Abstract
Polycomb group proteins Ring1b and Bmi1 (B-cell-specific Moloney murine leukaemia virus integration site 1) are critical components of the chromatin modulating PRC1 complex. Histone H2A ubiquitination by the PRC1 complex strongly depends on the Ring1b protein. Here we show that the E3-ligase activity of Ring1b on histone H2A is enhanced by Bmi1 in vitro. The N-terminal Ring-domains are sufficient for this activity and Ring1a can replace Ring1b. E2 enzymes UbcH5a, b, c or UbcH6 support this activity with varying processivity and selectivity. All four E2s promote autoubiquitination of Ring1b without affecting E3-ligase activity. We solved the crystal structure of the Ring–Ring heterodimeric complex of Ring1b and Bmi1. In the structure the arrangement of the Ring-domains is similar to another H2A E3 ligase, the BRCA1/BARD1 complex, but complex formation depends on an N-terminal arm of Ring1b that embraces the Bmi1 Ring-domain. Mutation of a critical residue in the E2/E3 interface shows that catalytic activity resides in Ring1b and not in Bmi1. These data provide a foundation for understanding the critical enzymatic activity at the core of the PRC1 polycomb complex, which is implicated in stem cell maintenance and cancer.
Keywords: Bmi1, crystal structure, E3 ligase, polycomb, Ring1b
Introduction
The Ring-domain proteins Ring1b and Bmi1 are important members of Polycomb group (PcG) proteins that adjust chromatin conformation and mediate target gene repression. During development, PcG proteins and their counteracting Trithorax-group (Trx-G) antirepressor complexes are responsible for stable maintenance of correct transcription patterns of specific key regulators such as homeotic genes. They possess an epigenetic transcriptional memory function during development and cell proliferation, which is not only required for correct cell behaviour and body plan but also later in life to maintain cell fate and to prevent oncogenic cell transformations (reviewed in Jacobs and van Lohuizen, 2002; Orlando, 2003; Lund and van Lohuizen, 2004a). Absence of Bmi1 leads to a defect in stem cell self-renewal (Park et al, 2003; Leung et al, 2004; Bruggeman et al, 2005), illustrating a crucial role in maintenance of cell fate also at the cellular level.
PcG proteins assemble into at least two biochemically and functionally distinct complexes. The initiation complex Polycomb repressive complex 2 (PRC2) and the maintenance complex PRC1 are involved in developmentally regulated and tissue-specific transcriptional silencing of Hox genes in flies and mammals (Lund and van Lohuizen, 2004b; Cernilogar and Orlando, 2005). In human cells, the PRC1 core complex contains stoichiometric amounts of RING1B (RNF2), RING1A (RING1), BMI1, EDR1 (HPH1) and CBX4 (HPC2), whereas the PRC2 core complex consists of EED, EZH2 and SUZ12 (Orlando, 2003; de Napoles et al, 2004; Lund and van Lohuizen, 2004b; Wang et al, 2004). Gene duplication has generated the RING1A and RING1B pair and the BMI1 and MEL18 pair. Although these proteins have very high sequence similarity, they appear to fulfil different functions. The PRC1 protein Bmi1 was originally identified as an oncogene, which, when overexpressed, collaborates with c-Myc to induce the formation of B-cell lymphomas (reviewed in Jacobs and van Lohuizen, 2002). Next to BMI1, it has become evident that deregulation of several other PcG genes may play a role in a variety of human tumours such as different types of leukaemia, medulloblastoma, breast cancer and non-small-cell lung cancer (Raaphorst, 2005). When compared to other PRC1 core proteins, Bmi1 appears to fulfil a unique role in that its levels are tightly regulated and it displays strong dosage sensitivity: even two-fold changes in protein expression have profound effects on biological outcome such as lymphoma formation (Jacobs et al, 1999). PcG complexes bind to chromatin and initiate and maintain gene repression, which is thought to be achieved by covalent modifications including methylation, deacetylation and ubiquitination of core histones (Francis et al, 2001; de Napoles et al, 2004; Wang et al, 2004; Cernilogar and Orlando, 2005).
Recent evidence supports the role of RING1A, RING1B and BMI1 in the ubiquitination pathway of histone H2A (de Napoles et al, 2004; Fang et al, 2004; Wang et al, 2004). Ubiquitination is a form of post-translational modification in which a small 76 amino-acid protein, ubiquitin, is conjugated through its C-terminus to lysines on cellular proteins, where it provides a signal for degradation (K48 polyubiquitin chains) or a regulatory function (monoubiquitination and other chain types). Ubiquitin conjugation is mediated by a conserved E1–E2–E3 cascade of enzymatic reactions (Weissman, 2001; Pickart and Eddins, 2004) in which the E3 ligase catalyses the last step of ubiquitin conjugation by forming an isopeptide bond between a target lysine and the C-terminus of ubiquitin. (Laney and Hochstrasser, 1999; Pickart, 2001; Glickman and Ciechanover, 2002).
RING1A, RING1B and BMI1 are essential for the function of the PRC1 complex, which acts as an ubiquitin E3 ligase that monoubiquitinates histone H2A at lysine 119 (Wang et al, 2004). Monoubiquitinated H2A and MacroH2A are enriched on the inactive female X-chromosome and are thought to be involved in its stable silencing (Fang et al, 2004; de Napoles et al, 2004; Hernandez-Munoz et al, 2005). Although both RING1A and RING1B have been implicated in monoubiquitination of H2A, RING1B appears to serve a predominant role in that loss of RING1B leads to major depletion of monoubiquitinated H2A, whereas loss of RING1A has only a minor effect. Interestingly, this also corresponds with the relative importance for development of these related Ring-domain proteins as Ring1a knockout mice only display minor skeletal aberrations, whereas loss of Ring1b leads to a full loss of PRC1 silencing resulting in early embryonic lethality (del Mar Lorente et al, 2000; Voncken et al, 2003).
Both Bmi1 and Ring1b contain an N-terminal Ring-domain. Ring-domains interact directly with the E2-ubiquitin thiolester and catalyse the ubiquitin transfer from the E2 to the target (Joazeiro and Weissman, 2000), most likely by positioning the E2 close to the target (Zheng et al, 2000, 2002). Ring-domains and another type of E3-ligase, the U-box domains, are structurally related, but whereas the latter is stabilised by a set of hydrogen bonding and salt bridges, the Ring-domain is stabilised by chelation of Zn2+ (Joazeiro and Weissman, 2000; Hatakeyama et al, 2001; Ohi et al, 2003). Ring-domains can function in the context of larger complexes, such as the SCF complex, in the context of larger proteins, such as Cbl, or in the context of Ring–Ring complexes such as BRCA1/BARD1 (Freemont, 2000; Joazeiro and Weissman, 2000; Brzovic et al, 2001b).
BMI1 and RING1A were shown to form a heterodimer both in vivo and in vitro (Satijn and Otte, 1999). Their Ring-domains promote this interaction and allows homodimerisation of each protein (Satijn and Otte, 1999), whereas the Ring-domain of Bmi1 is critical for its tumorigenic function (Alkema et al, 1997). Here we study the enzymatic activity, structure and complex-formation of the Ring1b/Bmi1 complex.
