Abstract
Eukaryotic topoisomerases I and II efficiently remove helical tension in naked DNA molecules. However, this activity has not been examined in nucleosomal DNA, their natural substrate. Here, we obtained yeast minichromosomes holding DNA under (+) helical tension, and incubated them with topoisomerases. We show that DNA supercoiling density can rise above +0.04 without displacement of the histones and that the typical nucleosome topology is restored upon DNA relaxation. However, in contrast to what is observed in naked DNA, topoisomerase II relaxes nucleosomal DNA much faster than topoisomerase I. The same effect occurs in cell extracts containing physiological dosages of topoisomeraseI and II. Apparently, the DNA strand-rotation mechanism of topoisomerase I does not efficiently relax chromatin, which imposes barriers for DNA twist diffusion. Conversely, the DNA cross-inversion mechanism of topoisomerase II is facilitated in chromatin, which favor the juxtaposition of DNA segments. We conclude that topoisomerase II is the main modulator of DNA topology in chromatin fibers. The nonessential topoisomerase I then assists DNA relaxation where chromatin structure impairs DNA juxtaposition but allows twist diffusion.
Keywords: chromatin dynamics, DNA transcription, DNA twist, DNA writhe, nucleosome dynamics
Introduction
Helical tension in double-stranded DNA occurs during most chromosomal transactions. During DNA transcription, the template rotates relative to RNA polymerase, causing overwinding ahead of the traversing complex and unwinding behind it. Similarly, rotation of the duplex entering a replication complex overwinds DNA ahead of replication forks. Chromatin assembly and remodeling also induce changes of helical tension, as nucleosomes constrain DNA supercoils. The scarcity of DNA free-ends inside the cell precludes simple winding to dissipate helical stress. The enzymes responsible for resolving this problem are the ubiquitous DNA topoisomerases, which produce temporary single- or double-strand DNA breaks (reviewed in Champoux, 2001; Wang, 2002).
Eukaryotic cells have two classes of topoisomerases, type-1B and type-2, to remove helical tension from DNA. The type-1B, including the eukaryotic topo I (encoded by TOP1) and poxvirus topo I, use an active-site tyrosine as a nucleophile to cleave one strand of duplex DNA, generating a covalent 3′-phosphotyrosyl bond. This reaction leaves a 5′-hydroxyl DNA end that can rotate in either direction around the uncleaved strand, allowing relaxation of (+) and (−) helical stress. In the rejoining reaction, the 5′-hydroxyl group acts as a nucleophile to attack the 3′-phosphotyrosyl linkage and restores the continuity of the double helix (Stewart et al, 1998; Krogh and Shuman, 2000). As this ‘strand rotation' mechanism of type-1B topoisomerases functions without energetic cofactor, only DNA torque and friction drive one or several integral rotations of the duplex (Koster et al, 2005). The type-2 topoisomerases, that is, the eukaryotic topo II (encoded by TOP2), are functional homodimers that use ATP to transport one DNA duplex through a transient double-strand break in another duplex (Wang, 1998). The gated duplex, termed G-segment, is cleaved with a four-base stagger generating a 5′-phosphotyrosyl bond with each dimer subunit of the enzyme. The transported duplex, termed T-segment, enters one side the dimer interface, crosses the transiently gated G-segment, and exits the topoisomerase through the opposite side (Roca et al, 1996). This ‘cross-inversion' mechanism allows topo II to remove (+) and (−) DNA supercoils, as well as knot–unknot or catenate–decatenate DNA molecules, depending on the intra- or intermolecular location of the G- and T-segments.
Despite the advances in understanding the DNA relaxation mechanism of eukaryotic topoisomerases I and II, their relative contributions to the supercoiling of intracellular DNA remain unclear. Topo I concentrates in the nucleolus (Muller et al, 1985; Zhang et al, 1988) and in highly transcribed regions (Fleischmann et al, 1984; Mao et al, 2002), whereas topo II is more evenly distributed throughout the interphase nucleus (Gasser et al, 1986; Klein et al, 1992; Christensen et al, 2002). These observations had supported a general view of topo I as the relaxase of helical tension generated during transcription and other DNA-tracking processes, and of topo II as devoted to unlinking replicated DNA duplexes and preparing chromosomes for segregation (Holm et al, 1985; Uemura et al, 1987; Adachi et al, 1991). However, numerous observations argue against this simple partition of roles and indicate that topo II participates also in DNA relaxation tasks. Although topo I is required during development in higher eukaryotes (Zhang et al, 2000; Takahashi et al, 2002), it is dispensable for cell viability in Saccharomyces cerevisiae and Schizosaccharomyces pombe (Uemura and Yanagida, 1984). In these cells, topo II relaxes DNA supercoils in the absence of topo I (Saavedra and Huberman, 1986; Brill and Sternglanz, 1988; Giaever and Wang, 1988). Accordingly, DNA replication can proceed in yeast cells as long as one of the two topoisomerases is active (Kim and Wang, 1989b). In addition, inactivation of either topo I or topo II does not significantly affect DNA transcription, whereas inactivation of both enzymes markedly reduces rRNA synthesis (Schultz et al, 1992) and, to a lesser extent, mRNA synthesis (Cavalli et al, 1996; Collins et al, 2001). Finally, yeast Δtop1 top2-ts double mutants, but not single mutants, show mitotic hyper-recombination (Christman et al, 1988), increased excision of rDNA as extrachromosomal rings (Kim and Wang, 1989a), and multimerization of circular minichromosomes (Trigueros and Roca, 2001).
The apparently exchangeable activities of topoisomerases I and II indicate that either enzyme can relax the helical tension of intracellular DNA. Yet, a deeper analysis of their normal contributions is impaired by the complexity of in vivo experimental systems and the multiple functions in which they are involved. So far, the catalytic efficiency of eukaryotic topoisomerases has been examined in vitro on naked DNA molecules, by using supercoiled plasmids and DNA single-molecule assemblies (Charvin et al, 2003; Koster et al, 2005). However, their activities on nucleosomal DNA, their natural substrate, have not been contrasted. Here, we report the first comparative analysis of the extent to which topoisomerases I and II relax (+) supercoiled DNA in native yeast minichromosomes. We show that histones do not dissociate from DNA under high (+) helical tension and that DNA relaxation restores the typical nucleosome topology. In contrast to what is observed with naked DNA, however, topoisomerase II relaxes nucleosomal DNA more efficiently than topoisomerase I. This finding highlights a differential effect of chromatin hydrodynamics on the DNA strand-passage mechanisms of topoisomerases I and II. Functional implications to the supercoiling of intracellular DNA are discussed.
Results
High DNA supercoiling density can be generated in eukaryotic chromatin
DNA relaxation by topoisomerases I and II prevent the accumulation of helical tension in circular minichromosomes in TOP1 TOP2 yeast cells. However, when Escherichia coli topo I, a type-1A topoisomerase that relaxes (−) supercoils by acting on unwound regions of the duplex, is expressed in a Δtop1 top2-ts mutant, (+) supercoils accumulate upon thermal inactivation of topo II (Giaever and Wang, 1988). We used this unbalanced relaxation of supercoils to determine a threshold of (+) DNA helical tension that transcription or other DNA-tracking processes can generate in vivo (Liu and Wang, 1987). We transformed the strain JCW28 (Δtop1 top2–4) with plasmid pJRW13, a vector for constitutive expression of E. coli topo I; and with plasmid Yp4.4, our reporter minichromosome. The cells were cultured at 26°C (permissive temperature) and shifted to 35°C to inactivate topo II. Native yeast minichromosomes were extracted at different times, and their DNA was displayed by two-dimensional gel electrophoresis to examine the linking number (Lk) of the Yp4.4 circles (Figure 1).
