Abstract
In the fungus Ustilago maydis, early endosomes move bidirectionally along microtubules (MTs) and facilitate growth by local membrane recycling at the tip of the infectious hypha. Here, we set out to elucidate the molecular mechanism of this process. We show that endosomes travel by Kinesin-3 activity into the hyphal apex, where they reverse direction and move backwards in a dynein-dependent manner. Our data demonstrate that dynein, dynactin and Lis1 accumulate at MT plus-ends within the hyphal tip, where they provide a reservoir of inactive motors for retrograde endosome transport. Consistently, endosome traffic is abolished after depletion of the dynein activator Lis1 and in Kinesin-1 null mutants, which was due to a defect in targeting of dynein and dynactin to the apical MT plus-ends. Furthermore, biologically active GFP-dynein travels on endosomes in retrograde and not in anterograde direction. Surprisingly, a CLIP170 homologue was neither needed for dynein localization nor for endosome transport. These results suggest an apical dynein loading zone in the hyphal tip, which ensure that endosomes reach the expanding growth region before they reverse direction.
Keywords: CLIP-170, dynactin, dynein, endosome motility, LIS1, plant pathogen
Introduction
Fungal pathogenicity is based on the ability to invade the host tissue by polarized growth of the hypha. It is thought that this process is based on motor-mediated transport of growth supplies along the cytoskeleton to the growing hyphal apex (Steinberg, 2000), where polarized exocytosis occurs (Gow, 1995). In addition, evidence exist that endosome-based membrane recycling supports hyphal tip growth (Wedlich-Söldner et al, 2000). Interestingly, in yeast-like cells and hyphae of the dimorphic plant pathogenic fungus Ustilago maydis, early endosomes (EE) show rapid microtubule (MT)-dependent motility (Wedlich-Söldner et al, 2000). In yeast-like cells, this bidirectional traffic is mediated by Kinesin-3 and cytoplasmic dynein (Wedlich-Söldner et al, 2002), and similar motors might mediate the motility of endosomes in vertebrates (Hoepfner et al, 2005).
Currently, there are several concepts for the activity of kinesins and dynein in bidirectional organelle traffic. Most experimental evidence from animal cells suggests that both motors interact on the same organelle, thus controlling each other's activity (Gross et al, 2002; Welte, 2004). This notion is supported by the finding that Kinesin-1 directly binds the dynein complex (Ligon et al, 2004). However, such physical interaction could also reflect that one motor is a cargo of the other. This notion is supported by recent results showing that fungal dynein, which accumulates at the plus-ends of MTs (Han et al, 2001; Lee et al, 2003; Sheeman et al, 2003), requires Kinesin-1 for its plus-ends targeting in Aspergillus nidulans (Zhang et al, 2003).
In Saccharomyces cerevisiae growing MT plus-ends take dynein to the cortex, where it becomes activated and ‘off-loaded' in order to migrate towards the minus-ends, thereby mediating spindle positioning (Lee et al, 2003; Sheeman et al, 2003). A key component in this process is Pac1 (Sheeman et al, 2003), which is an NudF/LIS1 homologue (Xiang et al, 1999). These factors are thought to activate dynein for minus-end directed motility (Faulkner et al, 2000; Smith et al, 2000; Coquelle et al, 2002; Zhang et al, 2003; Lansbergen et al, 2004; Mesngon et al, 2006). Another factor involved in dynein regulation is dynactin (Schroer, 2004), which also affects dynein activity and its MT affinity at plus-ends (Valetti et al, 1999; Zhang et al, 2003). Moreover, together with the cytoplasmic linker protein 170 (CLIP-170), dynactin is thought to recruit endosomes to MT plus-ends (Pierre et al, 1992; Vaughan, 2005), and similar mechanisms might also account for other cellular cargoes (Mimori-Kiyosue and Tsukita, 2003). Taken together, these findings emphasize the importance of MT plus-ends for intracellular organelle motility.
Based on these studies, it is tempting to speculate that plus-end located fungal dynein represents a reservoir for minus-end directed transport in interphase (Yamamoto and Hiraoka, 2003; Zhang et al, 2003). Here, we provide experimental evidence for such a concept. We show that cytoplasmic dynein and homologues of the dynein activators LIS1 and dynactin localize to plus-ends of MTs. A Kinesin-3 delivers EE to the hyphal apex, where they are loaded onto dynein for retrograde transport. This ensures that endosomes reach the hyphal tip, where they participate in cell expansion by endocytic membrane recycling.
Results
Dynein and a Kif1A-like kinesin support bidirectional endosome traffic in hyphae of U. maydis
It was previously shown that EE move along MTs in yeast-like cells and hyphae of U. maydis (Wedlich-Söldner et al, 2000). As a first step towards an understanding of this motility, we determined the directionality of MTs in hyphae by the observation of a YFP-tagged EB1-homologue Peb1 that binds only to growing MT plus-ends (Straube et al, 2003). Therefore, any moving signal indicates the orientation of a MT. Infectious hyphae of U. maydis consist of a single tip cell of ∼120–150 μm in length and 2–3 μm width (Figure 1A), with the nucleus being located close to the center of the cell (Figure 1B [II]; chromosomal DNA labeled by CFP-histone 4 is given in blue; strain AB33Peb1Y_H4C, for all strains see Table I; see Supplementary Movie 1). Hyphae contain long MTs that often form bundles (Steinberg et al, 2001). The majority of Peb1-YFP signals that, based on their consistent signal intensity, marked individual MT plus-ends moved towards the growing apex and the basal septum, suggesting that plus-ends are directed to the cell poles (Figure 1B [I]+[III] and Figure 1C; Peb1 in red). In the middle of the cell, Peb1-YFP signals grow towards each other (Figure 1B [II], 1C) and passed the nucleus in opposite directions, indicating that MTs have an antipolar orientation in the central region of the hypha. Consistent with previous results (Wedlich-Söldner et al, 2000), individual MTs and bundles of MTs served as tracks for bidirectional motility of EE that were tagged with the specific t-SNARE Yup1-GFP (Figure 2A; strain AB33RT_YG; see Supplementary Movie 2), but which also localizes to stationary vacuoles, albeit to a minor extend (Wedlich-Söldner et al, 2000). Interestingly, most EE reached the MT ends in the hyphal tip before they reversed direction (Figure 2B). This was a first indication that factors in the hyphal apex participate in retrograde transport. In yeast-like cells, EE traffic depends on Kinesin-3 and dynein (Wedlich-Söldner et al, 2002). Consistently, deletion of Kinesin-3 in hyphae almost abolished EE motility (Figure 2C, ΔKin3; strain AB33ΔKin3_YG; for ‘control' see Figure 2D, strain AB33YG; for Figure 2D–F; see Supplementary Movie 3 on EMBO web site) and led to clustering of EE around the nucleus in the middle of the shorter hyphae (Figure 2E), whereas EE were equally distributed in control hyphae (Figure 2D). Although these Δkin3 hyphae were bipolar (Figure 2E, Schuchardt et al, 2005), MT orientation was not altered (89% plus-ends towards tip; n=59; strain AB33ΔKin3_Peb1Y), indicating that EE are trapped at minus-ends of MTs near the cell center. In contrast, depletion of dynein shifted EE to the cell poles (Figure 2F, Dyn1↓; strain AB33rDyn1_YG), where MT plus-ends were concentrated. Unfortunately, Western analysis revealed that expression of dynein was not completely abolished in these mutants (not shown). Therefore, we analyzed EE traffic in pheromone-induced hyphae of temperature-sensitive dynein mutants (strain FB1Dyn2ts_YG). Such dynein mutant hyphae show growth defects (Fuchs et al, 2005) and contained longer MTs (not shown). However, MT orientation was not affected, as 95.5% of all Peb1-RFP signals were directed to the tip (n=40; strain FB1Dyn2tsPeb1Y). Again, EE were trapped at plus-ends in the hyphal tip (not shown) and almost all motility was abolished (Figure 2C, Dyn2ts). These observations are most consistent with the notion that a balance of Kinesin-3 and dynein activity underlies long-distance EE transport in U. maydis hyphae.
Figure 1.