Results and discussion
H2A ubiquitination by Ring1b is enhanced by Bmi1 in vitro
The importance of Ring1b for monoubiquitination of histone H2A by the core PRC1 complex was recently shown (Fang et al, 2004; Wang et al, 2004). We confirmed the ubiquitin-conjugating activity of Ring1b of H2A in vitro using purified mouse GST-Ring1b amino acids (aa 1–331) (Q9CQJ4) lacking eight residues at the C-terminus (Figure 1A) in a mixture of nucleosomal histones, recombinant ubiquitin, E1, UbcH5c as E2, ubiquitin and ATP (Figure 1B, lane 3). We then tested the effect of adding full-length Escherichia coli-expressed Bmi1 (P25916) to the reaction. Titration of Bmi1 into GST-Ring1b shows an activating effect of Bmi1 on histone H2A modification (Figure 1B, upper panel), whereas Bmi1 alone does not show any E3-ligase activity, as we observe no increase over background of monoubiquitinated histone H2A (Figure 1B, lower panel). This dose-dependent increase of Ring1b's target-specific E3-ligase activity is interesting as it may provide one possible explanation for the unique sensitivity of the cell to changes in the dose of Bmi1 in the PRC1 complex in vivo (Jacobs et al, 1999).
Figure 1.
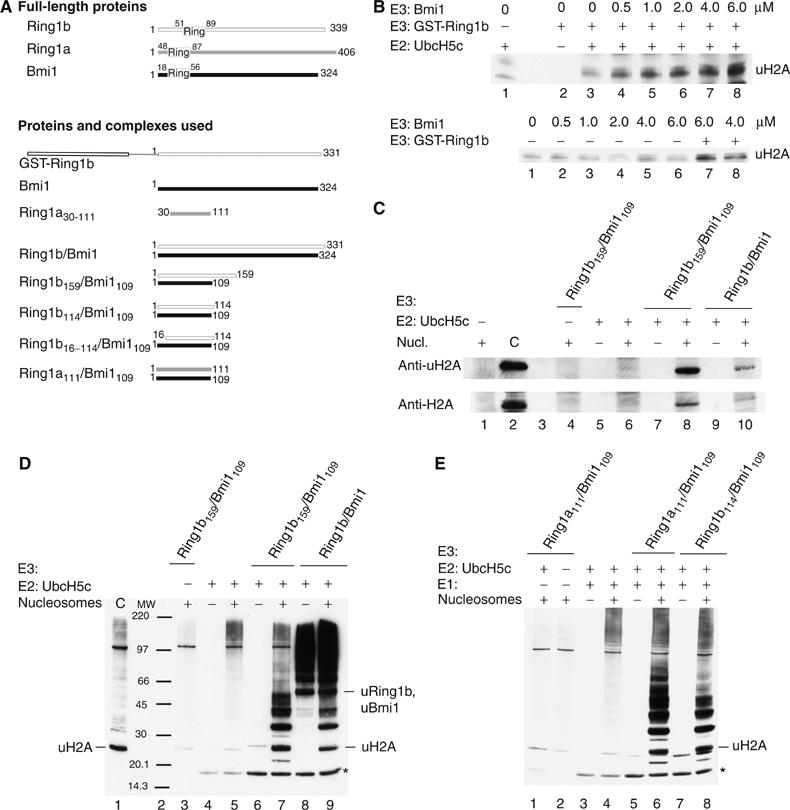
Ring1b/Bmi1 and the minimal Ring1b/Bmi1 complex are catalytically active on histone H2A. GST-Ring1b, full-length Ring1b/Bmi1 (2 μM) or different Ring-domain constructs of Ring1b and Ring1a in complex with Bmi1109 (2 μM) were incubated in ligase buffer in the presence of 28 nM human E1, 1.5 μM UbcH5c, 22 μM ubiquitin, 3 mM ATP and 4.8 μM H2A (18 μg nucleosomes). The reaction mixture was incubated at 30°C for 1 h. Samples were analysed for ubiquitination by SDS–PAGE and Western blot as described in Materials and methods. (A) Proteins and complexes used in this study. (B) Bmi1 enhances ligase activity of GST-Ring1b in vitro and is not active by itself. Upper panel: Bmi1 was titrated to the indicated final concentration into 2 μM GST-Ring1b, E1, MgATP, UbcH5c and nucleosomes. Lower panel: titration of Bmi1 with the indicated final concentrations in the presence of E1, MgATP, UbcH5c and nucleosomes and in the presence of 2 μM GST-Ring1b in lane 7 and 8, respectively. Western blots against ubiquitin. (C, D) The short Ring–Ring complex, Ring1b159/Bmi1109, is catalytically active in the presence of human E1 and UbcH5c. (C) Western blot against ubiquitinated histone H2A (uH2A) and histone H2A (H2A). (D) Western blot against ubiquitin. Ubiquitinated H2A, ubiquitinated Ring1b and Bmi1 are indicated. *Indicates diubiquitin. (E) Ring1a can substitute for Ring1b and the Ring1a111/Bmi1109 complex is equally active as the minimal Ring1b114/Bmi1109 complex against H2A; Western blot against ubiquitin. *Indicates diubiquitin.
We copurified a Ring1b/Bmi1 complex after individual expression of Ring1b (aa 1–331) and full-length murine Bmi1. This complex forms a heterotetramer, a dimer of heterodimers, as shown by gel filtration and multiangle static light scattering (MALLS) (138 kDa compared to a theoretical value of 150 kDa, data not shown). We confirmed that this complex is active in monoubiquitination of histone H2A with antibodies against ubiquitin, H2A and ubiquitin-H2A (Figure 1C). Besides the monoubiquitinated H2A, various ubiquitinated products are formed in these in vitro reactions. Antibodies against Ring1b and Bmi1 detect monoubiquitinated Ring1b and Bmi1, both running at ∼47 kDa (see Supplementary Figure 2). In addition, longer polyubiquitin chains are formed. The formation of these chains is not dependent on the presence of nucleosomes (Figure 1D, lane 8) and there is neither Ring1b nor Bmi1 in most of these products (Supplementary Figure 2), and also no Ubc5c (data not shown), indicating that these are unanchored polyubiquitin chains.
A minimal Ring1b/Bmi1 complex is sufficient for the observed E3-ligase activity
To define the E3-ligase activity within the Ring1b/Bmi1 complex, we used sequence alignment and secondary structure predictions aiming at a minimal catalytic domain. Domain boundaries were set by the end of predicted secondary structure. We coexpressed GST-fusions of either Ring1b159 or Ring1b114 together with untagged Bmi1109 in E. coli. The minimal Ring1b/Bmi1 complexes were copurified using the GST-tag (for Ring1b) and the GST-tag was removed. The proteins did not dissociate during purification or tag removal, indicating a stable complex. Both minimal heterodimers eluted from a gel-filtration column at the apparent size of the heterodimer and there was no evidence for higher order complexes. We could show that these Ring–Ring complexes are active in monoubiquitinating H2A (Figure 1C–E) and that the reaction is dependent on the presence of the E1 and E2 enzymes.
