Abstract
By controlling the subcellular localization of growth factor receptors, cells can modulate the activity of intracellular signal transduction pathways. During Caenorhabditis elegans vulval development, a ternary complex consisting of the LIN-7, LIN-2 and LIN-10 PDZ domain proteins localizes the epidermal growth factor receptor (EGFR) to the basolateral compartment of the vulval precursor cells (VPCs) to allow efficient receptor activation by the inductive EGF signal from the anchor cell. We have identified EGFR substrate protein-8 (EPS-8) as a novel component of the EGFR localization complex that links receptor trafficking to cell fate specification. EPS-8 expression is upregulated in the primary VPCs, where it creates a positive feedback loop in the EGFR/RAS/MAPK pathway. The membrane-associated guanylate kinase LIN-2 recruits EPS-8 into the receptor localization complex to retain the EGFR on the basolateral plasma membrane, and thus allow maximal receptor activation in the primary cell lineage. Low levels of EPS-8 in the neighboring secondary VPCs result in the rapid degradation of the EGFR, allowing these cells to adopt the secondary cell fate. Extracellular signals thus regulate EGFR trafficking in a cell type-specific manner to control pattern formation during organogenesis.
Keywords: Caenorhabditis elegans , epidermal growth factor, receptor, trafficking, vulval development
Introduction
The establishment of epithelial polarity involves the asymmetric segregation of cell surface proteins to different plasma membrane compartments (Knoblich, 2001). During this process, the epidermal growth factor receptor (EGFR) is localized to the basolateral plasma membrane compartment (He et al, 2002). The intracellular trafficking of receptor tyrosine kinases such as the EGFR is regulated at multiple steps (Burke et al, 2001). First, the EGFR is sorted from the trans-Golgi network into secretory vesicles that are transported to the basolateral compartment. The basolateral sorting of the EGFR depends on a 23 amino-acid segment located in the cytoplasmic juxtamembrane domain (Hobert et al, 1997). After reaching the basolateral compartment, the receptor is retained on the basolateral plasma membrane. The basolateral retention of the EGFR depends on a second, distinct signal near the cytoplasmic tail of the receptor (Hobert et al, 1997). If the retention signal is deleted, then the EGFR is initially sorted to the basolateral compartment, but it undergoes rapid endocytosis and accumulates on the apical side of the cells. The rate of EGFR endocytosis is greatly accelerated by ligand binding. Since the dephosphorylation of the EGFR occurs mainly in intracellular vesicles (Haj et al, 2002; Berset et al, 2005), ligand-induced receptor endocytosis may serve to attenuate the EGFR signal. Endocytosed EGFR can either be targeted to the lysosomal pathway leading to its degradation or it can be recycled and sent back to the plasma membrane.
The development of the Caenorhabditis elegans vulva, the egg-laying organ of the hermaphrodite, serves as an excellent model to study how intercellular signals control cell fate specification and pattern formation during organogenesis (Sternberg and Han, 1998; Sundaram, 2005). During C. elegans vulval development, the anchor cell (AC) in the somatic gonad secretes the EGF-like growth factor LIN-3 that activates the EGFR homolog LET-23 in the adjacent vulval precursor cells (VPCs) to specify the primary (1°) vulval cell fate in the nearest VPC P6.p. A lateral signal produced by the 1° cell P6.p then specifies the secondary (2°) vulval cell fate in the neighboring VPCs P5.p and P7.p via the LIN-12 NOTCH signaling pathway (Greenwald et al, 1983). In order to receive the AC signal, the EGFR must be kept on the basolateral surface of the VPCs facing the AC (Simske et al, 1996; Kaech et al, 1998). An evolutionary conserved tripartite protein complex consisting of the three PDZ domain proteins LIN-2 (termed CASK in vertebrates), LIN-7 (termed VELIs in vertebrates) and LIN-10 (termed Mint-1 or X-11α in vertebrates) is required for the basolateral localization of the EGFR in the VPCs (Kaech et al, 1998; Whitfield et al, 1999). The LIN-7 adaptor protein directly binds to the C-terminus of LET-23 EGFR, and the membrane-associated guanylate kinase LIN-2 serves as a scaffolding protein for LIN-7 and LIN-10. Loss-of-function mutations in lin-2, lin-7 or lin-10 result in the apical mislocalization of LET-23 EGFR, and thus prevent the efficient activation of the receptor by LIN-3 EGF secreted on the basal side of the epithelium (Hoskins et al, 1996; Simske et al, 1996). However, it has been unknown which components of the receptor localization complex control the sorting, basolateral retention or endocytosis of LET-23 EGFR.
Here, we report the identification of the C. elegans homolog of the mammalian EGFR substrate protein 8 (EPS-8) as a new component of the receptor localization complex. Mammalian EPS-8 binds to the EGFR and inhibits receptor endocytosis, possibly by activating the GTPase-activating protein RNtre that inhibits RAB-5 signaling (Fazioli et al, 1993; Lanzetti et al, 2000; Burke et al, 2001). In addition, EPS-8 links EGFR signaling to actin remodeling via a Rac signaling pathway and through a novel actin-capping activity (Lanzetti et al, 2000; Croce et al, 2004; Disanza et al, 2004). During C. elegans vulval induction, EPS-8 binds to LIN-2 to prevent the endocytosis and subsequent degradation of the EGFR in the 1° cell lineage, while low levels of EPS-8 in the neighboring 2° vulval cells lead to the rapid downregulation of LET-23 EGFR in the 2° cell lineage. EPS-8 thus links EGFR trafficking to cell fate specification during development.
Results
EPS-8 positively regulates vulval development
To identify additional factors that control EGFR localization, we searched the C. elegans interactome database (Walhout et al, 2000) for putative interactors with LIN-7, LIN-2 or LIN-10 and tested candidates by RNA interference (RNAi) and mutant analysis for their genetic interaction with the EGFR/RAS/MAPK signaling pathway (see below and Table I). This approach has pointed at the EPS-8 protein (Croce et al, 2004), which interacts with the LIN-2 protein in a yeast two-hybrid assay, as a positive regulator of EGFR signaling. For the subsequent analysis, we used the eps-8(by160) allele that carries a deletion of exons 8–12, which are common to all known eps-8 splice variants, and thus likely represents a null allele (Croce et al, 2004). Since the eps-8(by160) deletion causes an early larval lethal phenotype, we used a strain in which the larval lethality has been rescued by expressing a functional eps-8::gfp fusion under control of the gut specific opt-2 promoter (Croce et al, 2004). This strain allowed us to study the consequences of a loss of eps-8 function during vulval development, and we refer to the animals as eps-8(lf) henceforth. In eps-8(lf) single mutants, P5.p, P6.p and P7.p adopt the wild-type pattern of 2°–1°–2° cell fates generating 22 vulval cells, and the animals exhibit no other obvious vulval defects based on morphological criteria (Figure 1A and C and Table I, row 2). However, eps-8(lf) or RNAi against eps-8 enhances the vulvaless (Vul) phenotype caused by lin-3 egf or let-23 egfr reduction-of-function (rf) and lin-7 loss-of-function (lf) mutations (Table I, rows 4–6, 8, 9 and 11–14). Interestingly, eps-8(lf) does not significantly enhance the lin-2(lf) Vul phenotype (Table I, rows 16 and 17), suggesting that loss of EPS-8 function in the absence of LIN-2 does not lead to a further decrease in vulval induction. Moreover, eps-8(lf) or eps-8 RNAi suppress the multivulva (Muv) phenotype caused by a let-60 ras gain-of-function (gf) mutation or by overexpression of the wild-type MAP kinase mpk-1 together with activated mek-2 (hs::mpk-1) (Lackner and Kim, 1998) (Table I, rows 21–28). Since both the let-60 ras(gf) mutation as well as the hs::mpk-1 transgene are sensitive to a reduction of the inductive signal upstream of LET-60 RAS (Dutt et al, 2004), this analysis did not allow us to determine at which step EPS-8 regulates the EGFR/RAS/MAPK pathway.