Figure 1.
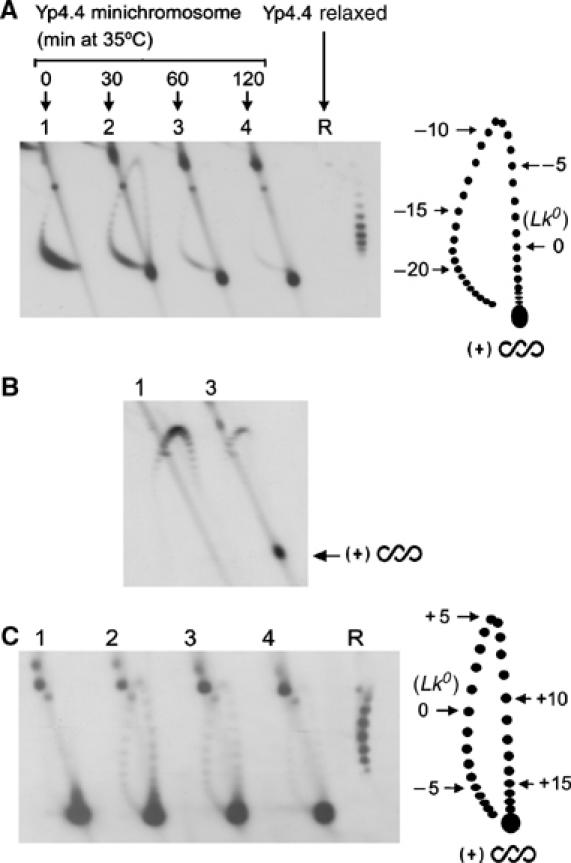
DNA supercoiling density in yeast circular minichromosomes. (A) Lanes1–4, DNA topology in Yp4.4 minichromosomes extracted from JCW28 cells that expressed of E. coli topo I, and were then shifted to 35°C to inactivate topo II for the indicated times (min). Lane R, Yp4.4 plasmid relaxed in vitro at 35°C with topoisomerase I. Two-dimensional electrophoresis of DNA was carried at 25°C in a 0.8% agarose gel in TBE buffer at 50 V for 14 h in the first dimension (top to bottom), and TBE buffer plus 2 μg/ml of chloroquine, at 60 V for 8 h in the second dimension (left to right). The gel-blot was probed for Yp4.4. The scheme depicts gel migrations of Yp4.4 topoisomers, indicating ΔLk values relative to relaxed DNA. (B) DNA samples (lanes 1 and 3 as in A) analyzed like above but in TBE buffer plus 3 μg/ml of chloroquine in the first dimension, and TBE buffer plus 15 μg/ml of chloroquine in the second dimension. (C) DNA samples (all as in A) analyzed by two-dimensional electrophoresis at 4°C in a 0.8% agarose gel in TBE buffer plus magnesium acetate 5 mM at 33 V for 40 h in the first dimension (top to bottom), and TBE buffer alone at 60 V for 4 h in the second dimension (left to right). The scheme depicts gel migrations of Yp4.4 topoisomers, indicating ΔLk values relative to relaxed DNA.
By the electrophoresis conditions described in Figure 1A, gel bands corresponding to Lk topoisomers of Yp4.4 are distributed in an arch, in which higher Lk values migrate clockwise. Yp4.4 minichromosomes extracted from cells cultured at 26°C (lane 1) had linking number difference (ΔLk) near –22 relative to the Lk0 position determined in the relaxed Yp4.4 plasmid (lane R). Since Lk0 is about 420 (4414/10.5), the DNA supercoiling density (σ) in these minichromosomes is about −0.052 (−22/420). After thermal inactivation of topo II (lanes 2–4), increasing amounts of the Yp4.4 circles migrated toward one end of the topoisomer arch. An electrophoresis in the presence of high chloroquine concentration (Figure 1B) confirmed that these circles were positively supercoiled (Lk>Lk0). To resolve their Lk distribution, we used electrophoresis at 4°C and in the presence of magnesium ions in the first gel dimension (Panyutin et al, 1989; Xu and Bremer, 1997). These conditions decrease the writhe of (+) supercoiled DNA and, as a result, the migration of Lk topoisomers shifts counter-clockwise (Bednar et al, 1994; Rybenkov et al, 1997). Although we achieved the largest shift with the gel depicted in Figure 1C, the Lk distribution of (+) supercoiled circles remained unresolved. Relative to Lk0 (lane R), these molecules had a ΔLk above +18, which translates into σ greater than +0.042 (18/420). Similar results were obtained for the yeast 2-μm circle and other yeast circular minichromosomes, for which the Lk distribution of (+) supercoiled circles could not be resolved. Yet, since E. coli topo I had to act on unwound regions of the duplex to accumulate (+) helical tension, local values of σ in vivo must be substantially higher than +0.042. Thus far, this σ value establishes the helical tension against which DNA-tracking ensembles still are able to move along eukaryotic chromatin.
Nucleosomal organization is preserved in DNA under positive helical tension
To examine whether DNA supercoiling generated in vivo was constrained by chromatin, we incubated native yeast minichromosomes, for which σ∼−0.05 and for which σ>+0.04, with catalytic amounts of type-1B topoisomerase (S. cerevisiae topo I or vaccinia virus topo I) and type-2 topoisomerase (S. cerevisiae topo II). All reactions contained also an excess of a (−) supercoiled plasmid, which had been added during minichromosome extraction. This plasmid served as internal control to calibrate the DNA relaxation activity of the topoisomerases, as well as to quench nonspecific protein–DNA interactions. DNA topology in minichromosomes and their accompanying control plasmids was then analyzed by two-dimensional gel electrophoresis (Figure 2A). Incubation of minichromosomes for which σ∼−0.05 (lane 1) with topo I (lane 2) or topo II (lane 3) did not significantly change their ΔLk value and merely induced slight broadening of the Lk distributions. In the same mixtures, however, the control plasmid was fully relaxed by the topoisomerases. Therefore, the ΔLk of these minichromosomes reflects the (−) supercoils constrained by typical chromatin structure, in which each nucleosome stabilizes roughly one (−) DNA supercoil. Incubation of Yp4.4 minichromosomes for which σ>+0.04 (lane 5) with topo I (lane 6) and topo II (lane 7) produced Lk distributions that, surprisingly, were nearly identical to those constrained by typical chromatin (compare lanes 2 and 3, with lanes 6 and 7). In the same reactions, the control plasmid had no constrained supercoils and was again fully relaxed by the topoisomerases. This complete recovery of (−) supercoils was observed in different yeast circular minichromosomes, was independent of the topoisomerase used, and persisted even when relaxations were carried out in the presence of a large excess of control plasmid (Figure 2B).
Figure 2.