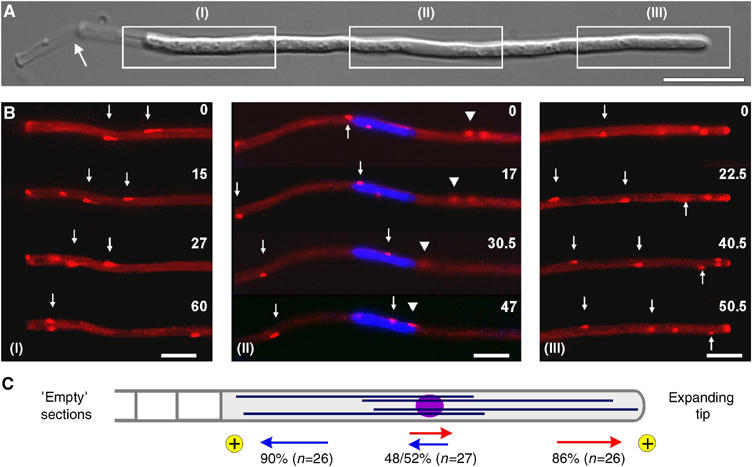
Microtubule orientation in hyphae of U. maydis. (A) Overview of a hypha of U. maydis. Boxed regions (I–III) correspond to series given in 1B. Note that tip cells leave ‘empty' cell wall section behind (arrow) while they grow at their tip (right end). Bar: 10 μm. (B) Peb1-YFP (given in red) marks MT plus-ends that elongate towards the basal septum (I, arrows), as well as the hyphal tip (III, arrows), indicating that the plus-ends are orientated towards both poles of the hyphal cell. Around the nucleus (II; labeled by histone 4-CFP is given in blue), Peb1-signals move in both directions (arrow and arrowhead), indicating that MTs have an antipolar orientation. Elapsed time is given in seconds. Bars: 5 μm. (C) Quantitative analysis of growing MT ends demonstrates that the plus-ends (indicated by ‘plus') are focused to the tip (indicated by red arrows) and the subapical septum (indicated by blue arrows). An antipolar MT orientation in the middle of the cell suggests that minus-ends are located near the nucleus. Numbers indicate the percentage of Peb1-YFP signals that move in the direction indicated. Supplementary movie for Panel B is given on the EMBO web site.
Table 1.
Strains and plasmids used in this study
| Strains/plasmid | Genotype | Reference |
|---|---|---|
| AB33RT_YG | a2 PnarbW2 PnarbE1, bleR/potefRFPTub1/potefYup1GFP | This study |
| AB33YG | a2 PnarbW2 PnarbE1, bleR/potefYup1GFP | This study |
| AB33ΔKin3_YG | a2 PnarbW2 PnarbE1 Δkin3∷hygR, bleR/potefYup1GFP | This study |
| FB1Dyn2ts_YG | a1b1 dyn2ts, natR/potefYup1GFP | This study |
| AB33ΔKin1_Peb1Y | a2 PnarbW2 PnarbE1 Δkin1∷hygR, Ppeb1-peb1-yfp, bleR, natR | This study |
| AB33ΔKin3_Peb1Y | a2 PnarbW2 PnarbE1 Δkin3∷hygR, Ppeb1-peb1-yfp, bleR, natR | This study |
| FB1Dyn2tsPeb1Y | a1b1 dyn2∷dyn2ts Ppeb1-peb1-yfp, natR, bleR | This study |
| AB33rDyn1_YG | a2 PnarbW2 PnarbE1 Pcrg1-dyn1, bleR, natR/potefYup1GFP | This study |
| AB33ΔKin1_YG | a2 PnarbW2 PnarbE1 Δkin1∷hygR/potefYup1GFP | This study |
| AB33Peb1Y_H4C | a2 PnarbW2 PnarbE1 Ppeb1-peb1-yfp, bleR, natR/pH4CFP | This study |
| AB33G3Dyn2 | a2 PnarbW2 PnarbE1 3xgfp-dyn2, bleR, natR | This study |
| AB33G3Dyn2_RT | a2 PnarbW2 PnarbE1 3xgfp-dyn2, bleR, hygR/potefRFPTub1 | This study |
| AB33ΔKin1_G3D2_RT | a2 PnarbW2 PnarbE1 Δkin1∷hygR 3xgfp-dyn2, natR, bleR/potefRFPTub1 | This study |
| AB33ΔKin1_G3D2_YR2 | a2 PnarbW2 PnarbE1 Δkin1∷hygR 3xgfp-dyn2, natR, bleR/potefYup1RFP2 | This study |
| AB33G3Dyn2_YR2 | a2 PnarbW2 PnarbE1 3xgfp-dyn2, hygR, bleR/potefYup1RFP2 | This study |
| FB2GLis1 | a2b2 PLis1-gfp-lis1, hygR | This study |
| FB2GLis1_RT | a2b2 PLis1-gfp-lis1, hygR/potefRFPTub1 | This study |
| FB2nGLis1 | a2b2 Pnar-gfp-lis1, natR | This study |
| FB2G3Dyn2_nLis1 | a2b2 Pdyn2-3xgfp-dyn2 Pnar-lis1, hygR natR | This study |
| FB2G3Dyn2 | a2b2 Pdyn2-3xgfp-dyn2, hygR | This study |
| FB2G3Dyn2_nLis1_RT | a2b2 Pdyn2-3xgfp-dyn2 Pnar-lis1, hygR natR/potefRFPTub1 | This study |
| FB2G3Dyn2_nLis1_YR2 | a2b2 Pdyn2-3xgfp-dyn2 Pnar-lis1, hygR natR/potefYup1RFP2 | This study |
| FB2YR2 | a2b2/potefYup1RFP2 | This study |
| FB2nLis1_Peb1R_GT | a2b2 Pnar-lis1 Ppeb1-peb1-rfp, natR, bleR/potefGFPTub1 | This study |
| FB2G3Lis1_YR2 | a2b2 Plis1-3xgfp-lis1, natR /potefYup1RFP2 | This study |
| FB2G3Dya1_RT | a2b2 Pdya1-3xgfp-dya1, bleR/potefRFPTub1 | This study |
| FB2rGDya1 | a2b2 Pcrg1-gfp-dya1, bleR | This study |
| FB2rDya1_G3D | a2b2 Pcrg1-dya1 Pdyn2-3xgfp-dyn2, bleR, hygR | This study |
| FB2rDya1_G3D_RT | a2b2 Pcrg1-dya1 Pdyn2-3xgfp-dyn2, bleR, hygR/potefRFPTub1 | This study |
| FB2rDya1_YG | a2b2 Pcrg1-dya1, bleR/potefYup1GFP | This study |
| FB2rDya1_Peb1R_GT | a2b2 Pcrg1-dya1 Ppeb1-peb1-rfp, natR, bleR/potefGFPTub1 | This study |
| FB2G3Dya1_YR2 | a2b2 Pdya1-3xgfp-dya1, bleR/potefYup1RFP2 | This study |
| FB2YG | a2b2/potefYup1GFP | This study |
| FB2ΔKin1_G3Dya1_RT | a2b2 Δkin1∷hygR Pdya1-3xgfp-dya1, bleR/potefRFPTub1 | This study |
| FB2rKin1_GLis1 | a2b2 Plis1-gfp-lis1 Pcrg1-kin1, hygR natR | This study |
| FB2GClip1_RT | a2b2 Pclip1-gfp-clip1, hygR/potefRFPTub1 | This study |
| FB2ΔClip1 | a2b2 Δclip1∷bleR | This study |
| FB2ΔClip1_YG | a2b2 Δclip1∷bleR/potefYup1GFP | This study |
| FB2ΔClip1_G3Dyn2_RT | a2b2 Δclip1∷bleR Pdyn2-3xgfp-dyn2, hygR/potefRFPTub1 | This study |
| FB2GT | a2b2/potefGFPTub1 | (Steinberg et al, 2001) |
| FB2ΔClip1_GT | a2b2 ΔClip1∷bleR/potefGFPTub1 | This study |
| FB2ΔClip1 _Peb1R_GT | a2b2 Δclip1∷bleR, Ppeb1-peb1-rfp, natR/potefGFPTub1 | This study |
| RWS9 | a1b1/pKin3YFP/pPX10-146CFP | (Wedlich-Söldner et al, 2002) |
| potefGFPTub1 | Potef-egfp-tub1, cbxR | (Steinberg et al, 2001) |
| potefRFPTub1 | Potef-mrfp-tub1, cbxR | (Straube et al, 2005) |
| potefYup1GFP | Potef-yup1-sgfp, cbxR | (Wedlich-Söldner et al, 2000) |
| potefYup1RFP2 | Potef-yup1-2xmrfp, cbxR | This study |
| pKin3YFP | Potef-kin3-yfp, cbxR | (Wedlich-Söldner et al, 2002) |
| pPX10-146CFP | Potef-yup1PX1–146-cfp, bleR | (Wedlich-Söldner et al, 2002) |
| PH4CFP | Phis4-h4-cfp, hygR | (Straube et al, 2005) |
| a, b, mating type loci; Δ, deletion; P, promoter; ∷, homologous replacement; -, fusion; hphR, hygromycin resistance; bleR, phleomycin resistance; natR, nourseothricin resistance; cbxR, carboxin resistance; /, ectopically integrated; otef, constitutive promoter; nar, conditional nitrate reductase promoter; crg1, conditional arabinose-induced promoter; E1, W2, genes of the b mating type locus; gfp, enhanced green fluorescent protein; yfp, yellow-shifted fluorescent protein; mrfp, monomeric red fluorescent protein; yup1: endosomal t-SNARE; kin1, kinesin-1 family member; kin3, kinesin-3 family member; dyn1: N-terminal half of the dynein heavy chain; dyn2: C-terminal half of the dynein heavy chain; dyn2ts: temperature-sensitive allele of dyn2; tub1, α-tubulin; lis1, LIS1 homologue; clip1, CLIP-170 homologue; dya1, p150glued homologue; yup1PX1−146, localization domain of Yup1; his4, histone 4. | ||
Figure 2.
