Abstract
We report an efficient, controllable, site-specific replication roadblock that blocks cell proliferation, but which can be rapidly and efficiently reversed, leading to recovery of viability. Escherichia coli replication forks of both polarities stalled in vivo within the first 500 bp of a 10 kb repressor-bound array of operator DNA-binding sites. Controlled release of repressor binding led to rapid restart of the blocked replication fork without the participation of homologous recombination. Cytological tracking of fork stalling and restart showed that the replisome-associated SSB protein remains associated with the blocked fork for extended periods and that duplication of the fluorescent foci associated with the blocked operator array occurs immediately after restart, thereby demonstrating a lack of sister cohesion in the region of the array. Roadblocks positioned near oriC or the dif site did not prevent replication and segregation of the rest of the chromosome.
Keywords: Escherichia coli, recombination, replication fork stalling, repressor binding, restart
Introduction
Bacterial genomes have evolved multiple levels of organisation that are shaped by replication. In Escherichia coli, replication proceeds bidirectionally from a unique origin, oriC, and terminates diametrically opposite in the ter region, which is flanked by ter sites that act as polar replication terminators (Hill et al, 1987). The genome is thus divided into two almost equal arms or replichores (Blattner et al, 1997). Furthermore, base composition bias and consequent organisation of specific sequence motifs relate directly to the direction of replication. In order to maintain constant gene concentration, highly expressed genes are frequently located close to oriC; these and essential genes are generally transcribed in the same direction as the replication forks. This replichore organisation is important for normal physiological function and for chromosome processing (e.g. see Rocha, 2004; Bigot et al, 2005; Lesterlin et al, 2005). Furthermore, the circular bacterial chromosome is highly organised within the bacterial cell, with specific genetic regions occupying specific cellular locations, and with organisation and gene position undergoing a defined choreography throughout the cell cycle (Niki et al, 2000; Lau et al, 2003; Sherratt, 2003; Viollier et al, 2004). Experiments in which two genetic markers were simultaneously tracked in living E. coli led to the proposal that replichores are spatially organised within a new-born cell, ‘left' replichore on the ‘left' of the cell and ‘right' replichore on the ‘right' with oriC in the middle (Wang et al, 2005). Bidirectional replication appears to be largely confined to mid-third of the E. coli cell, with progression of two sister forks not necessarily coordinated (Bates and Kleckner, 2005; Breier et al, 2005).
Complete DNA replication and its faithful segregation to daughter cells are central to the maintenance of genome integrity. Mechanisms for repairing and restarting stalled or broken replication forks are important for genome integrity and cell viability, and stalled or broken forks often induce checkpoints that serve to minimise the potential consequences of incomplete genome replication (reviewed in Kolodner et al, 2002; McGlynn and Lloyd, 2002; Lesterlin et al, 2004; Michel et al, 2004). It is now clear that recombination proteins, with or without homologous recombination, often participate at stalled or broken replication forks to enable replication restart. Furthermore, different types of arrest lead to different requirements for restart. For example, in E. coli, replication forks blocked at Ter sites bound by Tus protein remain stable but provoke recombination if a subsequent round of replication runs into the back of them to generate free double-strand ends (Bidnenko et al, 2002). Furthermore, replication arrest in E. coli owing to an impaired replisome requires recombination proteins for restart (reviewed in Michel et al, 2004). In contrast, fork stalling by impairment of DNA gyrase, which acts ahead of the replication fork to remove positive supercoiling as it accumulates during fork travel, could be reversed in a reaction that does not require recombination proteins (Grompone et al, 2003).
Although the possibility of adventitious stalling of replication forks by DNA-binding proteins that do not play specialised roles in replication and its termination has been considered, no detailed investigations of this, or of the requirements for replication restart once the protein block is removed, have been reported. Previously, the binding of fluorescent derivatives of the LacI and TetR repressors has been used to provide insight into gene localisation, duplication and segregation in living cells (Gordon et al, 1997; Viollier et al 2004; Fekete and Chattoraj, 2005). Our own work in E. coli has demonstrated that such binding can be achieved without detectably perturbing cell cycle parameters by having cognate inducers present, thereby reducing the affinity of repressor binding (Lau et al, 2003; Wang et al, 2005). As this work also indicated that high-level expression of repressor expression in the absence of inducer might interfere with DNA replication, we initiated a study to determine how tight repressor binding to operator arrays influences DNA metabolism. By using a combination of genetics, DNA analysis, microscopy, flow cytometry and microbiology, we have tracked efficient and persistent site-specific replication blocking and its rapid reversal when the avidity of repressor binding is reduced.
Results
Binding of fluorescent repressor to operator arrays prevents cell proliferation
Fluorescent fusion derivatives of the repressors, TetR (TetR-YFP) and LacI (LacI-CFP), were expressed tandemly from the arabinose (ara) inducible promoter of the multicopy plasmid pLau53 (Lau et al, 2003). E. coli strains were constructed with 240 copy arrays of their cognate binding sites, tetO or lacO, respectively, inserted independently at three possible positions: 15 kb anticlockwise of the replication origin (ori1), 200 kb (ter2), or 50 kb (ter3) clockwise of dif in the replication termination region, ter (Figure 1A), or with combinations of these positions. The arrays contain 10 bp of sequence heterology between each operator in order to minimise genetic instability.
Figure 1.