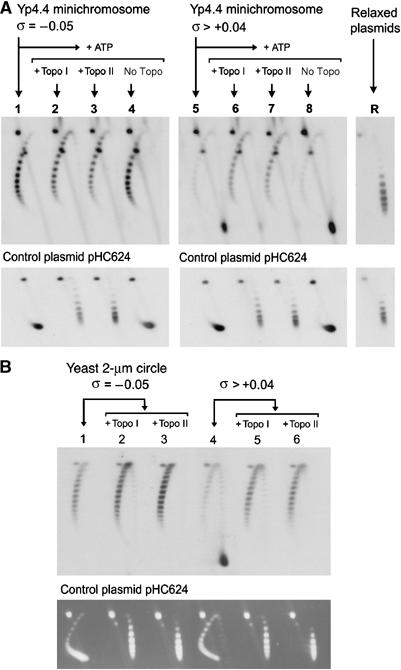
Relaxation of yeast minichromosomes by DNA topoisomerases. (A) Yeast minichromosomes, for which σ∼−0.05 (lane 1) and for which σ∼>+0.04 (lane 5) were incubated at 30°C for 30 min with catalytic amounts of vaccinia virus topo I (lanes 2 and 6), S. cerevisiae topo II (lanes 3 and 7) or no enzyme (7 and 8). Plasmids Yp4.4 and pHC624 were also relaxed in vitro (lane R). Two-dimensional electrophoresis of DNA was carried out at 25°C in a 0.8% agarose gel in TBE buffer plus 0.6 μg/ml of chloroquine at 50 V for 14 h in the first dimension (top to bottom), and TBE buffer plus 3 μg/ml of chloroquine, at 60 V for 8 h in the second dimension (left to right). Gel-blots were probed for Yp4.4 (upper panel) and for the control plasmid pHC624 (lower panel). (B) Yeast minichromosomes, for which σ∼−0.05 (lane 1) and for which σ>+0.04 (lane 4) were supplemented with an excess of control plasmid pHC624 (1 mg/ml), and incubated with catalytic amounts of S. cerevisiae topo I (lanes 2 and 5) or S. cerevisiae topo II (lanes 3 and 6) at 30°C for 30 min. After two-dimensional electrophoresis of DNA, gels were blotted and probed for the yeast 2-μm circle (upper panel) or ethidium stained to visualize pHC624 (lower panel).
To corroborate the recovery of the typical chromatin structure upon relaxation of (+) supercoiled minichromosomes, we used micrococcal nuclease to footprint nucleosome positions along the TRP1 ARS1 region of Yp4.4 (Figure 3). Minichromosomes for which σ∼−0.05 (B) showed preferential DNA cleavage sites that were consistent with the nucleosome positions determined by Thoma et al (1984). Minichromosomes for which σ>+0.04 (C) showed few additional cleavage sites, which indicated an alteration of chromatin structure. Following their incubation with topo I (D) or topo II (E), however, such additional sites disappeared and the footprints became nearly identical to those of the typical chromatin (B). Therefore, when intracellular DNA undergoes (+) helical tension, the bulk of histones seem to remain bound to DNA and conformational changes of chromatin apparently revert by DNA relaxation.
Figure 3.
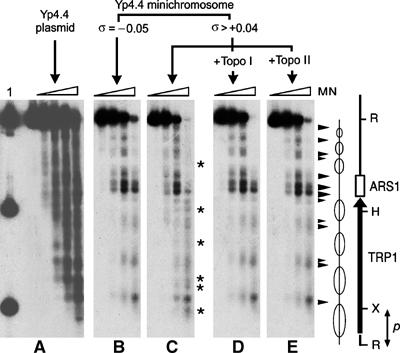
Chromatin structure upon relaxation of yeast minichromosomes. Preferential DNA cleavage sites by micrococcal nuclease (MN) digestion along the TRP1-ARS1 region of Yp4.4 were mapped on the following substrates: Yp4.4 plasmid (A), Yp4.4 minichromosome for which σ∼−0.05 (B), Yp4.4 minichromosomes for which σ>+0.04 (C), and after their relaxation by S. cerevisiae topo I (D) or S. cerevisiae topo II (E). Digested DNA samples were restricted with endonuclease EcoRI, separated on a 1% agarose gel, blotted and probed with the radio-labeled 186 bp EcoRI–XbaI TRP1 fragment (p). Lane 1, XbaI and HindIII site markers. Arrowheads denote main DNA cleavage sites visible in (B, C, D, E). Asterisks denote additional DNA cleavage sites visible in (C). The scheme depicts nucleosome positions determined by Thoma et al (1984) along the TRP1-ARS1 yeast DNA segment.
Topoisomerase II relaxes supercoiled chromatin more efficiently than topoisomerase I
In the above experiments, we had noticed that more type-1B than type-2 topoisomerase activity was required to completely relax the yeast minichromosomes. Hence, control plasmids added during minichromosome extraction allowed us to normalize the catalytic amount of topo I and of topo II needed to relax naked DNA at comparable rates and thereby compare their relaxation kinetics on supercoiled chromatin. In these conditions (Figure 4A), (+) supercoiled minichromosomes were relaxed by topo II faster than by topo I. The same result was found when comparing topo II with vaccinia virus topo I. This kinetic divergence was also observed in the yeast 2-μm circle and other yeast minichromosomes, and it persisted in a broad range of ionic conditions: KCl or NaCl from 80 to 200 mM, and MgCl2 from 2 to 12 mM. Therefore, the difference in the efficiency with which type-1B than type-2 topoisomerases relax supercoiled chromatin was probably not attributable to DNA sequences or reaction conditions.
Figure 4.
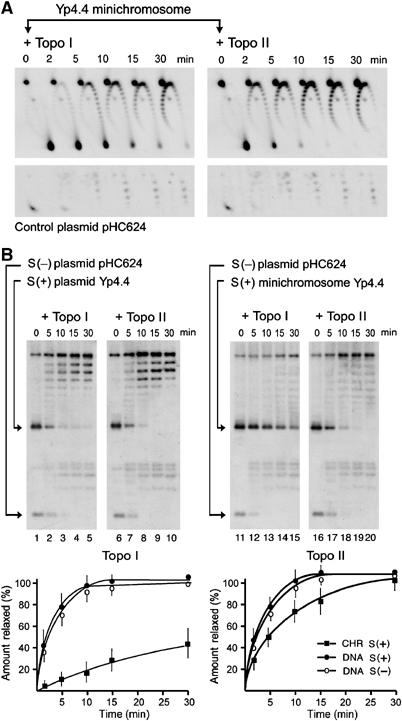
Relaxation kinetics of supercoiled chromatin. (A) Positively supercoiled Yp4.4 minichromosomes and their accompanying control plasmid pHC624 were incubated with catalytic amounts of topoisomerase I or topoisomerase II (as indicated). Reactions were quenched at the indicated periods (min). Following DNA electrophoresis, the gel-blots were probed for Yp4.4 (up) and for pHC624 (down). Note that topoisomerase I and II activities were adjusted to relax the control plasmid at comparable rates. (B) Mixtures containing (+) supercoiled Yp4.4 plasmid plus (−) supercoiled pHC624 plasmid, or containing (+) supercoiled Yp4.4 minichromosome plus (−) supercoiled pHC624 plasmid were relaxed with catalytic amounts of topoisomerase I or topoisomerase II (as indicated). Reactions were quenched at the indicated periods (min). DNA electrophoresis was carried at 25°C in a 0.9% agarose gel in TBE buffer at 40 V for 14 h. Gel-blots were probed for Yp4.4 plus pHC624. DNA relaxation rates, by topoisomerases I and II, for supercoiled minichromosomes (CHR S(+)) and supercoiled plasmids (DNA S(−), DNA S(+)) were determined by measuring the gain of relaxed topoisomers excluding nicked ones. Graphs represent the average of four experiments with error bars indicating s.d.'s from the mean.