Table 1.
eps-8 positively regulates EGFR/RAS/MAPK signaling
| Row | Genotype | Induced VPCsa | % Vulb | % Muvc | n | P-valued |
|---|---|---|---|---|---|---|
| 1 | Wild-type | 3.0 | 0 | 0 | Many | — |
| 2 | eps-8(lf) | 3.0 | 0 | 0 | 35 | — |
| 3 | [lin-31p::eps-8a] | 3.1 | 0 | 16 | 131 | <0.0001 (1) |
| 4 | lin-3(rf) | 0.8 | 95 | 0 | 75 | — |
| 5 | lin-3(rf); gfp RNAi | 0.5 | 88 | 0 | 128 | — |
| 6 | lin-3(rf); eps-8 RNAi | 0.2 | 98 | 0 | 97 | <0.005 (5) |
| 7 | lin-3(rf); [lin-31p::eps-8a] | 2.1 | 35 | 0 | 71 | <0.005 (4) |
| 8 | let-23(rf) | 1.1 | 80 | 0 | 106 | — |
| 9 | let-23(rf); eps-8(lf) | 0.4 | 97 | 0 | 32 | <0.0001 (8) |
| 10 | let-23(rf); [lin-31p::eps-8a] | 3.0 | 0 | 0 | 38 | <0.0001 (8) |
| 11 | lin-7(lf) | 0.9 | 94 | 0 | 72 | — |
| 12 | lin-7(lf); eps-8(lf) | 0.3 | 94 | 0 | 33 | <0.001 (11) |
| 13 | lin-7(lf); gfp RNAi | 0.9 | 95 | 0 | 73 | — |
| 14 | lin-7(lf); eps-8 RNAi | 0.3 | 98 | 0 | 71 | <0.001 (13) |
| 15 | lin-7(lf); [lin-31p::eps-8a] | 3.0 | 0 | 0 | 35 | <0.0001 (11) |
| 16 | lin-2(lf) | 0.6 | 92 | 3 | 38 | — |
| 17 | lin-2(lf); eps-8(lf) | 1.0 | 77 | 0 | 26 | <0.25 (16) |
| 18 | lin-2(lf); [lin-31p::eps-8a] | 1.8 | 73 | 3 | 30 | <0.0001 (16) |
| 19 | sem-5(rf) | 1.1 | 75 | 0 | 64 | — |
| 20 | sem-5(rf); [lin-31p::eps-8a] | 1.1 | 74 | 0 | 27 | <0.95 (19) |
| 21 | let-60(gf) | 4.5 | 0 | 84 | 44 | — |
| 22 | let-60(gf); eps-8(lf) | 3.3 | 0 | 25 | 20 | <0.0001 (21) |
| 23 | let-60(gf); gfp RNAi | 4.9 | 0 | 90 | 101 | — |
| 24 | let-60(gf); eps-8 RNAi | 3.7 | 0 | 64 | 75 | <0.0001 (23) |
| 25 | [hs::mpk-1]e | 4.5 | 0 | 69 | 24 | |
| 26 | [hs::mpk-1]; eps-8(lf)e | 3.0 | 0 | 0 | 28 | <0.0001 (25) |
| 27 | [hs::mpk-1]; gfp RNAie | 4.3 | 0 | 70 | 40 | — |
| 28 | [hs::mpk-1]; eps-8 RNAie | 3.4 | 0 | 23 | 49 | <0.0001 (27) |
| 29 | lin-12 (n302) | 0.4 | 92 | 0 | 37 | |
| 30 | lin-12 (n302); eps-8(lf) | 0.7 | 96 | 0 | 27 | <0.2 (29) |
| 31 | lin-12 (n379) | 0.5 | 90 | 0 | 42 | |
| 32 | lin-12 (n379); eps-8(lf) | 0.3 | 100 | 0 | 45 | <0.2 (31) |
| Alleles used: zhIs11[lin-31::eps-8a], eps-8(by160); Ex[opt-2::eps-8A], lin-3(e1417), let-23(sy1), lin-7(e1413), lin-12(n302); lin-12(n379), lin-2(n397), sem-5(n2019), let-60(n1046gf), gaIs36[hs::mpk-1(+), D-mek-2(gf)]. | ||||||
| aThe induction index indicates the average number of VPCs per animal that adopted 1° or 2° vulval fates as described (Berset et al, 2001). | ||||||
| b%Vul indicates the fraction of animals with fewer than three induced VPCs. | ||||||
| c%Muv indicates the fraction of animals with more than three induced VPCs. | ||||||
| dStatistical significance was tested with two-tailed Student's t-test. Numbers in parentheses indicate the row to which a data set was compared. | ||||||
| eTo induce MPK-1 expression, L1 larvae were heat-shocked for 30 min at 33°C every 24 h until the L4 larval stage. | ||||||
Figure 1.
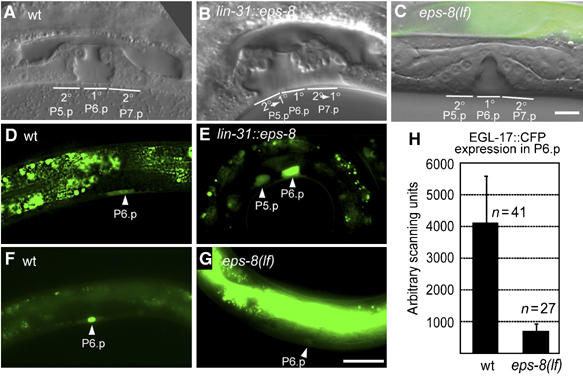
EPS-8 positively regulates vulval cell fate specification. (A) Morphology of a wild-type vulva at the L4 stage. The position of the 2° descendants of P5.p and P7.p and the 1° descendants of P6.p (out of focus) is indicated. Note that some of the 2° vulval cells remain attached to the cuticula while all 1° cells are detached. (B) Vulval morphology in a lin-31::eps-8a L4 larva showing a 2° towards 1° fate transformation of P5.p and P7.p descendants. (C) eps-8(lf) L4 larva with a wild-type vulva consisting of 22 cells. (D) Expression of the 1° cell fate marker EGL-17::GFP in a wild-type early L3 larva is restricted to P6.p. (E) Ectopic EGL-17::GFP expression in P5.p in a lin-31::eps-8a L3 larva (17% of the cases, n=24). (F) EGL-17::CFP expression in the nucleus of P6.p in a wild-type L3 larva. (G) Reduced EGL-17::CFP expression in P6.p in an eps-8(lf) L3 larva. Note in (C) and (G) the bright GFP expression in the gut due to the presence of the rescuing opt-2::eps-8::gfp transgene. Scale bar in (G) is 10 μm. (H) Quantification of EGL-17::CFP expression in P6.p of wild-type and eps-8(lf) larvae.
EPS-8 likely affects the intracellular trafficking of several proteins. In particular, EPS-8 could regulate the activity of the NOTCH signaling pathway, which requires the endocytosis of the Delta ligand in the signal producing cells (Wang and Struhl, 2005). For example, eps-8(lf) might promote LIN-12 NOTCH signaling by increasing the rate of endocytosis of a Delta ligand in the 1° vulval cells. However, eps-8(lf) neither enhances nor suppresses the Vul phenotype of two weak lin-12 notch gain-of-function mutants, in which no 1° fate is specified due to the absence of an AC and the induced VPCs always adopt the 2° cell fate (Table I, rows 29–32) (Greenwald and Seydoux, 1990). This observation indicates that at least during vulval induction eps-8(lf) does not significantly alter LIN-12 NOTCH signaling.
We thus conclude that EPS-8 positively regulates EGFR/RAS/MAPK signaling during vulval development, but EPS-8 is not required for vulval induction under normal conditions.