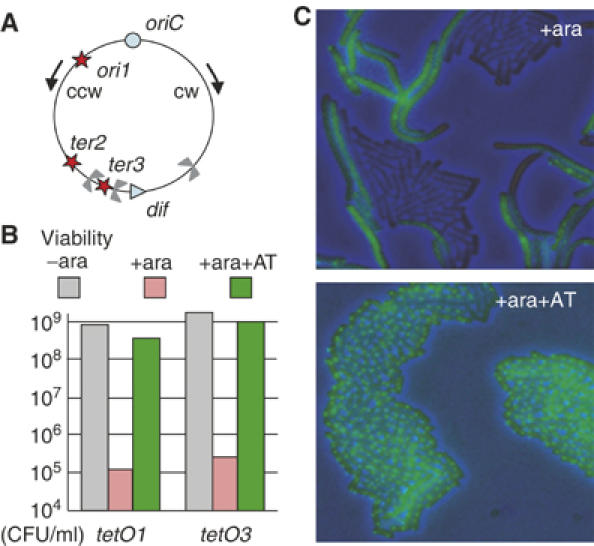
TetR binding to tetO array prevents cell proliferation. (A) E. coli chromosome map indicating position of operator arrays. Arrows indicate the ccw and cw replication forks. (B) Viability of tetO1 and tetO3 strains on LB+Ap after 4 h of the indicated treatment. (C) Microcolony formation (overnight room temperature growth) of a tetO1 strain on an LB agarose layer after 2 h of ara induction without (top) or with AT (bottom).
First, the effects of overexpression of fluorescent repressor on cell viability were examined. Cells were grown for 4 h in LB with ara-induced repressor expression, with or without the inducers anhydrotetracycline (AT), and isopropyl-β-D-thiogalactopyranoside (IPTG), which reduce the binding affinity of TetR and LacI, respectively. Cells with the indicated array were then plated on ara-free plates containing ampicillin (Ap), which selects for continued maintenance of pLau53. In the absence of ara induction, the plating efficiencies were the same, irrespective of whether Ap was present, thereby demonstrating that pLau53 is stably maintained (not shown). After induction of fluorescent repressor expression by 0.05% ara, the plating efficiency of strains tetO1, tetO2 or tetO3 containing the tetO array at ori1, ter2 or ter3, respectively, was dramatically reduced when AT was absent (Figure 1B). Similarly, repressor expression in a strain containing lacO array reduced plating efficiency, albeit to a lesser extent (not shown). When the appropriate inducer was present during ara-induction, TetR and LacI repressor overexpression did not lead to a reduction in plating efficiency.
Microcolony formation of a tetO1 strain was examined cytologically after overnight growth on LB agarose lacking ara, Ap and AT. From an ara-induced culture in the presence of AT, microcolonies contained normal-looking cells with the expected fluorescent foci. In contrast, cells in which TetR was induced in the absence of AT were heterogeneous in size and in fluorescence signal (Figure 1C). Very few cells had fluorescent foci because they had lost the plasmid during microcolony formation in the absence of Ap; those cells that had fluorescent foci were filamentous. We conclude that high occupancy and tight binding of repressor to operator arrays prevents viable cell proliferation.
Tight repressor binding to operator arrays leads to replication fork stalling and to associated cytological defects
In order to test whether tight repressor binding prevents replication or segregation, tetO1 cell samples harvested after various times of cultivation, without ara, with ara or with ara+AT, were analysed by three methods: (1) two-dimensional (2D) neutral–neutral gel electrophoresis of the tetO1 chromosomal region, a method that detects replication intermediates as a consequence of their nonlinear shape (Brewer and Fangman, 1987); (2) fluorescence microscopy, which allows direct visualisation of tetO1 focus position and duplication and (3) flow cytometry after completion of rounds of chromosomal replication in cells inhibited for cell division and replication reinitiation (‘run-out'), by using a combination of cephalexin and rifampicin treatment (Figure 2; Skarstad et al, 1985).
Figure 2.
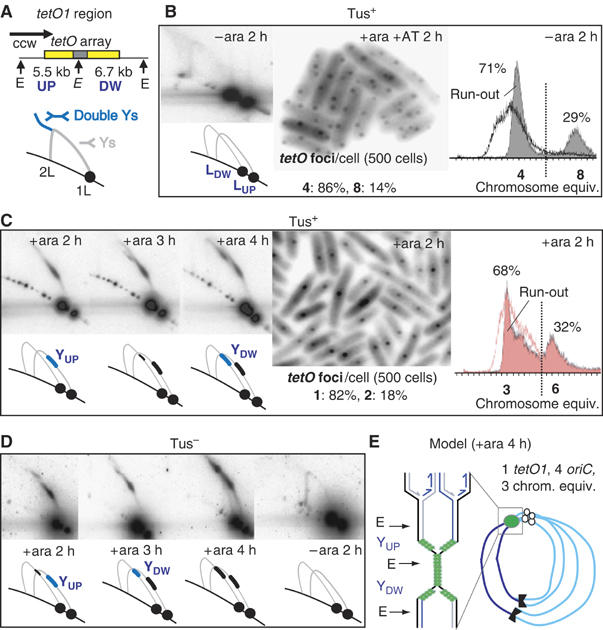
Persistent replication fork stalling at tetO1. (A) EcoRV (denoted E) restriction map of the tetO1 region. The two restriction fragments detected by tetO probe are named UP and DW in respect to the ccw replication fork (horizontal arrow). Replication fork progression at tetO1 in Tus+ (B, C) and Tus− (D) strains cultivated with the indicated additions and for the indicated timing, analysed by 2D gel (left B, left C and D), tetO1 foci (centre of B and C) and flow cytometry (right of B and C). A cartoon of each 2D gel representing the major species observed is shown underneath the respective gels. The light grey lines show the positions of the Y arcs as reference, even when not visible on the gel. L: linear fragment; Y, forked fragment. The percentages shown on the flow cytometry profiles refer to the number of cells grouped on either side of the vertical line. (E) Model representing the chromosome after 4 h ara induction of roadblock.
EcoRV digestion of genomic DNA prepared from cells cultivated in the absence of ara generated the expected 5.5 and 6.7 kb linear fragments (LUP and LDW) plus a very faint arc of Y-shaped replication intermediates when the tetO array was used as a probe (Figure 2A and B). A probe of the UP region alone was used to confirm the identity of UP fragments (not shown). DNA from cells grown in presence of ara+AT gave the same pattern (not shown).