As control plasmids were (−) supercoiled and minichromosomes were (+) supercoiled, we also ruled out an effect of DNA supercoiling handedness (Figure 4B). We deproteinized (+) supercoiled Yp4.4 minichromosomes to obtain (+) supercoiled Yp4.4 plasmid. This DNA was mixed with the (−) supercoiled pHC624 and adjusted to the reaction conditions used to relax minichromosomes. We then compared the relaxation rates of (−) supercoiled DNA, (+) supercoiled DNA, and (+) supercoiled chromatin by topoisomerase I and II. Topo I (lanes 1–5) and topo II (lanes 6–10) did not show any significant bias in relaxing (+) and (−) supercoiled plasmids. This result was consistent with that observed in previous studies on naked DNA (Charvin et al, 2003; Koster et al, 2005). However, in the same catalytic conditions, topo I relaxed the supercoiled minichromosomes (lanes 11–15) about 15 times slower than the control plasmids. This difference markedly contrasted with that of topo II, which relaxed the minichromosomes (lanes 16–20) only 1.5 times slower than the plasmids.
Intracellular topoisomerase II is the main relaxase of supercoils in nucleosomal DNA
To assess the biological relevance of the high efficiency with which topo II relaxes chromatin, we measured the specific DNA relaxation activities of intracellular topoisomerases I and II in yeast. We incubated (+) supercoiled minichromosomes and control plasmids with serial dilutions of extracts from Δtop1 TOP2 and TOP1 TOP2 yeast cells, and determined DNA relaxation rates in the absence or presence of ATP (Figure 5A). In the absence of ATP, topo I is the only cellular enzyme to relax supercoiled DNA efficiently. Accordingly, extracts from Δtop1 TOP2 cells showed no significant ATP-independent DNA relaxase activity; whereas, in similar conditions, extracts from TOP1 TOP2 cells relaxed the control plasmids and to a lesser extent the minichromosomes. In the presence of ATP, however, the minichromosomes and the control plasmids were relaxed with comparable efficiency by the Δtop1 TOP2 yeast extracts, as well as by the TOP1 TOP2 yeast extracts. This ATP-dependent DNA relaxase is topo II; thus, no other relaxase activity is detected in extracts from Δtop1 TOP2 cells containing the topo II inhibitor ICRF-193, or extracts from Δtop1 top2-ts cells inactivated at 35°C. Therefore, ATP-independent and ATP-dependent DNA relaxation rates allowed us to calculate relative specific activities of yeast topoisomerases I and II within their physiological dosages (Figure 5B). The results indicated that yeast topo I activity relaxed naked DNA twice as fast as that of topo II. However, yeast topo II activity relaxed chromatin five times as fast as that of topo I. These figures were consistent with the relaxation rates determined with the purified enzymes, in which topo II relaxed chromatin about 10-fold faster than topo I.
Figure 5.
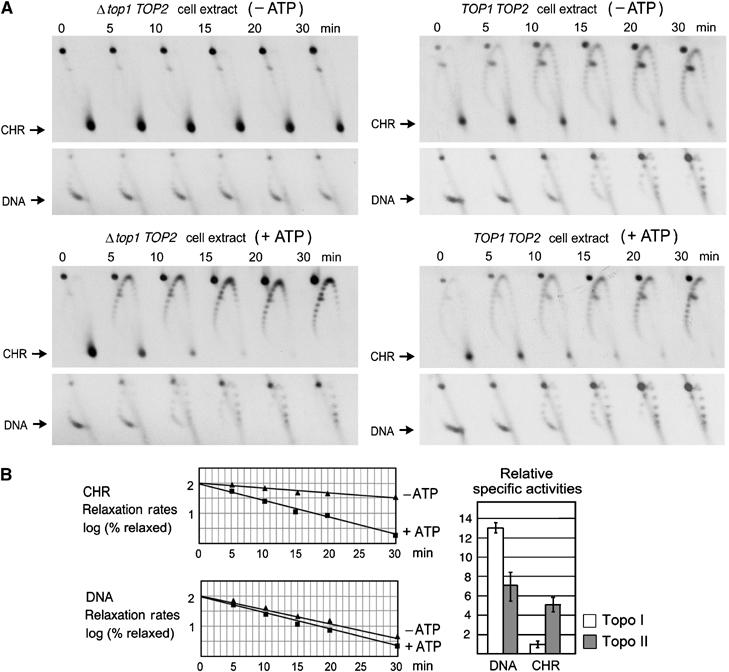
Relative relaxase activities of intracellular topoisomerases I and II. (A) Positively supercoiled Yp4.4 minichromosomes (CHR) and their accompanying control plasmid pHC624 (DNA) were supplemented with extracts from Δtop1 TOP2 or TOP1 TOP2 yeast cells, as indicated. Incubations proceeded at 30°C in the absence or presence of ATP, and were quenched at the indicated time periods (min). Two-dimensional electrophoresis of DNA was as in Figure 2A and the gel blots were probed for Yp4.4 and pHC624. (B) Right, rates of chromatin and DNA relaxation, in the presence and the absence of ATP, determined by measuring the gain of relaxed topoisomers excluding nicked ones. Left, relative specific activities of topo I (ATP-independent rate) and topo II (ATP- dependent minus ATP-independent rate). Error bars denote the s.d. from the mean of five different experiments.
Discussion
We have shown that DNA superhelical density can rise above +0.04 in eukaryotic chromatin without displacement of the histones or disruption of the nucleosome organization. This σ value establishes an helical tension below which DNA-tracking ensembles still operate in eukaryotic chromatin. We have also shown that, in contrast to what occurs in naked DNA, eukaryotic topo II is more efficient than topo I in relaxing nucleosomal DNA. This divergence has biological relevance, thus we found that topo II is the dominant relaxase of nucleosomal DNA in cell extracts containing physiological dosages of topoisomerase I and II. These findings reveal novel interactions between DNA supercoiling, chromatin dynamics and the functions of topoisomerases I and II.