Constitutive expression of eps-8 results in the specification of multiple 1° cells
To examine the consequences of EPS-8 overexpression on vulval development, we constitutively expressed EPS-8A, the longest splice variant derived from the Y57G11C.24g transcript, in all VPCs under control of the Pn.p cell-specific lin-31 promoter (lin-31p::eps-8a, Tan et al, 1998). lin-31p::eps-8a animals exhibit a weak Muv phenotype due to the ectopic induction of vulval cell fates in P3.p, P4.p and P8.p. In addition, the vulva of lin-31p::eps-8a animals shows morphological changes characteristic of a 2° to 1° cell fate transformation in P5.p and P7.p (Figure 1A and B and Table I, row 3). In 91% (n=131) of L4 larvae, the descendants of P5.p and P7.p, which normally adopt the 2° vulval fate, have detached from the cuticle and migrated inwards like the 1° descendants of P6.p (Katz et al, 1995). This morphological cell fate transformation is accompanied by the ectopic expression of the 1° cell fate marker egl-17::gfp, which is normally detectable only in P6.p and its descendants (Burdine et al, 1998; Cui and Han, 2003). In 17% of lin-31p::eps-8a animals (n=24), EGL-17::GFP is expressed in P5.p and/or P7.p in addition to P6.p (Figure 1D and E). On the other hand, in eps-8(lf) mutants, the levels of egl-17::cfp in P6.p are about 10-fold reduced when compared to wild-type animals (Figure 1F through H; here, we used the cfp version of the 1° fate marker because it is more sensitive than the egl-17::gfp marker used above). It is remarkable that despite the strong reduction in 1° marker expression, eps-8(lf) single mutants develop a wild-type vulva as shown above (Figure 1C and Table I, row 2).
To determine at which step of the inductive signaling pathway EPS-8 acts, we performed an epistasis analysis with the lin-31::eps-8a transgene and mutations that reduce the activity of the EGFR/RAS/MAPK pathway at different steps. lin-31p::eps-8a completely suppresses the Vul phenotype caused by a let-23 egfr(rf) or a lin-7(lf) mutation (Table I, rows 10 and 15), but only weakly suppresses the lin-3 egf and lin-2(lf) Vul phenotypes and does not affect an rf mutation in sem-5, which encodes a GRB2 adaptor protein that transduces the signal between LET-23 EGFR and SOS-1 (Table I, rows 7 and 20) (Clark et al, 1992). Taken together, these results indicate that EPS-8 positively regulates EGFR/RAS/MAPK signaling upstream of SEM-5 GRB2 and that the function of EPS-8 largely depends on the presence of LIN-2 and on the inductive LIN-3 EGF signal from the AC.
eps-8 expression is upregulated in the 1° vulval cell lineage
In order to study the expression pattern of eps-8, we generated transgenic animals carrying a transcriptional gfp reporter (eps-8p::nls::gfp). This construct contains 2.5 kb of the 5′ regulatory region that is sufficient to rescue the larval lethality of eps-8(lf) mutants when fused to eps-8a cDNA (Croce et al, 2004 and data not shown). The eps-8p::nls::gfp reporter is expressed in a variety of cell types including neurons, gut, muscle and seam cells as well as in the VPCs and their descendants (Figure 2A–D and data not shown). A time course analysis revealed a dynamic expression pattern of eps-8::nls:.gfp in the vulval cells. In mid-L2 larvae, eps-8p::nls::gfp is weakly expressed in all VPCs except for P3.p (Supplementay Figure S1A). Around 6 h later in early L3 larvae, before the first round of vulval cell divisions, expression is strongest in the 1° cell P6.p, lowest in the 2° cells P5.p and P7.p and intermediate in the 3° cells P3.p, P4.p and P8.p (Figure 2A and Supplementay Figure S1A). At the Pn.px stage, the proximal descendants of P5.p and P7.p (P5.pp and P7.pa) that are in direct contact with the two 1° descendants of P6.p and hence receive more lateral LIN-12 NOTCH signal express less eps-8p::nls::gfp than the distal descendants (P5.pa and P7.pp in Figure 2B and C and Supplementay Figure S1A). A similar asymmetric expression in the 2° lineage has previously been observed for the EGFR/RAS/MAPK target gene egl-17 (Burdine et al, 1998; Berset et al, 2005).
Figure 2.
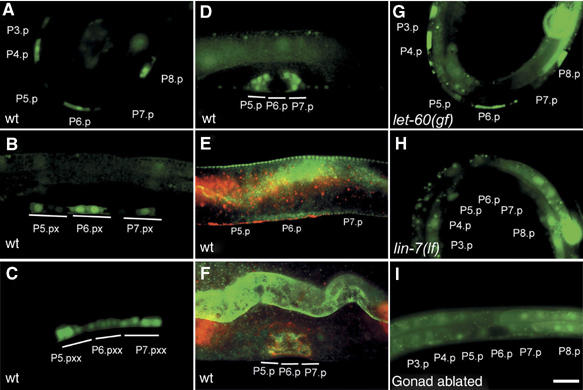
EPS-8 expression is upregulated in the 1° vulval cells. Expression of the eps-8p::gfp transcriptional reporter in the VPCs or their descendants of (A) a wild-type larva at the Pn.p cell stage (early L3), (B) at the Pn.px, (C) at the Pn.pxx stage (both mid L3) and (D) at the L4 stage. (E) Whole-mounts of wild-type larvae at the Pn.p cell stage and (F) at the L4 stage stained with polyclonal EPS-8 antibodies (in green). Adherens junctions are stained with MH27 in red. (G) eps-8p::gfp expression in a let-60(n1046gf) and (H) lin-7(e1413) mutant at the Pn.p stage (early L3). (I) eps-8p::gfp expression in a gonad-ablated L3 larva lacking the AC at the Pn.p stage (early L3). For a quantification of the expression patterns, see Supplementay Figure S1. Scale bar in (I) is 10 μm.
Immuno-staining of whole-mount L3 larvae with polyclonal antibodies against EPS-8 (Croce et al, 2004) detected endogenous EPS-8 protein in the same tissues expressing the eps-8p::nls::gfp reporter (data not shown). Among the vulval cells, EPS-8 protein was detectable only in P6.p and its descendants (Figure 2E), suggesting that either the expression observed with the transcriptional eps-8p::nls::gfp reporter in the 3° VPCs P3.p, P4.p and P8.p is too weak to be detected by antibody staining or that post-transcriptional downregulation of EPS-8 occurs in the 3° cells. At later stages during vulval morphogenesis, eps-8p::nls::gfp expression and antibody staining is found in all vulval cells including the descendants of P5.p and P7.p (Figure 2D and F; the 1° descendants of P6.p are in different focal planes).
The eps-8p::nls::gfp expression pattern suggested that eps-8 transcription may be activated in the 1° cell lineage by EGFR/RAS/MAPK signaling and repressed in the 2° cell lineage due to lateral inhibition of the EGFR/RAS/MAPK pathway by LIN-12 NOTCH (Greenwald et al, 1983; Berset et al, 2001; Yoo et al, 2004). We therefore asked whether increasing or decreasing EGFR/RAS/MAPK signaling alters eps-8p::nls::gfp expression pattern. In let-60 ras(gf) mutants, eps-8p::nls::gfp expression is enhanced in the distal VPCs P3.p, P4.p and P8.p (Figure 2G and Supplementay Figure S1B). Conversely, lin-7 (lf) mutants or gonad ablated animals that lack the AC show a strong reduction of eps-8p::nls::gfp expression in all VPCs (Figure 2H and I and Supplementay Figure S1C and D). Thus, eps-8 transcription is induced in the 1° vulval lineage, directly or indirectly, by the inductive AC signal.
EPS-8 retains LET-23 EGFR on the basolateral plasma membrane of the 1° cells
Since mammalian EPS-8 inhibits EGFR endocytosis (Lanzetti et al, 2000), we asked whether eps-8(lf) or constitutive expression of eps-8 in the VPCs change the subcellular localization or expression pattern of LET-23 EGFR. In wild-type mid-L2 larvae before vulval induction, LET-23 EGFR is weakly but uniformly expressed in all VPCs (Kaech et al, 1998). Around the time of induction in early L3 larvae, LET-23 EGFR expression is strongly upregulated in the 1° VPC P6.p, where it accumulates on the basolateral plasma membrane and near the adherens junctions (Figure 3A, A′ and Table II, row 1). At the same time, LET-23 EGFR disappears first in the 2° VPCs P5.p and P7.p and later in the 3° VPCs P3.p, P4.p and P8.p (Figure 3C).
Figure 3.