Under these same conditions, the number of tetO1 foci per cell (four in 86% and eight in 14% of the 500 cells examined) corresponds to the number of chromosome equivalents per cell measured after ‘run-out' (four in 71% and eight in 29% of the cell population). Hence, one fluorescent focus represents one chromosome copy. The parental strain, AB1157, has been reported to have similar proportions of four or eight origin cells under the same growth conditions (Markovitz, 2005), confirming that replication was apparently unperturbed in these cells in the absence of tight repressor binding.
In contrast, DNA from cells grown in ara for 2, 3 or 4 h showed substantial amounts of Y-shaped DNA molecules with branch points restricted to a specific region (Figure 2C). At 2 h, the ‘extended spot' (YUP) of blocked forks was derived from the blocked counter-clockwise (ccw) replication fork entering the array from the UP side, whereas at 4 h, a second spot (YDW spot) of comparable intensity is observed consistent with the clockwise (cw) fork eventually overcoming the block of the replication termination protein, Tus, bound to ter sites and entering the array from the DW side (Figure 2E). It is also possible, although unlikely, that some ccw forks slowly ‘crawl' through the array to generate a fraction of the observed YDW arc. At 3 h, only a small amount of blocked cw fork was seen, when compared to the blocked ccw fork. After 2 h and 3 h of ara induction in a tetO1 Tus− strain, which is impaired in Tus-dependent termination at ter sites, higher levels of the cw fork entered the array from the DW side as expected (Figure 2D). Estimation of the molecular weight of the blocked Y structures (YUP and YDW) from the EcoRV digest and a SacII digest (not shown) shows that the position of the replication block ranges from the beginning of the array to about ∼500 bp into the array. Although we believe the block to be complete throughout the repressor-producing population, the 2D gels always showed linear fragment as well as the Ys resulting from fork blockage. We suspect this is because a fraction of the Ys are nicked or branch migrate during isolation and thereby are converted to linear DNA.
Cytological analysis showed a failure to duplicate the tetO1 fluorescent focus after 2–4 h of ara induction. The number of foci per cell decreased from four or eight in unblocked cells to one or two after 2 h of TetR expression (compare Figure 2C with B).
Similarly, the flow cytometry analysis of the 2 h ara-induced ‘run-out' culture shows cells as having between three and six chromosome equivalents (Figure 2C), as compared to the four and eight of an unblocked culture (Figure 2B). After ara-induced blockage of the ccw forks, the cw forks will proceed as far as the ter sites, where they too will be halted. The arrival of another cw fork allows the bypass of the ter site and the cw fork will thus eventually arrive at the DW end of the tetO array. This will yield the observed three chromosome equivalent cells: with four copies of the right-hand half of the chromosome and two almost complete copies of the left-hand half (see Figure 2E). Similarly, cells that contained eight origins at the time of blockage initiation will yield six chromosome equivalents (simply double of the situation with three equivalents). Without run-out treatment, an ara-induced culture generated DNA profiles which over time approached those of Figure 2C (run-out), being consistent with the model in Figure 2E by 4 h (not shown). In addition, this scheme is supported by quantitative analysis of relative gene copy number by Southern hybridisation after ara induction. Probes to pheA and lacZ were used to report relative concentrations of the left and right chromosome arms, respectively. In a culture without ara, the pheA /lacZ ratio was close to 1 (0.85 after 2 h and 1.05 after 4 h), whereas it dropped to 0.35 in an ara-induced culture (0.38 after 2 h and 0.32 after 4 h). This is consistent with a heterogeneous population in which there has been an under-replication of the left arm.
We conclude that high occupancy tight binding of the tetO array by TetR produces an effective replication roadblock independent of the fork polarity. Furthermore, blockage of ccw forks 15 kb after initiation does not appear to prevent the eventual progression of some cw forks around the whole chromosome, which requires bypass of ter sites, a process that requires several hours to occur (Figure 2E).
Site-specific fork stalling in the ori region: segregation of the opposite replichore, colocalisation of SSB proteins with the blocked forks and cell division inhibition
We examined the replication-segregation of LacI-CFP bound loosely to a lacO2 array, 200 kb cw of dif, in the presence of IPTG, when tetO1, 15 kb ccw of oriC, was blocked by tightly bound TetR (Figure 3). After 2 or 4 h of ara induction, most of the cells exhibited one tetO1 focus (green) at midcell or two foci at the quarter positions, consistent with the expected replication block. By 4 h, cells are elongated with little evidence of septation-cell division. Simultaneous tracking of the lacO2 locus revealed a delay in its duplication, as at 2 h most of cells harboured a single lacO2 focus close to midcell. Nevertheless, by 4 h, most cells had duplicated and segregated the lacO2 foci away from midcell in a bipolar fashion; note that the replication of lacO2 requires overcoming the replication terminators terB and terC. This confirms that the stalling of a fork near oriC does not prevent the opposite fork from continuing replication of the ter region and its subsequent bipolar migration (Figure 3; cartoon). A small proportion of cells at 4 h had four lacO2 foci evenly distributed, suggesting that these cells have reinitiated replication, despite the block at tetO1, and replicated-segregated the ter region. Time-lapse analysis (not shown) supported this overall scheme.
Figure 3.
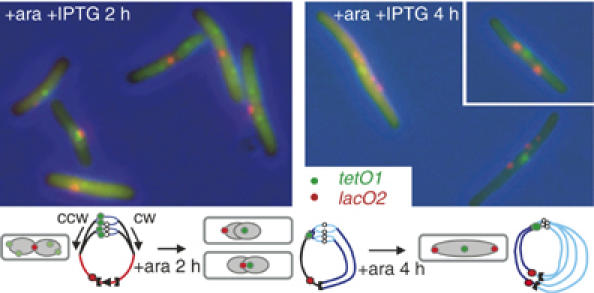
Segregation of lacO2 in tetO1-blocked cells. Images and cartoon of representative tetO1 (green)/lacO2 (red) cells cultivated as indicated.