The first question raised by our results is how DNA can preserve its nucleosome organization under (+) helical stress. Lee and Garrard (1991) had reported the presence of bound histones in (+) supercoiled circles extracted from yeast. We found that, indeed, the bulk of histones may remain bound to (+) supercoiled DNA, and that the native nucleosome organization is recovered by relaxation alone. Although histones can form nucleosome-like structures in (+) supercoiled DNA in vitro (Clark and Felsenfeld, 1991), we do not know whether such structures are comparable to those of the supercoiled minichromosomes generated in vivo. In canon nucleosomes, the core DNA (∼145 bp) has positive twist (ΔTwC∼+0.8) and negative writhe (ΔWrC∼−1.8), whereas linker DNA segments (30–90 bp) do not bear significant twist (ΔTwL) or writhe (ΔWrL) deformations. As ΔLk=ΔWr+ΔTw, a typical nucleosome stabilizes DNA helical tension equivalent to about −1 ΔLk. Given that a nucleosome-like organization is preserved at σ>+0.04, each nucleosome had to accommodate on average DNA deformations equivalent to ∼+0.8 ΔLk. Such deformations can hardly be confined to the linker regions. As [ΔWrC+ΔTwC] is ∼−1, [ΔWrL+ΔTwL] would be +1.8! Probably, torsion energy alters also the conformation of the core DNA, by changing the phase of histone–DNA superhelical interactions (ΔTwC), or the wrapping of DNA around the histone octamer (ΔWrC).
Our results demonstrate that chromatin adapts to (+) helical tension with a conformation that reverts upon relaxation. On that basis, we modeled plausible conformations that could support σ values >+0.04 while preserving most native histone–DNA interactions (Figure 6). In model a, entry and exit DNA segments in each nucleosome flip to form a (+) instead of the typical (−) crossing, as proposed by Sivolob and Prunell (2003). This model, however, could hardly support σ values greater than +0.05. In model b, core DNA unwraps by roughly one turn in each nucleosome and then folds in left-handed plectoneme branches. In model c, further unwrapping of DNA is associated with unfolding of the histone octamer and a switch in the helical handedness of the histone (H3–H4)2 tetramer, as proposed by Hamiche et al (1996). The presence of additional micrococcal nuclease cleavage sites in (+) supercoiled chromatin supports DNA unwrapping conformations b and c. However, a general model is difficult to assess experimentally. Intermediate states might occur, and individual nucleosomes may equilibrate differently according to their DNA sequences and histone modifications.
Figure 6.
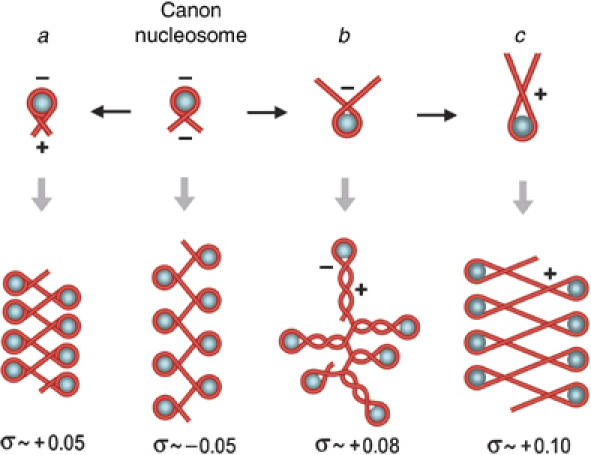
Plausible chromatin conformations that accommodate positive helical tension of DNA. The topology of DNA in the canon nucleosome and in models a, b, and c is explained in the text. To calculate approximate σ values supported in each model, the ratio of the change in writhe to the change in twist was fixed at ∼2.6:1 in histone-free DNA regions (Boles et al, 1990). For σ over +0.03, the twist regime tends to saturate and deformation occurs mainly by the writhe regime (Koster et al, 2005). We then took ΔTw values no higher than 1-U/200 bp and ΔWr values by averaging (−) and (+) crossings in several planar projections of a given conformation. For simplicity, we depicted histone octamers as spheres, although partial unfolding of octamers may occur in models b and c.
The second question raised by our results is why topo II relaxes chromatin more efficiently than topo I. This difference is unlikely to be related to DNA-binding hindrance. Both enzymes interact with DNA segments of comparable length (Figure 7A). If access to DNA were impaired by chromatin structure, one would expect topo II, which is larger and interacts with two DNA segments, to have more restrictions than topo I. The different efficiencies of topoisomerases I and II in relaxing nucleosomal DNA are more likely attributable to their mode of interaction and manipulation of DNA strands. In the first place, chromatin structure may differently affect their substrate availability. Torsional elasticity of chromatin could be smaller than for naked DNA, leading to a smaller driving torque and thus to a low activity of topo I. Conversely, chromatin might favor DNA transport activity of topo II by increasing the juxtaposition probability of DNA segments (Sun et al, 2005). Such increase may be significant in (+) supercoiled chromatin. Numerous (+) DNA crossings can occur if chromatin hydrodynamics deviates the normal partition between twist and writhe deformations of DNA (Boles et al, 1990). Moreover, as seen in Figure 6, many (+) DNA crossings may also happen to overcome twist saturation and residual wrapping of DNA around histone octamers. In the second place, chromatin structure may affect the efficiencies of topoisomerases I and II during their strand passage step. Topo I cleaves one DNA strand allowing the duplex to complete one or more rotations around the intact phosphodiester bond on the uncleaved strand. For naked DNA in free solution, axial rotation is fast, allowing the duplex to spin like a speedometer-cable. Only DNA bends increase the effective hydrodynamic volume of the duplex and, hence, the viscous drag against axial rotation (Nelson, 1999). However, in a nucleosomal DNA, axial rotation of the duplex comes up against friction barriers. Twist diffusion is delayed because the interaction of DNA with histone octamers converts chromatin into a bundle of DNA bends. Alternatively, rotation of entire nucleosomes implies large hydrodynamic volumes, especially when the angle between the entry and exit DNA segment in each nucleosome is small. Finally, other protein–DNA interactions or clashes may further increase viscous friction and restrict twist diffusion. Therefore, when DNA is folded in chromatin, topo I must encounter kinetic limitations to complete axial rotations of the cleaved duplex. Conversely, the DNA cross-inversion mechanism of topo II only involves a short and unidirectional translocation of the T-segment across the G-segment (Roca, 2004). This ATP-dependent process might be barely perturbed by friction outside the enzyme–DNA complex. Hence, we envisage that the spatial arrangement and hydrodynamics of DNA in chromatin fibers affect the efficiencies of type-1B and type-2 topoisomerases, but in opposite ways (Figure 7B).
Figure 7.
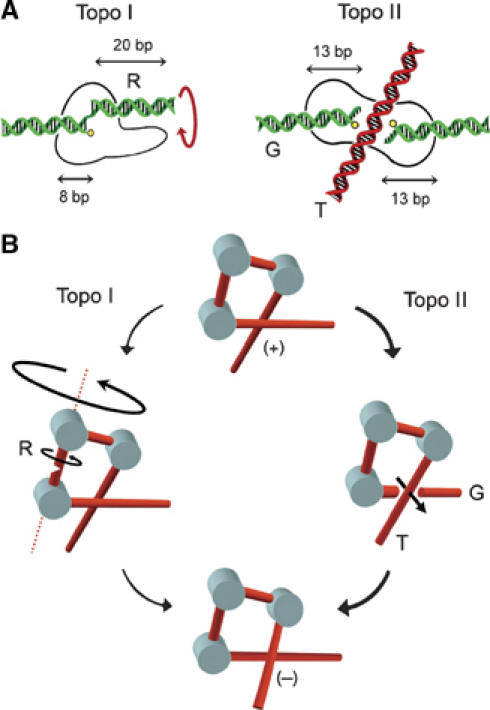
Comparative mechanics of topoisomerases I and II in relaxing nucleosomal DNA. (A) DNA segment lengths required for the function of eukaryotic topoisomerases are represented according to structural and biochemical data. Topo I clamps around 8 bp of DNA up to the cleavage site. Then, about 20 bp of duplex (R) should be free to rotate without colliding with the protruding domains of the enzyme (Stewart et al, 1998). Topo II interacts with the gated duplex (G) along 26 bp of DNA. The transported duplex (T) must cross the 50-Å wide dimer interface of the enzyme (Fass et al, 1999). (B) In nucleosomal DNA, bending of the duplex slows twist diffusion, and rotation of entire nucleosomes implies high viscous friction. Consequently, driving torque is small and a duplex cleaved by topo I comes up against kinetic limitations to complete axial rotations. Conversely, chromatin folding may favor the juxtaposition of DNA segments. Then, the cross-inversion mechanism of topo II involves a short translocation of the T-segment across the G-segment.
