Hsp70 to NEF was 2:1. Fluorescence excitation was measured at 334 nm, and a cutoff filter at 385 nm was used.
Luciferase refolding experiments
Firefly luciferase (100 nM) was incubated in HKM buffer supplemented with 1 mM DTT at 42°C for 12 min. To prevent aggregation, denaturation was performed in presence of 1 μM Hsc70 or 1 μM Hsp110, as described in the text and the figure legend. Refolding was initiated by addition of 2 mM ATP and additional chaperones. After dilution in refolding buffer, activity was determined in a luminometer (Berthold LB 9507) with 100% corresponding to the enzyme activity of native luciferase in refolding buffer. For chemical denaturation of luciferase, 6 M guanidinium chloride in the presence of 2 mM DTT was used. Denatured luciferase was added to a final concentration of 100 nM in HKM buffer containing the indicated combinations of chaperones. Activity was determined as for thermally denatured luciferase.
In vivo luciferase activity measurements
Strain SBY528 (sse1Δ) was constructed by PCR-mediated disruption of the SSE1 gene by the HIS3 marker in the wt S. cerevisiae strain, YPH499 (MATa ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52) (Sikorski and Hieter, 1989), according to the standard methods (Lorenz et al, 1995). Strain SBY526 (SSE1↑) was constructed by transforming pCM189-SSE1 into YPH499 using standard procedure (Rose et al, 1988). All strains used in this study additionally contain the plasmid, p415-GAL-Luc (Agashe et al, 2004), which has a c-myc tagged firefly luciferase gene under the control of a galactose-inducible promoter. For time course analyses, wt, the sse1Δ strain and the SSE1↑ strain were adjusted to 0.5 OD600/ml in the presence of 2% galactose to induce synthesis of luciferase, and were grown at 37°C. At different time points, luciferase activity was measured in vivo, as described (Agashe et al, 2004). For analysis of luciferase protein levels, cell samples were harvested, and spheroplasts were prepared by zymolyase treatment and lysed in 25 mM HEPES-KOH pH 7.5, 2 mM DTT, 0.1% Triton X-100, 1 mM PMSF, and Complete protease inhibitor cocktail (Roche). Yeast cell extracts were analyzed by SDS–PAGE and Western blotting using an antibody against the c-myc epitope (Santa Cruz).
Computational methods
All curve fitting was performed with the SX.18V software (Applied Photophysics) or with the KaleidaGraph 4.0 (Synergy) software package. Release curves were fitted using a single exponential model, Fl(t)=m1+m2 exp(−m3 t) with Fl, fluorescence, and m3=koff. Each curve represents the average of at least six identical measurements. koff dependence on NEF concentration (Figure 2) was fitted with a sigmoidal curve, koff(NEF conc.)=m1+(m2−m1)/(1+(NEF conc./m3)m4) with m1=koff max.
Supplementary Material
Supplementary Information
Acknowledgments
We would like to acknowledge Dr Dejana Mokranjac for critical reading of the manuscript.