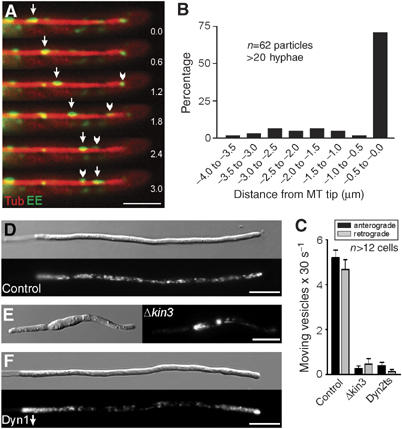
Endosome organization and motility in the Kinesin-3 and dynein mutants. (A) EE that are labeled by a fusion protein of GFP and the membrane receptor Yup1 (arrowhead, arrow; Wedlich-Söldner et al, 2000) migrate bidirectioally along MTs, which are visualized by expression of RFP-α-tubulin. Elapsed time is given in seconds. Bar: 3 μm. (B) A quantitative analysis of the behavior of individual EE confirmed that most organelles reach the MT plus-ends before they reverse direction. (C) Quantitative analysis of the EE motility frequencies at 10 μm distance to the tip of control and mutant hyphae. Almost identical numbers of vesicles move towards the tip (anterograde) and the basal nuclear region (retrograde) of control hyphae. This motility is abolished in Kinesin-3 mutants (Δkin3) and in dynein mutants (Dyn2ts). (D) In control hyphae, Yup1-GFP labeled EE are motile and equally distributed along the length of the hyphae. Occasionally, transient endosome concentrations are found in the apex. Bar: 10 μm. (E) Δkin3 mutant hyphae are usually bipolar and much shorter (Schuchardt et al, 2005). The EE cluster near the nucleus, where the minus-ends of MTs are concentrated. Note that the MT orientation in Δkin3 hyphae was confirmed by analysis of Peb1-YFP motility (not shown). Bar: 10 μm. (F) In the conditional dynein mutants, depletion of dynein strengthens anterograde EE motility and leads to accumulations of EE at the MT plus-ends near the cell poles. Bar: 10 μm. Supplementary movie for Panels A and D–F is given on the EMBO web site.
Dynein accumulates at MT plus-ends in the hyphal tip
Next, we set out to visualize dynein by fusing a triple GFP-tag to the N-terminus of the endogenous dyn2 gene (strain AB33G3Dyn2), which encodes the C-terminal half of the dynein heavy chain (Straube et al, 2001). The fusion protein did not cause any morphological phenotypes, indicating that it is fully functional. GFP3-Dyn2 formed comet-like motile structures (Figure 3A, arrowhead; see Supplementary Movie 5) and was significantly concentrated at MT ends in the tip of growing hyphae (Figure 3A, arrow and Figure 3B; strain AB33G3Dyn2_RT). From there, small dynein particles could be detected that move in retrograde direction (see below), suggesting that the dynein accumulation is a reservoir for retrograde motor proteins. Quantitative analysis of the fluorescence intensity revealed that the dynein signals in the first 3 μm of the hyphae were significantly stronger than those at MT tips in subapical regions (Figure 3C). A more detailed analysis revealed that the amount of dynein remained constant over long distances, but rapidly increased in the apical 5 μm and reached maximum values in the close proximity to the hyphal tip (Figure 3D), suggesting that dynein is actively concentrated in the apex. In vivo observation of EE in strain AB33G3Dyn2_YR2 revealed that EE reach the dynein accumulation before they reverse direction (Figure 3E, arrow; see Supplementary Movie 4).
Figure 3.
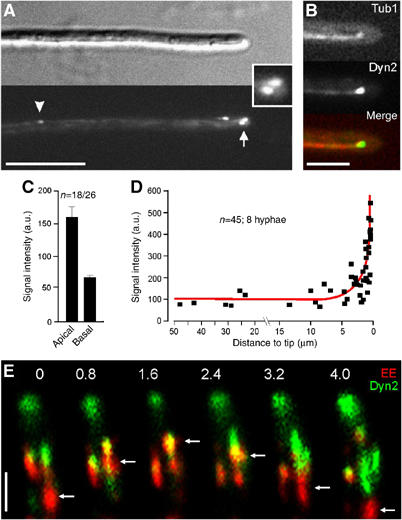
Dynein localization and EE movement. (A) In U. maydis hyphae dynein, visualized by a fusion of 3xGFP and the endogenous Dyn2 forms motile comet-like structures (arrowheads). The strongest signals are always found in the hyphal apex (arrow, inset). Bar: 10 μm. (B) Colocalization of GFP3Dyn2 (Dyn, green in overlay) and RFP-α-tubulin labeled MTs (red) in control hyphae. Dynein localizes to the MT plus-ends that are reaching into the hyphal apex. Bar: 5 μm. (C) A quantitative analysis of the GFP3-Dyn2 signal intensity at MT plus-ends in the first 3 μm of the hyphal tip (apical) and in at least 12 μm distance from the tip (basal) reveals that the dynein accumulations at the hyphal tip is more than twice as strong as in basal parts of the cell. Note that the signal intensity has been normalized to a single GFP. (D) The average intensity of dynein signals at MT plus-ends was measured in relation to distance to the tip, demonstrating that the amount of dynein remains constant over most of the length of the hypha, but rapidly increases at the hyphal apex. (E) Colocalization of GFP3Dyn2 (Dyn, green in overlay series) and Yup-RFP2 (EE, red in overlay series) demonstrates that EE (arrows) migrate into the dynein signal before they reverse direction (colocalization results in yellow). Elapsed time is given in seconds. Bar: 2 μm. Supplementary movie for Panel E is given on the EMBO web site.
Retrograde EE movement depends on activation of dynein by an LIS1 homologue
The accumulation of dynein in the hyphal tip suggested that the motor stays inactive until its cargo reaches the MT plus-ends. Therefore, we analyzed the mechanism of dynein activation and its impact on EE motility. A good candidate for dynein regulation is LIS1, which is thought to activate dynein (Faulkner et al, 2000; Smith et al, 2000; Zhang et al, 2003; Mesngon et al, 2006). The LIS1 homologue in U. maydis shares 47% amino-acid identity with its closest homologue NudF from A. nidulans (Xiang et al, 1999) and, like other LIS1-members, is predicted to contain an LisH motif, a coiled-coil region and seven WD40 domains (Figure 4A). We fused 3xGFP to the N-terminus of the endogenous lis1 gene, and this did not result in any defects in nuclear migration or growth (see below), indicating that the protein is fully functional. GFP3-Lis1 localized to MT tips (Figure 4B; strain FB2GLis1_RT), and concentrates at plus-ends in the hyphal tip (Figure 4B and C), suggesting that the putative dynein activator is part of the apical dynein complex. Next, we analyzed the role of Lis1 in dynein activation. In strain FB2G3Dyn2_nLis1, lis1 is expressed under the control of the inducible nar1-promoter (Brachmann et al, 2001). Upon growth in complete medium (CM) containing ammonium, this promoter is repressed and Lis1 is depleted (see Supplementary Figure 1 on EMBO web site), whereas it is induced in the presence of nitrate (NM). Reduction of Lis1 levels led to nuclear migration defects in yeast-like cells and longer MTs in hyphae (not shown). However, 87.5% of all plus-ends were still oriented to the hyphal tip (n=48; strain FB2nLis1_Peb1R_GT). Neither of these treatments affected dynein protein levels (see Supplementary Figure 1 on EMBO web site). However, the amount of dynein at MT plus-ends in the hyphal tip was significantly increased in the absence of Lis1, while it was slightly decreased upon overexpression of Lis1 (Figure 4D and E; Lis1↑: high protein levels, Lis1: no Lis1; FB2G3Dyn2_nLis1_RT; see Supplementary Movie 5). Consistent with the concept that EE are loaded onto dynein at the hyphal apex and that Lis1 activates dynein, EE were not able to leave the tip in the absence of Lis1 (Figure 4F; overlay of two time points, stationary organelles appear in yellow; FB2G3Dyn2_nLis1_YR2; for Figures 4F, 5F and 6E see also Supplementary Movie 1). Consequently, EE motility was significantly impaired (Figure 4G; control strain: FB2YR2; see Supplementary Movie 6). In summary, these results indicate that the LIS1-homologue Lis1 is crucial for the activation of dynein for retrograde EE transport.
Figure 4.