Then, we visualised simultaneously SSB-CFP with TetR-YFP in a tetO1 strain (Figure 4). Most cells grown in LB+Ap had two SSB-CFP foci near the quarter positions and four ori1 foci organised as pairs flanking each SSB-CFP focus consistent with two sister replisomes being associated with a single SSB focus in this medium, the focus being positioned close to the centre of each sister nucleoid (Figure 4A and D). After 2 h of ara induction, SSB foci were arranged as one pair per tetO1 focus, one SSB focus colocalising with tetO1 focus and the other close by in the midcell region (Figure 4B and D). After 4 h of ara induction, a central SSB signal was obvious in most of cells (Figure 4C and D), but the ori1 signal was diffuse and lacked distinct foci (not shown). Most cells should contain six forks either stalled at tetO1 and/or at ter sites (Figure 2E). We conclude that replisome-associated SSB-CFP persists at repressor-blocked forks for long periods.
Figure 4.
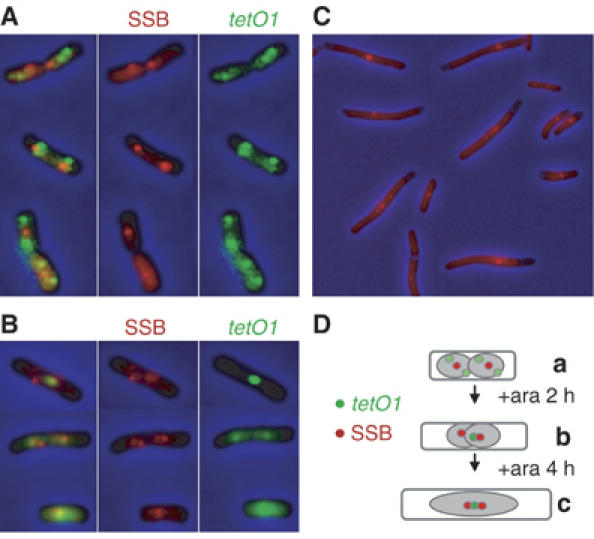
Colocalisation of the SSB signal and tetO1 roadblock. tetO1 cells expressing SSB-CFP were cultivated with ara and AT for 2 h (A), with only ara for 2 h (B) or 4 h (C). Green and red foci correspond to tetO1 loci and SSB proteins, respectively, with the merged image shown on the left where appropriate. (D) A cartoon represents the tetO1/ SSB cell patterns observed in (a), (b) and (c).
The replication roadblock at ori1, 15 kb ccw of oriC, delays but does not prevent replication-segregation of the complete chromosome, although cell division is inhibited. The inhibition of cell division almost certainly arises because the unsegregated nucleoid prevents placement of the FtsZ ring; we do not know if such cells initiate the SOS response.
Site-specific fork stalling in the ter region: biased uniparental inheritance and ter locus release after cell division
The consequences of a repressor-induced replication block in ter region was analysed in a strain containing tetO2, 50 kb clockwise of dif and a lacO1 reporter array 15 kb ccw of oriC. LacI and TetR expression in the presence of IPTG leads to replication block at tetO2, while allowing normal replication of lacO1. A tetO2-blocked culture was compared to an unblocked control using time-lapse analysis over three generations (Figure 5A). The unblocked cell shows a pattern of growth and duplication consistent with earlier analysis (Lau et al, 2003). In contrast, tracking of the tetO2 focus in the blocked cells revealed that the tetO2 focus did not duplicate during the three generation time course, whereas lacO1 duplication and cell division appeared to go on relatively normally (Figure 5B). At each generation, the tetO2 focus was transmitted only to one daughter cell, where it was ‘released' from the septum/new pole at division and localised to the new midcell position. This pattern of inheritance and cell division requires chromosome breakage. Daughter cells lacking a tetO2 focus failed to divide and formed filaments, presumably through induction of the SOS response in response to the broken chromosome. We also noticed that the segregation of the tetO2 focus at the second and third divisions was biased towards the daughter cell containing the newer of the two old poles (n1, then n2; Figure 5B; not shown), corresponding to the pole where tetO2 originates. This bias, which is similar to the bias of asymmetric positioning of the ter region reported from previous time-lapse analysis (Lau et al, 2003), suggests that the ter region is restrained to a cell compartment, where it oscillates between new pole and midcell positions. This pattern of positioning could result directly or indirectly from the DNA breakage required to complete cell division.
Figure 5.
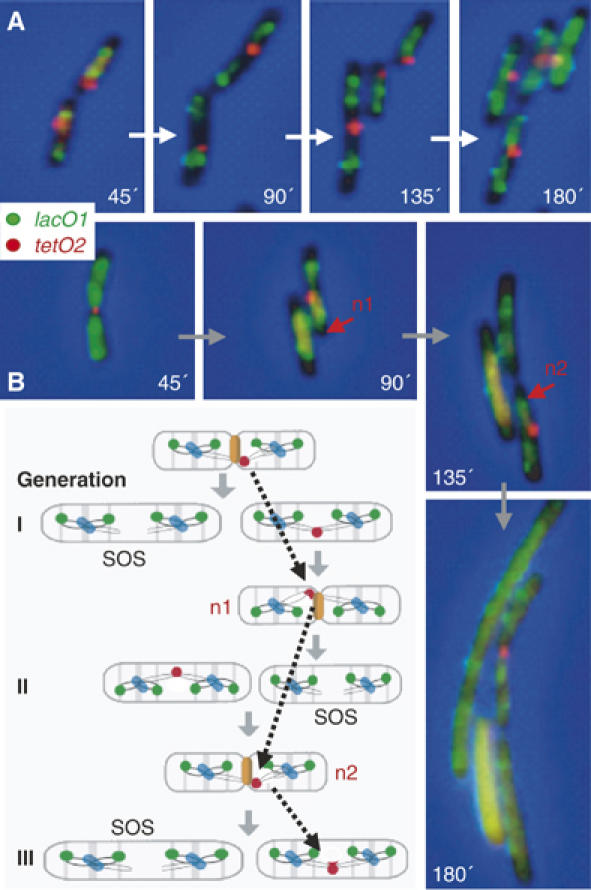
Biased inheritance of the tetO2 roadblock. After 1 h 30 min of ara-induced LacI-CFP and tetR-YFP expression, cells were placed on LB agar layer containing IPTG with (A) or without AT (B). lacO1 and tetO2 foci were tracked under the microscope (images taken every 45 min). A schematic of the time lapse observed in (B) shows a zigzag behaviour (broken arrows) of the tetO2 roadblock (indicated as n1–n2) with blue dots representing replisomes.