The fact that even these Ring–Ring complexes are fully active has implications for the binding site of the substrate. Traditionally, the Ring-domains are expected to bind to the E2 in ubiquitin conjugation, but in this case they must also be able to bind to target H2A in the nucleosome.
The Ring-domain of Ring1a is equally active as E3 ligase
Gene duplication in mammals has created a Ring1b homolog, the Ring1a protein, that is also present in the PRC1 complex in stoichiometric amounts (Satijn and Otte, 1999; Wang et al, 2004). We retrieved Ring1a sequences from several species that are up to 30 amino acids longer at their N-terminus than the commonly used murine sequence in the literature (Schoorlemmer et al, 1997). The alignment reveals that part of the novel N-terminal sequence (aa 11–30) of Ring1a is well conserved in Ring1b (Figure 3E).
Figure 3.
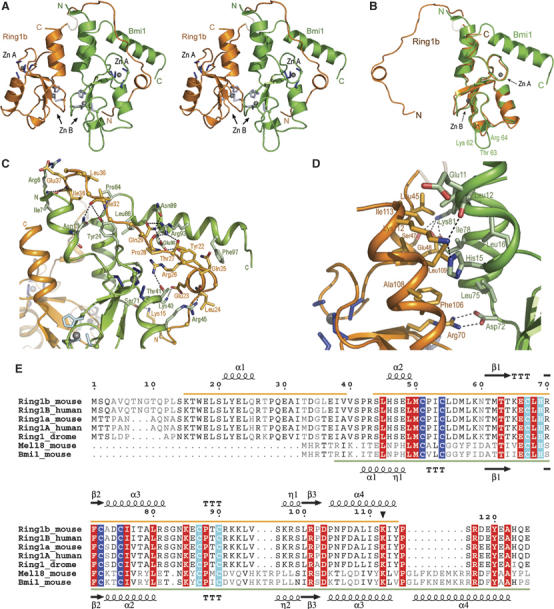
The crystal structure of the Ring1b/Bmi1 Ring–Ring complex. (A) Stereo ribbon representation of the crystal structure of the Ring1b159/Bmi1109 complex, with Ring1b in orange, Bmi1 in green. The Zn2+-ions and ligands are shown in grey. Site A is formed by Cys 51, 54, 72 and 75 in Ring1b and by Cys 18, 21, 39 and 42 in Bmi1. Site B is formed by Cys 67, 87, 90 and His 69 in Ring1b and by Cys 34, 53, 56 and His 36 in Bmi1. Transparent segments indicate disordered regions. (B) Both Ring motifs are very similar. Superposition of the Ring1b and Bmi1 Ring-domains. A three-residue insertion in the Bmi1 Ring-domain is indicated. (C, D) Details of the interface between Ring1b and Bmi1 (C) formed by the embracing arm of Ring1b and Bmi1's Ring-domain and kinked C-terminal helix, with important interactions indicated. (D) Close up view of the interface region formed by the helical bundle flanking the Ring-domains. Side chains of residues involved in the interaction according to analysis with PISA are shown. Arg 70 in Ring1b forms a salt bridge with Asp 72 in Bmi1, which is stabilised by lateral interactions with Phe 106 and Leu 75. (E) Sequence alignment of Ring-domain proteins in PRC1 with secondary structure indicated. Zn2+ binding site I is highlighted in blue and Zn2+ binding site II is highlighted in cyan. The autoubiquitination site in Ring1b is marked with a filled triangle. Sequence visible in the structure is underlined.
We coexpressed and copurified a complex of the Ring-domains of Ring1a and Bmi1 (Ring1a111/Bmi1109), including this novel N-terminus. This complex is active as E3 ligase and basically indistinguishable from the Ring1b114/Bmi1109 heterodimer in ubiquitin conjugation of histone H2A, polyubiquitin chain formation and autoubiquitination (Figure 1E). In vivo, however, Ring1a and Ring1b are functionally distinct, as most of the ubiquitinated H2A is depleted upon loss of Ring1b and only a low level of uH2A remains, which can be removed when Ring1a is additionally lost (Fang et al, 2004; de Napoles et al, 2004).
Only a subset of E2s promotes Ring1b/Bmi1 ligase activity
We tested Ring1b/Bmi1 ligase activity with a panel of available ubiquitin-conjugating E2 enzymes to see if the preference for UbcH5c that was reported by Wang et al (2004) for RING1B is conserved in the heterodimer. In Ring1b159/Bmi1109-dependent assays with nine different human E2s (UbcH5a, UbcH5b, UbcH5c, UbcH3, Rad6, E2-25K, UbcH6, UbcH7 and UbcH10), ubiquitination of histone H2A can only be observed in the presence of any UbcH5 subtype or UbcH6 (Figure 2). This is a common set of E2 enzymes that was also active in promoting E3-ligase activity of other proteins such as ARD1 (Vichi et al, 2005) or Topors (Rajendra et al, 2004). In the Ring1b/Bmi1 reactions, the processivity and selectivity of the different E2s varies: UbcH5c creates more polyubiquitin chains then UbcH5a and UbcH5b, whereas UbcH6 does not produce these at all.
Figure 2.
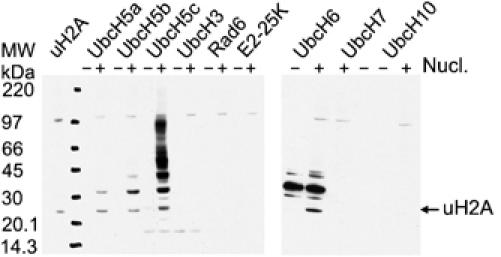
Specific E2 requirement for histone H2A ubiquitination by the Ring1b159/Bmi1109 complex. Western blots against ubiquitin showing the effect of different E2 enzymes on H2A ubiquitination and poly-ubiquitin chain formation. For conditions, see Figure 1 and Materials and methods.
A physical interaction of Ring1b with E2-25K/Hip2 had been reported (Lee et al, 2001). We confirmed the existence of this E2/E3 complex in analytical gel-filtration experiments with Ring1b159/Bmi1109 but saw no stimulation of E2 catalytic activity compared to the E2 alone. None of the other E2s tested (UbcH5c, UbcH3, Rad6, Ubc9) formed a stable complex with Ring1b159/Bmi1109 in gel-filtration experiments. Thus, neither absence nor presence of a tight physical interaction correlates with catalytic activity in these E2/E3 pairs. A similar discrepancy between catalytic efficiency and binding has been noted previously for UbcH7 in complex with BRCA1/BARD1 (Brzovic et al, 2003).