Further inference from the relaxation of chromatin by eukaryotic topoisomerases concerns the cleavage-religation equilibrium of DNA. Because transiently cleaved-DNA complexes can be converted into DNA lesions (Liu, 1989), the assignment of cellular functions of topo I and II must minimize the risk of genome damage. For topo II, ATP-driven conformational changes enforce a very transient DNA gating step (Roca, 2004). For topo I, however, cleavage-religation equilibrium is governed by DNA torque and friction (Koster et al, 2005). Then, the religation of DNA could be stalled owing to twist diffusion barriers imposed by chromatin. In our experiments, however, we could not detect an increase of cleaved-DNA intermediates when topo I interacted with supercoiled chromatin. Hence, we augur that topo I-DNA friction might create an energy landscape that prevents nonproductive releases of the cleaved strand when the duplex is unlikely to complete a full turn.
Given that nucleosomal DNA is an optimal substrate for relaxation by topoisomerase II, what is then the role of topoisomerase I? We believe that topo I contributes to DNA relaxation in chromatin regions where topo II is not proficient. As topo II activity depends on the juxtaposition probability of DNA segments (Roca and Wang, 1996), topo II will relax helical tension as long as there is substantial deformation of DNA by the writhe regime. In regions where the transcription rate is high, however, molecular crowding and DNA-pulling forces may preclude the local formation of supercoils. Helical tension would then deform DNA mostly by the twist regime, allowing topo I to be more efficient than topo II. This scheme is consistent with the preferential localization of topo I in highly transcribed regions (Fleischmann et al, 1984; Mao et al, 2002) and mainly in the nucleolus (Muller et al, 1985; Zhang et al, 1988), where topo I interacts directly with the RNA polymerase I holoenzyme (Christensen et al 2004). These interactions may induce topo I to function where full turn rotations of the duplex are fast, thereby increasing the overall efficiency of DNA relaxation and avoiding the stall of cleaved-DNA intermediates. Another scenario where topo I may be efficient is chromatin assembly. Either topo I or topo II can provide the relaxation activity required for replication-independent nucleosome assembly in budding yeast (Garinther and Schultz, 1997). Furthermore, nucleosome assembly in Xenopus egg extracts have suggested that in this system DNA relaxation is largely performed by topo I (Almouzni and Mechali, 1988). During chromatin assembly, histones are deposited on naked DNA and (+) helical tension rises locally to balance the (−) writhe of DNA in nucleosomes. Thus, global supercoiling density is never positive. Therefore, the initial conformation of this substrate is not equivalent to that of the (+) supercoiled chromatin (σ>+0.04) reported here. Hence, DNA relaxation by topo I may be efficient, especially in early stages of chromatin assembly when rotations of the duplex can be fast. Topo I, however, is dispensable for cell viability in budding and fission yeast (Uemura and Yanagida, 1984; Saavedra and Huberman, 1986; Brill and Sternglanz, 1988; Giaever and Wang, 1988). This observation was surprising because no other cellular topoisomerase uses a DNA strand-rotation mechanism. Hence, topo I, rather than topo II, was considered the proficient relaxase of DNA in eukaryotic cells. Now, our results clarify why topo II suffices to modulate supercoiling of DNA in the absence of topo I. The molar amount of topo I and topo II in S. cerevisiae had been estimated to about 3000 and 5000 copies per cell, respectively (Ghaemmaghami et al, 2003). These quantities are consistent with our results showing that the topo II activity relaxing nucleosomal DNA in yeast extracts dominates by a factor of five that of topo I. Therefore, topo II is not only more efficient than topo I in relaxing chromatin, but probably accounts for the bulk of this cellular activity.
Numerous observations had denoted that topo II might have cellular roles other than unlinking newly replicated DNA strands. For example, Topo II alleviates transcription repression by chromatin (Mondal and Parvin, 2001) and affects nucleosome positioning (Germe and Hyrien, 2005). Topo II, rather than topo I, cleaves DNA ahead of replication forks (Holm et al, 1989; D'Arpa et al, 1990). Topo II, rather than topo I, is active in post replicative spermatogenic cells that undergo the nucleo-histone nucleo-protamine transition (Roca and Mezquita, 1989). The β-isoform of topo II found in higher eukaryotes is not required for cell proliferation, but involved in the regulation of cell fate (Lyu and Wang, 2003). The topological interconversions of DNA underlying these molecular processes are largely unknown. Our results suggest that, in addition to relaxing DNA supercoils, topo II can be an efficient modulator of chromatin structure by altering the topology of nucleosomal DNA. This role of topo II may be as relevant for the topology of cellular DNA, as that of other type-2 topoisomerases in bacterial cells. In most bacteria, both topo IV and DNA gyrase modulate the supercoiling of DNA (Champoux, 2001; Wang, 2002). Remarkably, bacteria do not have a type-1B topoisomerase to assist the relaxation of (+) helical tension under the twist regime. However, unlike topo II, bacterial gyrase enforces the juxtaposition of DNA segments to invert (+) crossings. In light of our results, we anticipate that eukaryotic chromatin may configure optimal DNA crossings to be inverted by topo II. Further studies may decipher whether this prospect has deeper implications for the eukaryotic gene regulation.
Materials and methods
Strains, plasmids, and enzymes
S. cerevisiae strains JCW27 (Δtop1 TOP2) and JCW28 (Δtop1 top2–4), carrying the null mutation Δtop1 or the thermo-sensitive mutation top2–4, are derivatives of FY251 (TOP1 TOP2 MATa his3-Δ200 leu2-Δ1 trp1-Δ63 ura3–52) (Roca et al, 1992). Plasmid JRW13, a derivative of YEp13, carries the E. coli topA gene under constitutive pGPD yeast promoter. Plasmid Yp4.4 (4414 bp) carries the 1.4 kbp EcoRI TRP1ARS1 chromosomal fragment of S. cerevisiae. Plasmid pHC624 (2065 bp) is a derivative of pBR322 containing the ampR-oriC sequences. DNA Topoisomerase I of vaccinia virus was purified from E. coli cells harboring the expression clone pET11vvtop1 (Shuman et al, 1988). DNA topoisomerase I of S. cerevisiae was purified from yeast cells harboring the expression clone YCpGAL-TOP1 (Bjornsti and Fertala, 1999). DNA topoisomerase II of S. cerevisiae was purified from yeast cells harboring the expression clone YEpTOP2GAL1 (Worland and Wang, 1989).