The position and movement of two ter loci, lacO2 and tetO3, separated by 150 kb, were tracked simultaneously in cephalexin-induced filaments. In the presence of IPTG and AT, replication of lacO2 and tetO3 was not perturbed and the foci colocalised and segregated uniformly along the filament (Figure 6A). In absence of AT, tetO3 replication was blocked and the focus remained at the middle of the filament, whereas the upstream lacO2 foci duplicated and segregated normally (Figure 6B). The observation that chromosomal loci 150 kb apart can be separated by several cell lengths illustrates the flexibility of the chromosome organisation. The failure of the blocked locus to be ‘released' from midcell in the cephalexin filaments contrasts to the situation in blocked yet dividing cells; supporting the idea that the ‘release' may require DNA breakage, which is dependent on cell division.
Figure 6.
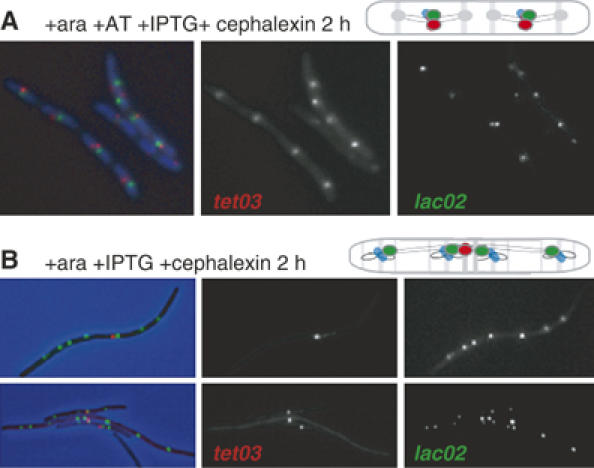
The tetO3 roadblock is not released from midcell in a cephalexin filament. Micrographs and cartoons of representative tetO3/lacO2 cells cultivated as indicated with (A) or without AT (B).
Rapid reversibility by AT of the tetO1 replication roadblock
We then tested if the addition of AT could reverse the replication roadblock. After 2 h of ara induction, the viability of a tetO1- and a tetO3-blocked culture fully recovered after AT addition for 2 h (Figure 7A and not shown). Similarly, a normal replication profile, as judged by ‘run-out', was restored after 2 h AT treatment (Figure 7B).
Figure 7.
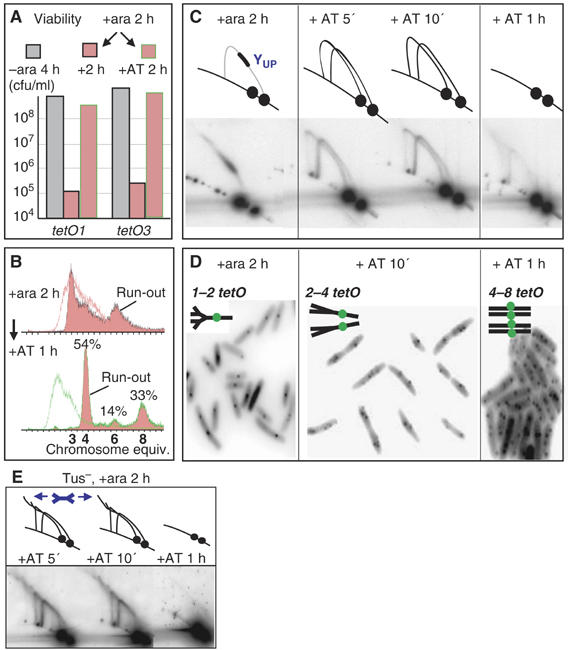
Tracking of AT-dependent replication restart. (A) Viability of tetO1 and tetO3 strains as determined in Figure 1, after cultivation as indicated. (B) Tracking of replication fork restart at tetO1 in Tus+ cells after the indicated conditions by flow cytometry. (C) 2D gel analysis of tetO1 block and restart (Tus+). (D) Microscopic analysis of tetO1 block and restart (Tus+). (E) Release of block at tetO1 in a Tus− strain visualised by 2D gel electrophoresis; the −AT control is shown in Figure 2D. The blue double-Y indicates the position of double-forked UP and DW fragment species.
The observation of this rapid and homogenous recovery of cell viability–physiology by AT addition led us to track the replication restart over time by using 2D gels and cytological analysis. AT was added to a tetO1 culture ara-induced for 2 h. Cell samples were harvested after 5, 10 min and 1 h of AT treatment. The effects of AT addition on replication were obvious by 2D DNA analysis after 5 min. The YUP spot, which was the only replication intermediate observed before AT addition, had disappeared and the entire YUP and YDW arcs became visible, consistent with a substantial number of forks moving through the array (Figure 7C). At the cytological level, pairs of adjacent sister foci were already observed in some cells, whereas only single foci were present before AT addition (not shown). After 10 min of AT treatment, these phenotypes were reinforced: two strong Y arcs were still evident and a large proportion of cells had ori1 focus doublets (Figure 7D). Some cells even had ori1 focus triplets or quadruplets, consistent with new rounds of replication entering the locus and the persistence of Y arcs. These observations demonstrate that replication can restart within 5 min of AT addition and that subsequent segregation is not subject to any significant period of cohesion. After 1 h of AT, the cell population had lost all obvious consequences of the roadblock: no replication intermediate accumulation was detectable and ori1 number and positioning were indistinguishable from cells that had never experienced the roadblock; the FACS profile had returned to normal.