Autoubiquitination of Ring1b
We observed monoubiquitination of both Ring1b and Bmi1, as confirmed by Western blot (Supplementary Figure 2). These products are also observed when nucleosomes are left out of the reaction (Figure 1D, lane 8). In the shorter Ring–Ring complexes Ring1b159/Bmi1109 (Figure 1D, lane 6) and Ring1b114/Bmi1109 (Figure 1E, lane 7), we could only detect modification of Ring1b, suggesting that Bmi1 is modified at a position outside of the Ring-domain. Mass-spectrometry after in-gel tryptic digestion identified Lys 112 of Ring1b as the site for auto-monoubiquitination of Ring1b159/Bmi1109 (data not shown). This site is found in the C-terminal α-helix of Ring1b that interacts with Bmi1 (see below, Figure 3D). The lysine residue is accessible for modification and the ubiquitin would not interfere with E2 interaction as predicted from known E2/E3 complexes (Zheng et al, 2000; Zhang et al, 2005).
We mutated Lys 112 to arginine in Ring1b to test whether the autoubiquitination has an effect on E3-ligase activity of the heterodimeric Ring–Ring complex. This mutant showed no difference in target modification on nucleosomes. The analysis is complicated by the fact that even the mutant complex still creates ubiquitinated Ring1b product (Supplementary Figure 2B), indicating that another lysine in the Ring1b Ring-domain now becomes ubiquitinated. However, one of our later constructs of Ring1b, Ring1b16–114 (lacking the 15 N-terminal residues), is not autoubiquitinated. This complex also shows no difference in target ubiquitination when compared to Ring1b114/Bmi1109, (not shown) or Ring1a111/Bmi1109 (Figure 5A), suggesting that autoubiquitination of the Ring1b Ring-domain has no influence on catalytic activity of the Ring–Ring complex.
Figure 5.

Role of the N-terminus of Ring1a and Ring1b. (A) Assays performed with isolated Ring1a30–111 and Bmi1 and complexes of Ring1b/Bmi1, Ring1b16–114/Bmi1109 and Ring1a111/Bmi1109 using conditions as described in the legend of Figure 1 and Materials and methods; Western blot against ubiquitin. (B) Bmi1 was titrated to the indicated final concentration into 2 μM Ring1a30–111, E1, MgATP, UbcH5c and nucleosomes; Western blot against ubiquitin. (C) Titration of Bmi1 to the indicated final concentrations into 2 μM Ring1a30–111 or Ring1a111/Bmi1109, respectively, using the same conditions as described in (B); Western blot against ubiquitin.
Only UbcH5c seems to promote polyubiquitination of Ring1b and Bmi1, whereas the other three E2s promote only monoubiquitination of the E3 proteins (see Supplementary Figure 2). This is most likely a function of the general processive nature of UbcH5c, as there seems to be no change in target modification compared to other E2s (Figure 2). Altogether these biochemical studies show no detectable effect of the autoubiquitination of either Ring1b or Bmi1, but the possibility remains that autoubiquitination of Ring1b at Lys 112 has a modulatory effect in vivo on the function of Ring1b within the larger Polycomb complex, for instance by affecting PRC1 complex formation.
Crystal structure of the heterodimeric Ring1b/Bmi1 Ring–Ring complex
We determined the crystal structure of the N-terminal Ring containing fragments of coexpressed Ring1b159/Bmi1109 that was active in histone H2A ubiquitination. The structure was determined at 2.0 Å using the anomalous diffraction of the four zinc atoms and one iodine from the solvent. The weak anomalous data collected at remote wavelengths gave sufficient phasing information to solve the structure and autotrace the model with ARP/wARP (see Supplementary Table 1 for data analysis and refinement statistics). The final refined model has an R-factor of 17.6 with Rfree of 21.3%. Electron density for the first 14 and the last 45 C-terminal residues of Ring1b is missing, most likely because this part of the protein is degraded during crystallisation, as revealed by SDS–PAGE.
The Ring1b and Bmi1 structures contain the usual Ring motif with flanking N- and C-terminal helices (Figure 3). Both have an extension that forms a helix at the C-terminus of Bmi1 and a long loop at the N-terminus in Ring1b. The Ring motif is composed of two large Zn2+-binding loops, a short three-stranded antiparallel β-sheet, a central α-helix and a 310-helix (Figure 3A). The overall structure of the central Ring motif in both subunits of the heterodimer is similar to other Ring-domain structures (Bellon et al, 1997; Brzovic et al, 2001b), characterised by two large Zn2+-binding loops, in which eight Cys and His residues bind two Zn2+ ions in a so-called cross-brace arrangement. The first and third pair of Zn2+ ligands coordinate the first Zn2+ ion, whereas the second and fourth pair coordinate the second Zn2+ ion (Borden, 2000). The overlay of the Bmi1 and Ring1b Ring motifs shows that both structures are very similar in this region (Figure 3B, r.m.s.: 0.58 Å on 54 Cα atoms). The only difference can be observed in a loop extension of Bmi1 after the 310-helix, where it has three additional residues (aa 62–64) (Figure 3B).
Comparison of the individual Bmi1 and Ring1b structures to the PDB using the DALI server (Holm and Sander, 1993) gives the highest similarity (z-score 10.6 for Bmi1, 9.6 for Ring1b) to the U-box protein CHIP (Zhang et al, 2005). High similarities were also identified for the U-box protein AtPub14 (Andersen et al, 2004), and for Ring-containing proteins such as the V(D)J recombination-activating protein Rag1 (Bellon et al, 1997), the breast and ovarian cancer tumour suppressor BRCA1 and its partner BARD1 (Brzovic et al, 2001b).
Heterodimer formation
The major part of the heterodimer interface between Ring1b and Bmi1 is formed by the N-terminal arm of Ring1b (residues 15–39) that bends away and loops around to embrace the Bmi1 Ring-domain, bending the C-terminal helix of Bmi1 (residues 85–103). It seems that this arrangement of the C-terminal helix in Bmi1 helps to position the N-terminus of Ring1b. The N- and C-terminal flanking regions of the central Ring motif (residues 50–103 of Ring1b and 17–72 of Bmi1) interact with each other and form a short four-helix bundle involving only 1–2 turns. Residues 45–49 and 104–114 of Ring1b form short antiparallel helices that interact with the two Ring-domain flanking helices of Bmi1 formed by residues 9–16 and 73–84. These interfaces agree with data that show that N- and C-terminal regions adjacent to the Ring-domains are critical for complex formation between RING1A (the closely related homologue of Ring1b) and BMI1 (Satijn and Otte, 1999). The total interface buries 2289 Å2 and is relatively hydrophilic. There are 15 salt bridges and 31 hydrogen bonds in the interface (Figure 3C and D) and Arg 70 in Ring1b forms a salt bridge with Asp 72 in Bmi1. Mutation of the equivalent arginine in dRing (R65C) caused loss of function in Drosophila (Fritsch et al, 2003) and abrogated the RING1B (R70C) enzymatic activity on H2A (Wang et al, 2004). This was interpreted as a loss of enzymatic activity, but the structure reveals that Arg 70 in Ring1b is buried in the dimer interface and is most likely critical for the interaction with Bmi1 (Figure 3D). This indicates how important the Ring1b interaction with Bmi1 is in vivo.