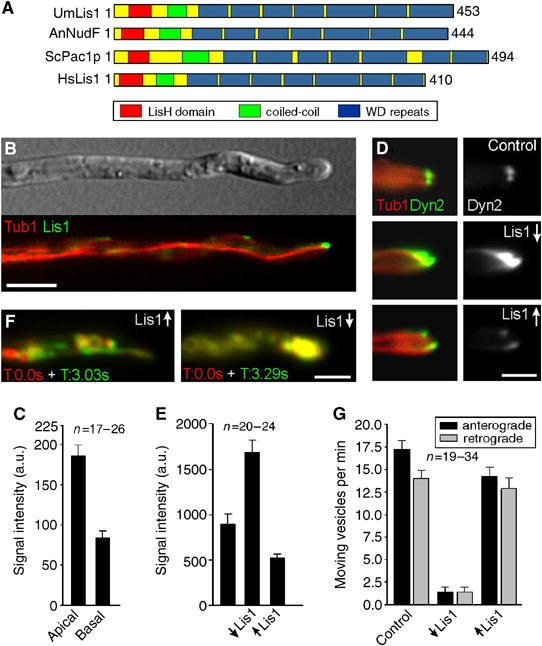
The role of a LIS1-homologue in dynein localization and endosome motility. (A) Lis1 from U. maydis (UmLis1, Accession Number: EAK84394) shares a similar domain structure with other LIS1-homologues, including those from A. nidulans (AnNudF, EAA57983), S. cerevisiae (ScPac1p, AAB00685) and human (HsLis1, NP 000421). (B) GFP-tagged endogenous Lis1 (green) localizes to plus-ends of RFP-α-tubulin containing MTs (red). Note that the tip contains the strongest signal. Bar: 5 μm. (C) Quantification of the GFP-Lis1 intensity at MT plus-ends in the apical 3 μm of the hyphal tip (apical) and in at least 12 μm distance from the tip (basal) demonstrates that Lis1 is concentrated at MT plus-ends in the tip. (D) The absence of Lis1 (Lis1↓) results in an increase of dynein at plus-ends within the hyphal tip. Overexpression of Lis1 (Lis1↑) reduces the apical dynein signal. Bar: 2 μm. (E) Quantitative analysis of GFP3Dyn2 intensity at MT plus-ends revealed that the absence of Lis1 (↓Lis1) significantly increases the amount of dynein at MT plus-ends. Consistently, Lis1 overexpression (↑Lis1) has the opposite effect. (F) Overlay of two images at different time points illustrates the motility of RFP2-tagged endosomes. Moving organelles appear in green or red, while stationary signals result in yellow color. Depletion of Lis1 led to an immobile accumulation of EE in the hyphal apex (strain FB2G3Dyn2_nLis1_YR2). Bar: 5 μm. Supplementary movies for Panel D and F are given on the EMBO web site. (G) A quantitative analysis of EE traffic reveals that overexpression of Lis1 (↑Lis1) affects EE motility only slightly, while nearly all motility is abolished in the absence of Lis1 (↓Lis1).
Figure 5.
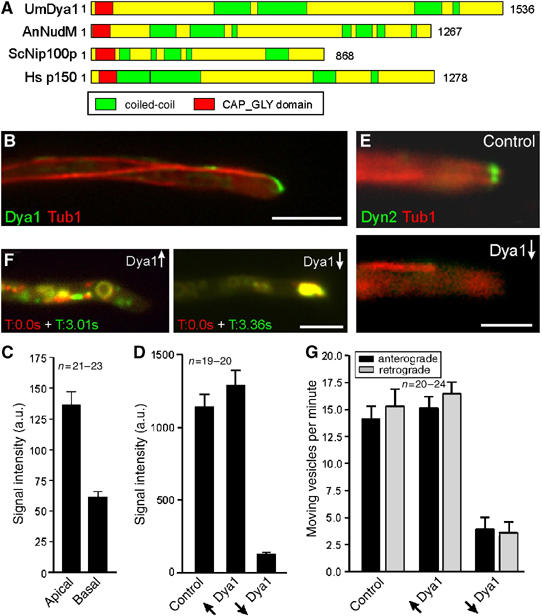
The role of a p150glued homologue in dynein localization and endosome motility. (A) The p150glued homologues from U. maydis (UmDya1, Accession Number: EAK84712) shares a similar domain structure with other p150glued homologues from A. nidulans (AnNudM, EAA58707), S. cerevisiae (ScNip100p, NP 015151) and human (Hsp150, NP 004073). (B) GFP3-tagged endogenous Dya1 (green) accumulates at the plus-ends of RFP-α-tubulin labeled MTs (red). The strongest signal is present in the hyphal apex. Bar: 5 μm. (C) Fluorescence intensity measurements of GFP3Dya1 at MT plus-ends in the first 3 μm of the hyphal tip (apical) and in basal parts of the cell (in >12 μm distance from the tip) reveals that dynactin was ∼2-times more concentrated at MT plus-ends in the hyphal tip than in subapical regions. (D) Quantification of the GFP3Dyn2 signals at MT plus-ends and in the apical cytoplasm shows that high levels of Dya1 (↑Dya1) has no significant effect on dynein localization (P=0.306), whereas dya1 repression (↓Dya1) leads to a dramatic decrease of the dynein signal. (E) No dynein is present at apical MT plus-ends after depletion of Dya1 (Dya1↓). Bar: 2.5 μm. (F) Overlay of two images at different time points illustrates the motility of GFP-tagged endosomes in strain FB2rDya1_YG. Moving organelles appear in green or red, while stationary signals result in yellow. Depletion of Dya1 leads to an immobile accumulation of EE in the hyphal apex. Bar: 5 μm. Supplementary movies for Panels E and F are given on the EMBO web site. (G) The expression of Dya1 (↑Dya1) has no effect on EE transport, whereas the absence of Dya1 (↓Dya1) abolishes almost all EE motility.
Figure 6.
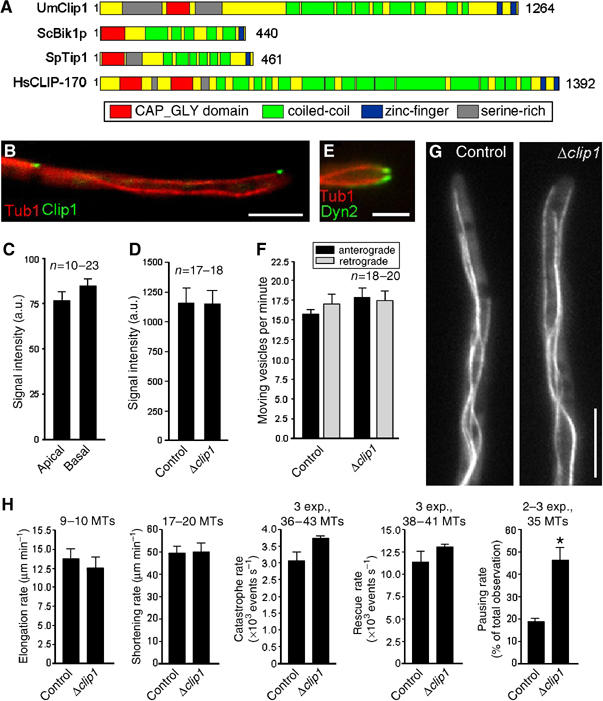
The role of a CLIP-170 homologue in dynein and endosome targeting. (A) The CLIP-170 homologues from U. maydis (UmClip1, Accession Number: EAK82811) shares a similar domain structure with other CLIP-170 members from S. cerevisiae (ScBik1p, NP 009901), Schizosaccharomyces pombe (SpTip1, P79065) and human (HsCLIP170, A43336). (B) GFP-tagged endogenous Clip1 (green) localizes to plus-ends of RFP-α-tubulin labeled MTs (red). Bar: 5 μm. (C) In contrast to dynein, dynactin and Lis1 intensities GFP-Clip1 fluorescence signals at MT plus-ends in the first 3 μm of the hyphae (apical) do not differ from those in basal parts of the cells (in >12 μm distance from tip). (D) In Δclip1 cells, the intensity of the dynein signal at MT plus-ends in the tip is not different from those of control cells. (E) In the clip1 deletion strain, GFP3-tagged dynein (green) still accumulates at plus-ends of RFP-labeled MTs (red). Bar: 2.5 μm. (F) Deletion of clip1 does not affect bidirectional EE motility. (G) In Δclip1 mutants, the organization of GFP-α-tubulin labeled MTs is not altered compared to control cells (strain FB2GT). Bar: 5 μm. Supplementary movies for Panels E and G are given on the EMBO web site. (H) Parameters of MT dynamic instability in control cells and Δclip1 mutant hyphae.