Replication restart at tetO1 was also analysed in Tus− mutant (Figure 7E) after 2 h of ara induction, after which both ccw (and to a lesser extent cw) forks are blocked at the repressor-bound array (see Figure 2D). After 5 min of AT addition, the two spots corresponding to blocked forks had disappeared and two Y arcs appeared with a trailing tails at their top. This additional tailing, which is not observed in the Tus+ strain, likely corresponds to double Ys arising from restart on at least one side of an array bound by blocked forks of opposite polarity.
Direct replication restart without recombination
The ability to block and restart replication at tetO1 was tracked by 2D gels and microscopy in recA and recB mutant strains, in order to test whether homologous recombination or recombination proteins are required for replication restart at repressor-blocked arrays. After cultivation without ara or for 2 h with ara+AT, no replication intermediates were observed (Figure 8). After 2 h of ara induction, a spot migrating along the Y arc curve of the UP fragment (YUP) was observed for both recA and recB mutant strains. The blocked fork accumulated after 3 and 4 h of ara induction at a position similar to that for the wild-type strain (not shown). Note that in the recB mutant, the level of block is somewhat reduced and there is a higher background Y arc. We believe this is a consequence of plasmid loss and plasmid copy number heterogeneity in the recB mutant, leading to a fraction of cells that are not blocked, and some that have ongoing restart at the locus because of lower levels of repressor. Cytological analysis supports this interpretation. The pattern of fluorescent foci in the recA mutant was essentially indistinguishable to that of Rec+ cells. In contrast, only a small proportion of recB cells showed clear fluorescent foci, but those cells with fluorescent foci exhibited the same pattern as Rec+ cells (not shown).
Figure 8.
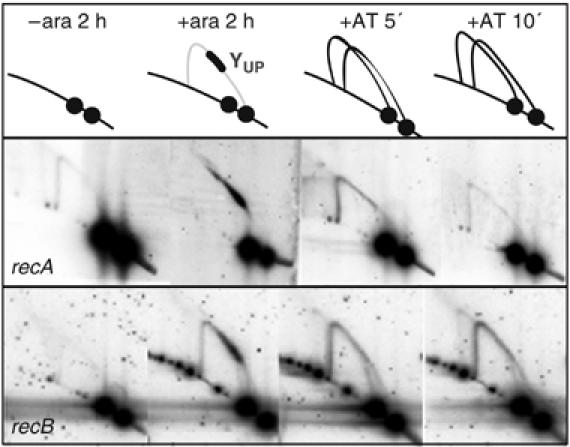
Direct restart in recA and recB mutants. The RecA+ control is shown in Figure 7C. 2D gel electrophoresis after the indicated growth conditions.
After 5 or 10 min of AT treatment, the YUP spot had disappeared in both the recA and recB mutants, just as in the Rec+ strain. Furthermore, increased Y arc intensity was evident, as in Rec+ cells, consistent with restarted forks moving through the locus. After 30 min of AT treatment, the pattern was indistinguishable from unblocked cells (not shown).
In the recA mutant, the apparent normal restart after AT addition was correlated with a recovery of viability to a level comparable to that in Rec+ cells (Figure 1; not shown). In contrast, recovery of viability in the recB mutation was very low (∼1%), despite the apparently normal kinetics of restart. We do not know the reason for this inviability, but it appears not to be associated directly with the block, or with any fork block at Tus-ter, as recovery of viability was no higher in a Tus− recB strain. Perhaps the nuclease activity of RecBCD is required to remove toxic DNA ends that accumulate after blockage (B Michel, personal communication), or the moderate plasmid instability in a recB mutant is exacerbated after the replication fork block is applied to cells.
Discussion
This is the first report of a reversible site-specific replication roadblock that prevents viable cell proliferation. Natural replication barriers, for example the E. coli Tus protein binding to 30 bp ter sites, or the yeast Schizosaccharomyces pombe Rtf1/2 complex that binds four repeated RTS1 sites, cause a transient pause rather than a persistent blockage that does not affect cell viability (Bidnenko et al, 2002; Ahn et al, 2005). In contrast, we have shown that TetR repressor-bound operator DNA efficiently and persistently stalls replication forks of both polarities. E. coli cells apparently cannot overcome this barrier, either by running a second fork into it, or through the activity of any housekeeping enzymes. Positioning of the roadblock close to oriC or within ter did not impede progress of the other replication fork, which could reach the opposite side of the blocked array, thereby reinforcing the conclusion that the two replication forks act independently (Breier et al, 2005). Furthermore, blocking one fork did not prevent either the segregation of the opposite replichore, or a locus on the same replichore just 150 kb upstream. In cells blocked at ori1, or in cephalexin-treated tetO2-blocked cells, the blocked array persisted at midcell for many hours, presumably trapped adjacent to potentially active replication machinery. This persistence is dependent on the failure to undergo cell division, because once a cell divides, a blocked tetO2 array moves away to the new midcell position. Whether the release is dependent on replisome disassembly and/or cleavage of DNA by guillotining is not known. Addition of the cognate inducer, anhydrotetracyline, leads to an almost instantaneous release of the replication block and a return to the cell's normal physiological state within 60 min. As release of tight repressor binding leads to immediate duplication-segregation of fluorescent foci, we can rule out any extensive period of sister cohesion.