Comparison to other dimeric Ring–Ring or U-box complexes
The heterodimeric arrangement of the Ring1b/Bmi1 heterodimer strongly resembles the heterodimeric BRCA1/BARD1 complex (Brzovic et al, 2001b) (Figure 4A), whereas it is very different from the RAG1 dimer (Bellon et al, 1997). Alignment of Ring1b/Bmi1 on the four-helix bundle of BRCA1/BARD1 reveals that the Ring motifs have a very similar local arrangement and that the interaction is mainly defined by structural elements that flank the Ring motifs. However, whereas the main BRCA1/BARD1 dimerisation interface is formed by a four-helical antiparallel bundle, the Ring1b/Bmi1 heterodimer utilises a much shorter helical bundle and the embracing N-terminal loop of Ring1b to create the main interface with Bmi1. Nevertheless, both interfaces bury similar solvent-accessible surface areas (Brzovic et al, 2001b). In BRCA1/BARD1, the heterodimer interface is functionally extremely important, as many of the BRCA1 point mutations that are linked to breast cancer predisposition are found in the interface between the helices (Brzovic et al, 2001a). This correlates well with the positive effect that the dimerisation with Bmi1 has on the Ring1b activity and with the importance of the R65C mutation in dRing.
Figure 4.
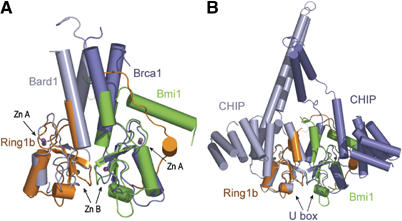
Structural comparison of Ring1b/Bmi1 with Ring and U-box dimers. (A) Superposition of Ring1b/Bmi1 onto the four-helix bundle of BRCA1/BARD1 (blue). The Zn2+ atoms in Ring1b/Bmi1 (grey) and BRCA1/BARD1 (magenta) are also depicted. (B) Superposition of Ring1b/Bmi1 onto the U-box domain of the ChIP homodimer (blue). The interface in these dimers is made by the regions that flank the Ring-domains.
The heterodimeric Ring–Ring complex of the tumour suppressor BRCA1 and BARD1 is also an E3 ligase and also monoubiquitinates histone H2A (Lorick et al, 1999; Hashizume et al, 2001; Mallery et al, 2002), but in contrast to the Ring1b/Bmi1 complexes the minimal interacting Ring-domains BRCA1 (1–109) and BARD1 (26–119) were not active in H2A ubiquitination (Mallery et al, 2002) although they form a stable complex (Brzovic et al, 2001b). Much larger Ring-containing fragments of BRCA1 (1–250) and BARD1 (14–186) were necessary for ubiquitination of H2A (Mallery et al, 2002).
Another difference between these two Ring–Ring heterodimers is that the polyubiquitination of BRCA1/BARD1 at multiple sites of both proteins stimulates the ubiquitin E3-ligase activity against H2A more than 20-fold (Mallery et al, 2002), whereas we do not observe such an activation for Ring1b/Bmi1.
The Ring1b/Bmi heterodimer also greatly resembles the dimer arrangement in the U-box proteins CHIP (Zhang et al, 2005) and Atpub14 (Andersen et al, 2004) (Figure 4B). In the case of CHIP, the critical interaction Arg70/Asp72 is conserved and considered important for the homodimerisation. The CHIP homodimer has a surprising structure because it is intrinsically asymmetric, much more so than the heterodimer of BRCA1/BARD1. The Ring1b/Bmi1 heterodimer reflects this type of asymmetry by the N-terminal arm of Ring1b that embraces the Bmi1 protein.
Role of the N-terminus of Ring1 proteins for complex formation and E3-ligase activity
In order to study the role of the N-terminal arm of Ring1a and Ring1b for E3-ligase activity, we used a variant of Ring1a that lacks the N-terminus (Ring1a30–111) (Figure 5A). In contrast to the Ring1a111/Bmi1109 heterodimer, this N-terminally truncated protein cannot be purified as stable complex with Bmi1109. A variant of Ring1b that starts where the N-terminus becomes visible in the structure (Ring1b16–114) still purifies as a heterodimer, indicating that the part of the N-terminal arm that is visible in the crystal structure (15–32 in Ring1b) is crucial and sufficient for complex formation of Bmi1 with the Ring1 proteins.
Both dimerizing forms, Ring1a111/Bmi1109 and Ring1b16–114/Bmi1109 heterodimers, are equally active as E3 ligases in ubiquitin conjugation of histone H2A (Figure 5A), whereas Ring1a30–111 or Bmi1 alone is not active, indicating that the dimer formation is important for the activity of Ring1 proteins. Gradual additions of Bmi1 to Ring1a30–111 restore some activity indicating that weak complex formation may be induced and that this restores some activity (Figure 5B), although not to the level of Ring1a111/Bmi1109 (Figure 5C).
The importance of this type of dimer formation has consequences for Polycomb assembly. Several types of interaction are possible by Ring1a or Ring1b and Bmi1 or Mel18, but they are likely to occur in pairs, where the Ring protein embraces the Bmi1 homologue. As Mel18 does not appear to have the activating role of Bmi1 (Cao et al, 2005), fine tuning the assembly is apparently of great physiological significance.
E2/E3 interaction
The presence of the two Ring-domains in the heterodimer generates two putative E2 interaction sites in the Ring–Ring heterodimer and we wondered whether both are active. We modelled the possible interactions of our structure with UbcH5c, based on E2/E3 complexes of CHIP with UbcH13 (Zhang et al, 2005) and Cbl with UbcH7 (Zheng et al, 2000) (Figure 6A). This modelling shows no clashes between the E2 and the Ring1b/Bmi1 complex. The N-terminus of Ring1b would show side-chain interactions with the E2 on the Bmi1 side, but it would not noticeably interfere with E2 interaction. However, it is theoretically possible that the very N-terminal 15 residues that are absent in our structure have an interfering effect on the Bmi1/E2 interaction, although we did not see a significant increase in activity when we remove these residues (Figure 5A).
Figure 6.

The E2 interaction of Ring1b/Bmi1. (A) Model of the E2/E3 interaction of Ring1b/Bmi1 with UbcH5c, based on the crystal structure of the ChIP U-box complexed to UbcH13. The Ser-Pro-Ala motif important for interaction with the U-box is indicated in UbcH5 (cyan). (B) Mutation of E2-interacting residues in Ring1b and in Bmi1 only shows effect on activity in Ring1b. Ubiquitination assay (Western blot against ubiquitin) with Ring1b159/Bmi1109 (wt/wt), Ring1b159 I53A/Bmi1109 (mt/wt) or Ring1b159/Bmi1109 L20A (wt/mt), as indicated with UbcH5c or UbcH6 as E2. For conditions, see Figure 1 and Materials and methods.