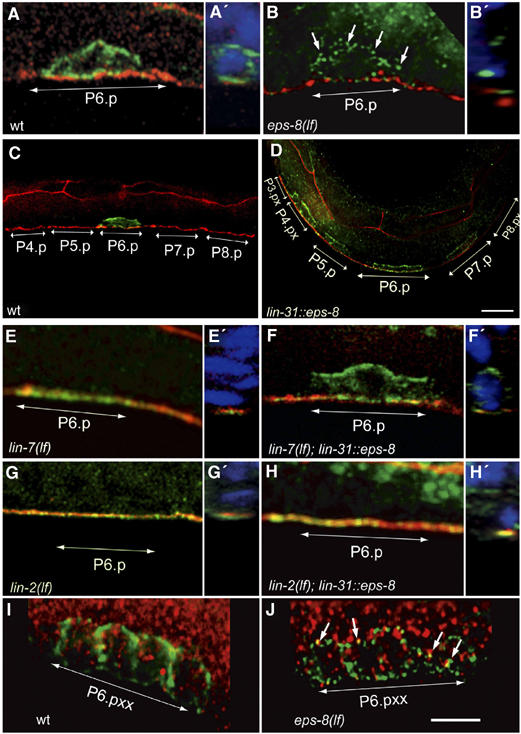
EPS-8 regulates LET-23 EGFR trafficking in the VPCs. LET-23 EGFR staining (green) in whole-mount L3 larvae using a polyclonal LET-23 antibody (see Materials and methods). The adherens junctions (red) were stained with the MH27 antibody in (A) through (H). The basal side of the VPCs is up in all panels. (A) Basolateral localization of LET-23 EGFR in P6.p of a wild-type early L3 larva. (B) Intracellular accumulation of LET-23 EGFR in an eps-8(lf) larva (arrows point at LET-23 punctae). (C) In late L2/early L3 larvae LET-23 EGFR is downregulated in all VPCs except for P6.p. (D) lin-31::eps-8a animals show persisting LET-23 EGFR expression in all VPCs. For a quantification of this phenotype, see Supplementay Figure S2. (E) Apical mislocalization of LET-23 EGFR in a lin-7(e1413) larva. (F) Partial relocalization of LET-23 EGFR to the basolateral compartment in a lin-7(e1413) larva caused by the lin-31::eps-8a transgene. (G) Apical mislocalization of LET-23 EGFR in a lin-2(n397) mutant. (H) lin-31::eps-8a fails to rescue the mislocalization of LET-23 EGFR in lin-2(n397) mutants. (I) Wild-type L3 larva (Pn.pxx stage) stained with antibodies against LET-23 (green) and the early endosomal marker EEA1 (red). (J) Partial co localization of LET-23 EGFR with EEA1 (arrows) in an eps-8(lf) larva. Panels A′, B′ and E′ to H′ show z–y sections in which DAPI stained nuclei are shown in blue color. Scale bars in (D) and (J) are 5 μm.
Table 2.
The first L27 domain in LIN-2 is required for basolateral retention of LET-23 EGFR
| Row | Genotype | Line #a | induced VPCs | % Vul (n) | % apicalb | % basolateralb | % intracellularb | n |
|---|---|---|---|---|---|---|---|---|
| 1 | Wild type | — | 3.0 | 0 | 0 | 100 | 0 | 46 |
| 2 | eps-8(lf) | — | 3.0 | 0 (58) | 0 | 16 | 89 | 19 |
| 3 | lin-2(lf) | — | 0.5 | 92 (38) | 100 | 0 | 0 | 37 |
| 4 | lin-2(lf); [lin-2 w.t.] | 1 | 3.0 | 0 (22) | 0 | 91 | 18 | 11 |
| 5 | lin-2(lf); [lin-2 w.t.] | 2 | 3.0 | 0 (29) | 17 | 67 | 33 | 6 |
| 6 | lin-2(lf); [lin-2 w.t.] | 3 | 3.0 | 0 (17) | 18 | 65 | 18 | 17 |
| 7 | lin-2(lf); [lin-2 L407S] | 1 | 3.0 | 0 (18) | 50 | 8 | 83 | 12 |
| 8 | lin-2(lf); [lin-2 L407S] | 2 | 3.0 | 0 (22) | 56 | 11 | 72 | 18 |
| 9 | lin-2(lf); [lin-2 L407S] | 3 | 3.0 | 0 (23) | 58 | 8 | 67 | 12 |
| 10 | lin-2(lf); [lin-2 I439S] | 1 | 0.3 | 100 (19) | 100 | 0 | 0 | 9 |
| 11 | lin-2(lf); [lin-2 I439S] | 2 | 0.3 | 95 (19) | 100 | 0 | 0 | 15 |
| 12 | lin-2(lf); [lin-2 I439S] | 3 | 0.4 | 93 (15) | 100 | 0 | 0 | 11 |
| Vulval induction was scored as described in the legend to Table I and Materials and methods. | ||||||||
| aFor each construct, three independent transgenic lines were scored. | ||||||||
| bLET-23 EGFR localization in P6.p was scored in whole-mount larvae stained with LET-23 and MH27 antibodies and classified as apical (ventral to the adherens junctions), basolateral (on the plasma membrane dorsal to the adherens junctions) and intracellular as shown in Figure 5. Note that LET-23 staining within a cell is often detected in more than one compartment. Alleles used: eps-8(by160), lin-2(n397), zhEx129.1[lin-2 w.t., sur-5::gfp] through zhEx129.3, zhEx165.1[lin-2 L407S, sur-5::gfp] through zhEx165.3, zhEx166.1[lin-2 I439S, sur-5::gfp] through zhEx166.3. | ||||||||
However, in eps-8(lf) animals, LET-23 EGFR accumulates in P6.p in intracellular punctae that partially colocalize with the early endosome marker EEA1 (Figure 3B, I and J and Table II, row 2) (Mills et al, 1998; Wilson et al, 2000). The altered subcellular localization of LET-23 EGFR in eps-8(lf) animals suggests that EPS-8 is required to keep LET-23 on the basolateral plasma membrane, either by inhibiting receptor endocytosis or by stimulating receptor recycling.
lin-31p::eps-8a animals, on the other hand, show persisting LET-23 EGFR staining in additional VPCs besides P6.p (Figure 3D; and Supplementay Figure S2). Thus, constitutive overexpression of EPS-8 prevents the downregulation of LET-23 EGFR in those VPCs that normally adopt the 2° or 3° cell fate. We also tested if EPS-8 can compensate for a loss of LIN-2 or LIN-7 function. To this aim, we introduced the lin-31p::eps-8a transgene into a lin-2(lf) and lin-7(lf) mutant background and determined the subcellular localization of LET-23 EGFR. Loss-of-function mutations in lin-2 or lin-7 cause a mislocalization of LET-23 EGFR to the apical VPC compartment (Figure 3E and G and Table II, row 3) (Simske et al, 1996; Kaech et al, 1998). lin-31p::eps-8a partially restores the basolateral localization of LET-23 EGFR in lin-7(lf) mutants (50% of the cases, n=18), but it does not alter the apical mislocalization of LET-23 EGFR in lin-2(lf) mutants (Figure 3F and H). Thus, EPS-8 can only regulate LET-23 EGFR localization in the presence of LIN-2.
EPS-8 binds to the first L27 domain in LIN-2
To further characterize the interaction between EPS-8 and LIN-2, we narrowed down the binding domains in C. elegans EPS-8 and LIN-2 using a yeast two-hybrid assay, and then confirmed the data by performing GST pull-down experiments with the mammalian homologs. The LIN-2 binding domain in EPS-8 maps between amino acids 462 and 511, a region that shows no significant homology to known protein domains (Figure 4A). Interestingly, LIN-7 and EPS-8 bind to distinct domains in LIN-2. As reported previously, LIN-7 binds to the second LIN-2/-7 interaction (L27) domain of LIN-2 (amino acids 425–555, Figure 4B; Harris et al, 2002), while EPS-8 binds to the first L27 domain of LIN-2 (amino acids 368–426, Figure 4B). Similar results were obtained in GST pull-down experiments using human LIN-2 CASK fused to GST as a bait and MDCK cell lysates as source of mammalian (m) LIN-7A and mEPS-8 (Figure 4C). Moreover, LIN-2 CASK from MDCK cells could be co-immunoprecipitated with mEPS-8 or mLIN-7A antibodies, and LIN-2 CASK immuno-precipitates contained mEPS-8 and mLIN-7A (Figure 4D). Finally, all three proteins were partially co-localized at the lateral plasma membrane of MDCK cells (Supplementary Figure S3).