Extraction of minichromosomes
Yeast cells were grown at 26°C in synthetic selective media. Thermal inactivation of topo II was carried out during exponential growth (OD ∼0.8) by shifting cell cultures to 35°C. Cells from a 100 ml culture were harvested and washed in Tris–HCl 10 mM (pH 8), EDTA 1 mM, at 4°C. Cells were resuspended at 4°C in 1 ml of buffer L (Tris–HCl 10 mM pH 8.0, EDTA 1 mM, EGTA 1 mM, NaCl 150 mM, DTT 1 mM, Triton –X-100 0.1%, pepstatin 1 μg/ml, leupeptin 1 μg/ml, PMSF 1 mM, and 10 μg/ml of supercoiled plasmid pHC624). About 1 ml of glass beads was added and the suspension was stirred six times by 30-s pulses at 4°C. Supernatants were recovered after two successive centrifugations (20 000 g at 4°C) and then loaded on a Sephacryl S-300 column equilibrated with buffer L at 4°C. Yeast circular minichromosomes and supercoiled plasmids were eluted in the first filtration volume.
Micrococcal nuclease digestions
Plasmids and minichromosomes were solubilized in buffer L plus CaCl2 adjusted to 5 mM. Following preincubation at 37°C for 5 min, micrococcal nuclease was added at 2–250 U/ml and digestions proceeded at 37°C for 2 min. Reactions were terminated by the addition of one volume of buffer K (EDTA 40 mM, SDS 1%, proteinase K, RNAse A). After 1 h at 60°C, samples were extracted by phenol and DNA was recovered by EtOH precipitation.
Topoisomerase reactions
Mixtures containing yeast minichromosomes and control plasmids were adjusted to 8 mM MgCl2 and 1 mM ATP (when indicated), preincubated at 30°C for 5 min, and then supplemented with catalytic amounts of topoisomerases. Mixtures were also supplemented with serial dilutions of cell extracts of JCW27 or FY251 obtained by physical cell disruption, as described above. Following incubations at 30°C, reactions were quenched at indicated times by adding one volume of buffer K (EDTA 40 mM, SDS 1%, proteinase K, RNAse A). Following 1 h incubation at 60°C, samples were extracted by phenol and DNA was recovered by EtOH precipitation.
DNA electrophoresis and topology analysis
DNA was analyzed by agarose gel electrophoresis in the conditions specified in figure legends. Gel-blot hybridization was carried out using 32P-labeled DNA probes obtained by random priming on purified DNA sequences. DNA linking number (Lk) distributions was analyzed by quantifying the amount of every given topoisomer by phosphorimaging the probed gel-blots. DNA supercoiling density (σ) was calculated with σ=ΔLk/Lk0 (Wang et al, 1982). Linking number difference (ΔLk) was determined with ΔLk=Lk−Lk0, in which Lk0=N/h0, where N is the DNA circle size (in bp) and h0 (10.5 bp/turn) the most probable helical repeat of DNA in the relaxation conditions used (Horowitz and Wang, 1984).
Acknowledgments
This work was supported by Grants from the Plan Nacional I+D+I (Ministerio de Educación y Ciencia de España) and DURSI (Generalitat de Catalunya). JS was recipient of a FPU fellowship from the Spanish Government.
References
- Adachi Y, Luke M, Laemmli UK (1991) Chromosome assembly in vitro: topoisomerase II is required for condensation. Cell 64: 137–148 [DOI] [PubMed] [Google Scholar]
- Almouzni G, Mechali M (1988) Assembly of spaced chromatin involvement of ATP and DNA topoisomerase activity. EMBO J 7: 4355–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednar J, Furrer P, Stasiak A, Dubochet J, Egelman EH, Bates AD (1994) The twist, writhe and overall shape of supercoiled DNA change during counterion-induced transition from a loosely to a tightly interwound superhelix. J Mol Biol 235: 825–847 [DOI] [PubMed] [Google Scholar]
- Bjornsti MA, Fertala J (1999) Overexpression and purification of DNA topoisomerase I from yeast. Methods Mol Biol 94: 179–186 [DOI] [PubMed] [Google Scholar]
- Boles TC, White JH, Cozzarelli NR (1990) Structure of plectonemically supercoiled DNA. J Mol Biol 213: 931–951 [DOI] [PubMed] [Google Scholar]
- Brill SJ, Sternglanz R (1988) Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell 54: 403–411 [DOI] [PubMed] [Google Scholar]
- Cavalli G, Bachmann D, Thoma F (1996) Inactivation of topoisomerases affects transcription-dependent chromatin transitions in rDNA but not in a gene transcribed by RNA polymerase II. EMBO J 15: 590–597 [PMC free article] [PubMed] [Google Scholar]
- Clark D, Felsenfeld G (1991) Formation of nucleosomes on positively supercoiled DNA. EMBO J 10: 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins I., Weber A, Levens D (2001) Transcriptional consequences of topoisomerase inhibition. Mol Cell Biol 21: 8437–8451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J (2001) DNA Topoisomerases: structure, function, and mechanism. Annu Rev Biochem 70: 369–413 [DOI] [PubMed] [Google Scholar]
- Charvin G, Bensimon D, Croquette V (2003) Single-molecule study of DNA unlinking by eukaryotic and prokaryotic type-II topoisomerases. Proc Natl Acad Sci USA 100: 9820–9825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen MO, Krokowski RM, Barthelmes HU, Hock R, Boege F, Mielke C (2004) Distinct effects of topoisomerase I and RNA polymerase I inhibitors suggest a dual mechanism of nucleolar/nucleoplasmic partitioning of topoisomerase I. J Biol Chem 279: 21873–21882 [DOI] [PubMed] [Google Scholar]
- Christensen MO, Larsen MK, Barthelmes HU, Hock R, Andersen CL, Kjeldsen E, Knudsen BR, Westergaard O, Boege F, Mielke C (2002) Dynamics of human DNA topoisomerases IIalpha and IIbeta in living cells. J Cell Biol 157: 31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman MF, Dietrich FS, Fink GR (1988) Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell 55: 413–425 [DOI] [PubMed] [Google Scholar]
- D'Arpa P, Beardmore C, Liu LF (1990) Involvement of nucleic acid synthesis in cell killing mechanisms of topoisomerase poisons. Cancer Res 50: 6919–6924 [PubMed] [Google Scholar]
- Fass D, Bogden CE, Berger JM (1999) Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat Struct Biol 6: 322–326 [DOI] [PubMed] [Google Scholar]
- Fleischmann G., Pflugfelder G, Steiner EK, Javaherian K, Howard GC, Wang JC, Elgin SC (1984) Drosophila DNA topoisomerase I is associated with transcriptionally active regions of the genome. Proc Natl Acad Sci USA 81: 6958–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garinther W, Schultz M (1997) Topoisomerase function during replication-independent chromatin assembly in yeast. Mol Cell Biol 17: 3520–3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser SM, Laroche T, Falquet J, Boy de la Tour E, Laemmli UK (1986) Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol 188: 613–629 [DOI] [PubMed] [Google Scholar]
- Germe T, Hyrien O (2005) Topoisomerase II–DNA complexes trapped by ICRF-193 perturb chromatin structure. EMBO Rep 6: 729–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Giaever GN, Wang JC (1988) Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell 55: 849–856 [DOI] [PubMed] [Google Scholar]