The interaction sites of Bmi1 or Ring1b with the E2 are very similar to those seen in the CHIP/UbcH13 interface, with important contacts of the Ser-Pro-Ala region in the E2 interacting with Ile 53 and Pro 88 in Ring1b and Leu 20 and Pro 54 in Bmi1. In analogy to experiments performed on BRCA1/BARD1 (Brzovic et al, 2003), a mutation of Ile 53 in Ring1b or Leu 20 in Bmi1 would be expected to destroy the E2/E3 interface, without effecting the folding of the E3 itself. In practice, the Ring1b159/Bmi1109 complex with an I53A mutation in Ring1b lost almost all catalytic activity, but the heterodimer with the equivalent L20A mutation in Bmi1 was fully active (Figure 6B). We found the same results for either UbcH5c or UbcH6, indicating that the E2/E3 interaction for monoubiquitination of H2A in the Ring1b/Bmi1 complex is located in the Ring1b protein. Thus, Bmi1 does not recruit an E2 in this interaction but plays its role as a stimulator in the ubiquitination of H2A and partner in the assembly of the PRC1 complex.
Conclusion
Here, we have characterised the ubiquitin conjugation activity of the Ring1b/Bmi1 and Ring1a/Bmi1 heterodimers. We show that the E2s that are active in nucleosomal histone H2A ubiquitin conjugation do not form a stable complex with the Ring–Ring complex, whereas E2-25K/Hip2 makes a stable complex but shows no catalytic activity against this target. We found that Bmi1 can promote ubiquitin conjugation by Ring1b in vitro and we showed that autoubiquitination of this E3 takes place, but does not seem to affect target-specific ubiquitination.
Importantly, we found that in vitro there is no difference in ubiquitin conjugation activity on histone H2A between the long constructs and the minimal construct for which we determined the crystal structure. This implies that not only the E2 interaction site but also the H2A recognition site is contained within this region. It also implies that the multimerisation that we observe in the longer forms of the proteins, which form heterotetramers, apparently is not important for the catalytic rate on H2A. This is in contrast to the initial assembly of the heterodimer between Ring1b and Bmi1 Ring-domains. In the absence of this heterodimerisation, ubiquitin conjugation activity is lost, unless homodimers are made by, for example, GST-Ring1b.
As these proteins function in the cell in the context of the larger PRC1 complex, these assembly points are of great relevance. Regulation of multicomponent systems can depend critically on local and temporal regulation of the assembly of the system involved. Here we show the atomic detail of the core enzymatic components of the PRC1 complex and we establish the effect of the Ring1a or Ring1b complexation to Bmi1 for catalytic activity in chromatin. It is quite likely that the higher order complexation that we observe for the longer constructs is equally important for the final functioning within the Polycomb complex.
Within the PRC1 complex, the Bmi1 protein has a unique role that is critical for stem cell maintenance and cancer formation. Its strong dose-dependent phenotype, where overexpression can lead to B- and T-cell lymphomas and where partial reduction of Bmi1 leads to significant reduction in lymphoma formation (Jacobs et al, 1999) and brain tumour formation (Bruggeman et al, 2005) indicate the importance of tight and controlled Bmi1 regulation. In this respect, our current observation that Bmi1 acts as a dose-dependent regulator of the E3 ligase Ring1b is of particular relevance. The detailed insight in the catalytic function and the atomic structure of the Ring1b/Bmi1 catalytic domains may therefore provide a valuable basis for drug design.
Materials and methods
Antibodies, plasmids and enzymes
Mouse monoclonal anti-ubiquitin (Santa Cruz Biotechnology), anti-ubiquityl-histone H2A (Upstate Biotechnology), anti-Ring1b (H Koseki, RIKEN Research Center for Allergy and Immunology, Yokohama, Japan), anti-Bmi1 (Upstate Biotechnology), rabbit polyclonal anti-UbcH5 (Boston Biochem) and secondary antibodies were obtained from Biorad Laboratories and Biosource. Mammalian ubiquitin E1 was obtained from Affinity or Boston Biochem. Human UbcH5c plasmid was a gift from P Jackson (Stanford University School of Medicine) and plasmids of human UbcH5b and UbcH6 were a gift from B Winkler (Bijvoet Center for Biomolecular Research). Purified recombinant ubiquitin, E2-25K and Rad6 were generous gifts from WJ van Dijk, P Knipscheer and V Notenboom, respectively (The Netherlands Cancer Institute). UbcH3, UbcH5a, UbcH7 and His-UbcH10 were obtained from Boston Biochem. All E2 enzymes were active as tested by the ability to form di-ubiquitin.
Protein expression
GST-fusion proteins of Ring1b (residues 1–331), full-length Bmi1, UbcH5b, UbcH5c and UbcH6 were expressed separately from the pGEX6P-1 vector in E. coli Rosetta (DE3)/pLysS. The Ring1b1-331/Bmi1 complex was made by copurification. Ring1a was expressed in pGEX6P-1 after PCR amplification with Ring1a IMAGE clone (ID:6315481) as template.
In contrast, the shorter Ring-domain complexes were made by coexpression. Ring1b (residues 1–159 or 1–114 or 16–114) and Bmi1 (1–109) or Ring1a (1–111 or 30–111) and Bmi1 (1–109), respectively, were coexpressed in E. coli Rosetta (DE3)/pLysS from a modified vector pGEX6P (a gift from Anna De Antoni) with both genes on a single promoter and a glutathione-S-transferase (GST) fusion tag on the Ring fragments. Mutants of the Ring1b Ring-domain were generated using Quick Change Site-Directed Mutagenesis (Stratagene) as described by the manufacturer's protocol, and expressed and purified like the wild-type proteins.
Protein purification
Cells grown to OD600 of 0.8 in LB medium with 50 μg/ml carbenicillin and 34 μg/ml chloramphenicol were induced at 16°C with 0.1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) and cultivated for 8–12 h. After homogenizing the cells with an EmulsiFlex-C5 (Avestin, Ottawa, CA), the suspension was centrifuged at 30 000 g for 1.5 h.
GST-Ring1b and GST-Ring1a fragments in complex with Bmi1109 were purified on glutathione sepharose (Hitrap column, Pharmacia). After application of the sample, the column was washed extensively with buffer A (50 mM Tris/HCl pH 7.5, 100 mM NaCl, 1 μM ZnCl2, 2 mM DTT), followed by buffer B (50 mM Tris/HCl pH 7.5, 100 mM NaCl, 50 mM KCl, 10 mM MgCl2, 1 μM ZnCl2, 2 mM ATP, 2 mM DTT) and again with buffer A. The proteins were eluted with buffer A that contained 20 mM glutathione. After overnight cleavage with GST-PreScission (Amersham), the proteins were further purified by ion exchange chromatography (MonoS, pH 7.0). GST and the excess of GST-Ring1b did not bind to the column. The Ring1b/Bmi1 complex was eluted with 300–350 mM NaCl and was further purified by gel-filtration chromatography (Superdex 75, Pharmacia) with buffer A, which resulted in the Ring1b constructs (residues 1–159 or 1–114 or 16–114) and Ring1a construct (residues 1–111) that contain six additional amino acids (Gly, Pro, Leu, Gly, Ser, His) at the N-terminus. Fractions containing the complex were pooled and concentrated to 7 mg/ml. We obtained about 0.1 mg of complex per gram of cells.