Figure 4.
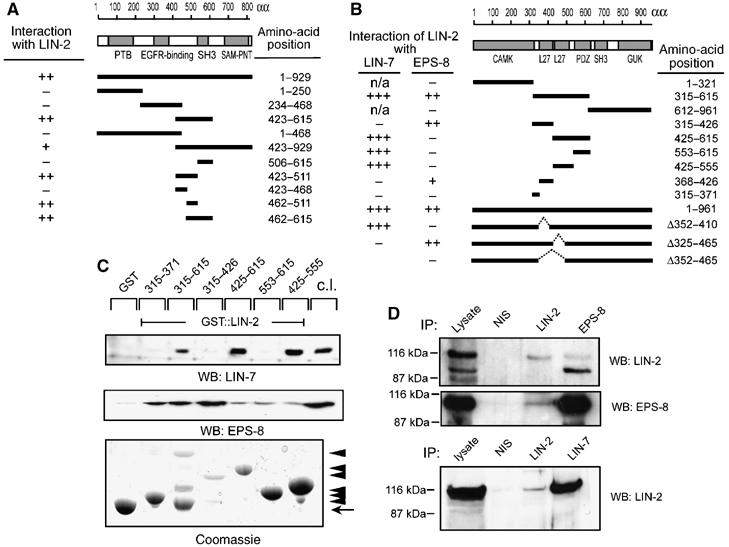
EPS-8 associates with the LIN-2/LIN-7/LIN-10 complex. (A) The indicated fragments of C. elegans EPS-8 were tested in a yeast two-hybrid assay using amino acid 315–615 of C. elegans LIN-2 as bait. (B) The indicated fragments of C. elegans LIN-2 were tested in a yeast two-hybrid assay for interaction with full-length LIN-7 and EPS-8, respectively. Interactions were tested both by His− growth and lacZ activity (+++ strong, ++ medium, + week interaction, defined by lacZ filter staining). (C) GST pull-down assays using the indicated fragments of human LIN-2 CASK cDNA fused to GST and MDCK cell lysates as source of mLIN-7A and mEPS-8 (p97 form). Bound proteins were detected on Western blots with mLIN-7A (upper panel) or mEPS-8 (middle panel) antibodies. The GST::LIN-2 fusion proteins were detected by Coomassie staining (bottom panel). Arrowheads point at GST::LIN-2 fusions, the arrow at GST. (D) Coimmunoprecipitation of mammalian LIN-2 CASK, mEPS-8 and mLIN-7A from MDCK cells. NIS indicates control precipitations with pre-immunoserum. Precipitated proteins were detected on Western blots with polyclonal LIN-2 CASK (upper panel), and monoclonal mEPS-8 antibodies (middle panel). Binding of LIN-2 CASK to mLIN-7A was detected with monoclonal LIN-2 CASK antibodies (lower panel).
Thus, EPS-8 interacts with the EGFR localization complex via the first L27 domain in LIN-2. The physiological function of the first L27 domain in LIN-2 has so far not been characterized.
The first L27 domain in LIN-2 is required for basolateral membrane localization of LET-23 EGFR
The binding experiments described above predict that disrupting the interaction between LIN-7 and LIN-2 will result in the apical mislocalization of LET-23 EGFR, while blocking the binding of EPS-8 to LIN-2 should lead to the intracellular accumulation of LET-23 EGFR. To test these predictions, we introduced into a rescuing lin-2 minigene (Hoskins et al, 1996) point mutations into each of the two L27 domains and tested the mutant constructs for rescue of the lin-2(lf) Vul and receptor mislocalization phenotypes. Analogous to the inactivating mutations described for the SAP97 L27 domain (Feng et al, 2004), mutations of residues Leu407 to Ser in the first and Ile439 to Ser in the second L27 domain of LIN-2 disrupt the yeast two-hybrid interaction of LIN-2 with EPS-8 and LIN-7, respectively (Figure 5D). A lin-2 minigene carrying the Leu407 Ser mutation in the first L27 domain completely rescues the lin-2(lf) Vul phenotype, yet LET-23 EGFR staining is detected predominantly in intracellular punctae similar to the mislocalization observed in eps-8(lf) animals (Figure 5B and Table II, rows 7–9). In contrast, a lin-2 minigene carrying the Ile439 Ser mutation in the second L27 domain neither rescues the lin-2(lf) Vul phenotype nor the apical mislocalization of LET-23 EGFR (Figure 5C and Table II, rows 10–12). Thus, inactivation of the first L27 domain in LIN-2 results in the intracellular accumulation of LET-23 EGFR as observed in eps-8(lf) animals, while the second L27 domain is necessary for the basolateral localization of LET-23 EGFR via LIN-7.
Figure 5.
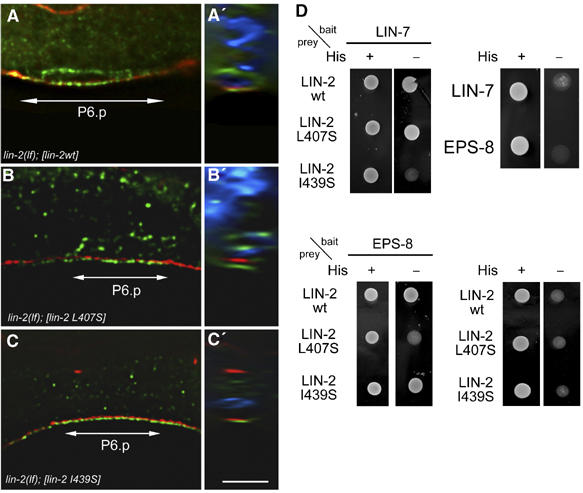
The first L27 domain in LIN-2 is required for EPS-8 activity. (A) Basolateral localization of LET-23 EGFR (green) in P6.p of an early L3 lin-2(lf) larva carrying the wild-type lin-2 minigene. (B) Punctate and intracellular staining of LET-23 EGFR in an L3 larva carrying a lin-2 minigene with the Leu407 to Ser mutation in the first L27 domain. (C) Apical mislocalization of LET-23 EGFR in an L3 larva carrying a lin-2 minigene with the Ile439 to Ser mutation in the second L27 domain. Adherens junctions are stained with MH27 in red and nuclei stained with DAPI (in the z–y sections in A′, B′ and C′) are shown in blue. Scale bar in C is 10 μm. (D) Yeast two-hybrid interaction of LIN-2 carrying the indicated point mutations with LIN-7 and EPS-8, respectively, as described in Figure 4.
Discussion
EPS-8 is a new component of the EGF receptor localization complex
Here, we show that during C. elegans vulval development the intracellular trafficking of the EGFR homolog LET-23 is regulated in a cell fate-specific manner. The EPS-8 protein is a new component of the LIN-2/LIN-7/LIN-10 receptor localization complex that plays a key role in regulating the plasma membrane localization of LET-23 EGFR in the 1° cell lineage. The following molecular model might account for the cell lineage-specific differences in EGFR trafficking we observed (Figure 6). EPS-8 function largely depends on its interaction with the membrane-associated guanylate kinase LIN-2 as the inactivation of the EPS-8 binding site in LIN-2 has the same effect on LET-23 EGFR localization as an eps-8(lf) mutation. Thus, LIN-2 appears to act as a scaffolding protein that recruits EPS-8 into the EGF receptor localization complex. Consistent with the protein interaction experiments, EPS-8 acts in parallel with the LIN-7 adaptor protein, as elevated levels of EPS-8 can partially rescue the apical mislocalization of LET-23 EGFR in lin-7(lf) mutants. Nevertheless, EPS-8 and LIN-7 are not functionally equivalent, but they rather regulate LET-23 EGFR trafficking at different steps in a sequential manner. A LIN-7::GFP reporter is uniformly expressed in all VPCs already before induction (data not shown), and loss of LIN-7 function equally affects LET-23 EGFR localization in all VPCs (Simske et al, 1996; Kaech et al, 1998). EPS-8, on the other hand, is expressed at highest level in the 1° cell lineage, and it regulates LET-23 EGFR trafficking predominantly in P6.p and its descendants. Loss of EPS-8 function leads to the intracellular accumulation of LET-23 causing a reduction in the activity of the inductive signaling pathway to a level that is still sufficient for normal induction. Loss of LIN-7, on the other hand, results in the mislocalization of LET-23 EGFR to the apical plasma membrane and an almost complete block in the inductive signaling pathway because LIN-3 EGF secreted from the AC cannot activate the mislocalized LET-23 EGFR.