Dynactin is essential for dynein plus-ends localization and retrograde EE transport
In order to further characterize the regulation of dynein, we investigated the role of the dynein activator dynactin (Schroer, 2004) in EE transport. We identified dya1, a putative component of the dynactin complex that shares 19% amino acid identity with its homologue from A. nidulans. Similar to other p150glued homologues, Dya1 is predicted to contain a CAP-Gly motif, as well as numerous coiled-coil regions (Figure 5A). We fused a triple GFP-tag to the N-terminus of the endogenous dya1 gene and found that the fusion protein is functional, as it did not cause the dya1 mutant phenotype (see below). The fusion protein localized to the plus-ends of MTs (Figure 5B; strain FB2G3Dya1_RT), with most dynactin being concentrated at the apical MT ends (Figure 5B and C), which corresponds well with the data raised for dynein and Lis1 (see above). Next, we generated conditional mutants that expressed dya1 under the control of the strong crg1-promoter (Bottin et al, 1996). In the presence of arabinose, this promoter allowed expression of GFP-Dya1, whereas in the presence of glucose it was repressed and no fusion protein was detected (see Supplementary Figure on EMBO web site; strain FB2rGDya1). These growth conditions neither affected the orientation of MTs (86.2% of plus-ends grow towards the hyphal tip, n=29; strain FB2rDya1_Peb1R_GT) nor changed the levels of dynein in cell extracts of strain FB2rDya1_G3D (see Supplementary Figure on EMBO web site). Depletion of Dya1 abolished dynein accumulation in the tip (Figure 5D and E; Dya1↓: no Dya1, Dya1↑: high levels of Dya1; strain FB2rDya1_G3D_RT; see Supplementary Movie 5) and this led to an EE aggregation in the hyphal apex (Figure 5F; overlay of two time points, stationary signals appear in yellow) and a significant reduction in EE motility (Figure 5G; strain FB2rDya1_YG; see Supplementary Movie 6). These results demonstrate that the dynactin subunit Dya1 is essential for setting up a dynein reservoir at the hyphal tip, which in turn is required for retrograde EE traffic. Hence, these results add further support to our concept that organelles are loaded onto dynein in the hyphal tip.
Clip1 is not necessary for dynein plus-ends binding and EE movement
In vertebrates, the cytoplasmic linker protein CLIP-170 links endocytic membranes (Pierre et al, 1992) and the dynactin complex, and thereby indirectly dynein to MT plus-ends (Valetti et al, 1999; Lansbergen et al, 2004). Thus, we investigated the role of Clip1, the U. maydis CLIP-170 homologue, in EE traffic and dynein targeting. Although sequence similarity between Clip1 and human CLIP-170 is low (15% amino acid identity), Clip1 is predicted to have approximately the same size and contains a CAP-Gly motif flanked by two Serine-rich regions, as well as numerous coiled coil regions and two C-terminal zinc-finger like domains (Figure 6A). In addition, a fusion protein of GFP and Clip1 localized to MT plus-ends (Figure 6B; strain FB2GClip1_RT), which is consistent with its predicted role in targeting organelles or dynein/dynactin to MT tips. However, in contrast to dynein, Lis1 and Dya1, the CLIP-170 homologue showed similar signal intensities at apical and subapical MT plus-ends (Figure 6C), which argues against a role in dynein/dynactin anchoring. In order to characterize the role of Clip1 at the plus-ends, we constructed a clip1 deletion mutant (strain FB2ΔClip1) and confirmed the absence of the gene by Southern blot and RT–PCR (not shown). The absence of Clip1 had no effect on the amount of dynein at MT plus-ends in the tip (Figure 6D and E; strain FB2ΔClip1_G3Dyn2_RT; see Supplementary Movie 5) and neither anterograde nor retrograde EE traffic was inhibited (Figure 6F; FB2ΔClip_YG; see Supplementary Movie 6). Surprisingly, deletion of Clip1 also did not have a significant effect on MT organization and polarity either (Figure 6G; strains FB2ΔClip_GT; FB2ΔClip_Peb1R_GT) and only pausing rates of MTs were affected (Figure 6H). Taken together, these data show that the CLIP-170 homologue is neither involved in dynein anchoring nor in endosome targeting. In addition, Clip1 has only minor roles in MT dynamic instability.
Kinesin-1-based targeting of dynein/dynactin to MT plus-ends promotes minus-end directed endosome motility
The results presented so far were consistent with the notion that EE are loaded onto dynein at the apex. Therefore, we next asked how this apical dynein loading zone is established. In A. nidulans Kinesin-1 participates in plus-ends targeting of dynein (Zhang et al, 2003), and we considered it possible that U. maydis Kinesin-1 (Lehmler et al, 1997) also delivers dynein/dynactin to MT plus-ends. In Kinesin-1 null mutants, MT orientation was not altered (91% of all plus-ends directed towards hyphal apex; n=131; AB33ΔKin1_Peb1Y). However, no GFP-tagged dynein was found at the MT tips in the apex of these hyphae, but concentrated at subapical regions (Figure 7A, inset shows hyphal tip, strain AB33ΔKin1_G3D2_RT; see Supplementary Movie 7). A similar distribution was found for dynactin (Figure 7B; strain FB2ΔKin1_G3Dya1_RT), indicating that Kinesin-1 delivers a complex of dynein and dynactin to MT plus-ends in the hyphal tip. In contrast, the Lis1 accumulation at MT tips was only partially reduced in Kinesin-1 null mutants (not shown), suggesting that Lis1 is targeted by other mechanisms, which might also involve Kinesin-1. Consistent with the concept that dynein has to accumulate at the hyphal tip for retrograde transport, EE were trapped at apical MT plus-ends in Δkin1 hyphae (Figure 7C; strain AB33ΔKin1_YG), and almost no EE motility was observed (Figure 7D). This demonstrates that the apical localization of dynein/dynactin is required for retrograde EE transport.
Figure 7.
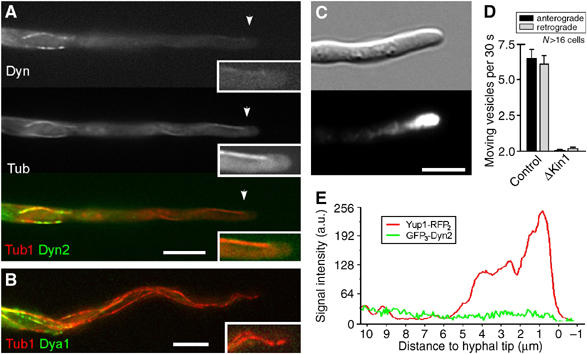
Kinesin-1-based targeting of dynein and dynactin to MT plus-ends in the hyphal tip. (A) Colocalization of GFP3Dyn2 (Dyn, green in overlay) and RFP-α-tubulin labeled MTs (Tub, red in overlay) in Δkin1 mutants reveals that dynein colocalizes with MTs in the subapical region. However, dynein does not reach the plus-ends in the hyphal tip (insets). Bar: 5 μm. (B) No GFP3Dya1 (green) appears at apical MT plus-ends (red) in Kinesin-1 null mutants, demonstrating that dynactin requires Kinesin-1 for long-distance plus-end targeting. Bar: 5 μm. (C) In Δkin1 hyphae EE cluster in the hyphal tip, indicating that deletion of Kinesin-1 strengthened plus-end directed transport, which is due to a lack of dynein in the hyphal tip. Bar: 5 μm. (D) Quantitative analysis of EE motility in the apical 10 μm of control and Kinesin-1 null mutant hyphae. Almost all motility is abolished in the absence of Kinesin-1. (E) Linescan analysis of intensities of GFP3-Dyn2 and Yup1-RFP2 in Δkin1 mutant hyphae (strain AB33ΔKin1_G3D2_YR2). In the absence of Kinesin-1, EE travel to the tip but are trapped there. Dynein is not enriched in the endosome cluster, which argues against a direct binding of the motor to anterogradely moving organelles. Supplementary movies for Panels A and C are given on the EMBO web site.
Dynein locates only on retrogradely moving endosomes
In a final set of experiments, we set out to obtain direct evidence for our hypothesis that EE are loaded onto dynein in the hyphal tip. Consistent with our hypothesis that dynein does not travel on EE towards the hyphal apex, we found no increase in dynein signals in the apical cluster of EE that are trapped in the hyphal tip of Δkin1 hyphae (Figure 7E). In addition, we did colocalization studies of EE and the functional GFP-fusion proteins. We analyzed the existing strains AB33G3Dyn2_YR2 and RWS9, which contains Kinesin-3YFP and CFP-labeled EE (Wedlich-Söldner et al, 2002). In addition, we generated mutant strains for Dya1 and Lis1 (FB2G3Dya1_YR2 and FB2G3Lis1_YR2; see Table I). The experiments demonstrated that 88.5% of all faint GFP3-dynein signals (n=52) leave the dynein accumulation for retrograde traffic (Figure 8A, colored arrows indicate individual moving signals; see Supplementary Movie 8). Consistently, dynein and dynactin were found to move on retrogradely travelling EEs (Figure 8B; Dynein, Dynactin). A quantitative analysis demonstrated that 100% of the backward moving EE carried dynein (n=20; 10 cells), while GFP3-Dyn2 was not found on anterogradely moving EE (n=14; 12 cells; Figure 8C). In contrast, GFP3-labeled Lis1 was found on 10% of the tipward moving EE, but localized to only 18.2% of the retrograde EE (Figure 8C) and often left the EE immediately after it started moving (Figure 8B; Lis1). Finally, we found that YFP-labeled Kinesin-3 colocalized with all moving EE in both directions (Figure 8C). Taken together, these results support our hypothesis and strongly suggest that dynein and dynactin bind to endosomes at the MT plus-ends and move organelles retrogradely. In contrast, Lis1 appears to have only transient contact with retrograde endosomes.