GST-fusion proteins of Ring1b (1–331), Ring1a (30–111), full-length Bmi1, UbcH5c, UbcH5b and UbcH6 were also loaded on glutathione sepharose and washed with buffer A, buffer B and again with buffer A. After cleavage with GST-PreScission overnight on the column, UbcH5b, UbcH5c and UbcH6 were eluted with buffer A. The proteins were further purified by gel-filtration chromatography (Superdex 75, Pharmacia). GST-Ring1b and GST-Bmi1 were eluted with buffer A that contained 20 mM glutathione. Solutions were combined and after cleavage with GST-PreScission (Amersham) overnight, the Ring1b1–331/Bmi1 complex was further purified as described for the Ring–Ring complexes of Ring1b and Bmi1. For size exclusion chromatography, a Superdex 200 gel-filtration column (Pharmacia) with running buffer A was used.
Nucleosome preparation
Nucleosomes were prepared from HEK293 cells, digested with micrococcal nuclease, according to Hernandez-Munoz et al (2005). The quality of the purified nucleosomes was analysed by gel electrophoresis.
Size and mass determination
MALLS experiments were performed at 4°C on a Mini-Dawn light scattering detector (Wyatt Technology) online with a Superdex S200 10/30 column using buffer A. Refractive index and light scattering detectors were calibrated using BSA (Sigma). Gel-filtration columns were calibrated with Pharmacia marker sets. Analytical gel filtration was carried out on a Smart® System (Pharmacia Biotech). A Superdex™ 200 PC 3.2/30 column was equilibrated with buffer A. For analysing complex formation between E2s and the Ring1b159/Bmi1109 E3, 50 μl solutions with two-fold molar excess of the E2 protein were used. Fractions (50 μl) were collected and analysed by SDS–PAGE. Mass spectrometry was performed according to Pichler et al (2005)).
In vitro ubiquitin ligase assay
Ligase activity was measured by assaying ubiquitination of histone H2A and by ubiquitin chain formation. Reactions were carried out by incubating human E1 (28 nM), human UbcH5c (E2, 1.5 μM), GST-Ring1b, Ring1a or Ring1b/Bmi complex (E3, 2.0 μM monomer), ubiquitin (22 μM), ATP (3 mM) and 18 μg nucleosomes (H2A final concentration: 4.8 μM) in buffer C (50 mM Tris/HCl (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 1 μM ZnCl2, 1 mM DTT) at 30°C for 1 h. The reactions were stopped by addition of LDS sample buffer (NuPAGE®, Invitrogen). Positive controls were enriched histones from acid-lysed 293 cells. Reaction products were separated by NuPAGE® in 4–12% Bis–Tris precast gels (Invitrogen), transferred to nitrocellulose membrane and probed with an anti-ubiquitin antibody (Santa Cruz) unless otherwise indicated and developed by chemiluminescence with ECL (Amersham). Signals were detected with a Fujifilm LAS-3000 Imager (FUJIFILM Medical Systems).
Crystallisation
Crystals were grown at room temperature by vapour diffusion from sitting nano-drops, formed by equal volumes (100+100 nl) of protein (7 mg/ml) and crystallisation buffer (10% PEG 3350, 0.1 M NaI, 0.05 M Bis–Tris-propane pH 7.5), suspended over a reservoir of 75 μl, as described by Newman et al (2005). Small crystalline needles (average size 100 × 10 × 5 μm3) appeared after 7–9 days. Crystals were optimised in hanging drops (1+1 μl) with macroseeding protocols and reached a typical size of 600 × 30 × 20 μm3. Crystallisation only occurred after C-terminal degradation of Ring1b. Crystals belong to the hexagonal space group P63 with cell dimensions a=b=120 Å, c=27 Å, α=β=90°, γ=120° and contain one Ring1b/Bmi1 Ring–Ring complex in the asymmetric unit with a solvent content of 48%.
Data collection
Crystals were transferred to stabilisation buffer (15% PEG 3350, 0.1 M NaI, 0.05 M Bis–Tris-propane, pH 7.5) containing 20% (v/v) glycerol as a cryoprotectant. A vitrified crystal diffracted X-rays to 3.0 Å resolution in our home rotating anode CuKα source with Montel mirrors (Bruker), and a full data set was collected. Data from two additional crystals were collected at ESRF beamline ID14-EH1 to 2.5 and 2.0 Å resolution, respectively. Intensities were integrated with Mosflm (Leslie, 1991) and scaled with Scala (CCP4, 1994) (see Supplementary Table 1).
Structure determination and refinement
The two synchrotron data sets were collected at the fixed wavelength ID14-EH1 beamline (0.934 Å), where Zn2+ has a weak anomalous signal (2.3 e−). That signal was sufficient to give good quality anomalous data that allowed SHELXD (Schneider and Sheldrick, 2002) to locate five atoms in a straightforward manner, in each synchrotron data set independently. As only four Zn2+ ions were expected, the lowest occupancy site was assigned as an I− (originating from the NaI in the crystallisation solution), which displays 2.94 anomalous e− at this wavelength. No sites were detectable in the rotating anode data set, despite an expected signal of 0.69 and 6.9 e− from Zn2+ and I−, respectively. The rotating anode data set was included in phasing for its useful dispersive differences when combined with the synchrotron data, which contributed 1.51 and 0.24 e− for Zn2+ and I− respectively. Phase probability distributions using these three data sets and the five heavy atom sites were calculated with the SHARP program (de la Fortelle and Bricogne, 1997). Solvent flattening was performed with SOLOMON (Abrahams and Leslie, 1996) and DM (CCP4, 1994) as implemented in SHARP. The quality of the derived phases allowed most of the Ring1b/Bmi1 model (residues 7–101 in chain A, residues 16–37 and 46–108 in chain B) to be automatically built with ARP/wARP (Perrakis et al, 1999) and to be manually completed (residues 6–103 in chain A and residues 15–114 in chain B, 173 waters molecules, four Zn2+ and two I− atoms) using O (Jones et al, 1991) and COOT (Emsley and Cowtan, 2004). Refinement was carried out with REFMAC (Murshudov et al, 1999). Inspection of the model with PROCHECK did not show any unfavourable geometries. Interface analysis was performed with PISA at the EBI website (Krissinel and Henrick, 2005). Structure figures were generated using PyMOL (http://www.pymol.org/).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Information
Acknowledgments
We thank Jesper Velgaard Olsen and Matthias Mann for mass spectrometry, Puck Knipscheer, Maria Hernandez-Munoz, Valerie Notenboom, Pim van Dijk, Erwin Boutsma and group members for discussion; beam line scientists at the ESRF and Patrick Celie for assistance with data collection. Funding was provided by EU-Spine (grant QLG2-CT-2002-00988) and NWO-CW Pionier to TKS. PvdS was supported by a Centre for Biomedical Genetics grant to MvL. Coordinates and structure factors have been deposited with the PDB, accession number 2ckl, for the structure factors r2cklsf.