Figure 6.
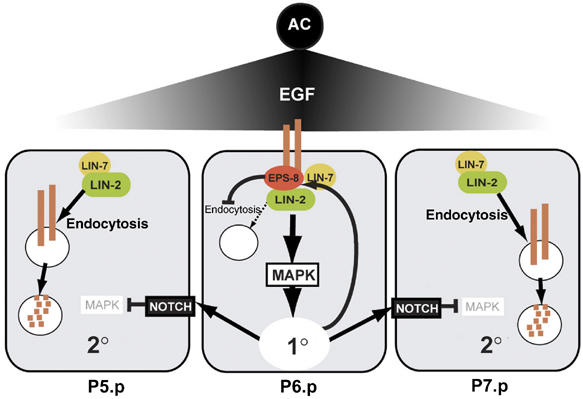
A model for EPS-8 function during vulval fate specification. The inductive AC signal upregulates EPS-8 expression in the 1° VPC (P6.p), where EPS-8 associates with LIN-2 to retain LET-23 EGFR on the basolateral plasma membrane. In the adjacent 2° VPCs (P5.p and P7.p) EPS-8 levels are low due to the lateral inhibition of the EGFR/RAS/MAPK signaling pathway by LIN-12 NOTCH, and LET-23 is therefore rapidly internalized and degraded in the 2° lineage.
Taken together, these observations suggest that LIN-7 probably serves together with LIN-2 and LIN-10 to transport or localize LET-23 EGFR to the basolateral compartment of the VPCs before the AC produces the inductive LIN-3 EGF signal. During vulval induction, however, LIN-7 may not be sufficient to block the ligand-induced endocytosis of LET-23 EGFR. Thus, eps-8 transcription is induced, directly or indirectly, in the 1° cell lineage by the inductive AC signaling pathway to maintain high levels of LET-23 EGFR on the basolateral plasma membrane of the 1° cells. EPS-8 either inhibits ligand-induced EGF receptor endocytosis, as it has been proposed for the mammalian EPS-8 homologs (Lanzetti et al, 2000), or alternatively, EPS-8 is required for the recycling of endocytosed receptor molecules. Our data are consistent with the two possible modes of EPS-8 function. In both scenarios, EPS-8 is part of a positive feedback loop that ensures the continuous activation of LET-23 EGFR in the 1° cell lineage. In the adjacent 2° VPCs P5.p and P7.p, on the other hand, eps-8 expression is downregulated because lateral signaling by LIN-12 NOTCH inactivates the EGFR/RAS/MAPK pathway in these cells (Berset et al, 2001; Yoo et al, 2004). The rapid disappearance of LET-23 EGFR first in the 2° and shortly thereafter in the 3° cells may be due to an accelerated rate of receptor endocytosis followed by degradation in the lysosomes. Supporting this idea, LET-23 EGFR downregulation in P5.p, P7.p and also in the distal VPCs can be blocked by constitutive EPS-8 overexpression.
EPS-8 positively regulates inductive signaling at the level of the EGFR
eps-8(lf) animals develop a wild-type vulva despite the strong reduction in 1° fate marker expression. This observation seems at first surprising, yet vulval induction under laboratory conditions appears to exhibit a high buffering capacity to compensate for a wide range of changes in the activity of the inductive signaling pathway. Hence, single mutations in many conserved regulators of LET-23 EGFR such as sli-1, ark-1 or dep-1 do not change the normal pattern of vulval cell fates, and like eps-8 their functions manifest only in sensitized genetic backgrounds (Yoo et al, 2004; Berset et al, 2005).
EPS-8 probably affects the intracellular trafficking of many other proteins besides LET-23 EGFR in different tissues. EPS-8 might, for example, inhibit LIN-12 NOTCH signaling by preventing the endocytosis of the DSL Delta ligands in the 1° vulval cells. However, our data point at a rather specific function of EPS-8 in regulating LET-23 EGFR activity. In addition to the changes in LET-23 EGFR localization in eps-8(lf) mutants, epistasis analysis indicates that EPS-8 regulates vulval induction at the level of LET-23 EGFR. Overexpression of EPS-8 suppresses the Vul phenotype of LET-23 EGFR reduction-of-function mutants, but it cannot rescue a mutation in sem-5, which encodes a GRB2 adaptor molecule that transduces the signal downstream of LET-23 EGFR (Clark et al, 1992). Moreover, a loss-of-function mutation in lin-2 is epistatic to eps-8 overexpression, indicating that EPS-8 acts via LIN-2. Finally, we found no genetic interaction between eps-8(lf) and two weak lin-12 notch alleles that hyper-activate the NOTCH pathway, indicating that at least during vulval induction loss of EPS-8 does not significantly alter LIN-12 NOTCH signaling. We thus propose that EPS-8 regulates vulval induction primarily by controlling LET-23 EGFR activity.
Regulation of EGFR trafficking is critical for pattern formation
The lineage specific regulation of receptor trafficking provides an additional level of control in the EGFR pathway that is used to modulate the sensitivity of initially equivalent precursor cells towards an extracellular growth factor signal. The inductive AC signal ensures that P6.p, which has to adopt the 1° fate, presents the highest levels of LET-23 EGFR, while the adjacent cells P5.p and P7.p become unresponsive to the inductive signal and hence adopt the 2° fate. Thus, perturbing receptor trafficking not only changes EGFR expression and localization but it also disrupts normal vulval cell fate specification and pattern formation.
In humans, the expression of the EGFR is upregulated in a large fraction of epithelial tumors, either due to gene amplification, increased gene expression or by altered turnover of the protein (Rao et al, 2003; Holbro and Hynes, 2004). Accordingly the expression of mammalian EPS-8 is upregulated by oncogenes such as v-src, and EPS-8 itself exhibits transforming activity when overexpressed in cell lines (Matoskova et al, 1995; Gallo et al, 1997). It will therefore be interesting to see if regulated EGFR trafficking is linked to development also in higher organisms, and if deregulated EGFR trafficking contributes to tumor formation in humans.
Materials and methods
General methods and strains used
Standard methods were used for maintaining and manipulating C. elegans (Brenner, 1974). The C. elegans Bristol strain, variety N2, was used as the wild-type reference strain in all experiments. Unless noted otherwise, the mutations used have been described previously (Riddle and National Center for Biotechnology Information (US), 2001) and are listed below by their linkage group. LGII: let-23(sy1), rrf-3(pk1426) (Simmer et al, 2002), lin-7(e1413); LGIII: unc-119(e2498) (Maduro and Pilgrim, 1995); lin-12(n302); lin-12(n379) LGIV: eps-8(by160) (Croce et al, 2004), let-60(n1046gf), lin-3(e1417); LGX: sem-5(n2019), lin-2(n397). Integrated transgenic arrays: arIs92[egl-17::cfp,tax-3::gfp] (Yoo et al, 2004), zhIs13[eps-8p::nls::gfp] (this study), zhIs11[lin-31p::eps-8a] (this study), ayIs4 (Burdine et al, 1998; Cui and Han, 2003), gaIs36[hs::mpk-1] (Lackner and Kim, 1998), Ex[opt-2::eps-8] (Croce et al, 2004). Extrachromosomal transgenic arrays: zhEx129.1[lin-2 w.t., sur-5::gfp] through zhEx129.3, zhEx165.1[lin-2 L407S, sur-5::gfp] through zhEx165.3, zhEx166.1[lin-2 I439S, sur-5::gfp] through zhEx166.3 (all this study).