Figure 8.
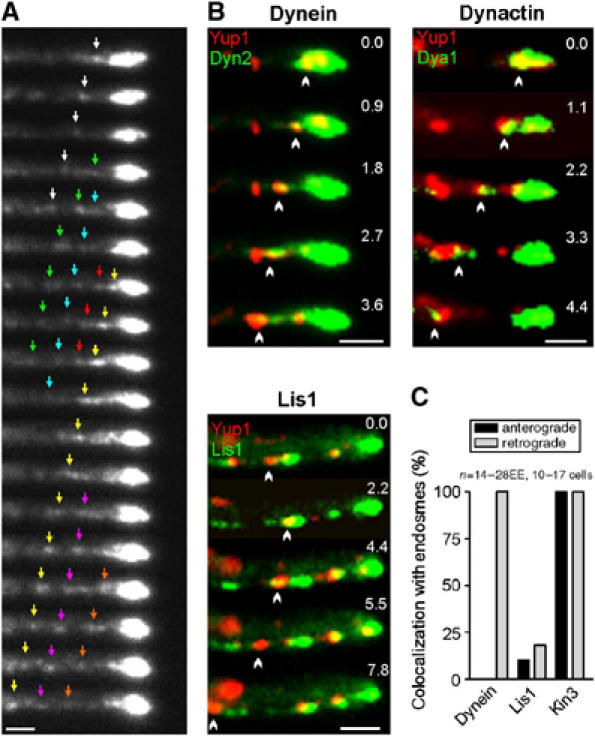
Dynein, dynactin, Lis1 on moving early endosomes. (A) In vivo observation of functional GFP3-Dyn2 at the hyphal tip demonstrates that dynein is continuously released from its apical accumulation. Individual signals marked by colored arrows. Interval between frames is 1 s. Bar: 1 μm. (B) Colocalization studies on strains expressing Yup1-RFP2 and GFP-tags fused to dyn2 (dynein), dya1 (dynactin) and lis1 (Lis1) demonstrate that the dynein/dynactin complex localizes to retrogradely moving organelles, whereas Lis1 only transiently appears on EE. Bars: 1 μm. (C) Quantitative analysis of EE that move for at least 3 μm. Kinesin-3 locates on all EE, whereas dynein was only found on retrogradely moving organelles. Supplementary movie for Panel B is given on the EMBO web site.
Discussion
Bidirectional motility of organelles is a very common phenomenon, which was found in various organelles, including endosomes, secretory vesicles, lysosomes and mitochondria (Gross 2004; Welte, 2004). Bidirectional motility was also described for endosomes in yeast-like cells of U. maydis (Wedlich-Söldner et al, 2000, 2002). In this plant pathogenic fungus EE participate in hyphal growth (Wedlich-Söldner et al, 2000), and hyphal growth is essential for host invasion and pathogenic development (Weber et al, 2003). Thus, we set out to understand the molecular mechanism of bidirectional EE motility in hyphae. We used the tSNARE Yup1, which was found to be located on rapidly moving organelles that were initially identified as EE based on their affinity to the endocytic dye FM4-64 (Wedlich-Söldner et al, 2000). Recent studies demonstrated that endosome specific Rab5-GTPases colocalize with Yup1-GFP in U. maydis (U Fuchs, I Schuchardt, G Steinberg, unpublished), which confirms that the Yup1-RFP2 positive organelles are EE.
The role of plus-end binding proteins in regulation of dynein function
The results presented here support the idea that dynein at MT plus-ends is inactive until an organelle reaches the tip (Figure 9). A well-studied dynein interacting protein is NudF/Lis1 (Xiang et al, 1999), which is thought to regulate dynein activity in various cell types (Faulkner et al, 2000; Smith et al, 2000; Coquelle et al, 2002; Zhang et al, 2003; Lansbergen et al, 2004; Mesngon et al, 2006). Here, we provide in vivo evidence for such a function. High levels of Lis1 reduced the dynein signal at the tip, whereas the depletion of Lis1 significantly increased the amount of dynein at apical MT plus-ends, which coincided with an inhibition of retrograde EE motility. Interestingly, Lis1 accumulates at the plus-ends in amounts similar to that of dynein, indicating that Lis1 is part of the inactive dynein complex. This suggests that the effects of Lis1 on dynein are regulated by other upstream factors. Such components could travel on the arriving EE and trigger the retrograde transport machinery (Figure 9), which might involve phosphorylation of Lis1 (Coquelle et al, 2002).
Figure 9.
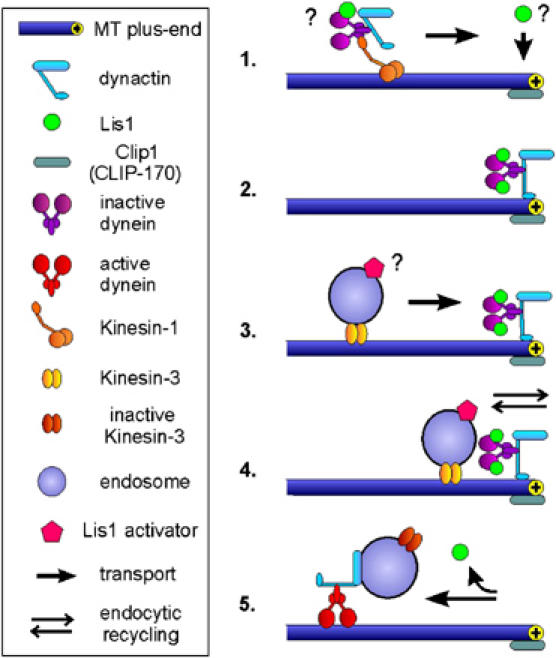
Dynein in minus-end directed traffic of early endosomes. (1) In the current model, Kinesin-1 takes dynein/dynactin to the MT plus-ends in the hyphal tip. Preliminary evidence indicates that a portion of Lis1 might hitchhike on the transported dynein/dynactin complex (not shown), but other mechanisms are also likely. (2) An inactive complex of dynein, dynactin and Lis1 accumulates at the plus-ends close to the growth region of the hypha. (3) Kinesin-3 transports EE along MTs to the inactive dynein/dynactin/Lis1 complex, thereby delivering an unknown activator of Lis1. (4) When EE reach the plus-ends at the hyphal apex they quickly exchange material with the growing tip. This might involve the uptake of material for transport towards the cell body as well as local membrane recycling processes. (5) Subsequently, the unknown activator triggers Lis1-dependent activation of the dynein/dynactin complex, which results in retrograde motility of the EE.
Another activator of dynein is the dynactin complex (Schroer, 2004). Initially discovered as a factor involved in dynein-dependent organelle traffic in vitro (Gill et al, 1991), it recently emerged as a factor that increases the processivity of dynein (King and Schroer, 2000) and participates in linking organelles to MTs via its MT binding region (Vaughan et al, 2002). In mammalian cells, dynactin concentrates at MT plus-ends (Valetti et al, 1999; Vaughan et al, 1999), where it is thought to mediate loading of endosomes to dynein (Vaughan, 2005). Indeed, our results demonstrate that an intact dynactin complex is required for the accumulation of dynein at plus-ends and, consequently, for retrograde EE transport. In contrast, dynactin is not required for plus-ends targeting of dynein in S. cerevisiae (Sheeman et al, 2003). This contradiction might result from a different targeting mechanism, as both dynein and dynactin are delivered to the hyphal apex by Kinesin-1, whereas this kinesin is not even present in bakers yeast. However, further studies on the mechanism of plus-ends targeting of both proteins are required to elucidate the molecular details of delivery of dynein/dynactin to MT plus-ends.