DNA-bound proteins in normal cells must be frequently encountered by a replisome, whose progression must not be compromised in the long term if a cell is to proliferate. Although the repressor-bound operator arrays used here to block replication do not occur naturally, we are confident that in normal cells, replication forks encounter large nucleoprotein complexes that block replication in a manner similar to that described here, and that replication restart can occur quickly and efficiently once the block is removed without the participation of recombination proteins. For example, combinations of proteins bound to regulatory regions, or stalled transcription–translation complexes may stop replication, with restart occurring by the mechanism described here after release of nucleoprotein complex. Some proteins can form extensive nucleoprotein complexes extending for many kilobases. For example, the parS-parB plasmid and chromosomal segregation system leads to ParB spreading over at least an 11 kb region, where it interferes with transcription (Rodionov et al, 1999) and apparently replication when present in a plasmid (G Ebersbach and DJ Sherratt, unpublished). Additionally, several DNA-binding proteins bind generally to the E. coli nucleoid, covering on average about 60% of the chromosomal DNA, as ∼40% of the chrosmosome exhibits unconstrained supercoiling; a high density of such nucleoprotein complexes in one location could well interfere with replication. Whereas some of these proteins are present at all growth phases (e.g., H-NS, HU), others show cell cycle dependence (Azam and Ishihama, 1999). For example, the Dps protein increases from 6000 to 180 000 molecules/cell in stationary phase, when it protects against stress. When such stationary phase cells resume DNA synthesis, replication blockage may occur in regions that are not immediately freed of bound Dps protein. Viability will require that any such blocks are reversible. The Rep protein has been proposed to facilitate removal of bound proteins in E. coli in vivo (Matson et al, 1994; Uzest et al, 1995), and the related helicase Rrm3 appears to have the same function in Saccharomyces cerevisiae (Ivessa et al, 2003). Consistent with the existence of systems to remove blocking complexes, replication fork progression appears to be more easily impaired by bound proteins in vitro than in vivo. For example, wild-type LacI repressor can prevent replication fork progression in vitro when as few as three operators are present, although in vivo at least 22 are required to interfere with progression, as judged by 2D DNA analysis (P McGlynn, personal communication). However, other proteins may participate in removal in vivo, as in vitro studies did not show an ability of Rep to alleviate the fork stalling by operator-bound LacI (P McGlynn, personal communication). Our observation that replication restarts immediately after release of tight repressor binding, without the participation of recombination proteins, and that replisome-associated proteins SSB persists for many hours at a repressor-bound array, favours the view that a stable stationary replisome can persist in the vicinity of a blocking nucleoprotein complex. The rescue of this block via direct replication restart leads us to suggest that such reversible blocks could have been previously gone unnoticed, and may be the predominant way that cells handle nucleoprotein blocks to replication. The ability to cope efficiently with transient blocks in this manner is both economical and avoids possible mutagenic outcomes of dealing with a collapsed replication fork. In contrast, when replication forks encounter DNA damage, when the replisome is impaired, or when specific replication termination sites are bound by their cognate termination proteins, recombination proteins and/or homologous recombination are required before restart (Michel et al, 2004). We note that inhibiting DNA gyrase seems to result in a similar mechanism of recombination protein-free restart to that described here (Grompone et al, 2003). Indeed protein blocks like those here could conceivably mediate their effect by preventing gyrase access, thus leading to replication fork stalling.
It is clear from our analysis of tetO1 strains, using multiple approaches, that the replisome can overcome the Tus-ter blocks that naturally occur and that a replication fork could eventually continue around almost the whole chromosome. However, to see a significant signal from 2D gel analysis that gave evidence of this required 4 h of growth. Even in a tus mutant derivative of tetO1, the appearance of the second fork was slow, and a strong blocked fork signal required 3–4 h of growth. A replication fork is able to replicate half the chromosome of E. coli in 50–60 min under our growth conditions (Wang et al, 2005). Therefore, the slow appearance of the second Y-arc signal emphasizes just how much influence chromosome organisation exerts over the replication process. Once the replication fork progresses past the ter region, it appears to require at least 2 h further to replicate the second half of the chromosome, two-three times longer than it took for the first half. We imagine this is because of collisions with convergent transcription complexes (e.g. see Rocha, 2004).
The work here emphasises the need to exercise caution when specific DNA-binding proteins are used to monitor gene position and number in living cells; it is particularly important to ensure that the conditions of use do not perturb normal DNA metabolism. Nevertheless, the controllable and site-specific replication roadblock characterized here is a powerful tool with broad applicability for investigations of replication stalling and restart at different loci and in different situations, and for studies of replisome stability.
Materials and methods
Bacterial strains and plasmids
Derivatives of E. coli K12 AB1157 were used (Bachmann, 1972). The strains carrying 240 copies of the lacO and tetO arrays and the repressor producing plasmid pLau53 (Lau et al, 2003). The SSB-CFP allele inserted in place of the E. coli lamB gene was provided by A Wright (G Leung et al, unpublished).
Microscopy and flow cytometry
Cells were cultivated in LB medium containing Ap, with other indicated reagents where appropriate. Expression of fluorescent repressors was induced by addition of 0.05% ara. The gratuitous inducers, IPTG (1 mM) and AT (100 ng/ml), were used to prevent tight repressor binding where indicated. For microscopy, cells were grown to A600 of 0.1 and transferred to a 1.0% agarose layer containing phosphate-buffered saline for snapshot microscopy or LB for time lapse. Cells were visualized with a × 100 objective on a Nikon Eclipse TE2000-U microscope, equipped with a Photometrics CoolSNAP HQ CCD camera and a temperature-controlled incubation chamber. The images were taken, analyzed and processed by MetaMorph 6.2 and Adobe Photoshop. Flow cytometry was performed on a FACSCalibur flow cytometer (Becton Dickinson). Samples for flow cytometry were grown, fixed and the DNA was stained with Syto16 (Molecular Probes). Replication ‘run-out' was performed by treatment of a culture with rifampicin (300 μg/ml) and cephalexin (100 μg/ml), which inhibit replication initiation at oriC and cell division, respectively, thereby allowing only completion of ongoing rounds of replication (Skarstad et al, 1985). If chromosome replication is not blocked, after ‘run-out', the DNA content per cell should correspond to 2n chromosome equivalents, reflecting the number of oriC origins per cell at the time that treatment had started. Normalisation was done from one chromosome-containing cells (Wang et al, 2005). After a persistent replication roadblock, partially replicated chromosomes will lead to a non-2n chromosome number.