Unless noted in the tab. legends, all experiments were conducted at 20°C. Transgenic lines were generated by injecting the DNA at a concentration of 100 ng/μl into both arms of the syncytial gonad as described (Mello et al, 1991). pUNC-119 (20 ng/μl) or pTG96 (100 ng/μl) were used as a transformation markers (Maduro and Pilgrim, 1995; Yochem et al, 1998). Gonads were ablated by removing the Z1 and Z4 precursors of the somatic gonad at the L1 stage (Kimble, 1981). eps-8 RNAi experiments were performed by feeding worms with double-stranded RNA producing bacteria as described (Kamath et al, 2001).
Vulval induction
Vulval induction was scored by examining worms at the L4 stage under Nomarski optics as described (Berset et al, 2001). The number of VPCs that had adopted a 1° or 2° vulval fate was counted for each animal, and the induction index was calculated by dividing the number of 1° or 2° induced cells by the number of animals scored. Statistical analysis was performed using two-tailed Student‘s t-test for independent samples.
Plasmid constructs
The eps-8p::nls::gfp transcriptional reporter was generated by PCR amplification of a genomic fragment containing 2.4 kb of 5′ promoter sequence and 18 bp of the open reading frame using the primers (CTCGAGTAACGTCAGAGATGTGC and GGATCCACCTCGAC GCATC) and cloned into the SalI and BamHI sites of pPD95.69 expression vector (kind gift of A Fire). The lin-31p::eps-8a construct was generated by cloning worm eps-8 cDNA isolated by PCR amplification with the primers (AGATCTATGCGTCGAGGTGGATCGATG and GTCGACCTAGAGAATTGGGTTAATAGTG) into the BglII and SalI sites of the pB253 vector (Tan et al, 1998). To generate GST::LIN-2 or GST::EPS-8 fusion proteins, mammalian cDNA fragments of both proteins covering the regions shown in Figure 1 were isolated by PCR amplification and subcloned in frame into the bacterial expression vector pGEX-5X3 (Pharmacia). Point mutations were introduced into the lin-2 minigene as described (Hoskins et al, 1996).
Yeast two-hybrid assays
Fragments of C. elegans lin-2 cDNA shown in Figure 1 were fused in-frame to the Gal-4AD in the vector pGAD GH (BD Bioscience) and the indicated fragments of C. elegans eps-8 or lin-7 cDNAs were fused to the LexA DNA-binding domain in the vector pBTM-116 (Invitrogen). DNA was introduced into the L40 yeast strain using the lithium-acetate method, and interactions were monitored both by growth on His- medium and by LacZ activity on filter assays.
Cell culture and GST pull-down experiments
MDCK cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum. Ninety percent confluent cells were starved for 24 h in DMEM supplemented with 0.5% BSA, treated for 10 min with 20 ng/ml TGFα where indicated, washed and lysed in a buffer containing 50 mM HEPES, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 10% glycerol, 10 mM sodium pyrophosphate, 2 mM sodium vanadate, 10 mM sodium fluoride, and protease inhibitor cocktail (Roche). Lysates were cleared by centrifugation at 13 000 r.p.m. (10 000 g) for 5 min at 4°C and used for immunoprecipitation or GST pull-down experiments. GST fusion proteins were produced in the BL-21DE3 bacterial strain (Stratagene), purified and eluted from glutathione-sepharose beads, as recommended by the manufacturer (Pharmacia). For immunoprecipitation, 500 μg total cell lysate was incubated with the indicated antibodies together with protein-A agarose beads, washed three times and subjected to SDS gel-electrophoresis. Antibodies used for Western blot were mouse monoclonal EPS-8 (Transduction Laboratories, 1:1000 dilution) rabbit polyclonal EGFR (clone 1005, Santa Cruz, 1:1000 dilution), mouse monoclonal LIN-2 (Transduction Laboratories, 1:500 dilution) or rabbit polyclonal LIN-2 (Santa Cruz, 1:250 dilution), rabbit polyclonal LIN 7 (see Perego et al, 1999; 1:500 dilution) and mouse monoclonal GST (Biomol, 1:2000 dilution).
Whole mount staining and microscopy of C. elegans
Animals were fixed using a modified Finney–Ruvkun protocol as described (Kaech et al, 1998). Antibody incubations were carried out overnight in 200 μl of ABA buffer (PBS containing 0.05% Triton X-100 and 3% BSA) with affinity-purified rabbit anti-LET-23 antibodies (1:100 dilution) raised against a recombinant protein encoding the C-terminal 196 amino acids and the monoclonal MH27 (see Kaech et al, 1998) or EEA1 antibodies (Transduction Laboratories, 1:200 dilution) at 4°C. Secondary donkey anti-rabbit-Alexa488 and donkey anti-mouse-Cy5 antibodies (Jackson labs) were applied for 2 h at room temperature and nuclei were stained with DAPI (1:1000 dilution of saturated solution). For EPS-8 immunostaining, animals were fixed using Bouin's fixative (0.75 ml of saturated picric acid, 0.25 ml of formalin, 0.05 ml of glacial acetic acid, 0.25 ml of methanol, and 0.01 ml of β-mercaptoethanol) for 30 min (Nonet et al, 1997), washed in BTB (1 × borate-buffer, 0.5% Triton X-100, 2% β-mercaptoethanol) for 3 h and incubated with MH27 and polyclonal anti-EPS-8 antibodies (S85 or K49 1:200 dilution; (Croce et al, 2004) overnight in 200 μl of ABA buffer.
MDCK cells were grown on coverslips to confluency, fixed with methanol for 5 min rehydrated in PBS and blocked for 30 min with ABA. Antibody incubations were carried out for 2h in ABA buffer with rabbit polyclonal (1:50 dilution) (Santa Cruz) or mouse monoclonal (1:100 dilution) LIN-2 CASK (Transduction Laboratories), rabbit polyclonal mLIN-7A (1:100 dilution), or mouse monoclonal EPS-8 antibodies (1:200 dilution) (Transduction Laboratories). Fluorescent images were recorded with a Leica TCS4 confocal microscope or a Leica DMRA wide-field microscope equipped with a cooled CCD camera (Hamamatsu ORCA-ER) controlled by the Openlab 3.0 software package (Improvision). For the time course analysis of eps-8::nls::gfp expression shown in Figure 2, larvae were synchronized at the L1 stage by food starvation, then grown for 18 h until the mid-L2 stage, and expression was observed thereafter every hour until the L4 stage. For the quantification of the EGL-17::CFP expression (Figure 1H), the maximal intensities were measured in P6.p and normalized to the background outside the worms. The images shown in Figures 3 and 5 were processed by iterative deconvolution (Improvision) to remove out-of-focus light.
Supplementary Material
Supplementay Information
Acknowledgments
We thank the members of our group for critical discussion and Konrad Basler and Urs Greber comments on the manuscript. We are grateful to Andrew Fire for GFP vectors, the C. elegans genetics centre and Ralf Baumeister for providing strains and Yuji Kohara for EST clones. We would also thank Urs Greber for cell culture facilities and EEA1 antibody. This research was supported by grants from the SNF and Oncosuisse to AH and the Kanton of Zürich.