We found that Clip1, a homologue of mammalian cytoplasmic linker protein 170 (CLIP-170), is neither involved in dynein targeting nor in EE traffic. This finding is most surprising, as CLIP-170 takes part in anchoring of dynein/dynactin in mammals (Coquelle et al, 2002; Lansbergen et al, 2004) and yeast (Sheeman et al, 2003), but also anchors endocytic membranes to MT plus-ends (Pierre et al, 1992; Vaughan, 2005). Members of the CLIP-170 protein family have also been shown to regulate MT dynamics as anticatastrophe factors in fungi (Berlin et al, 1990; Brunner and Nurse, 2000; Efimov et al, 2006), and are rescue factors in mammalians (Komarova et al, 2002). However, the deletion of Clip1 had no effect on MT organization and only increased pausing rates of MTs in U. maydis hyphae. Thus, it appears that CLIP-170 like proteins perform various cellular roles in the different systems, and it is tempting to speculate that this variation reflects the complex interaction of the numerous plus-ends binding proteins in organizing or regulating the plus-ends complex.
A dynein-loading site at the plus-ends of MTs in the hyphal tip
We previously reported that EE travel to the tip of U. maydis hyphae, where they are thought to support tip growth by membrane recycling (Wedlich-Söldner et al, 2000). The major finding of this study is that EE reach the plus-ends in the hyphal tip by the activity of Kinesin-3, where they are apparently loaded onto an inactive dynein/dynactin/Lis1 complex that becomes activated. The results presented here demonstrate that dynein/dynactin takes endosomes, and presumably inactive Kinesin-3, towards the cell body, while most Lis1 does not bind endosomes or leaves early after reversal of transport direction (Figure 9). This model is supported by several lines of evidence: (1) MT plus-ends in the hyphal tip carry the strongest dynein, dynactin and Lis1 signals; (2) most EE move into the dynein/dynactin/Lis1 accumulation at MT plus-ends before they reverse direction; (3) EE are trapped at plus-ends in the hyphal tip when dynein is not activated by Lis1; (4) EE cluster at plus-ends in the tip when dynein targeting to the tip is impaired in Kinesin-1 null mutants or when dynactin is depleted; and (5) dynein/dynactin colocalizes with retrogradely but not anterogradely moving EE. Taken together, these results strongly suggest that dynein and associated factors are targeted to the hyphal tips by Kinesin-1, where dynein binds to Kinesin-3 delivered EE (Figure 9). Recent evidence from Drosophila oocytes provide good indication that a similar mechanism of dynein targeting exists in animal cells. Here, dynein/dynactin localization and function also depends on the activity of Kinesin-1 (Duncan and Warrior, 2002; Januschke et al, 2002). In mammals, plus-ends located dynactin is thought to mediate targeting of membranous cargoes to the MT tip, which is followed by the recruitment of dynein and initiation of retrograde transport (Vaughan et al, 2002; Vaughan, 2005). Thus, mammalian dynein is usually absent from MT plus-ends (Vaughan et al, 1999), whereas an already assembled complex of dynein/dynactin/Lis1 is waiting at plus-ends of interphase MTs in U. maydis (this study) and A. nidulans (Han et al, 2001; Zhang et al, 2003). These results suggest that some of the basic principles of bidirectional organelle traffic differ between cell types. The reason for the situation in fungi might lie in the way fungal hyphae expand.
The role of the dynein loading zone in hyphal tip growth
In U. maydis, a strong dynein accumulation is found in the growing tip of the hypha that waits for a Kinesin-3-delivered organelle in order to be activated and ‘off-loaded' onto the organelle. In contrast, in mammalian systems, kinesin and dynein motors are often located on the same organelle (Gross, 2004; Ligon et al, 2004; Welte, 2004). Having both motors on a single organelle appears to be an economic way to mediate bidirectional motility, which raises the question why filamentous fungi have established an apical dynein loading zone. The answer to this question might be the key to an understanding of fungal tip growth. Hyphae are surrounded by a rigid cell wall that is synthesized and modified by polarized exocytosis (Gow, 1995) and local membrane recycling (Wedlich-Söldner et al, 2000) at the growing tip. One way to ensure that organelles reach the tip is that retrograde motors wait there to take organelles backwards. If this is true, hyphal tip growth and the existence of an apical dynein loading zone are intimately linked and most likely co-evolved. During evolution, bidirectional transport along MTs was lost in the yeast S. cerevisiae (Madden and Snyder, 1998), but the same principal of dynein waiting at plus-ends to be activated in order to move towards the minus-end is still used for nuclear migration (Lee et al, 2003; Sheeman et al, 2003). Taken together, we speculate that the establishment of a dynein loading zone in the hyphal apex was a crucial step in fungal evolution, which is essential for tip growth and, therefore, the evolutionary success of this important organismic group.
Materials and methods
Strains and plasmids
MTs were labeled with ectopically expressed RFP-α-tubulin or GFP-α-tubulin (Steinberg et al, 2001), while EE were marked by a fusion protein of GFP or a 2xRFP protein fused to an endosome specific t-SNARE (Wedlich-Söldner et al, 2000), respectively. Dynein and dynactin were visualized with a 3xGFP-tag fused to the N-terminus of the endogenous dyn2 or dya1 gene, while the endogenous Clip1 and Lis1 were labeled by a single GFP. In order to deplete dya1 or dyn1, both genes were expressed under the control of the crg1 promoter that is repressed in glucose (Bottin et al, 1996), while lis1 was expressed under control of the nar1 promoter (Brachmann et al, 2001). Deletion of clip1 was performed by homologous replacement of the whole open reading frame with a phleomycin resistance cassette. Further details on U. maydis strains and plasmids are listed in Table I.
Growth condition
Strains were grown over night at 28°C in CM (Holliday, 1974) supplemented with 1% glucose (CM-G) or 1% arabinose (CM-A). Solid medium contained 2% (w/v) bacto-agar. Induction of hyphal growth in AB33-derivatives, in which the filament-inducing bE/bW transcription factor is under control of the nitrate inducible nar1 promoter, was done as previously described (Fuchs et al, 2005). Hyphal growth of strains derived from FB1 or FB2 was induced by synthetic pheromone (Fuchs et al, 2005). AB33rDyn1_YG was grown over night in CM-A, and the crg1-promoter was repressed and hyphal growth induced in glucose containing nitrate minimal medium (Holliday, 1974) for 16–22 h at 28°C, 200 r.p.m. Strains that allowed controlled expression of lis1 (FB2nGLis1 and FB2G3Dyn2_nLis1 and derivates) and dya1 (FB2rDya1_G3D and derivates, FB2rDya1_YG and FB2rGDya1) were grown over night in CM-G, washed in fresh medium, followed by resuspension in 0.5 ml NM-G or CM-A, respectively. Hyphal growth was induced by synthetic pheromone 6–8 h at 22°C and 200 r.p.m.
Sequence analysis
U. maydis homologues of LIS1, CLIP-170 and p150glued were identified in the public databases by using PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi). Domain analysis was performed using SMART, Coils and MotifScan (all accessible on ExPASy web site http://us.expasy.org/).
Microscopy, image processing and quantitative analysis
Logarithmically growing cells were embedded in low melt agarose or placed on top of a 2% agar cushion, and were immediately observed using a Zeiss Axioplan II imaging microscope (Oberkochen, Germany) and filter sets for YFP (BP500/20, FT515, BP535/30), CFP (BP436, FT455, BP480-500), FITC (BP470/20, FT510, BP515-565), and DsRed (BP565/30, FT585, BP620/60). Image aquisition was carried out using a CoolSNAP-HQ CCD camera (Photometrics, Tocsen, USA) controlled by the imaging software MetaMorph (Universal Imaging, Downing Town, USA). All image processing, including adjustment of brightness, contrast and gamma-values, was performed with MetaMorph and Photoshop (Adobe). Two-tailed t-tests were carried out using Prism (GraphPad). Signal intensities of GFP3-Dyn2, GFP-Lis1, GFP3-Dya1 and GFP-Clip1 were measured using MetaMorph. The average intensity within at MT plus-ends was corrected for the cytoplasmic background or the extracellular background. Endosome motility was analyzed in image sequences of 20–60 frames, taken with an exposure time of 500–1000 ms. The anterograde and retrograde EE movements were counted in a distance of about 10 μm from the hyphal tip. Only organelles that were moving for at least 3 μm were considered. Quantitative analysis of MT dynamics was performed essentially as described (Steinberg et al, 2001). The pausing rate was defined as the time in percent that an MT plus-ends does neither elongate nor shorten. MT orientation in different areas of the hyphae was determined essentially as described (Straube et al, 2003).
Supplementary Material
Supplementary Figure on EMBO web site
Supplementary Figure on EMBO web site; strain FB2rGDya
Movie 1
Movie 2
Movie 3
Movie 4
Movie 5
Movie 6
Movie 7
Movie 8
Movie Legends
Acknowledgments
We thank R Wedlich-Söldner and U Fuchs for generating strains. We are grateful to L Stevermann and U Fuchs for helpful comments on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft (STE 799/4-2).