- Hamiche A, Carot V, Alilet M, De Lucia F, O'Donohe M, Revet B, Prunell A (1996) Interaction of the histone (H3-H4)2 tetramer of the nucleosome with positively supercoiled DNA minicircles: potential flipping of the protein from a left- to a right-handed superhelical form. Proc Natl Acad Sci USA 93: 7588–7593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C, Covey JM, Kerrigan D, Pommier Y (1989) Differential requirement of DNA replication for the cytotoxicity of DNA topoisomerase I and II inhibitors in Chinese hamster DC3F cells. Cancer Res 49: 6365–6368 [PubMed] [Google Scholar]
- Holm C, Goto T, Wang JC, Botstein D. (1985) DNA topoisomerase II is required at the time of mitosis in yeast. Cell 41: 553–563 [DOI] [PubMed] [Google Scholar]
- Horowitz DS, Wang JC (1984) Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J Mol Biol 173: 75–91 [DOI] [PubMed] [Google Scholar]
- Kim RA, Wang JC (1989a) A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell 57: 975–985 [DOI] [PubMed] [Google Scholar]
- Kim RA, Wang JC (1989b) Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J Mol Biol 208: 257–267 [DOI] [PubMed] [Google Scholar]
- Klein FT, Laroche ME, Cardenas J, Hofmann F, Schweizer D, Gasser SM (1992) Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol 117: 935–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH (2005) Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature 434: 671–674 [DOI] [PubMed] [Google Scholar]
- Krogh BO, Shuman S (2000) Catalytic mechanism of DNA topoisomerase IB. Mol Cell 5: 1035–1041 [DOI] [PubMed] [Google Scholar]
- Lee M, Garrard W (1991) Positive DNA supercoiling generates a chromatin conformation characteristic of highly active genes. Proc Natl Acad Sci USA 88: 9675–9679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF (1989) DNA topoisomerase poisons as antitumor drugs. Annu Rev Biochem 58: 351–375 [DOI] [PubMed] [Google Scholar]
- Liu LF, Wang JC (1987) Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA 84: 7024–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu YL, Wang JC (2003) Aberrant lamination in the cerebral cortex of mouse embryos lacking DNA topoisomerase IIbeta. Proc Natl Acad Sci USA 100: 7123–7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Mehl IR, Muller MT (2002) Subnuclear distribution of topoisomerase I is linked to ongoing transcription and p53 status. Proc Natl Acad Sci USA 99: 1235–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal N, Parvin JD (2001) DNA topoisomerase IIalpha is required for RNA polymerase II transcription on chromatin templates. Nature 413: 435–438 [DOI] [PubMed] [Google Scholar]
- Muller MT, Pfund WP, Mehta VB, Trask DK (1985) Eukaryotic type I topoisomerase is enriched in the nucleolus and catalytically active on ribosomal DNA. EMBO J 4: 1237–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P (1999) Transport of torsional stress in DNA. Proc Natl Acad Sci USA 96: 14342–14347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyutin IG, Kovalsky OI, Budowsky EI (1989) Magnesium-dependent supercoiling-induced transition in (dG)n (dC)n stretches and formation of a new G-structure by (dG)i strand. Nucleic Acid Res 17: 8257–8271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca J (2004) The path of the DNA along the dimer interface of topoisomerase II. J Biol Chem 279: 25783–25788 [DOI] [PubMed] [Google Scholar]
- Roca J, Berger JM, Harrison SH, Wang JC (1996) DNA transport by type II DNA topoisomerases: direct evidence of a two gate mechanism. Proc Natl Acad Sci USA 93: 4057–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca J, Gartenberg M, Oshima Y, Wang JC (1992) A hit-and-run system for targeted genetic manipulations in yeast. Nucleic Acids Res 20: 4671–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca J, Mezquita C (1989) DNA topoisomerase II activity in nonreplicating, transcriptionally inactive, chicken late spermatids. EMBO J 8: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca J, Wang JC (1996) The probabilities of supercoil removal and decatenation by yeast DNA topoisomerase II. Genes Cells 1: 17–27 [DOI] [PubMed] [Google Scholar]
- Rybenkov VV, Vologodskii AV, Cozzarelli NR (1997) The effect of ionic conditions on DNA helical repeat, effective diameter and free energy of supercoiling. Nucleic Acids Res 25: 1412–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra RA, Huberman JA (1986) Both DNA topoisomerases I and II relax 2 micron plasmid DNA in living yeast cells. Cell 45: 65–70 [DOI] [PubMed] [Google Scholar]
- Schultz MC, Brill SJ, Ju Q, Sternglanz R, Reeder RH (1992) Topoisomerases and yeast rRNA transcription: negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev 6: 1332–1341 [DOI] [PubMed] [Google Scholar]
- Shuman S, Golder M, Moss B (1988) Characterization of vaccinia virus DNA topoisomerase I expressed in Escherichia coli. J Biol Chem 263: 16401–16407 [PubMed] [Google Scholar]
- Sivolob A, Prunell A (2003) Linker histone-dependent organization and dynamics of nucleosome entry/exit DNAs. J Mol Biol 331: 1025–1040 [DOI] [PubMed] [Google Scholar]
- Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux J (1998) A model for the mechanism of human topoisomerase I. Science 279: 1534–1541 [DOI] [PubMed] [Google Scholar]
- Sun J, Zhang Q, Schlick T (2005) Electrostatic mechanism of nucleosomal array folding revealed by computer simulation. Proc Natl Acad Sci USA 102: 8180–8185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Matsuhara S, Abe M, Komeda Y (2002) Disruption of a DNA topoisomerase I gene affects morphogenesis in Arabidopsis. Plant Cell 14: 2085–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F, Bergman LM, Simpson RT (1984) Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol 177: 715–733 [DOI] [PubMed] [Google Scholar]
- Trigueros S, Roca J (2001) Circular minichromosomes become highly recombinogenic in topoisomerase-deficient yeast cells. J Biol Chem 276: 2243–2249 [DOI] [PubMed] [Google Scholar]
- Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M (1987) DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell 50: 917–925 [DOI] [PubMed] [Google Scholar]
- Uemura T, Yanagida M (1984) Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J 3: 1737–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC (1998) Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q Rev Biophys 31: 107–144 [DOI] [PubMed] [Google Scholar]
- Wang JC (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3: 430–440 [DOI] [PubMed] [Google Scholar]
- Wang JC, Peck LJ, Becherer K (1982) DNA supercoiling and its effects on DNA structure and function. Cold Spring Harbor Symp Quan Biol 47: 85–91 [DOI] [PubMed] [Google Scholar]
- Worland ST, Wang JC (1989) Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J Biol Chem 264: 4412–4416 [PubMed] [Google Scholar]
- Xu YC, Bremer H (1997) Winding of the DNA helix by divalent metal ions. Nucleic Acids Res 25: 4067–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CX, Chen AD, Gettel NJ, Hsieh TS (2000) Essential functions of DNA topoisomerase I in Drosophila melanogaster. Dev Biol 222: 27–40 [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang JC, Liu LF (1988) Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc Natl Acad Sci USA 85: 1060–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]