References
- Berset T, Hoier EF, Battu G, Canevascini S, Hajnal A (2001) Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science 291: 1055–1058 [DOI] [PubMed] [Google Scholar]
- Berset TA, Hoier EF, Hajnal A (2005) The C. elegans homolog of the mammalian tumor suppressor Dep-1/Scc1 inhibits EGFR signaling to regulate binary cell fate decisions. Genes Dev 19: 1328–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine RD, Branda CS, Stern MJ (1998) EGL-17(FGF) expression coordinates the attraction of the migrating sex myoblasts with vulval induction in C. elegans. Development 125: 1083–1093 [DOI] [PubMed] [Google Scholar]
- Burke P, Schooler K, Wiley HS (2001) Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol Biol Cell 12: 1897–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SG, Stern MJ, Horvitz HR (1992) C. elegans cell-signalling gene sem-5 encodes a protein with SH2 and SH3 domains. Nature 356: 340–344 [DOI] [PubMed] [Google Scholar]
- Croce A, Cassata G, Disanza A, Gagliani MC, Tacchetti C, Malabarba MG, Carlier MF, Scita G, Baumeister R, Di Fiore PP (2004) A novel actin barbed-end-capping activity in EPS-8 regulates apical morphogenesis in intestinal cells of Caenorhabditis elegans. Nat Cell Biol 6: 1173–1179 [DOI] [PubMed] [Google Scholar]
- Cui M, Han M (2003) Cis regulatory requirements for vulval cell-specific expression of the Caenorhabditis elegans fibroblast growth factor gene egl-17. Dev Biol 257: 104–116 [DOI] [PubMed] [Google Scholar]
- Disanza A, Carlier MF, Stradal TE, Didry D, Frittoli E, Confalonieri S, Croce A, Wehland J, Di Fiore PP, Scita G (2004) Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat Cell Biol 6: 1180–1188 [DOI] [PubMed] [Google Scholar]
- Dutt A, Canevascini S, Froehli-Hoier E, Hajnal A (2004) EGF signal propagation during C. elegans vulval development mediated by ROM-1 rhomboid. PLoS Biol 2: e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazioli F, Minichiello L, Matoska V, Castagnino P, Miki T, Wong WT, Di Fiore PP (1993) Eps8, a substrate for the epidermal growth factor receptor kinase, enhances EGF-dependent mitogenic signals. EMBO J 12: 3799–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Long JF, Fan JS, Suetake T, Zhang M (2004) The tetrameric L27 domain complex as an organization platform for supramolecular assemblies. Nat Struct Mol Biol 11: 475–480 [DOI] [PubMed] [Google Scholar]
- Gallo R, Provenzano C, Carbone R, Di Fiore PP, Castellani L, Falcone G, Alema S (1997) Regulation of the tyrosine kinase substrate Eps8 expression by growth factors, v-Src and terminal differentiation. Oncogene 15: 1929–1936 [DOI] [PubMed] [Google Scholar]
- Greenwald I, Seydoux G (1990) Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature 346: 197–199 [DOI] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, Horvitz HR (1983) The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell 34: 435–444 [DOI] [PubMed] [Google Scholar]
- Haj FG, Verveer PJ, Squire A, Neel BG, Bastiaens PI (2002) Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science 295: 1708–1711 [DOI] [PubMed] [Google Scholar]
- Harris BZ, Venkatasubrahmanyam S, Lim WA (2002) Coordinated folding and association of the LIN-2, -7 (L27) domain. An obligate heterodimerization involved in assembly of signaling and cell polarity complexes. J Biol Chem 277: 34902–34908 [DOI] [PubMed] [Google Scholar]
- He C, Hobert M, Friend L, Carlin C (2002) The epidermal growth factor receptor juxtamembrane domain has multiple basolateral plasma membrane localization determinants, including a dominant signal with a polyproline core. J Biol Chem 277: 38284–38293 [DOI] [PubMed] [Google Scholar]
- Hobert ME, Kil SJ, Medof ME, Carlin CR (1997) The cytoplasmic juxtamembrane domain of the epidermal growth factor receptor contains a novel autonomous basolateral sorting determinant. J Biol Chem 272: 32901–32909 [DOI] [PubMed] [Google Scholar]
- Holbro T, Hynes NE (2004) ErbB receptors: directing key signaling networks throughout life. Annu Rev Pharmacol Toxicol 44: 195–217 [DOI] [PubMed] [Google Scholar]
- Hoskins R, Hajnal AF, Harp SA, Kim SK (1996) The C. elegans vulval induction gene lin-2 encodes a member of the MAGUK family of cell junction proteins. Development 122: 97–111 [DOI] [PubMed] [Google Scholar]
- Kaech SM, Whitfield CW, Kim SK (1998) The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell 94: 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J (2001) Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2: RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz WS, Hill RJ, Clandinin TR, Sternberg PW (1995) Different levels of the C. elegans growth factor LIN-3 promote distinct vulval precursor fates. Cell 82: 297–307 [DOI] [PubMed] [Google Scholar]
- Kimble J (1981) Alterations in cell lineage following laser ablation of cells in the somatic gonad of Caenorhabditis elegans. Dev Biol 87: 286–300 [DOI] [PubMed] [Google Scholar]
- Knoblich JA (2001) Asymmetric cell division during animal development. Nat Rev Mol Cell Biol 2: 11–20 [DOI] [PubMed] [Google Scholar]
- Lackner MR, Kim SK (1998) Genetic analysis of the Caenorhabditis elegans MAP kinase gene mpk-1. Genetics 150: 103–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetti L, Rybin V, Malabarba MG, Christoforidis S, Scita G, Zerial M, Di Fiore PP (2000) The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature 408: 374–377 [DOI] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D (1995) Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141: 977–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoskova B, Wong WT, Salcini AE, Pelicci PG, Di Fiore PP (1995) Constitutive phosphorylation of eps8 in tumor cell lines: relevance to malignant transformation. Mol Cell Biol 15: 3805–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V (1991) Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J 10: 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills IG, Jones AT, Clague MJ (1998) Involvement of the endosomal autoantigen EEA1 in homotypic fusion of early endosomes. Curr Biol 8: 881–884 [DOI] [PubMed] [Google Scholar]
- Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, Horvitz HR, Jorgensen EM, Meyer BJ (1997) Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci 17: 8061–8073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego C, Vanoni C, Villa A, Longhi R, Kaech SM, Frohli E, Hajnal A, Kim SK, Pietrini G (1999) PDZ-mediated interactions retain the epithelial GABA transporter on the basolateral surface of polarized epithelial cells. EMBO J 18: 2384–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao DS, Bradley SV, Kumar PD, Hyun TS, Saint-Dic D, Oravecz-Wilson K, Kleer CG, Ross TS (2003) Altered receptor trafficking in Huntingtin Interacting Protein 1-transformed cells. Cancer Cell 3: 471–482 [DOI] [PubMed] [Google Scholar]
- Riddle DL, National Center for Biotechnology Information (US) (2001) C. elegans II. Ncbi, Plainview, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH (2002) Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol 12: 1317–1319 [DOI] [PubMed] [Google Scholar]
- Simske JS, Kaech SM, Harp SA, Kim SK (1996) LET-23 receptor localization by the cell junction protein LIN-7 during C. elegans vulval induction. Cell 85: 195–204 [DOI] [PubMed] [Google Scholar]
- Sternberg PW, Han M (1998) Genetics of RAS signaling in C. elegans. Trends Genet 14: 466–472 [DOI] [PubMed] [Google Scholar]
- Sundaram MV (2005) The love–hate relationship between Ras and Notch. Genes Dev 19: 1825–1839 [DOI] [PubMed] [Google Scholar]
- Tan PB, Lackner MR, Kim SK (1998) MAP kinase signaling specificity mediated by the LIN-1 Ets/LIN-31 WH transcription factor complex during C. elegans vulval induction. Cell 93: 569–580 [DOI] [PubMed] [Google Scholar]
- Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M (2000) Protein interaction mapping in C. elegans using proteins involved in vulval development. Science 287: 116–122 [DOI] [PubMed] [Google Scholar]
- Wang W, Struhl G (2005) Distinct roles for mind bomb, neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development 132: 2883–2894 [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Benard C, Barnes T, Hekimi S, Kim SK (1999) Basolateral localization of the Caenorhabditis elegans epidermal growth factor receptor in epithelial cells by the PDZ protein LIN-10. Mol Biol Cell 10: 2087–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JM, de Hoop M, Zorzi N, Toh BH, Dotti CG, Parton RG (2000) EEA1, a tethering protein of the early sorting endosome, shows a polarized distribution in hippocampal neurons, epithelial cells, and fibroblasts. Mol Biol Cell 11: 2657–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J, Gu T, Han M (1998) A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics 149: 1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Bais C, Greenwald I (2004) Crosstalk between the EGFR and LIN-12/Notch pathways in C. elegans vulval development. Science 303: 663–666 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementay Information