References
- Abrahams JP, Leslie AG (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D 52: 30–42 [DOI] [PubMed] [Google Scholar]
- Alkema MJ, Jacobs H, van Lohuizen M, Berns A (1997) Pertubation of B and T cell development and predisposition to lymphomagenesis in Emu Bmi1 transgenic mice require the Bmi1 RING finger. Oncogene 15: 899–910 [DOI] [PubMed] [Google Scholar]
- Andersen P, Kragelund BB, Olsen AN, Larsen FH, Chua NH, Poulsen FM, Skriver K (2004) Structure and biochemical function of a prototypical Arabidopsis U-box domain. J Biol Chem 279: 40053–40061 [DOI] [PubMed] [Google Scholar]
- Bellon SF, Rodgers KK, Schatz DG, Coleman JE, Steitz TA (1997) Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat Struct Biol 4: 586–591 [DOI] [PubMed] [Google Scholar]
- Borden KL (2000) Ring-domains: master builders of molecular scaffolds? J Mol Biol 295: 1103–1112 [DOI] [PubMed] [Google Scholar]
- Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, Hulsman D, Leung C, Arsenijevic Y, Marino S, van Lohuizen M (2005) Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev 19: 1438–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D III, Fukuda M, Ohta T, Klevit R (2003) Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin–ligase complex. Proc Natl Acad Sci USA 100: 5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Meza JE, King MC, Klevit RE (2001a) BRCA1 Ring-domain cancer-predisposing mutations. Structural consequences and effects on protein–protein interactions. J Biol Chem 276: 41399–41406 [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE (2001b) Structure of a BRCA1-BARD1 heterodimeric RING–RING complex. Nat Struct Biol 8: 833–837 [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y (2005) Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell 20: 845–854 [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Cernilogar FM, Orlando V (2005) Epigenome programming by Polycomb and Trithorax proteins. Biochem Cell Biol 83: 322–331 [DOI] [PubMed] [Google Scholar]
- de la Fortelle E, Bricogne G (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multi-wavelength anomalous diffraction methods. In Methods in Enzymology, Carter CW and Sweet RM (eds.), Vol. 276, pp 472–494, Macromolecular Crystallography Part A [DOI] [PubMed] [Google Scholar]
- del Mar Lorente M, Marcos-Gutierrez C, Perez C, Schoorlemmer J, Ramirez A, Magin T, Vidal M (2000) Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 127: 5093–5100 [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fang J, Chen T, Chadwick B, Li E, Zhang Y (2004) Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem 279: 52812–52815 [DOI] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE (2001) Reconstitution of a functional core polycomb repressive complex. Mol Cell 8: 545–556 [DOI] [PubMed] [Google Scholar]
- Freemont PS (2000) RING for destruction? Curr Biol 10: R84–R87 [DOI] [PubMed] [Google Scholar]
- Fritsch C, Beuchle D, Muller J (2003) Molecular and genetic analysis of the Polycomb group gene Sex combs extra/Ring in Drosophila. Mech Dev 120: 949–954 [DOI] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T (2001) The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 276: 14537–14540 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI (2001) U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem 276: 33111–33120 [DOI] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M (2005) Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci USA 102: 7635–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M (1999) Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev 13: 2678–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJ, van Lohuizen M (2002) Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim Biophys Acta 1602: 151–161 [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell 102: 549–552 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Part 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2005) Detection of protein assemblies in crystals. In CompLife 2005, Berthold MR, Glen R, Diederichs K, Kohlbacher O, Fischer I (eds), Vol. LNBI 3695, pp 163–174. Berlin: Springer-Verlag [Google Scholar]
- Laney JD, Hochstrasser M (1999) Substrate targeting in the ubiquitin system. Cell 97: 427–430 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Choi JY, Sung YM, Park H, Rhim H, Kang S (2001) E3 ligase activity of RING finger proteins that interact with Hip-2, a human ubiquitin-conjugating enzyme. FEBS Lett 503: 61–64 [DOI] [PubMed] [Google Scholar]
- Leung C, Lingbeek M, Shakhova O, Liu J, Tanger E, Saremaslani P, van Lohuizen M, Marino S (2004) Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature 428: 337–341 [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA 96: 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund AH, van Lohuizen M (2004a) Epigenetics and cancer. Genes Dev 18: 2315–2335 [DOI] [PubMed] [Google Scholar]
- Lund AH, van Lohuizen M (2004b) Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol 16: 239–246 [DOI] [PubMed] [Google Scholar]
- Mallery DL, Vandenberg CJ, Hiom K (2002) Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J 21: 6755–6762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D 55 (Part 1): 247–255 [DOI] [PubMed] [Google Scholar]
- Newman J, Egan D, Walter TS, Meged R, Berry I, Ben Jelloul M, Sussman JL, Stuart DI, Perrakis A (2005) Towards rationalization of crystallization screening for small- to medium-sized academic laboratories: the PACT/JCSG+ strategy. Acta Crystallogr D 61: 1426–1431 [DOI] [PubMed] [Google Scholar]
- Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL (2003) Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol 10: 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V (2003) Polycomb, epigenomes, and control of cell identity. Cell 112: 599–606 [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF (2003) Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302–305 [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Pichler A, Knipscheer P, Oberhofer E, van Dijk WJ, Korner R, Olsen JV, Jentsch S, Melchior F, Sixma TK (2005) SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat Struct Mol Biol 12: 264–269 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ (2004) Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta 1695: 55–72 [DOI] [PubMed] [Google Scholar]
- Raaphorst FM (2005) Of mice, flies, and man: the emerging role of polycomb-group genes in human malignant lymphomas. Int J Hematol 81: 281–287 [DOI] [PubMed] [Google Scholar]
- Rajendra R, Malegaonkar D, Pungaliya P, Marshall H, Rasheed Z, Brownell J, Liu LF, Lutzker S, Saleem A, Rubin EH (2004) Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J Biol Chem 279: 36440–36444 [DOI] [PubMed] [Google Scholar]
- Satijn DP, Otte AP (1999) RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity. Mol Cell Biol 19: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM (2002) Substructure solution with SHELXD. Acta Crystallogr D 58: 1772–1779 [DOI] [PubMed] [Google Scholar]
- Schoorlemmer J, Marcos-Gutierrez C, Were F, Martinez R, Garcia E, Satijn DP, Otte AP, Vidal M (1997) Ring1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boundaries in the mouse hindbrain. EMBO J 16: 5930–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vichi A, Payne DM, Pacheco-Rodriguez G, Moss J, Vaughan M (2005) E3 ubiquitin ligase activity of the trifunctional ARD1 (ADP-ribosylation factor domain protein 1). Proc Natl Acad Sci USA 102: 1945–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M (2003) Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci USA 100: 2468–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878 [DOI] [PubMed] [Google Scholar]
- Weissman AM (2001) Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol 2: 169–178 [DOI] [PubMed] [Google Scholar]
- Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH (2005) Chaperoned ubiquitylation—crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP–Ubc13–Uev1a complex. Mol Cell 20: 525–538 [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP (2002) Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 416: 703–709 [DOI] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP (2000) Structure of a c-Cbl–UbcH7 complex: Ring-domain function in ubiquitin–protein ligases. Cell 102: 533–539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Information