2D DNA gels
Samples of cells were taken at the indicated time points, 0.1% sodium azide was added and cells were put on ice. Cells were harvested and then embedded in 0.4% agarose plugs at a density of 5 × 109 cells/ml. Lysis was carried out in the plugs which were then washed extensively. DNA was digested either with EcoRV or SacII as appropriate. 2-D gel conditions and subsequent transfer to nylon membranes were as described by Lopes et al (2001); DNA was detected using radiolabelled tetO array or the UP fragment (Figure 2) as probe.
Southern hybridisation
Relative gene dosage was measured by Southern hybridisation using 32P probes to the lacZ and pheA genes (chromosomal position 363 and 2736 kb, respectively). Duplicate blots onto Hybond N+ membrane (Amersham) of two independent experiments were analysed by phosphorImaging using a Fuji FLA3000 and Image Gauge software.
Acknowledgments
We thank A Wright for providing the SSB-CFP strain, M Whitby and B Michel for helpful discussions. We thank P McGlynn for communicating unpublished results and invaluable discussion. We thank A Whitely for assistance in flow cytometry. CP and SRF were supported by EMBO long-term fellowships. SRF was also supported by a fellowship from the Fundação para a Ciência e Tecnologia, MCT, Portugal. The work was supported by the Wellcome Trust.
References
- Ahn JS, Osman F, Whitby MC (2005) Replication fork blockage by RTS1 at an ectopic site promotes recombination in fission yeast. EMBO J 24: 2011–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam TA, Ishihama A (1999) Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J Biol Chem 274: 33105–33113 [DOI] [PubMed] [Google Scholar]
- Bachmann BJ (1972) Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev 36: 525–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Kleckner N (2005) Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121: 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidnenko V, Ehrlich SD, Michel B (2002) Replication fork collapse at replication terminator sequences. EMBO J 21: 3898–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot S, Saleh OA, Lesterlin C, Pages C, El Karoui M, Dennis C, Grigoriev M, Allemand JF, Barre FX, Cornet F (2005) KOPS: DNA motifs that control E. coli chromosome segregation by orienting the FtsK translocase. EMBO J 24: 3770–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y (1997) The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1474 [DOI] [PubMed] [Google Scholar]
- Breier AM, Weier HU, Cozzarelli NR (2005) Independence of replisomes in Escherichia coli chromosomal replication. Proc Natl Acad Sci USA 102: 3942–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471 [DOI] [PubMed] [Google Scholar]
- Fekete RA, Chattoraj DK (2005) A cis-acting sequence involved in chromosome segregation in Escherichia coli. Mol Microbiol 55: 175–183 [DOI] [PubMed] [Google Scholar]
- Gordon GS, Sitnikov D, Webb CD, Teleman A, Straight A, Losick R, Murray AW, Wright A (1997) Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell 90: 1113–1121 [DOI] [PubMed] [Google Scholar]
- Grompone G, Ehrlich SD, Michel B (2003) Replication restart in gyrB Escherichia coli mutants. Mol Microbiol 48: 845–854 [DOI] [PubMed] [Google Scholar]
- Hill TM, Henson JM, Kuempel PL (1987) The terminus region of the Escherichia coli chromosome contains two separate loci that exhibit polar inhibition of replication. Proc Natl Acad Sci USA 84: 1754–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A, Lenzmeier B, Bessler J, Goudsouzian L, Schnakenberg S, Zakian V (2003) The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein–DNA complexes. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Putnam CD, Myung K (2002) Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552–557 [DOI] [PubMed] [Google Scholar]
- Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ (2003) Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol 49: 731–743 [DOI] [PubMed] [Google Scholar]
- Lesterlin C, Barre FX, Cornet F (2004) Genetic recombination and the cell cycle: what we have learned from chromosome dimers. Mol Microbiol 54: 1151–1160 [DOI] [PubMed] [Google Scholar]
- Lesterlin C, Mercier R, Boccard F, Barre FX, Cornet F (2005) Roles for replichores and macrodomains in segregation of the Escherichia coli chromosome. EMBO Rep 6: 557–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Pellicioli A, Cotta-Ramusino C, Liberi G, Plevani P, Muzi-Falconi M, Newlon C, Foiani M (2001) The checkpoint response stabilizes stalled DNA replication forks. Nature 412: 599–602 [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG (2002) Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol 3: 859–870 [DOI] [PubMed] [Google Scholar]
- Markovitz A (2005) A new in vivo termination function for DNA polymerase I of Escherichia coli K12. Mol Microbiol 55: 1867–1882 [DOI] [PubMed] [Google Scholar]
- Matson SW, Bean DW, George JW (1994) DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. BioEssays 16: 13–22 [DOI] [PubMed] [Google Scholar]
- Michel B, Grompone G, Flores MJ, Bidnenko V (2004) Multiple pathways process stalled replication forks. Proc Natl Acad Sci USA 101: 12783–12788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S (2000) Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev 14: 212–223 [PMC free article] [PubMed] [Google Scholar]
- Rocha EP (2004) The replication-related organization of bacterial genomes. Microbiology 150: 1609–1627 [DOI] [PubMed] [Google Scholar]
- Rodionov O, Lobocka M, Yarmolinsky M (1999) Silencing of genes flanking the P1 plasmid centromere. Science 283: 546–549 [DOI] [PubMed] [Google Scholar]
- Sherratt DJ (2003) Bacterial chromosome dynamics. Science 301: 780–785 [DOI] [PubMed] [Google Scholar]
- Skarstad K, Steen HB, Boye E (1985) Escherichia coli DNA distributions measured by flow cytometry and compared with theoretical computer simulations. J Bacteriol 163: 661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzest M, Ehrlich SD, Michel B (1995) Lethality of rep recB and rep recC double mutants of Escherichia coli. Mol Microbiol 17: 1177–1188 [DOI] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L (2004) Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA 101: 9257–9262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Possoz C, Sherratt DJ (2005) Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev 19: 2367–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]