References
- Agashe VR, Guha S, Chang H-C, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM (2004) Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell 117: 199–209 [DOI] [PubMed] [Google Scholar]
- Anttonen AK, Mahjneh I, Hamalainen RH, Lagier-Tourenne C, Kopra O, Waris L, Anttonen M, Joensuu T, Kalimo H, Paetau A, Tranebjaerg L, Chaigne D, Koenig M, Eeg-Olofsson O, Udd B, Somer M, Somer H, Lehesjoki AE (2005) The gene disrupted in Marinesco–Sjogren syndrome encodes SIL1, an HSPA5 cochaperone. Nat Genet 37: 1309–1311 [DOI] [PubMed] [Google Scholar]
- Brehmer D, Rudiger S, Gassler CS, Klostermeier D, Packschies L, Reinstein J, Mayer MP, Bukau B (2001) Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Biol 8: 427–432 [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92: 351–366 [DOI] [PubMed] [Google Scholar]
- Chen X, Easton D, Oh HJ, Lee-Yoon DS, Liu X, Subjeck J (1996) The 170 kDa glucose regulated stress protein is a large HSP70-, HSP110-like protein of the endoplasmic reticulum. FEBS Lett 380: 68–72 [DOI] [PubMed] [Google Scholar]
- Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, Patterson C (2001) The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol 3: 93–96 [DOI] [PubMed] [Google Scholar]
- Craven RA, Egerton M, Stirling CJ (1996) A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J 15: 2640–2650 [PMC free article] [PubMed] [Google Scholar]
- Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ (1998) JPred: a consensus secondary structure prediction server. Bioinformatics 14: 892–893 [DOI] [PubMed] [Google Scholar]
- DeLuca-Flaherty C, McKay DB, Parham P, Hill BL (1990) Uncoating protein (hsc70) binds a conformationally labile domain of clathrin light chain LCa to stimulate ATP hydrolysis. Cell 62: 875–887 [DOI] [PubMed] [Google Scholar]
- Easton DP, Kaneko Y, Subjeck JR (2000) The hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones 5: 276–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- Gari E, Piedrafita L, Aldea M, Herrero E (1997) A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13: 837–848 [DOI] [PubMed] [Google Scholar]
- Gässler CS, Wiederkehr T, Brehmer D, Bukau B, Mayer MP (2001) Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J Biol Chem 276: 32538–32544 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Goeckeler JL, Stephens A, Lee P, Caplan AJ, Brodsky JL (2002) Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1-151 thermosensitivity and restores Hsp90-dependent activity. Mol Biol Cell 13: 2760–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gragerov A, Zeng L, Zhao X, Burkholder W, Gottesman ME (1994) Specificity of DnaK-peptide binding. J Mol Biol 235: 848–854 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Nishiyama H, Nonoguchi K, Higashitsuji H, Kishishita M, Fujita J (1997) A novel hsp110-related gene, apg-1, that is abundantly expressed in the testis responds to a low temperature heat shock rather than the traditional elevated temperatures. J Biol Chem 272: 2640–2645 [DOI] [PubMed] [Google Scholar]
- Lee-Yoon D, Easton D, Murawski M, Burd R, Subjeck JR (1995) Identification of a major subfamily of large hsp70-like proteins through the cloning of the mammalian 110-kDa heat shock protein. J Biol Chem 270: 15725–15733 [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M (1991) Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA 88: 2874–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XD, Morano KA, Thiele DJ (1999) The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J Biol Chem 274: 26654–26660 [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Muir RS, Lim E, McElver J, Weber SC, Heitman J (1995) Gene disruption with PCR products in Saccharomyces cerevisiae. Gene 158: 113–117 [DOI] [PubMed] [Google Scholar]
- Lu Z, Cyr DM (1998) Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem 273: 27824–27830 [DOI] [PubMed] [Google Scholar]
- McClellan AJ, Scott MD, Frydman J (2005) Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121: 739–748 [DOI] [PubMed] [Google Scholar]
- Minami Y, Hohfeld J, Ohtsuka K, Hartl FU (1996) Regulation of the heat-shock protein 70 reaction cycle by the mammalian DnaJ homolog, Hsp40. J Biol Chem 271: 19617–19624 [DOI] [PubMed] [Google Scholar]
- Mukai H, Kuno T, Tanaka H, Hirata D, Miyakawa T, Tanaka C (1993) Isolation and characterization of SSE1 and SSE2, new members of the yeast HSP70 multigene family. Gene 132: 57–66 [DOI] [PubMed] [Google Scholar]