References
- Berlin V, Styles CA, Fink GR (1990) BIK1, a protein required for microtubule function during mating and mitosis in Saccharomyces cerevisiae, colocalizes with tubulin. J Cell Biol 111: 2573–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottin A, Kamper J, Kahmann R (1996) Isolation of a carbon source-regulated gene from Ustilago maydis. Mol Gen Genet 253: 342–352 [DOI] [PubMed] [Google Scholar]
- Brachmann A, Weinzierl G, Kamper J, Kahmann R (2001) Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol Microbiol 42: 1047–1063 [DOI] [PubMed] [Google Scholar]
- Brunner D, Nurse P (2000) CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102: 695–704 [DOI] [PubMed] [Google Scholar]
- Coquelle FM, Caspi M, Cordelieres FP, Dompierre JP, Dujardin DL, Koifman C, Martin P, Hoogenraad CC, Akhmanova A, Galjart N, De Mey JR, Reiner O (2002) LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol Cell Biol 22: 3089–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JE, Warrior R (2002) The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Curr Biol 12: 1982–1991 [DOI] [PubMed] [Google Scholar]
- Efimov VP, Zhang J, Xiang X (2006) CLIP-170 homologue and NUDE play overlapping roles in NUDF localization in Aspergillus nidulans. Mol Biol Cell 17: 2021–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O'Connell CB, Wang Y, Vallee RB (2000) A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol 2: 784–791 [DOI] [PubMed] [Google Scholar]
- Fuchs U, Manns I, Steinberg G (2005) Microtubules are dispensable for the initial pathogenic development but required for long-distance hyphal growth in the corn smut fungus Ustilago maydis. Mol Biol Cell 16: 2746–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Schroer TA, Szilak I, Steuer ER, Sheetz MP, Cleveland DW (1991) Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J Cell Biol 115: 1639–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NAR (1995) Tip growth and polarity. In: The Growing Fungus, Gow and Gadd (eds) London, Glasgow, Weinheim, New York, Melbourne, Madras: Chapman & Hall [Google Scholar]
- Gross SP (2004) Hither and yon: a review of bi-directional microtubule-based transport. Phys Biol 1: R1–R11 [DOI] [PubMed] [Google Scholar]
- Gross SP, Welte MA, Block SM, Wieschaus EF (2002) Coordination of opposite-polarity microtubule motors. J Cell Biol 156: 715–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G, Liu B, Zhang J, Zuo W, Morris NR, Xiang X (2001) The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr Biol 11: 719–724 [DOI] [PubMed] [Google Scholar]
- Hoepfner S, Severin F, Cabezas A, Habermann B, Runge A, Gillooly D, Stenmark H, Zerial M (2005) Modulation of receptor recycling and degradation by the endosomal kinesin KIF16B. Cell 121: 437–450 [DOI] [PubMed] [Google Scholar]
- Holliday R (1974) Ustilago maydis. In: Handbook of Genetics, King RC (ed), Vol. 1, pp 575–595. New York, USA: Plenum Press [Google Scholar]
- Januschke J, Gervais L, Dass S, Kaltschmidt JA, Lopez-Schier H, St Johnston D, Brand AH, Roth S, Guichet A (2002) Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr Biol 12: 1971–1981 [DOI] [PubMed] [Google Scholar]
- King SJ, Schroer TA (2000) Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol 2: 20–24 [DOI] [PubMed] [Google Scholar]
- Komarova YA, Akhmanova AS, Kojima S, Galjart N, Borisy GG (2002) Cytoplasmic linker proteins promote microtubule rescue in vivo. J Cell Biol 159: 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbergen G, Komarova Y, Modesti M, Wyman C, Hoogenraad CC, Goodson HV, Lemaitre RP, Drechsel DN, van Munster E, Gadella TW Jr, Grosveld F, Galjart N, Borisy GG, Akhmanova A (2004) Conformational changes in CLIP-170 regulate its binding to microtubules and dynactin localization. J Cell Biol 166: 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Oberle JR, Cooper JA (2003) The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol 160: 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmler C, Steinberg G, Snetselaar KM, Schliwa M, Kahmann R, Bölker M (1997) Identification of a motor protein required for filamentous growth in Ustilago maydis. EMBO J 16: 3464–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligon LA, Tokito M, Finklestein JM, Grossman FE, Holzbaur EL (2004) A direct interaction between cytoplasmic dynein and kinesin I may coordinate motor activity. J Biol Chem 279: 19201–19208 [DOI] [PubMed] [Google Scholar]
- Madden K, Snyder M (1998) Cell polarity and morphogenesis in budding yeast. Ann Rev Microbiol 52: 687–744 [DOI] [PubMed] [Google Scholar]
- Mesngon MT, Tarricone C, Hebbar S, Guillotte AM, Schmitt EW, Lanier L, Musacchio A, King SJ, Smith DS (2006) Regulation of cytoplasmic dynein ATPase by Lis1. J Neurosci 26: 2132–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Tsukita S (2003) ‘Search-and-capture' of microtubules through plus-ends-binding proteins (+TIPs). J Biochem (Tokyo) 134: 321–326 [DOI] [PubMed] [Google Scholar]
- Pierre P, Scheel J, Rickard JE, Kreis TE (1992) CLIP-170 links endocytic vesicles to microtubules. Cell 70: 887–900 [DOI] [PubMed] [Google Scholar]
- Schroer TA (2004) Dynactin. Annu Rev Cell Dev Biol 20: 759–779 [DOI] [PubMed] [Google Scholar]
- Schuchardt I, Assmann D, Thines E, Schuberth C, Steinberg G (2005) Myosin-V, Kinesin-1, and Kinesin-3 cooperate in hyphal growth of the fungus Ustilago maydis. Mol Biol Cell 16: 5191–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D (2003) Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol 13: 364–372 [DOI] [PubMed] [Google Scholar]
- Smith DS, Niethammer M, Ayala R, Zhou Y, Gambello MJ, Wynshaw-Boris A, Tsai LH (2000) Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat Cell Biol 2: 767–775 [DOI] [PubMed] [Google Scholar]
- Steinberg G (2000) The cellular roles of molecular motors in fungi. Trends Microbiol 8: 162–168 [DOI] [PubMed] [Google Scholar]
- Steinberg G, Wedlich-Söldner R, Brill M, Schulz I (2001) Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J Cell Sci 114: 609–622 [DOI] [PubMed] [Google Scholar]
- Straube A, Brill M, Oakley BR, Horio T, Steinberg G (2003) Microtubule organization requires cell cycle-dependent nucleation at dispersed cytoplasmic sites: polar and perinuclear microtubule organizing centers in the plant pathogen Ustilago maydis. Mol Biol Cell 14: 642–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A, Enard W, Berner A, Wedlich-Söldner R, Kahmann R, Steinberg G (2001) A split motor domain in a cytoplasmic dynein. EMBO J 20: 5091–5100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube A, Weber I, Steinberg G (2005) A novel mechanism of nuclear envelope break-down in a fungus: nuclear migration strips off the envelope. EMBO J 24: 1674–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valetti C, Wetzel DM, Schrader M, Hasbani MJ, Gill SR, Kreis TE, Schroer TA (1999) Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol Biol Cell 10: 4107–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan KT (2005) Microtubule plus ends, motors, and traffic of Golgi membranes. Biochim Biophys Acta 1744: 316–324 [DOI] [PubMed] [Google Scholar]
- Vaughan KT, Tynan SH, Faulkner NE, Echeverri CJ, Vallee RB (1999) Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J Cell Sci 112 (Part 10): 1437–1447 [DOI] [PubMed] [Google Scholar]
- Vaughan PS, Miura P, Henderson M, Byrne B, Vaughan KT (2002) A role for regulated binding of p150(Glued) to microtubule plus ends in organelle transport. J Cell Biol 158: 305–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I, Gruber C, Steinberg G (2003) A class-V myosin required for mating, hyphal growth, and pathogenicity in the dimorphic plant pathogen Ustilago maydis. Plant Cell 15: 2826–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R, Bölker M, Kahmann R, Steinberg G (2000) A putative endosomal t-SNARE links exo- and endocytosis in the phytopathogenic fungus Ustilago maydis. EMBO J 19: 1974–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Söldner R, Straube A, Friedrich MW, Steinberg G (2002) A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J 21: 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA (2004) Bidirectional transport along microtubules. Curr Biol 14: R525–R537 [DOI] [PubMed] [Google Scholar]
- Xiang X, Zuo W, Efimov VP, Morris NR (1999) Isolation of a new set of Aspergillus nidulans mutants defective in nuclear migration. Curr Genet 35: 626–630 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Hiraoka Y (2003) Cytoplasmic dynein in fungi: insights from nuclear migration. J Cell Sci 116: 4501–4512 [DOI] [PubMed] [Google Scholar]
- Zhang J, Li S, Fischer R, Xiang X (2003) Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol Biol Cell 14: 1479–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure on EMBO web site
Supplementary Figure on EMBO web site; strain FB2rGDya
Movie 1
Movie 2
Movie 3
Movie 4
Movie 5
Movie 6
Movie 7
Movie 8
Movie Legends