- Netzer WJ, Hartl FU (1997) Recombination of protein domains facilitated by co-translational folding in eukaryotes. Nature 388: 343–349 [DOI] [PubMed] [Google Scholar]
- Oh HJ, Chen X, Subjeck JR (1997) Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J Biol Chem 272: 31636–31640 [DOI] [PubMed] [Google Scholar]
- Oh HJ, Easton D, Murawski M, Kaneko Y, Subjeck JR (1999) The chaperoning activity of hsp110. Identification of functional domains by use of targeted deletions. J Biol Chem 274: 15712–15718 [DOI] [PubMed] [Google Scholar]
- Packschies L, Theyssen H, Buchberger A, Bukau B, Goody RS, Reinstein J (1997) GrpE accelerates nucleotide exchange of the molecular chaperone DnaK with an associative displacement mechanism. Biochemistry 36: 3417–3422 [DOI] [PubMed] [Google Scholar]
- Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B (2006) Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J [E-pub ahead of print: 11 May 2006; doi:10.1038/sj.emboj.7601139] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P (1988) Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Senderek J, Krieger M, Stendel C, Bergmann C, Moser M, Breitbach-Faller N, Rudnik-Schoneborn S, Blaschek A, Wolf NI, Harting I, North K, Smith J, Muntoni F, Brockington M, Quijano-Roy S, Renault F, Herrmann R, Hendershot LM, Schroder JM, Lochmuller H, Topaloglu H, Volt T, Weis J, Ebinger F, Zerres K, Anttonen AK, Mahjneh I, Hamalainen RH, Lagier-Tourenne C, Kopra O, Waris L, Anttonen M, Joensuu T, Kalimo H, Paetau A, Tranebjaerg L, Chaigne D, Koenig M, Eeg-Olofsson O, Udd B, Somer M, Somer H, Lehesjoki AE, Zhao L, Longo-Guess C, Harris BS, Lee JW, Ackerman SL (2005) Mutations in SIL1 cause Marinesco–Sjogren syndrome, a cerebellar ataxia with cataract and myopathy. Nat Genet 37: 1312–1314 [DOI] [PubMed] [Google Scholar]
- Shaner L, Trott A, Goeckeler JL, Brodsky JL, Morano KA (2004) The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related hsp70 family. J Biol Chem 279: 21992–22001 [DOI] [PubMed] [Google Scholar]
- Shaner L, Wegele H, Buchner J, Morano KA (2005) The yeast HSP110 SSE1 functionally interacts with the HSP70 chaperones SSA and SSB. J Biol Chem 280: 41262–41269 [DOI] [PubMed] [Google Scholar]
- Shomura Y, Dragovic Z, Chang HC, Tzvetkov N, Young JC, Brodsky JL, Guerriero V, Hartl FU, Bracher A (2005) Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell 17: 367–379 [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel GJ, Fullerton DM, Tyson JR, Stirling CJ (2004) Coordinated activation of Hsp70 chaperones. Science 303: 98–101 [DOI] [PubMed] [Google Scholar]
- Theyssen H, Schuster HP, Packschies L, Bukau B, Reinstein J (1996) The second step of ATP binding to DnaK induces peptide release. J Mol Biol 263: 657–670 [DOI] [PubMed] [Google Scholar]
- Wang XY, Chen X, Oh HJ, Repasky E, Kazim L, Subjeck J (2000) Characterization of native interaction of hsp110 with hsp25 and hsc70. FEBS Lett 465: 98–102 [DOI] [PubMed] [Google Scholar]
- Weiner MP, Costa GL, Schoettlin W, Cline J, Mathur E, Bauer JC (1994) Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151: 119–123 [DOI] [PubMed] [Google Scholar]
- Yam AY, Albanese V, Lin HT, Frydman J (2005) HSP110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J Biol Chem 280: 41252–41261 [DOI] [PubMed] [Google Scholar]
- Yamagishi N, Ishihara K, Hatayama T (2004) Hsp105alpha suppresses Hsc70 chaperone activity by inhibiting Hsc70 ATPase activity. J Biol Chem 279: 41727–41733 [DOI] [PubMed] [Google Scholar]
- Yasuda K, Nakai A, Hatayama T, Nagata K (1995) Cloning and expression of murine high molecular mass heat shock proteins, HSP105. J Biol Chem 270: 29718–29723 [DOI] [PubMed] [Google Scholar]
- Zhang C, Guy CL (2005) Co-immunoprecipitation of Hsp101 with cytosolic Hsc70. Plant Physiol Biochem 43: 13–18 [DOI] [PubMed] [Google Scholar]
- Zhao L, Longo-Guess C, Harris BS, Lee JW, Ackerman SL (2005a) Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP. Nat Genet 37: 974–979 [DOI] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, Boone C, Emili A, Houry WA (2005b) Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120: 715–727 [DOI] [PubMed] [Google Scholar]
- Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA (1996) Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272: 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


